Abstract
Leptin is an adipocyte-derived cytokine that regulates food intake and body weight via interaction with its Ob receptor (ObR). Serum leptin levels are chronically elevated in obese humans, suggesting that obesity may be associated with leptin resistance and the inability to generate an adequate ObR response. Evidence suggests that transcriptional activation of target genes by STAT3 (signal transducer and activator of transcription) in the hypothalamus is a critical pathway that mediates leptin’s action. Herein we report that activation of ObR induces the tyrosine phosphorylation of the tyrosine phosphatase SH2-containing phosphatase 2 (SHP-2) and demonstrate that Tyr986 within the ObR cytoplasmic domain is essential to mediate phosphorylation of SHP-2 and binding of SHP-2 to ObR. Surprisingly, mutation of Tyr986 to Phe, which abrogates SHP-2 phosphorylation and binding to the receptor, dramatically increases gene induction mediated by STAT3. Our findings indicate that SHP-2 is a negative regulator of STAT3-mediated gene induction after activation of ObR and raise the possibility that blocking the interaction of SHP-2 with ObR could overcome leptin resistance by boosting leptin’s weight-reducing effects in obese individuals.
Leptin, the product of the obese gene, is secreted by adipocytes and functions as a peripheral signal to the brain to regulate food intake and energy metabolism (1). Leptin is thought to exert its action in the hypothalamus through its Ob receptor (ObR). Rodents with mutations that prevent normal expression of either leptin or full-length ObR are profoundly obese and diabetic and have a reduced metabolic rate. However, human obesity does not generally appear to be associated with mutations in the genes encoding leptin or ObR (2, 3). Although mice with a mutant obese gene can be returned to normal weight by administration of recombinant leptin (4–6), it seems that this approach may not succeed in the majority of obese humans because their serum leptin levels are chronically elevated (7, 8). Obese humans, therefore, appear to be “leptin resistant” (7) in that they do not generate a signal commensurate with their serum leptin levels, perhaps because of defective transport of leptin across the blood–brain barrier (9) or an inadequate ObR response. Analysis of ObR signaling pathways may reveal alternative therapeutic approaches of boosting ObR responses to overcome leptin resistance and reverse obesity.
Leptin and ObR are members of the four-helical bundle cytokine and receptor superfamilies, respectively (10). ObR is most closely related to the gp130 and leukemia inhibitory factor receptor β (LIFRβ) signal transducing receptors that are activated by cytokines such as interleukin 6 and ciliary neurotrophic factor (CNTF), whose signaling pathways have been intensively studied (11). Ligand binding induces either homodimerization of gp130 or heterodimerization of gp130 with related signal transducing receptors such as LIFRβ, leading to activation of the receptor-associated Janus kinases (Jaks). The Jaks then phosphorylate gp130 on cytoplasmic tyrosine residues, forming phosphotyrosine-based motifs that recruit specific src homology domain 2 (SH2)-containing signaling molecules such as STAT3 (signal transducer and activator of transcription) and the protein tyrosine phosphatase SH2-containing phosphatase 2 (SHP-2) (12). Removal or mutation of the phosphotyrosine motifs in gp130 or LIFRβ eliminates activation of the corresponding SH2 target molecule (12). Cytoplasmic deletions appear to affect ObR in a similar manner: there are multiple isoforms of ObR corresponding to alternatively spliced products with different cytoplasmic domains (10, 13, 14), but only one isoform with several potential phosphotyrosine motifs, known as the long form or ObRb, appears capable of mediating leptin’s weight controlling effects (13–16). Obese diabetic mice have a mutation in ObR that prevents expression of the long ObR splice isoform that renders them incapable of appropriately mediating leptin’s actions (13, 14). The finding that only the long form of ObR contains the sequence YXXQ (10), which is a motif that specifies STAT3 activation (12), raised the possibility that STAT3 is critical for mediating leptin responses. Recent results verify that STAT3 is activated both in cultured cells (15, 16) and in vivo (17) by the long form of ObR and not by a truncated ObR or the long form of ObR with a mutant YXXQ motif (16, 18). Although leptin-induced activation of overexpressed STAT1 and STAT5b is also observed in transfected cells (15, 16), only activation of STAT3 has been detected in vivo upon stimulation of hypothalamic ObR by administration of leptin (17). Leptin may also have direct effects on peripheral tissues (19), although the contribution of these effects on leptin’s ability to regulate energy metabolism is unclear. Thus it appears likely, but unproven, that transcriptional activation of target genes by STAT3 in the hypothalamus is a critical pathway that mediates leptin’s regulation of food intake and energy metabolism.
The role that SHP-2 plays in signal transduction has been examined for several cytokine and growth factor receptors. SHP-2 is ubiquitously expressed and generally acts to positively regulate signaling, as exemplified by the prolactin (20), epidermal growth factor, and platelet-derived growth factor (21, 22) receptor systems. For the platelet-derived growth factor receptor, tyrosine phosphorylated SHP-2 acts as an adaptor molecule that recruits Grb2 and Sos, members of the signaling pathway leading to activation of Ras and the mitogen-activated protein kinase extracellular signal-regulated kinase (ERK). Likewise, gp130 can apparently use SHP-2 as one means to activate ERK2 by coupling to Grb2 and Sos, leading to stimulation of mitogenesis in BAF cells (23).
