Abstract
The fission yeast Schizosaccharomyces pombe has many biological characteristics that make it an ideal model organism for the study of meiosis. A nearly synchronous meiosis is one of the most important. Under certain environmental and genetic conditions, large cultures of S. pombe can be induced to undergo meiosis in a timely and predictable manner that allows for changes in the DNA to be observed and analyzed by gel electrophoresis. Initiation of meiotic recombination via programmed DNA double-strand breaks, the formation of joint molecule recombination intermediates, and the resolution of these intermediates into crossover DNA products can all be seen with consistent timing during the progression of a synchronous meiotic induction. The timing of recombination events, the genetic requirements for the formation and disappearance of recombination intermediates, and the analysis of the DNA structures of those intermediates allow a comparison of meiotic recombination in fission yeast with that in the only other species similarly studied, the budding yeast Saccharomyces cerevisiae.
Keywords: fission yeast, Schizosaccharomyces pombe, meiotic induction, DNA double-strand breaks (DSBs), joint molecules (JMs), Holliday junctions (HJs), intersister (IS), interhomolog (IH), crossover, pulsed-field gel electrophoresis (PFGE), two-dimensional (2D) gel electrophoresis
1. Introduction
Synchronous meiosis in S. pombe can be studied in two genetic backgrounds, with slightly different methods. (i) A diploid heterozygous both at the mating-type locus (h+/h-) and for complementing auxotrophic markers is cultured in minimal medium to mid-log phase and then starved of nitrogen. This starvation initiates meiosis in the cells (1), though not in a very synchronous manner. (ii) Greater synchrony can be achieved with a temperature-sensitive mutation (pat1-114) in the pat1 gene, which encodes a protein kinase that represses meiosis (2, 3). Even haploid pat1-114 cells initiate meiosis, form DSBs with the same timing and position, and proceed at least as far as the formation of joint molecules (JMs). S. pombe pat1-114 haploid or diploid cells (homozygous for h, to prevent starvation-induced meiosis) are grown to mid-log phase in minimal medium and then starved for nitrogen to arrest the cells in G1 of the cell cycle. The reintroduction of nitrogen concurrent with a shift to high temperature allows the arrested cells to progress through a highly synchronous meiosis. This enables the study of meiotic events in an entire population. Although the sites of DSBs are the same in pat1+ and pat1-114 (4), many of the meiotic events happen at such a low frequency that they cannot be visualized in a pat1+ (starvation-induced meiosis) background. Most studies have, therefore, been done with pat1-114 mutants.
Shortly after meiosis begins (at 2 - 3 h after induction) a round of DNA replication takes place (5). The branched DNA replication intermediates can be visualized at this time using two-dimensional (2D) gel electrophoresis, which separates DNA primarily by mass in the first dimension and shape in the second (6). After replication is complete (at 3 - 4 h), meiotic DNA double-strand breaks (DSBs) are introduced by the Rec12 protein, a Spo11 homolog that cuts the DNA and becomes covalently bound to the 5′ ends (7, 8). The DSBs are repaired and are no longer visible 5 - 6 h after meiosis begins (9). A mutation in the rad50 gene, called rad50S, prevents Rec12 from being removed from the DNA; hence, the DSBs cannot be repaired, and they accumulate (10). During the repair of the DSBs (at 4 - 5 h in rad50+ strains) a different set of branched DNA intermediates are formed - meiotic JMs called Holliday junctions (HJs). HJs are formed following the invasion of one homolog (or sister) by the other during DSB repair mediated by homologous recombination and are visualized using 2D gel electrophoresis and electron microscopy (11). Particularly useful for visualizing these intermediates are strains with a deletion of the HJ resolvase gene mus81, as accumulation of HJs in these mutants allows for easier analysis. Resolution of the HJs (at 5 - 6 h in mus81+ strains) leads to the formation of crossover DNA products.
We describe here the analysis, by gel electrophoresis and Southern blot hybridization, of each of these intermediates - DSBs, intersister and interhomolog JMs (HJs), and final crossover products.
2. Materials
2.1. S. pombe Cell Culture
Yeast extract liquid (YEL) medium: 5 g of yeast extract (Difco 212750), 30 g of glucose. Make to 1 liter with water and autoclave. For yeast extract agar (YEA) add 2% agar (Difco 214530) before autoclaving.
