Abstract
The fission yeast Schizosaccharomyces pombe is well-suited for studying meiotic recombination. Methods are described here for culturing S. pombe and for genetic assays of intragenic recombination (gene conversion), intergenic recombination (crossing-over), and spore viability. Both random spore and tetrad analyses are described.
Keywords: fission yeast, Schizosaccharomyces pombe, meiosis, intragenic recombination (gene conversion), intergenic recombination (crossing-over), spore viability, tetrad analysis
1. Introduction
Genetic analysis of meiotic recombination is especially facile in the fission yeast Schizosaccharomyces pombe for several reasons. Many independent meioses (>108) can be easily analyzed in one experiment. Having only three chromosomes, S. pombe produces many viable meiotic products, called spores, even if the cells are recombination-deficient: strains completely deficient for the early steps of meiotic recombination (DNA double-strand break formation) produce about 10 – 20% as many viable spores as wild type (e.g., 1). This feature has aided the analysis of mutations in more than 50 genes with documented effects on meiotic recombination. These and other studies have led to the formulation of a pathway of meiotic recombination for S. pombe(2). The commonly used strains are isogenic, which facilitates exchange of alleles and comparison of results from different labs. Over 1600 of the ~5000 genes have been experimentally characterized, which allows crosses between markers covering nearly all of the genome to be analyzed genetically. The genome sequence is essentially complete (3) (http://www.sanger.ac.uk/Projects/S_pombe/), which greatly facilitates physical analysis of DNA intermediates as described in the accompanying article (Hyppa and Smith, this book). Cytological analysis of meiotic nuclei, chromosomes, and proteins is described in other articles in this book (Loidl, Hiraoka, this book). This article describes the genetic analysis of meiosis and recombination. Additional genetic methods for S. pombe are described in refs. (4, 5) and on the website of Susan Forsburg at the University of Southern California (http://www-rcf.usc.edu/~forsburg/).
1.1. Life cycle of S. pombe
S. pombe typically grows as haploid cells, which express one of two mating types, called plus and minus; only cells of opposite mating type can fuse to form diploids and proceed into meiosis. Homothallic (h90) cells switch between plus and minus approximately once per cell division, by copying information from the unexpressed mat2 (plus) or mat3 (minus) locus into the expressed mat1 locus (5). Thus, cultures of an h90 strain contain cells of both mating types, which can mate and produce spores at high frequency; on appropriate media about 90% of the cells mate and complete meiosis. The mat loci are closely linked, and aberrant rearrangements, such as deletions and fusions of mat2 and mat3, can produce heterothallic cells that are stably plus (denoted with the genotype h+) or stably minus (h−). h+ and h− strains can mate with each other at high frequency, but strains of the same type do so about 10−4 as frequently.
Unlike the budding yeast Saccharomyces cerevisiae, S. pombe cells mate only when starved for nitrogen. This condition is also needed for entry into meiosis. Thus, when cells of opposite mating type are placed on starvation (sporulation) medium, they mate and undergo meiosis with no intervening mitotic divisions. If mated cells are returned to growth medium before the onset of meiosis, the diploid cells can be propagated indefinitely. Diploids are usually selected by use of closely linked, complementing markers, such as those conferring auxotrophy, and maintained by growth with selection, as diploids give rise to more stable haploids at about 10−4 per cell division.
Three kinds of meiosis are recognized. Zygotic meiosis results from cells mating and entering meiosis without intervening mitotic division, and azygotic meiosis results from established diploids entering meiosis. The corresponding asci have different shapes (Fig. 1), but otherwise meiosis is, to my knowledge, largely the same in the two types (see Note 1). In addition, the temperature-sensitive mutant pat1-114 initiates meiosis, even from the haploid state, when the temperature is raised (6). This mutant is widely used to induce nearly synchronous meiosis, which is critical for physical analysis of DNA intermediates during meiosis (see accompanying chapter by Hyppa and Smith). Chromosome segregation is slightly deficient during the first meiotic division in pat1-114 mutants; this deficiency is partially suppressed by ectopic expression of genes from both mat loci (7). Otherwise, meiosis seems similar in wild-type and pat1-114 meiosis.