Herein we report that SHP-2 becomes tyrosine-phosphorylated in response to leptin stimulation of ObR. However, in contrast to its actions described above, SHP-2 appears to negatively regulate STAT3-mediated transcriptional activation upon leptin stimulation of ObR.
MATERIALS AND METHODS
Expression Constructs and Cell Transfection.
The cytoplasmic domain and full-length human ObR was obtained by PCR using human fetal liver and adipocyte cDNA (CLONTECH) and cloned into the expression vectors pCMX and pMT21, respectively. The cytoplasmic domain of ObR was expressed as a chimeric receptor, constructed by fusing the extracellular domain and transmembrane domain of TrkC (amino acids 1–458) to the cytoplasmic domain of ObR (amino acids 865–1,165). A triple myc epitope tag was added at the C terminus of all ObR constructs (12). Truncation and point mutant constructs were generated by PCR, and the sequences of all constructs were verified. Expression constructs were transfected into COS-7 cells by using LipofectAMINE (Life Technologies) for ≈18 hr. Thirty-six to 48 hr after transfection, cells were starved for 2 hr for chimeric receptors or overnight for full-length receptors before stimulation.
Immunoprecipitation and Western Blotting.
Cells were stimulated with human neurotrophin 3 (NT-3; 50 ng/ml) for 15 min or human leptin (R & D Systems; 100 ng/ml) for 10 min. Cells were then lysed in a 1% Brij 96 buffer and receptor was immunoprecipitated with anti-myc antibody 9E10 (12) followed by protein G-Sepharose (Pharmacia). Unbound lysate was then immunoprecipitated with anti-phosphotyrosine (Upstate Biotechnology) or agarose-conjugated anti-SHP-2 antibody (Santa Cruz Biotechnology). For analysis of the STAT3 tyrosine phosphorylation state, STAT3 was first immunoprecipitated with anti-STAT3 antibody (Transduction Laboratories, Lexington, KY), and the unbound lysate was then immunoprecipitated with anti-myc antibody. Immunoprecipitated proteins were separated by SDS/PAGE; immunoblotted with antibodies to SHP-2, STAT3, and Jak1 (all from Transduction Laboratories), Jak2 (Upstate Biotechnology), phospho-STAT3 (P-STAT3, New England Biolabs), or myc; and then visualized by chemiluminescence (DuPont/NEN). Endogenous STAT3, SHP-2, Jak1, and Jak2 is detected in all immunoblotting experiments.
Glutathione S-Transferase (GST) and Thioredoxin (Trx) Fusion Proteins and in Vitro SHP2 Binding Assay.
cDNA encoding SHP-2 (amino acids 2–224) was fused downstream of either Schistosoma japonicum GST or Escherichia coli Trx. SHP-2 was cloned into pGEX-3X (Pharmacia) for the GST-SHP2-SH2 fusion or into pET-32c (Novagen) for the Trx-SHP2-SH2 fusion and expressed in E. coli. pY986 (ObR) phosphopeptide (Research Genetics, Huntsville, AL), which has the sequence QPFVKpYATLISNDYKDDDDK, includes a flag epitope tag. pY440 (interferon γ receptor α, IFN-γRα) phosphopeptide has the sequence TSFGpYDKPHVLVDDYKDDDDK. The ObR cytoplasmic domain (amino acids 865–1,165) possessing a C-terminal triple myc epitope tag was fused downstream of GST, cloned into pGEX-5X-1 (Pharmacia), and expressed in E. coli. To obtain phosphorylated GST-ObR cytoplasmic domain fusion protein, the GST-ObR construct was expressed in E. coli (strain TKX1, Stratagene), which possesses a plasmid encoding an inducible tyrosine kinase, Elk. Expression of Elk resulted in tyrosine phosphorylation of the GST-ObR cytoplasmic domain fusion proteins. Binding assays were performed as immunoprecipitations; 15 nM Trx-SHP2-SH2 and 15 nM GST-ObR-cytoplasmic domain or 200 nM GST-SHP2-SH2 and 100 nM flag-tagged phosphopeptide were mixed together in TBST (20 mM Tris⋅HCl, pH 7.4/150 mM NaCl/0.1% Tween 20) overnight at 4°C. Antibodies against the flag (M2; Kodak) or myc epitope tags were added, and the proteins were precipitated with protein-G Sepharose. Immunoprecipitated proteins were separated by SDS/PAGE, detected by Western blotting using either polyclonal anti-GST or anti-Trx antibodies (Sigma), and visualized by chemiluminescence.
Luciferase Reporter Gene Assay.
Expression constructs for ObR, ObRY1141F, ObRY1078,1079F, ObRY986F, and ObRY986,1141F were cotransfected with the luciferase reporter 3(G3)-Cy6-LUC and pCMX.LacZ into COS-7 cells (six-well dishes) by using LipofectAMINE for ≈18 hr. After transfection, medium was changed to serum-free OptiMEM (Life Technologies), and either murine (PeproTech, Boston; 100 ng/ml) or human (R & D Systems; 100 ng/ml) leptin was added for 24 hr. Expression constructs for TObR and TObRY986F were cotransfected with 3(G3)-Cy6-LUC and pCMX.LacZ into neuroblastoma (NBFL) cells by calcium phosphate precipitation for 6 hr. Cells were subsequently treated with NT-3 (20 ng/ml) for 36 hr. Luciferase and β-galactosidase activities were measured in cell lysates as described (24); luciferase activity was normalized to β-galactosidase activity to control for transfection differences.