EMM2* (modified Edinburgh minimal medium 2): 50 mL of 20 × EMM2 salts, 25 mL of 20% (w/v) NH4Cl, 25 mL of 0.40 M Na2HPO4, 12.5 mL of 40% glucose (w/v), 1 mL of 1000 × vitamins, 0.1 mL of 10,000 × trace elements. Make to 1 liter with water and filter sterilize.
20 × EMM2 salts: 30.6 g of potassium phthalate (monobasic), 10 g of KCl, 5 g of MgCl2, 0.1 g of Na2SO4, 0.1 g of CaCl2. Make to 500 mL with water and autoclave.
1000 × vitamins: 1 mg of biotin, 10 mg of calcium pantothenate, 1 g of nicotinic acid, 1 g of myoinositol. Make to 100 mL with water and autoclave.
10,000 × trace elements: 0.5 g of H3BO3, 0.4 g of MnSO4, 0.4 g of ZnSO4·7H2O, 0.2 g of FeCl3·6H2O, 0.15 g of Na2MoO4, 0.1 g of KI, 0.04 g of CuSO4·5H2O, 1 g of citric acid. Make to 100 mL with water and filter sterilize.
Pombe minimal (PM) medium: as EMM2 but with 20 mL of 0.40 M Na2HPO4 and 50 mL of 40% glucose per liter.
2.2. Meiotic Induction and DNA Extraction
50 mM ethylenediamine tetraacetic acid (EDTA) pH 8.0, kept at 4°C during the induction.
Spheroplasting Buffer (CPES): 0.40 M EDTA pH 8.0, 120 mM Na2HPO4, 40 mM citric acid (free acid), 1.2 M sorbitol, 10 mM sodium azide. Store at room temperature.
Lysing Enzymes, from Trichoderma harzianum (Sigma #L1412).
Lyticase, from Arthrobacter luteus, crude (Sigma #L4025).
1.0 M DTT.
Lysis Buffer: 0.5 M EDTA pH 8.0, 10 mM Tris-HCl pH 7.5, 10 mM sodium azide, 1% N-Lauroylsarcosine sodium salt.
Proteinase K (Invitrogen #25530-015): 20 mg/mL in 10 mM Tris-HCl pH 7.5, 20 mM CaCl2, 50% glycerol. Store at -20°C.
2% Low Melting Point agarose (BioRad #161-3111) in 0.25 M EDTA pH 8.0, 1.2 M sorbitol.
Plastic plug molds and ejectors (BioRad #170-3622).
TE: 10 mM Tris-HCl pH 7.5, 1 mM EDTA pH 8.0.
100 mM PMSF (phenylmethylsulfonyl fluoride, Sigma #P7626). Store at -20°C.
5 × psoralen stock: 0.5 mg/mL Trioxalen (4,5′,8 Trimethylpsoralen, Sigma #T6137) in 100% ethanol, stored at 4°C and wrapped in foil.
Working psoralen solution: 1 × psoralen stock, 50 mM Tris-HCl pH 8.0, 50 mM EDTA pH 8.0.
50 mM trisodium citrate.
Propidium iodide: 4 mg/mL in 50 mM trisodium citrate.
70% ethanol.
1.5 mL microcentrifuge tubes.
2.3. Gel Electrophoresis of Recombination Intermediates
CHEF Mapper (BioRad) or other pulsed-field gel electrophoresis (PFGE) unit.
Chromosomal Grade Agarose (BioRad #161-3109).
PFGE gel casting platform, 21 × 14 cm (BioRad #170-3704).
PFGE markers (New England Biolabs #N3551S, N0350S, and N0340S).
Agarose (Sigma #A0169).
β-agarase (New England Biolabs #M0392L).
5 × TBE concentrated stock: 54 g of Tris base, 27.5 g of boric acid, 20 mL of 0.50 M EDTA pH 8.0. Make to one liter with water.
50 × TAE concentrated stock: 242 g of Tris base, 57.1 mL of glacial acetic acid, 100 mL of 0.50 M EDTA pH 8.0 in one liter of water.
Ethidium bromide (EtBr): 10 mg/mL. Store in metal foil-wrapped tube or bottle at 4°C.
6 × gel loading buffer: 0.25% bromophenol blue, 0.25% xylene cyanol, 30% glycerol in water
3.0 M sodium acetate pH 5.2.
100% ethanol.
70% ethanol.