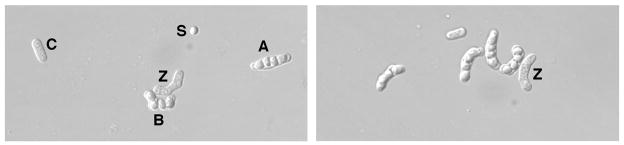
Fig. 1.
Asci from h90 meiosis.
Cells of an h90 strain were washed and spotted on a supplemented SPA plate, which was incubated for 3 d at 25°. A sample of the cell-ascus mixture was observed under a differential interference contrast microscope and photographed. (Left) The linear ascus (A) has the appearance of an azygotic ascus, whereas the curved ascus (B) has that of a zygotic ascus. A zygote (Z), an unmated cell (C), and a spore (S) from spontaneous lysis of an ascus are also visible in this rare field. (Right) A more typical field of asci, one of which (Z) is immature.
2. Materials
2.1. Culture media (see Note 2)
YEL (yeast extract liquid): 5 g of yeast extract (Difco 212750), 30 g of glucose. Make to 1 liter with water and autoclave. The corresponding solid medium containing 2% (w/v) agar is called YEA (yeast extract agar).
EMM2 (Edinburgh minimal medium 2): 50 mL of 20× EMM2 salts, 25 mL of 20% (w/v) NH4Cl, 25 mL of 0.40 M Na2HPO4, 25 mL of 40% (w/v) glucose, 1 mL of 1000× vitamins, 0.1 mL of 10,000× trace elements. Make to 1 liter with water.
20× EMM2 salts: 30.6 g of potassium phthalate (monobasic), 10 g of KCl, 5.0 g of MgCl2, 100 mg of Na2SO4, 100 mg of CaCl2. Make to 500 mL with water and autoclave.
1000× vitamins: 1.0 mg of biotin, 10 mg of calcium pantothenate, 1.0 g of nicotinic acid, 1.0 g of myoinositol. Make to 100 mL with water and autoclave.
10,000× trace elements: 0.50 g of H3BO3, 0.40 g of MnSO4, 0.40 g of ZnSO4·7H2O, 0.20 g of FeCl3·6H2O, 0.15 g of Na2MoO4, 0.10 g of KI, 0.040 g of CuSO4·5H2O, 1.0 g of citric acid. Make to 100 mL with water and filter sterilize.
NBL (nitrogen base liquid): 50 mL of 20× NB + Am2SO4, 25 mL of 40% (w/v) glucose, water to 1 liter. The corresponding solid medium is called NBA (nitrogen base agar).
20× NB + Am2SO4: 17 g of yeast nitrogen base without amino acids or ammonium sulfate (Difco 233520), 50 g of (NH4)2SO4. Make to 500 mL with water and filter sterilize. Store at 4°C.
SPA (sporulation agar): 25 mL of 40% (w/v) glucose, 10 mL of 10% (w/v) KH2PO4, 1 mL of 1000× vitamins. Add to 750 mL of 2.7% (w/v) agar (Difco 214530; autoclaved and tempered). Make to 1 liter with water.
MEA (malt extract agar): 30 g of malt extract (Difco 218630), 20 g of agar (Difco 214530). Make to 1 liter with water and autoclave.
YEA + G (see Note 3): To 1 L of molten YEA, add 4.0 mL of guanine-HCl solution (1.0 g of guanine-HCl plus 17.5 mL of 1.0 N NaOH in 50 mL total) and then 1.2 ml of 1.0 N HCl. pH should be that of YEA (5.5).
2.2. Spore preparation and analysis
Sterile distilled water.
Sterile flat toothpicks.
Glusulase (New England Nuclear, NEE-154).
60% (v/v) ethanol in water.
Sterile 1.5 mL microcentrifuge tubes.
Sterile velveteen replicating cloths, ~13 cm x 13 cm (Fig. 2).
70% (v/v) acetone in water.
Glass Petri dish cover with inner plastic ring and Whatman 3MM (or equivalent) filter paper inserts; replicating block (Fig. 2).
Iodine crystals.
Microscope with micromanipulator, such as Singer model MSM 300 or Leitz model MTR-27.
Microscope and counting chamber (hemacytometer).
Fig. 2.