RESULTS
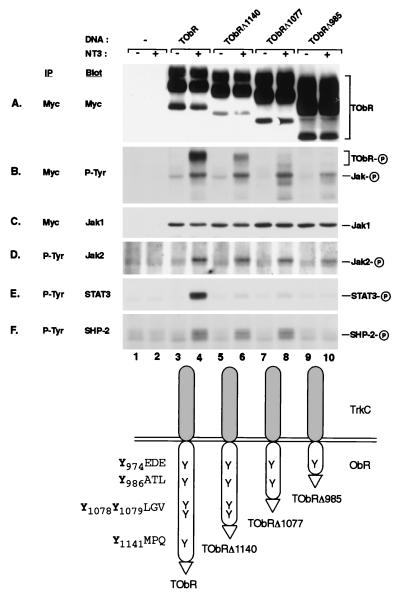
We first examined ObR signaling by creating a series of epitope-tagged chimeric receptors consisting of the extracellular and transmembrane domains of the receptor tyrosine kinase TrkC fused to the cytoplasmic domain of the long form of human ObR, thus allowing the use of a surrogate ligand to specifically activate the ObR signaling pathway. Analogous chimeric receptors have been used to investigate gp130 signaling and showed activation of the identical pathways as native gp130 (12). After transient transfection of COS cells with a vector expressing the TrkC-ObR chimera (TObR), receptor dimerization with NT-3 resulted in ligand-dependent tyrosine phosphorylation of TObR and downstream signaling molecules (Fig. 1). As first observed for gp130 (25), and subsequently noted for many other cytokine receptors, preassociation of Jak with the ObR cytoplasmic domain was observed in the absence of ligand (Fig. 1C, lane 3), and receptor activation resulted in tyrosine phosphorylation of the associated Jak (Fig. 1B, lane 4). The receptor-associated Jak is most likely Jak1 because immunoblots of myc immunoprecipitates show comigration of Jak1, but not Jak2, with the tyrosine-phosphorylated Jak (Fig. 1 B and C, lane 4, and data not shown). Jak2 also becomes tyrosine-phosphorylated on receptor activation (Fig. 1D, lane 4) but is not found associated with ObR after detergent lysis of the cells. Direct immunoprecipitation of Jak1 followed by immunoblotting with anti-phosphotyrosine revealed that Jak1 becomes tyrosine-phosphorylated when the cells were stimulated by NT-3 (data not shown). In addition, receptor was found to coimmunoprecipitate with Jak1 independent of ligand stimulation (data not shown). Ligand-dependent tyrosine phosphorylation of STAT3 was observed (Fig. 1E, lane 4), consistent with previous observations that STAT3 is activated by ObR (15–17). We also found that activation of ObR mediates tyrosine phosphorylation of the protein tyrosine phosphatase SHP-2 (Fig. 1F, lane 4), which has not been previously reported to be a target of leptin action.
Figure 1.
NT-3 stimulation of TrkC-ObR chimeras leads to phosphorylation of receptor, Jaks, STAT3, and SHP-2. myc (A–C) and phosphotyrosine (P-Tyr) (D–F) immunoprecipitates (IP) from unstimulated (−) and stimulated (+) cells transfected with TrkC-ObR (TOBR) expression constructs were immunoblotted (Blot) with antibodies to myc (A), P-Tyr (B), Jak1 (C), Jak2 (D), STAT3 (E), and SHP-2 (F). Truncation constructs (TOBRΔ1140, lanes 5 and 6; TOBRΔ1077, lanes 7 and 8; TOBRΔ985, lanes 9 and 10) are designated by the number of the last amino acid of the ObR cytoplasmic domain present before the myc epitope tag (indicated in the diagram by a triangle), and the amino acids surrounding each tyrosine are also indicated (diagram at bottom). The phosphorylated form of each protein is indicated by a circled P on the right.
To identify the region of ObR that is required to direct phosphorylation of SHP-2, we created a series of chimeras containing C-terminal truncations of TObR that successively eliminate tyrosine residues from the receptor cytoplasmic domain (Fig. 1). For gp130 and other cytokine receptors, phosphorylated tyrosines along with their surrounding amino acids constitute modular tyrosine-based motifs that recruit specific downstream targets through interaction with their SH2 domains (12, 26). Stimulation of cells expressing the truncated receptors TObRΔ1140 and TObRΔ1077 result in SHP-2 phosphorylation (Fig. 1F, lanes 6 and 8), whereas cells expressing TObRΔ985 can no longer mediate SHP-2 phosphorylation (Fig. 1F, lane 10). This truncated receptor can still confer phosphorylation of Jak2 and receptor-bound Jak1 (Fig. 1 B and D, lane 10) although total receptor tyrosine phosphorylation is substantially reduced (Fig. 1B, lane 10). TObRΔ985 lacks the sequence YATL, which is similar to other tyrosine-based motifs that interact with the SH2 domains of SHP-2 (27), and thus may specify SHP-2 interaction with ObR. Stimulation-dependent STAT3 phosphorylation is not observed for the construct TObRΔ1140, in which the sequence YMPQ, a tyrosine-based motif capable of specifying STAT3 activation (12), has been deleted (Fig. 1E, lane 6). This observation is consistent with previous studies demonstrating the importance of YMPQ in the activation of STAT3 upon stimulation of ObR (16–18).