UV box with camera.
2.4. Southern Blotting, Hybridization, and Analysis
Alkaline Transfer Solution: 1.5 M NaCl, 0.50 M NaOH.
0.25 N HCl.
UV Stratalinker 1800 (Stratagene) or equivalent.
Zeta-probe GT Genomic Tested blotting membrane (BioRad #162-0197) or equivalent.
20 × SSC: 3.0 M sodium chloride, 300 mM trisodium citrate pH 7.0.
1.0 M Tris-HCl pH 7.5.
Church Buffer: 7% SDS, 0.5 M NaH2PO4 pH 7 - 7.2, 1% BSA, 2 mM EDTA pH 8.0.
5 × labeling Buffer (Promega #U115A).
dATP, dGTP, dTTP mix (0.5 mM each).
[α-32P] dCTP, 3000 Ci/mmol.
10 × BSA: 1 mg/mL in water. Store at -20°C.
Klenow, DNA PolI Large Fragment (New England Biolabs #M0210S).
Microspin S-200 HR columns (GE Healthcare #27-5120-01).
Hybridization oven and bottles.
10% sodium dodecyl sulfate (SDS).
Plastic wrap (Saran or equivalent).
X-Ray film cassettes.
BioMax MS-Film (Kodak #829-4985).
Film processor.
Typhoon Trio variable mode imager (GE Healthcare) or equivalent.
Storage phosphor screens (GE Healthcare).
Image Quant software (GE Healthcare).
Microsoft Excel.
3. Methods
When inducing an S. pombe strain for meiosis, it is important to keep in mind the timing of events and the focus of the induction. This will dictate how much culture to induce, what time points to take, and what strains to use. Most meiotic events can be observed in a 0 - 6 h induction, time points being taken in hourly intervals. Half-hour time points can be taken for finer temporal analysis. This is useful when looking for replication and recombination intermediates (adding the 2.5 and 4.5 h points, respectively) or for the earliest sign of DSBs (adding the 3.5 h point). When mutants have delayed DSB initiation or repair, induction time points past 6 h should be taken. The induction is time-consuming, and it is important to have all tubes, plug molds, and reagents prepared and labeled ahead of time.
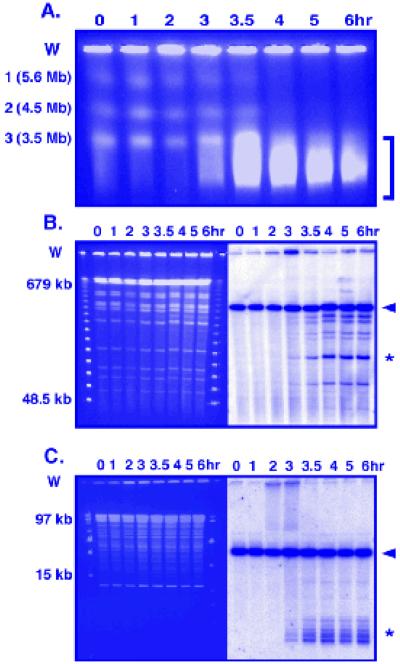
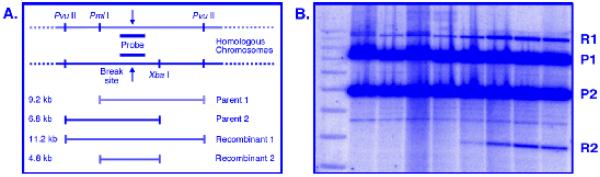
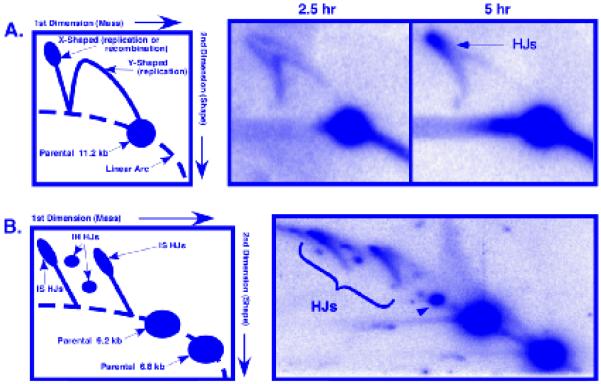
DNA from a meiotic induction can be analyzed in several ways for physical recombination intermediates. Meiotic DSBs can be visualized from DNA molecules in a large size-range. Pulsed-field gel electrophoresis (PFGE) reveals DSBs genome-wide on whole chromosomes (Fig. 1A), on DNA hundreds of kb long after cutting with rare restriction enzymes (Fig. 1B), and, with more frequently cutting enzymes, on DNA tens of kb long to reveal DSBs with higher resolution (Fig. 1C). Standard electrophoresis can be used to achieve separation of DNA up to about 20 kb long for fine scale mapping. The most intense meiotic DSB hotspots, those with more than a few percent DNA breakage, can be used to observe the formation of both recombination intermediates and products by the creation or destruction of restriction sites in heterozygous condition to form DNA with different strand lengths, as has been described for the DSB hotspot mbs1 (11) (Fig. 2A). Gel electrophoresis of meiotic DNA digested with the chosen restriction enzymes will separate the crossover DNA formed via meiotic recombination (Fig. 2B); isolation of the JM intermediates - interhomolog and intersister - requires electrophoresis in a second dimension (Fig. 3A and B).