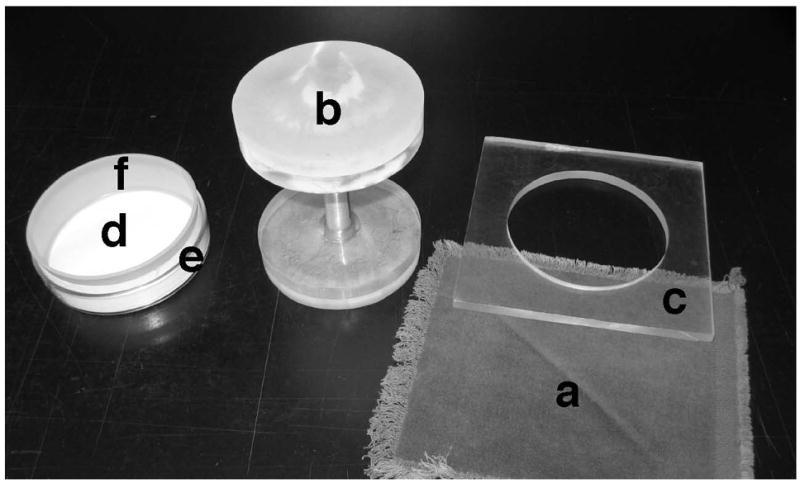
Assembly of Petri dish with acetone-soaked paper for killing vegetative cells.
A replicating velveteen (a) is stretched over the elevated plastic block (b) and held in place with the clear plastic ring (c). A master plate with colonies is replicated onto the velveteen. Acetone is pipetted onto the filter paper (d) held in the glass Petri dish (e) with an internal plastic ring (f). This assembly is then inverted over the velveteen. After exposure to the acetone vapors, the cell-ascus mixtures on the velveteen are replicated onto a fresh YEA plate.
3. Methods
3.1. Analysis of meiosis and viable spore yield on plates
This method is useful to identify rare heterozygous diploid spore colonies that could be confused with recombinants (Subheading 3.3.4). It is also useful for screening colonies from heterothallic strains to identify homothallic variants, or vice versa, and for screening homothallic colonies for mutants that produce few viable spores, as expected for mutants that make DSBs but do not repair them (“late” Rec mutants).
Use toothpicks to place isolates to be tested on a grid on YEA (Fig. 3). Incubate 1–2 d at 32°C (see Note 4). Replicate sequentially to supplemented SPA and to supplemented EMM2. Randomly placed colonies on SPA or EMM2 can also be used.
Incubate the SPA plate at 25°C for 2 d (see Note 5). Pour iodine crystals, about 25 g, into a large glass Petri dish, and invert the SPA plate over the crystals to expose the cell-ascus patches to iodine vapors for about 5–10 min. The ascus-spore mixture stains dark brown to black. Patches of cells that do not mate or that do not enter meiosis stain yellow.
Incubate the EMM2 plate at 25°C for 2 d (see Note 5). Transfer the cell-ascus mixture from the EMM2 plate onto velveteen (as for ordinary replica plating). Pipette 2.5 mL of 70% acetone onto the filter paper in the glass Petri dish assembly (Fig. 2), to thoroughly saturate the paper (see Note 6). Immediately place the glass Petri dish assembly over the velveteen, taking care that the velveteen does not touch the acetone-soaked paper. After 15 min at room temperature (about 22°C), remove the glass Petri dish assembly. Replicate the cell-ascus mixture onto YEA and incubate at 32°C. Isolates that produce viable spores will yield confluent growth on this YEA plate after 1–2 d. Isolates that do not mate, that do not complete meiosis, or that make inviable spores yield few or no colonies on this YEA plate.
Fig. 3.