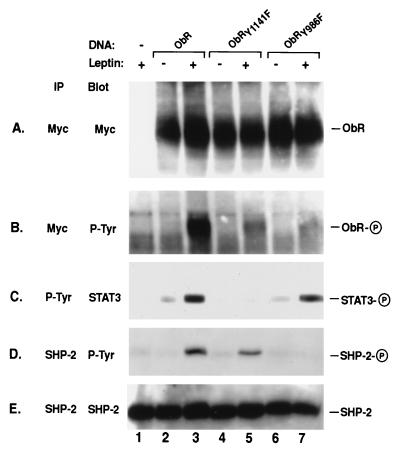
To confirm our assertion that the specific tyrosine residue within the YATL motif of ObR is critical for specifying SHP-2 phosphorylation, we created an epitope-tagged version of the long form of ObR containing a Tyr → Phe mutation within the YATL motif and examined its ability to activate SHP-2. Additionally, we constructed an ObR mutant with a Tyr → Phe substitution in the YMPQ motif that is required for activation of STAT3. Transfection of native and mutant ObR in COS cells resulted in similar levels of expression and, upon leptin stimulation, receptor tyrosine phosphorylation was observed (Fig. 2 A and B). Activation of native ObR after leptin stimulation results in tyrosine phosphorylation of both STAT3 and SHP-2 (Fig. 2 C and D, lane 3), which corroborates the observations made with the chimeric receptors. Mutation of the YATL motif in ObRY986F eliminates leptin-stimulated SHP-2 phosphorylation (Fig. 2D, lane 7) but does not affect STAT3 phosphorylation (Fig. 2C, lane 7). In contrast, mutation of the tyrosine within the YMPQ motif in ObRY1141F results in SHP-2 phosphorylation upon leptin stimulation (Fig. 2D, lane 5); however, as expected, STAT3 phosphorylation is no longer observed (Fig. 2C, lane 5). Stimulation of ObR possessing Phe substitutions at Tyr1078,1079 results in phosphorylation of both STAT3 and SHP-2, whereas ObR carrying a double mutation, ObRY986,1141F, cannot direct phosphorylation of either STAT3 or SHP-2 (data not shown). These results demonstrate that Tyr986 within ObR is required to specifically mediate the phosphorylation of SHP-2.
Figure 2.
SHP-2 phosphorylation is dependent on Tyr986. Native ObR (lanes 2 and 3) and mutant ObR containing phenylalanine (F) substitutions made at the indicated tyrosines (Y), ObRY1141F (lanes 4 and 5) and ObRY986F (lanes 6 and 7) were expressed in COS cells. Myc (A and B), phosphotyrosine (P-Tyr) (C) or SHP-2 (D and E) immunoprecipitates (IP) from unstimulated (−) and leptin stimulated (+) cells were immunoblotted (Blot) with antibodies to myc (A), P-Tyr (B and D), STAT3 (C), or SHP-2 (E). The phosphorylated form of each protein is indicated by a circled P on the right. ObR migrates at approximately 190 kDa.
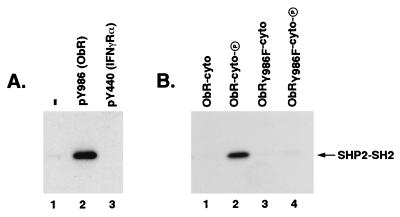
The phosphatase SHP-2 has been shown to bind to tyrosine-phosphorylated receptors, such as platelet-derived growth factor and epidermal growth factor receptors, via its SH2 domains (21, 22). We therefore explored whether SHP-2 could directly interact with ObR at Tyr986 through its SH2 domains by using either a GST or Trx fusion protein containing the N- and C-terminal SH2 domains of SHP-2. Indeed, immunoprecipitation experiments revealed that GST-SHP2-SH2 was able to specifically bind a synthetic phosphopeptide spanning the sequence surrounding Tyr986 and containing the tyrosine-based motif YATL (Fig. 3A, lane 2) but was unable to bind an unrelated phosphopeptide derived from IFN-γRα (Fig. 3A, lane 3). To confirm that SHP-2 binding to an intact ObR cytoplasmic domain showed the same requirement for Tyr986, we created constructs encoding GST fusions with native or mutant ObR cytoplasmic domains (GST-ObR-cyto) to use in binding studies with Trx-SHP2-SH2. Tyrosine-phosphorylated versions of the GST-ObR-cyto were obtained by expressing fusion proteins in E. coli TKX1 coexpressing the tyrosine kinase Elk. Specific binding of Trx-SHP2-SH2 to the cytoplasmic domain of ObR was observed, and this interaction required prior tyrosine phosphorylation of the recombinant ObR (Fig. 3B, lane 2). In contrast, Trx-SHP2-SH2 binding to the mutant ObRY986F was significantly reduced (Fig. 3B, lane 4) despite the existence of similar tyrosine phosphorylation levels of the native and mutant GST-ObR-cyto proteins (data not shown). These results show that SHP-2 can directly bind ObR, and this interaction is largely dependent on Tyr986 within the phosphotyrosine motif YATL.
Figure 3.