Fig. 1.
Pulsed-field gel electrophoresis (PFGE) of S. pombe meiotic DNA to assay DSBs. A pat1-114 rad50S haploid strain was induced for meiosis (Section 3.1.1 and 3.2). (A) PFGE of whole chromosomes (labeled 1, 2, and 3) (Section 3.3.1.1), stained with EtBr (0.5 μg/mL). The bracket on the right highlights the broken chromosomal DNA. (B) PFGE of DNA from the same induction digested with NotI, stained with EtBr (left panel) (Section 3.3.1.2). The size markers are Lambda Ladder PFG markers (NEB). A Southern blot of the gel (right panel) was hybridized with a 1 kb probe specific to the left end of the 501 kb NotI fragment J (arrowhead). DNA fragments broken at the meiotic DSB hotspot mbs1 are indicated with an asterisk. (C) PFGE of DNA from the same induction digested with EagI, stained with EtBr (left panel) (Section 3.3.1.2). The size markers are Mid Range 1 (left) and Low Range PFG markers (right) (NEB). A Southern blot of the gel (right panel) was hybridized with a 1 kb probe specific to the left end of the 60.3 kb EagI fragment (arrowhead) of the NotI fragment J. DNA fragments broken at the meiotic DSB hotspot mbs1 are indicated with an asterisk. In each panel W indicates the wells into which the plugs with DNA were placed.
Fig. 2.
Standard gel electrophoresis of S. pombe meiotic DNA to assay crossover DNA. (A) The DNA construct to assay JMs and crossover DNA at mbs1. The PmlI and XbaI sites are heterozygous in the diploid strain and flank the DSB hotspot mbs1. Digestion at the flanking PvuII sites in combination with PmlI and XbaI digestion produces parental DNA 9.2 kb long (parent 1; P1) and 6.8 kb long (parent 2; P2). Recombination (meiotic crossing-over or conversion of one marker) produces DNA 11.2 kb long (recombinant 1; R1) and 4.8 kb long (recombinant 2; R2). (B) Southern blot of the gel described in Section 3.3.2, probed as depicted in Fig. 2A. Note that incomplete digestion produces a fragment identical to R1. Therefore, quantitation of recombination is 2(R2)/[P1+P2+2(R2)]. Leftmost lane contains a 1 kb ladder; the bottom marker is 4 kb. (Modified from Ref. (11).)
Fig 3.
2D gel electrophoresis of DNA recombination intermediates at mbs1. (A) The left cartoon is a representation of the migration of different DNA forms during 2D electrophoresis, digested with a single restriction enzyme (Section 3.3.3.1). The left image is a Southern blot of DNA from a pat1-114 mus81Δ diploid strain, which was meiotically induced for 2.5 h. The branched intermediates are replication structures. The right image is from the same induction at 5 h. The X-shaped spikes are HJs arising from meiotic recombination (indicated by arrow). Both time points were digested with PvuII and probed as in Fig. 2A. (B) The left cartoon depicts the migration of different DNA forms from a diploid strain with heterozygous flanking restriction markers (Fig. 2A) triply digested and run in a 2D gel. The right image is of DNA prepared at 4.5 h after induction of the strain in (A), digested with PvuII, PmlI, and XbaI, and assayed by 2D gel electrophoresis (Section 3.3.3.2). The IH HJs run as two species (due to their asymmetric structure) at a position between the two IS HJ spikes (highlighted by bracket). See Ref. (11) for further explanation. The black arrowhead points to a partial digestion fragment. (Modified from Ref. (11)).