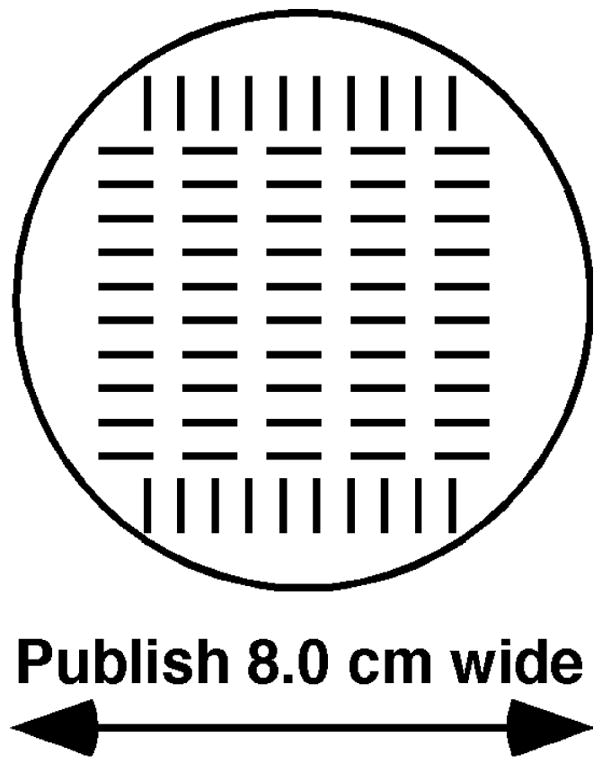
Grid (full size) for picking colonies for replication to test media.
A fresh YEA plate is centered over this grid. Cells are transferred from colonies onto the YEA plate, one over each grid mark. After incubation, this plate is replicated to test media.
3.2. Growing cells and conducting crosses
Streak cells of each parent separately on YEA and incubate at 32°C. After 2–3 d, pick a colony to 5 mL of YEL (see Note 7) and incubate on a roller drum at 32°C until saturated (overnight for healthy strains). Mix 0.05 mL of each parent in a 1.5 mL microcentrifuge tube. (For h90 × heterothallic crosses, use 0.05 mL of the h90 parent and 0.5 mL of the heterothallic parent.) Centrifuge 13,000 rpm (16,000g) for 10 s. Pour off or aspirate off supernatant. Wash cells twice in 1 mL of water. Suspend cells in the trace of water (about 20 μL) left in the tube. Deposit on a sporulation plate (supplemented SPA, EMM2, or MEA; see Note 8), making a spot about 5–7 mm in diameter. Incubate at 25°C (see Note 9). Sporulation is usually complete after 2 d.
If quantitation of the viable spore yield is desired, determine the number of viable cells per mL of saturated culture by diluting and plating on YEA (see Note 10). Incubate at 32°C for 2–4 d. Count the number of colonies and, with the dilution factors, calculate the concentration of viable cells per mL of saturated culture. Use this result in Subheading 3.3.2.
With a sterile toothpick remove a small amount of mating mixture from the sporulation plate, and examine it in a water mount under a phase contrast microscope. See Fig. 1 for examples of zygotic and azygotic asci and cells. If asci are not abundant, continue to incubate.
3.3. Analysis of random spores
With a sterile toothpick remove the entire cell-ascus mixture from each spot on the sporulation plate (Subheading 3.2). Suspend in 0.5 mL of glusulase (1:200 in water) in a 1.5 mL microcentrifuge tube. Vortex gently but thoroughly to make a homogeneous suspension of the cell-ascus mixture. Incubate at 32°C overnight. (See Note 11) Alternatively, use glusulase at 1:100 for 3–6 h, with occasional mixing. Add 0.5 mL of 60% ethanol, and incubate at room temperature (~22°C) for 10 min. Immediately centrifuge at 13,000 rpm (16,000g) for 10 s. Wash twice in 1 mL of water. Suspend in 1 mL of water and store at 4°C. This suspension is stable for many months.
3.3.1. Analysis of spore viability
Determine the concentration of visible spores in the suspension using a counting chamber (hemacytometer) under a phase contrast microscope.
Plate appropriate dilutions of the spore suspension on YEA (see Note 10) and incubate at 32°C for 3–4 d. Count the number of colonies and, with the dilution factors, calculate the concentration of viable spores per mL of spore suspension. Divide the concentration of viable spores (colony counts) by the concentration of visible spores (microscope counts) to obtain the fraction of spores that are viable.
As an alternative to steps 1 and 2, plate ~105 spores on a YEA plate and incubate at 32°C overnight. With a dissecting microscope determine, for a sample of the spores, the fraction of spores that germinate and produce a microcolony of at least 4 cells (see Note 12). Spore viability can also be determined from tetrad analysis (Subheading 3.4).