SHP-2 specifically binds ObR and this binding is dependent on Tyr986. (A) GST-SHP2-SH2 was incubated with no phosphopeptide (−; lane 1), a phosphopeptide containing the sequence surrounding Tyr986 in ObR [pY986 (ObR); lane 2], or a phosphopeptide derived from the cytoplasmic domain of the IFN-γRα (pY440, lane 3). Phosphopeptide was immunoprecipitated with an antibody to the flag epitope tag and coprecipitating GST-SHP2-SH2 was visualized with an anti-GST antibody. (B) Trx-SHP2-SH2 was incubated with native ObR (lanes 1 and 2) or mutant ObRY986F (lanes 3 and 4) ObR cytoplasmic domain-GST fusions, either unphosphorylated (ObR-cyto, lane 1; ObRY986F-cyto, lane 3) or phosphorylated (ObR-cyto-P, lane 2; ObRY986F-cyto-P, lane 4). ObR-cytoplasmic domain fusion proteins were immunoprecipitated with an antibody to the myc epitope tag and coprecipitating Trx-SHP2-SH2 was visualized with an anti-Trx antibody. The position of the Trx-SHP2-SH2 is indicated.
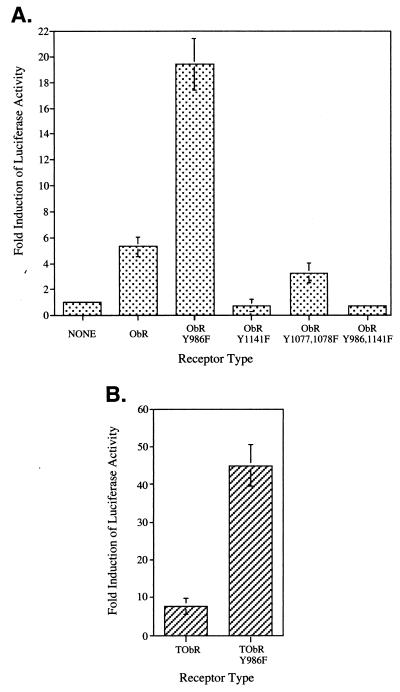
To assess whether SHP-2 plays a role in regulating leptin-stimulated gene induction, we cotransfected COS cells with expression vectors encoding ObR and the luciferase reporter 3(G3)-Cy6-LUC, which contains three copies of a STAT3-responsive DNA element from the promoter that drives expression of the neuropeptide vasoactive intestinal peptide (24). A 5-fold induction of luciferase activity was observed upon leptin stimulation of cells expressing native ObR (Fig. 4A). There was no induction of luciferase activity upon stimulation of cells expressing the mutant construct ObRY1141F (Fig. 4A), in which the mutated tyrosine motif is no longer competent to mediate activation of STAT3 (Fig. 2C, lane 5), while activation of the double mutant ObRY1078,1079F induced luciferase levels similar to that of the native receptor (Fig. 4A). Surprisingly, leptin stimulation of cells expressing the mutant receptor ObRY986F, which can mediate phosphorylation of STAT3 but not SHP-2 (Fig. 2 C and D, lane 7), results in ≈20-fold increase in luciferase activity (Fig. 4A). Similar results were obtained by using the TrkC–ObR chimeric receptors transiently transfected into neuroblastoma cells (NBFL). Activation of TObR resulted in an ≈8-fold increase in luciferase activity whereas expression of TObRY986F gave an ≈45-fold increase (Fig. 4B). Thus, abrogation of SHP-2 activation by mutating Tyr986 in ObR results in an up-regulation of STAT3-mediated signaling upon ObR activation.
Figure 4.
Mutation of Tyr986 in ObR leads to an enhancement of leptin-stimulated STAT3-mediated luciferase gene induction. (A) COS cells were transiently transfected with the indicated ObR receptor construct and 3(G3)-Cy6-LUC reporter. The fold induction of luciferase activity of stimulated compared with nonstimulated cells is indicated as the mean ± SEM for luciferase (LUC) alone (n = 8), ObR (n = 9), ObRY986F (n = 9), ObRY1141F (n = 4), or the average of two determinations for ObRY1077,1078F and ObRY986,1141F with the range indicated. (B) Neuroblastoma (NBFL) cells were transiently transfected with the indicated TrkC-ObR receptor constructs and 3(G3)-Cy6-LUC reporter. The fold induction of luciferase activity (mean ± SEM; n = 3) is indicated.