3.3.2. Analysis of viable spore yield
Divide the total number of viable spores in the spore suspension (Subheading 3.2) by the total number of cells (not the concentration) of the less-numerous parent put into the mating mixture. This result is the viable spores per viable cell. In theory, this is 2 for cells that, upon mating and meiosis, produce 4 viable spores per diploid cell (2 per one haploid cell). In practice, the number may be higher, if there is residual mitotic growth on the sporulation plate, or lower, if mating is inefficient. Thus, the number relative to a control, such as wild type, is most meaningful.
3.3.3. Intragenic (gene conversion) recombinant frequency (see Note 13)
Serially dilute spores appropriately in water (see Note 10) and plate on an appropriate medium for total viable spore determination and on a medium selective for the measured property. For example, plate on YEA for total viable spores and on YEA + G for Ade+ spores (see Note 3), or on EMM2 with uracil for total viable spores and on EMM2 without uracil for Ura+ spores (see Note 14).
Incubate plates at 32°C for 2–4 d, when colonies are usually large enough for all to be readily visible. Count colonies and, with the dilution factors, calculate the concentration of selected types per mL of spore suspension and the concentration of total viable spores per mL. Divide the concentration of the selected type by the concentration of total viable spores to obtain the recombinant frequency.
3.3.4. Intergenic (crossover) recombinant frequency (see Note 13)
Serially dilute the spore suspension and plate on YEA, as in Subheading 3.3.3, step 1. Incubate at a temperature that allows all genotypes to grow. Plate the parents or other appropriate strains as controls.
After 2–4 d, pick individual, well-isolated colonies to a gridded pattern (Fig. 3) on YEA (the “master plate”). Include each parent, or other appropriate strains, as controls. Incubate 1 or 2 d.
Using sterile velveteen, replicate from the master plate to plates that will allow a distinction of the two parental types for each marker to be scored. For example, replicate to EMM2 with and without adenine to score Ade+ and Ade−, or to YEA at 25°C and 37°C to score temperature-sensitive mutants (see Note 15). Incubate at the appropriate temperature(s) for 1–2 d. Also, replicate to SPA (see Note 16), incubate, and score for I2-reaction (Subheading 3.1) to identify diploids heterozygous for mating type; these diploids should be removed from the analysis if complementing diploids would have the same phenotype as recombinants.
Score each spore colony for each phenotype. This is conveniently done with spread sheet software, such as Excel, or by marking a scoring sheet, a paper with many rows (one for each colony) and columns (one for each marker). Count the number of recombinant types and divide by the number of colonies tested. To convert the resultant recombinant fraction into genetic distance, Use Haldane’s formula (13), x = −50 ln(1 – 2R), where x is the distance in centimorgans, and R = the recombinant fraction (a value between 0 and 0.5) (see Note 17).
3.4 Analysis of meiotic tetrads
Transfer a small amount of cell-ascus mixture (from a 2–3 day-old sporulation plate; Subheading 3.2, step 1) to 1 mL of water. Spread about 20 μL on the edge of a YEA plate (see Note 18). Under a microscope with a micromanipulator, place an isolated ascus (Fig. 1) at each of several isolated points on the plate. Mark the plate so it can be repositioned on the microscope stage. Incubate at 37°C for a few hours, or overnight at room temperature, to allow the ascal walls to dissolve (see Note 19).
With the micromanipulator, move each of the four spores from an ascus onto well-defined points separated by about 7 mm on the plate.
Incubate the plate for 2–4 d. Score the colony phenotypes by inspection (for example, for the red color of ade6 or ade7 mutants) or by transferring the colonies to a grid on YEA (the master plate) and, after incubation for 1–2 d, replicating to plates appropriate to score each phenotype (as in Subheading 3.3.4, step 3). Alternatively, replicate the dissection plate without transferring to a master plate (see Note 20).
Score each spore colony in each tetrad for each marker. Tetrads giving 1:3 segregation for a marker had a gene conversion for that marker. Single colonies with mixed phenotype indicate post-meiotic segregation (PMS; see Note 20). For each pair of linked markers, tetrads with two reciprocal recombinant types and two parental types (a tetratype tetrad) had one crossover (or an odd number of crossovers) between those markers. Tetrads with four reciprocal recombinant types (a non-parental ditype tetrad) had two crossovers (or an even number of crossovers) between the markers. Tetrads with all parental types had no or an even number of crossovers.