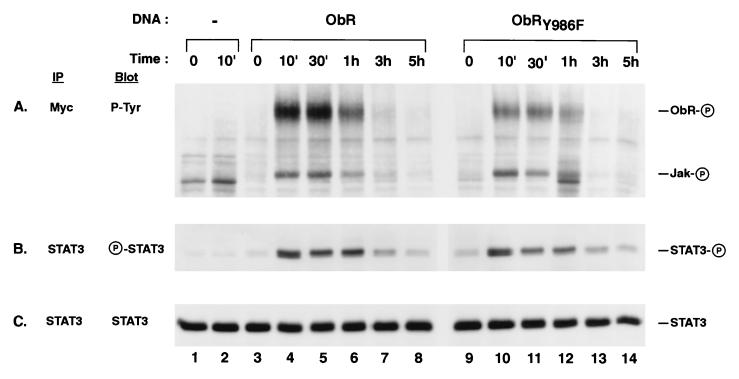
Up-regulation of STAT3-mediated gene transcription in cells expressing ObRY986F could be explained if SHP-2 normally functioned to directly dephosphorylate STAT3, and the receptor mutation preventing SHP-2 activation resulted in elevated or prolonged levels of STAT3 tyrosine phosphorylation. We therefore examined a time course of STAT3 tyrosine phosphorylation, by using an antibody specific for the phosphotyrosine at position 705, after leptin activation of native ObR or mutant ObRY986F. At 10 min, STAT3 is tyrosine-phosphorylated to similar levels in cells expressing either native ObR or mutant ObRY986F (Fig. 5B, lanes 4 and 10, respectively) and shows comparable rates of disappearance over 5 hr for both receptors (Fig. 5B, lanes 8 and 14, respectively). The total amount of receptor tyrosine phosphorylation is lower for ObRY986F than ObR despite similar levels of receptor expression (data not shown) due to the replacement of Tyr986 with Phe. However, the relative rates of disappearance of receptor tyrosine phosphorylation for ObR and ObRY986F were similar, with a maximum at 10 min (Fig. 5A, lanes 4 and 11, respectively) and detection of phosphorylated receptor for up to 1 hr (Fig. 5A, lanes 6 and 12, respectively). Tyrosine phosphorylation of receptor-associated Jak was also observed (Fig. 5A) and followed a similar rate of disappearance as receptor phosphorylation, although the overall amount appears slightly lower at 30 min for ObRY986F (Fig. 5A, compare lanes 5 and 11). These results indicate that there is no large difference in the tyrosine phosphorylation state or duration for either receptor, Jak or STAT3, between cells expressing native ObR and mutant ObRY986F. Thus, direct dephosphorylation of receptor, receptor-associated Jak, or STAT3 does not appear to be the mechanism by which SHP-2 down-regulates STAT3-mediated gene transcription upon leptin stimulation of ObR.
Figure 5.
Time course of STAT3 tyrosine phosphorylation is unchanged in cells expressing ObRY986F. COS cells expressing native ObR or mutant ObRY986F were left unstimulated (0) or stimulated with leptin for 10 min, washed once in PBS, and lysed immediately (10′) or incubated in medium (without leptin) for the indicated times and then lysed (the time equals the total incubation interval from addition of leptin). Myc (A) and STAT3 (B and C) immunoprecipitates (IP) were immunoblotted (Blot) with antibodies to phosphotyrosine (P-Tyr) (A), phospho-STAT3 (P-STAT3) (B), and STAT3 (C). The phosphorylated form of each protein is indicated by a circled P on the right.
DISCUSSION
Analysis of ObR signaling has provided evidence that the protein tyrosine phosphatase SHP-2 plays a negative role in STAT3-mediated gene induction. Upon leptin stimulation, SHP-2 becomes tyrosine-phosphorylated, and this phosphorylation is dependent on Tyr986 within the ObR cytoplasmic domain (Fig. 2). In addition, Tyr986 is required to mediate the direct binding of SHP-2 to recombinant tyrosine-phosphorylated ObR (Fig. 3). Thus Tyr986, which is part of the tyrosine-based motif YATL, appears to specify SHP-2 interaction with ObR. Importantly, activation of ObRY986F gave rise to a markedly increased induction of luciferase activity relative to that observed for native ObR in a STAT3-mediated transcriptional activation assay (Fig. 4). This effect is not restricted to ObR; an analogous finding is observed for the related receptor gp130, where a Tyr759 → Phe substitution, which prevents phosphorylation of SHP-2 upon activation of gp130, also gives rise to an enhanced level of gene induction by STAT3 (28). Moreover, overexpression of the N-terminal SH2 domain of SHP-2, which acts as dominant negative regulator that binds to phosphorylated gp130 and blocks native SHP-2 phosphorylation, can enhance the CNTF-mediated up-regulation of neuropeptides associated with cholinergic differentiation of sympathetic neurons (29).
The role of SHP-2 as a negative regulator of ObR and gp130 signaling is in contrast to its role as a positive regulator in several other types of receptor systems (20–22), including a report that SHP-2 is a mediator of ERK activation for gp130 itself (23). SHP-2, however, is not exclusively a positive regulator of signaling, as it associates with CTLA-4 and functions to down-regulate T cell receptor signaling (30). It appears that for ObR and gp130, SHP-2 plays a role more like its relative, SHP-1. SHP-1 generally functions as a negative regulator of receptor signaling in hematopoietic cells, where it is expressed. Recruitment of SHP-1 to the activated erythropoietin receptor causes inactivation of receptor-bound Jak2 and dampens proliferation (31). SHP-1 has been shown to similarly suppress signaling from the FcγRIIB1 receptor (32). Thus, SHP-2 appears to play a dual regulatory role for gp130, and possibly for ObR, in that it is required to mediate activation of ERK (and mitogenesis in a BAF cell), but functions as a negative regulator to suppress STAT3-mediated gene induction. The mechanism by which SHP-2 acts to down-regulate STAT3-mediated signaling is unclear but may differ from that observed for SHP-1. Overall levels of tyrosine phosphorylation of STAT3 and receptor-associated Jak appear equal (Fig. 5) in native and mutant ObRY986F that cannot recruit and activate SHP-2. In addition, there was also no difference in the duration of tyrosine phosphorylation of the receptor, STAT3, or receptor-associated Jak between native and mutant ObRY986F (Fig. 5). Therefore, in contrast to the role of SHP-1 in erythropoietin receptor signaling, SHP-2 may signal through a downstream effector, perhaps lying in the pathway leading to ERK activation, that can influence STAT3-mediated gene transcription. Alternatively, it is also possible that SHP-2 signaling involves the cytokine-inducible inhibitors of STAT signaling (33–35) to regulate levels of transcription upon ligand stimulation.