Calculate the frequency of gene conversion (or PMS) tetrads by dividing the number of tetrads with a conversion (or PMS) event by the total number of four-spore viable tetrads. Calculate the frequency of crossing-over between two linked markers using an appropriate formula that takes account of spores with two recombination events but unseen with the markers used. Perkins’s formula (14) is often used: x = 50 (T + 6N)/Σ, where x is the distance in cM, T is the number of tetratype tetrads, N = the number of non-parental ditype tetrads, and Σ = the total number of four-spore viable tetrads. This formula assumes that only single and double crossovers occur at significant frequency and that each N tetrad, which results from a four-strand (four-chromatid) double crossover event, is accompanied in the population on a statistical basis by two three-strand doubles, which are seen as tetratype tetrads, and one two-strand double, which is non-recombinant. Alternatively, one can treat the data as random spores and use the formula R = (2T + 4N)/Σ, where R is the fraction of spores that are recombinant. Then, one can use Haldane’s formula (13) x = −50 ln(1 - 2R), which assumes no crossover interference, as is the case for S. pombe (15).
4. Notes
In tht1 and tht2 mutants nuclear fusion (karyogamy) is much delayed. Consequently, the mutants have essentially no recombination in zygotic meiosis but normal recombination in azygotic meiosis (8,9).
Sterile distilled water is used throughout. To make solid medium for plates, use 2% agar (Difco 214530). For EMM2 agar and NBA it is convenient to make 2.7% agar in water; after melting, add concentrated components and make to proper volume with water (for example, 300 mL of 2.7% agar is made to 400 mL total). All media, including rich (YEL and YEA) media, are supplemented with all growth requirements (amino acids, purines, and pyrimidines) at 100 μg per mL, except as noted in Note 3.
Guanine inhibits the uptake of adenine, which is present in YEA at about 5 – 10 μg/mL. Thus, Ade− cells cannot grow on YEA + G. On YEA without added adenine, ade6 and ade7 mutants make red colonies; the color is enhanced after overnight incubation at 4°C. On YEA with added adenine (>20 μg/mL) Ade+ and Ade− cells make white colonies.
For convenience, our lab uses 32°C, but 30°C is more commonly used.
For reasons unclear SPA gives a stronger I2-reaction than EMM2, but EMM2 gives a better discrimination of viable spore yield. For the latter, 34°C is better for some situations, such as those with the temperature-sensitive rad50S mutation.
If more than one plate is to be treated, replace the filter paper and soak it with 70% acetone, as the acetone preferentially evaporates and killing would be inefficient if the paper were reused. The paper can, however, be dried and reused.
YEL liquid cultures can be kept in the refrigerator for about a month. In this case, dilute such a culture 1:50 into 5 mL of YEL and incubate overnight. Freshly saturated cultures mate more efficiently and give higher yields of asci than do old cultures.
An alternative method is to transfer with a toothpick a small amount of cells of each parent from a fresh colony or patch on a growth plate to about 20 μL of water on the sporulation plate and mix well. This less-quantitative method is suitable for making diploids or for constructing strains. Different laboratories use one or another sporulation medium (SPA, MEA, or EMM2), without obvious differences in results (but see Note 5). EMM2 with glutamate replacing NH4Cl is used by some investigators (4).
Meiosis is successful up to ~35°C, but the efficiency is lower. Recombinant frequencies differ less than a factor of ~2 at 35°C or 20°C compared to that at 25°C (10).
It is convenient to make 1:100 dilutions by pipetting 10 μL into 1.0 mL of water, or 0.05 mL into 5.0 mL, and to make 1:10 dilutions by pipetting 10 μL into 0.090 mL, or 0.1 mL into 0.90 mL. Plating 0.05, 0.1, or 0.2 mL of an appropriate dilution on each plate makes calculations easy.
Glusulase digests the ascal wall, to release spores, and digests vegetative cell walls, to kill most of the vegetative cells. At the indicated dose ethanol kills any remaining cells but leaves spores intact.
Some aneuploid spores can germinate and begin mitotic growth but soon cease, although chromosome III disomes make slow-growing colonies (11). A larger number of cell divisions, or longer incubation, may be needed for an appropriate distinction between viable and non-viable spores.