Activation of ObR and gp130 result in the recruitment and activation of common signaling molecules, most notably STAT3 (15–17) and SHP-2. Thus, for both receptor systems STAT3 appears to be critical for mediating the individual ligand’s biological actions. The receptor for CNTF, a neurocytokine, consists of the signal transducing components gp130 and LIFRβ, along with a specificity determining component CNTF receptor α (CNTFRα). Interestingly, functional CNTFRs are found in the hypothalamus, and administration of CNTF to humans mimics leptin’s ability to reduce food intake and cause weight loss (36). Activation of CNTFR by systemic administration of CNTF to obese mice that lack either functional leptin (ob/ob) or ObR (db/db) results in the reversal of their obesity phenotypes, including reduction of adiposity and hyperphagia (37). Both CNTF and leptin were shown to similarly activate STAT3 in a neuronal cell line and, moreover, mediate the induction of the STAT3 responsive tis-11 gene in the hypothalamus of ob/ob mice (37). The potent and similar effects of CNTF and leptin on the reduction of food intake and the increase in energy expenditure in mutant obese mice are most likely due to common signaling pathways, i.e., STAT3 activation, in the brain. Indeed, only STAT3 was activated upon leptin stimulation of hypothalamic ObR in vivo (17).
An exciting, but speculative, implication of the findings presented in this report is that specifically blocking the interaction of SHP-2 with ObR may enhance leptin’s STAT3-mediated effects. Enhancing leptin-induced gene responses could possibly overcome leptin resistance by increasing the effectiveness of the endogenous leptin signal, thereby boosting leptin’s weight-reducing effects in obese individuals. This possibility is analogous to one existing naturally for another cytokine: humans that express a truncated mutant of erythropoietin receptor that lacks the tyrosine-based motif that recruits SHP-1 display enhanced erythropoietin action and increased levels of red blood cells that result from the failure to down-regulate the activated receptor (38).
Acknowledgments
We thank Len Schleifer and our other colleagues at Regeneron, as well as Jim Inglese, for their interest and support. We are also grateful to R. Rossman, D. Hintz, and T. Chaiprasert for cell culture; E. Burrows and C. Murphy for graphics expertise; and R. Flowers for manuscript assistance.
ABBREVIATIONS
- ObR
Ob receptor
- SHP
SH2-containing phosphatase 2
- STAT
signal transducer and activator of transcription
- LIFRβ
leukemia inhibitory factor receptor
- CNTF
ciliary neurotrophic factor
- Jak
Janus kinase
- SH2
src homology 2 domain
- ERK
extracellular signal-regulated kinase
- NT-3
neurotrophin 3
- GST
glutathione S-transferase
- Trx
thioredoxin
- IFN-γRα
interferon γ receptor α
References
- 1.Leibel R L, Chung W K, Chua S C J. J Biol Chem. 1997;272:31937–31940. doi: 10.1074/jbc.272.51.31937. [DOI] [PubMed] [Google Scholar]
- 2.Considine R V, Considine E L, Williams C J, Nyce M R, Magosin S A, Bauer T L, Rosato E L, Colberg J, Caro J F. J Clin Invest. 1995;95:2986–2988. doi: 10.1172/JCI118007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Considine R V, Considine E L, Williams C J, Hyde T M, Caro J F. Diabetes. 1996;19:992–994. doi: 10.2337/diab.45.7.992. [DOI] [PubMed] [Google Scholar]
- 4.Pelleymounter M A, Cullen M J, Baker M B, Hecht R, Winters D, Boone T, Collins F. Science. 1995;269:540–543. doi: 10.1126/science.7624776. [DOI] [PubMed] [Google Scholar]
- 5.Halaas J L, Gajiwala K S, Maffei M, Cohen S L, Chait B T, Rabinowitz D, Lallone R L, Burley S K, Friedman J M. Science. 1995;269:543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- 6.Campfield L A, Smith F J, Guisez Y, Devos R, Burn P. Science. 1995;269:546–549. doi: 10.1126/science.7624778. [DOI] [PubMed] [Google Scholar]
- 7.Maffei M, Halaas J, Ravussin E, Pratley R E, Lee G H, Zhang Y, Fei H, Kim S, Lallone R, Ranganathan S, et al. Nat Med. 1995;1:1155–1161. doi: 10.1038/nm1195-1155. [DOI] [PubMed] [Google Scholar]
- 8.Considine R V, Sinha M K, Heiman M L, Kriauciunas A, Stephens T W, Nyce M R, Ohannesian J P, Marco C C, McKee L J, Bauer T L, Caro J F. N Engl J Med. 1996;334:292–295. doi: 10.1056/NEJM199602013340503. [DOI] [PubMed] [Google Scholar]
- 9.Caro J F, Kolaczynski J W, Nyce M R, Ohannesian J P, Opentanova I, Goldman W H, Lynn R B, Zhang P-L, Sinha M K, Considine R V. Lancet. 1996;348:159–161. doi: 10.1016/s0140-6736(96)03173-x. [DOI] [PubMed] [Google Scholar]