-
Most markers convert at <1% of chromatids; the mean for 31 markers is ~0.07% (P. Munz, pers. comm. in ref. 12). Thus, if markers are separated by more than a few cM, few of the recombinants arise from conversion; i.e., most of the events are reciprocal (crossovers). Conversely, in the few studies done, intragenic recombination is exclusively the result of conversion.
Note that selection methods will give genuine recombinant frequencies only if the markers are non-complementing. While this is generally true for markers in the same gene, intragenic complementation does occur, for example in the ade6 gene. Diploid spores arise from wild-type meiosis at about 10−3, and more frequently from meiosis of many mutants deficient in recombination or chromosome segregation. Selected colonies can be tested for their I2-reaction (Subheading 3.1), which will reveal some classes of diploids.
For all mitotic growth, EMM2 can be replaced with NBA, which is easier to prepare. Note that mating and meiosis occur frequently on EMM2 but infrequently on NBA and even less frequently on YEA.
Phloxin B (magdala red; 20 μg/mL) added to YEA can enhance scoring of temperature-sensitive mutants, since even a few dead cells in a patch of a temperature-sensitive mutant will stain with this dye and give a deeper red color than that for wild-type cells.
The SPA plate is not needed if a supplemented EMM2 plate that allows growth of all segregants is included, since it can be used for I2-staining.
Since S. pombe does not have crossover interference (15), Haldane’s formula is appropriate.
For this plate to have uniform thickness, it should be poured on a surface verified to be level.
After ascal wall dissolution the plate can be placed at 4°C if the spores cannot be immediately separated on the plate.
Sectored colonies, characteristic of post-meiotic segregation, may be more easily seen by direct replication (i.e., without transfer with toothpicks to a master plate. Unlike S. cerevisiae, however, S. pombe does not make half-sectored PMS colonies; rather, the two phenotypes are intermingled in streaks emanating from the center.
Acknowledgments
I am grateful to Sue Amundsen, Gareth Cromie, and Luther Davis for helpful comments on the manuscript. My laboratory is supported by research grants GM031693 and GM032194 from the National Institutes of Health.
References
- 1.Davis L, Smith GR. Non-random homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics. 2003;163:857–874. doi: 10.1093/genetics/163.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cromie GA, Smith GR. In: Genome Dynamics and Stability. Egel R, editor. Springer-Verlag; Berlin: in press . [Google Scholar]
- 3.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 4.Moreno S, Klar AJS, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 5.Egel R, editor. The molecular biology of Schizosaccharomyces pombe. Springer-Verlag; Berlin: 2004. [Google Scholar]
- 6.Iino Y, Yamamoto M. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol Gen Genet. 1985;198:416–421. doi: 10.1007/BF00332932. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto A, Hiraoka Y. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 2003;22:2284–2296. doi: 10.1093/emboj/cdg222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tange Y, Horio T, Shimanuki M, Ding DQ, Hiraoka Y, Niwa O. A novel fission yeast gene, tht1+, is required for the fusion of nuclear envelopes during karyogamy. J Cell Biol. 1998;140:247–258. doi: 10.1083/jcb.140.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martín-Castellanos C, Blanco M, Rozalén AE, Pérez-Hidalgo L, García AI, Conde F, Mata J, Ellermeier C, Davis L, San-Segundo P, Smith GR, Moreno S. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Genet. 2005;15:2056–2062. doi: 10.1016/j.cub.2005.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li YF, Smith GR. The Schizosaccharomyces pombe rec16 gene product regulates multiple meiotic events. Genetics. 1997;146:57–67. doi: 10.1093/genetics/146.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niwa O, Yanagida M. Triploid meiosis and aneuploidy in Schizosaccharomyces pombe: an unstable aneuploid disomic for chromosome III. Curr Genet. 1985;9:463–470. [Google Scholar]
- 12.Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
- 13.Haldane JBS. The combination of linkage values, and the calculation of distances between the loci of linked factors. J Genet. 1919;8:299–309. [Google Scholar]
- 14.Perkins DD. Biochemical mutants in the smut fungus Ustilago maydis. Genetics. 1949;34:607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]