- 10.Tartaglia L A, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards G J, Campfield L A, Clark F T, Deeds J, et al. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 11.Stahl N, Yancopoulos G D. Growth Factors and Cytokines in Health and Disease. 1997;2B:777–809. [Google Scholar]
- 12.Stahl N, Farruggella T J, Boulton T G, Zhong Z, Darnell J E, Yancopoulos G D. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 13.Lee G-H, Proenca R, Montez J M, Carroll K M, Darvishzadeh J G, Lee J I, Friedman J M. Nature (London) 1996;379:632–635. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Charlat O, Tartaglia L A, Woolf E A, Weng X, Ellis S J, Lakey N D, Culpepper J, Moore K J, Breitbart R E, et al. Cell. 1996;84:491–495. doi: 10.1016/s0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 15.Ghilardi N, Ziegler S, Wiestner A, Stoffel R, Heim M H. Proc Natl Acad Sci USA. 1996;93:6231–6235. doi: 10.1073/pnas.93.13.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumann H, Morell K K, White D W, Dembski M, Bailon P S, Kim H, Lai C-F, Tartaglia L A. Proc Natl Acad Sci USA. 1996;93:8374–8378. doi: 10.1073/pnas.93.16.8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaisse C, Halaas J L, Horvath C M, Darnell J E, Stoffel M, Friedman J M. Nat Genet. 1996;14:95–97. doi: 10.1038/ng0996-95. [DOI] [PubMed] [Google Scholar]
- 18.White D W, Kuropatwinski K K, Devos R, Baumann H, Tartaglia L A. J Biol Chem. 1997;272:4065–4071. doi: 10.1074/jbc.272.7.4065. [DOI] [PubMed] [Google Scholar]
- 19.Shimabukuro M, Koyama K, Chen G, Wang M Y, Trieu F, Lee Y, Newgard C B, Unger R H. Proc Natl Acad Sci USA. 1997;94:4637–4641. doi: 10.1073/pnas.94.9.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali S, Chen Z, Lebrun J-J, Vogel W, Kharitonenkov A, Kelly P A, Ullrich A. EMBO J. 1996;15:135–142. [PMC free article] [PubMed] [Google Scholar]
- 21.Lechleider R J, Freeman R M, Neel B G. J Biol Chem. 1993;268:13434–13438. [PubMed] [Google Scholar]
- 22.Li W, Nishimura R, Kashishian A, Batzer A G, Kim W J, Cooper J A, Schlessinger J. Mol Cell Biol. 1994;14:509–517. doi: 10.1128/mcb.14.1.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukuda T, Hibi M, Yamanaka Y, Takahasi-Tezuka M, Fujitani Y, Tamaguchi T, Nakajima K, Hirano T. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 24.Symes A, Lewis S, Corpus L, Rajan P, Hyman S E, Fink J S. Mol Endocrinol. 1994;8:1750–1763. doi: 10.1210/mend.8.12.7708062. [DOI] [PubMed] [Google Scholar]
- 25.Stahl N, Boulton T G, Farruggella T, Ip N Y, Davis S, Witthuhn B, Quelle F W, Silvennoinen O, Barbieri G, Pellegrini S, et al. Science. 1994;263:92–95. doi: 10.1126/science.8272873. [DOI] [PubMed] [Google Scholar]
- 26.Greenlund A C, Farrar M A, Viviano B L, Schreiber R D. EMBO J. 1994;13:1591–1600. doi: 10.1002/j.1460-2075.1994.tb06422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, et al. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 28.Symes A, Stahl N, Reeves S A, Farruggella T, Servidei T, Gearan T, Yancopoulos G D, Fink J S. Curr Biol. 1997;7:697–700. doi: 10.1016/s0960-9822(06)00298-3. [DOI] [PubMed] [Google Scholar]
- 29.Servidei T, Aoki Y, Lewis S E, Symes A, Fink J S, Reeves S A. J Biol Chem. 1998;273:6233–6241. doi: 10.1074/jbc.273.11.6233. [DOI] [PubMed] [Google Scholar]
- 30.Marengere L E M, Waterhouse P, Duncan G S, Mittrucker H-W, Feng G-S, Mak T W. Science. 1996;272:1170–1173. doi: 10.1126/science.272.5265.1170. [DOI] [PubMed] [Google Scholar]
- 31.Klingmuller U, Lorenz U, Cantley L C, Neel B G, Lodish H F. Cell. 1995;80:729–738. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- 32.D’Ambrosio D, Hippen K L, Minskoff S A, Mellman I, Pani G, Siminovitch K A, Cambier J C. Science. 1995;268:293–297. doi: 10.1126/science.7716523. [DOI] [PubMed] [Google Scholar]
- 33.Endo T A, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, et al. Nature (London) 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 34.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, et al. Nature (London) 1997;387:924–928. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 35.Starr R, Willson T A, Viney E M, Murray L J L, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, Hilton D J. Nature (London) 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 36.ALS CNTF Treatment Study Group. Neurology. 1996;46:1244–1249. doi: 10.1212/wnl.46.5.1244. [DOI] [PubMed] [Google Scholar]
- 37.Gloagen I, Costa P, Demartis A, Lazzaro D, Di Marco A, Graziani R, Paonessa G, Chen F, Rosenblum C I, Van der Ploeg L H T, et al. Proc Natl Acad Sci USA. 1997;94:6456–6461. doi: 10.1073/pnas.94.12.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de la Chapelle A, Traskelin A-L, Juvonen E. Proc Natl Acad Sci USA. 1993;90:4495–4499. doi: 10.1073/pnas.90.10.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]