3.1. S. pombe Cell Culture for Meiotic Induction
3.1.1. Conditions for pat1-114 Strains
Streak an S. pombe pat1-114 strain from a -80°C glycerol stock onto a YEA plate with appropriate supplements (100 μg/mL). Incubate the plate at 25°C.
When grown (about 4 - 5 d), pick an isolated colony to 5 mL of YEL with appropriate supplements (100 μg/mL; see Note 1) and incubate on a roller drum at 25°C until saturated (2 - 3 d).
Use the starter culture to inoculate (1:100) a 50 mL EMM2* culture with supplements (75 μg/mL). Grow at 25°C with moderate shaking until the OD600 is between 1.5 and 2.0 (2 - 3 d). Do not let the OD600 become much higher than this, as subsequent growth may be affected.
Dilute the EMM2* culture into 500 mL of EMM2* to an OD600 of 0.10 in a 2-liter flask. This needs to be done approximately 14 - 16 h before starvation is to occur (see Note 2). Incubate at 25°C with moderate shaking.
When the culture reaches an OD600 of 0.3 - 0.4, centrifuge the cells for 5 min at 4000 rpm (2800g) and wash once in sterile water. Resuspend the cells in 500 mL of EMM2* without NH4Cl and with supplements limited (10 μg/mL). Grow at 25°C with moderate shaking for 18 - 22 h.
The OD600 will have approximately doubled after starvation, to about 1 × 107 cells/mL. Add 12.5 mL of 20% NH4Cl and supplements to 75 μg/mL, and place the culture in a 34°C water bath with moderate shaking. This time marks the initiation of meiotic induction.
3.1.2. Conditions for pat1+ Strains
Streak an S. pombe pat1+/ pat1+ h+/h- diploid strain from a glycerol stock to an unsupplemented YEA plate (see Note 3) and incubate at 30°C.
When grown (3 - 4 d), pick an isolated colony to 10 mL of YEL (add guanine to 80 μg/mL if the diploid is maintained by ade6-M210/ade6-M216 complementation; only Ade+ cells can grow in the presence of guanine). Incubate on a roller drum at 30°C until nearly saturated (1 - 2 d).
Use the starter culture to inoculate (1:100) a 500 mL PM culture in a 2-liter flask (see Note 4) and incubate at 30°C with moderate shaking for 12 - 14 h.
When the OD600 has reached 0.5 - 0.7, centrifuge the cells for 5 min at 4000 rpm (2800g) and wash once in sterile water. Resuspend the cells in 500 mL of PM without NH4Cl and with glucose reduced to 1.0%. Place at 34°C and shake vigorously (see Note 5). This time marks the initiation of meiotic induction. The timing of a pat1+ induction is 3 - 4 h slower than that of pat1-114 (Fig. 1B), so allow for the extra time.
3.2. Meiotic Induction and DNA Extraction
Starting at 0 h - and at appropriate time points thereafter - centrifuge 30 mL of culture at 5000 rpm (3000g) at 4°C for 5 min. While this is spinning, put 1 mL of culture into a 1.5 mL microcentrifuge tube, spin the cells down at 13,000 rpm (16,000g) for 15 s, wash once in water, and resuspend in cold 70% ethanol. Store at -20°C until use for flow cytometric analysis (see Step 10 below). Wash the cell pellet from the 30 mL sample with 30 mL of cold 50 mM EDTA pH 8.0 and spin at 5000 rpm (3000g) at 4°C for 5 min. Decant and put on ice (see Note 6).
In the same tube, resuspend the cells in 300 μl of spheroplasting buffer (CPES) with DTT (5 mM), Lysing Enzymes (5 mg/mL), and Lyticase (1 mg/mL). Mix these components just prior to induction and store on ice.
Place the tube containing the cells in CPES in a 50°C water bath for 1 min. Quickly add 400 μl of 2% low melting point agarose (previously melted and kept at 50°C) and rapidly pipette up and down to mix thoroughly and quickly. Keep the tubes in the water bath while mixing to prevent the agarose from solidifying. Pipette the cell-agarose mixture into prepared plug molds and cool at 4°C for 3 - 5 min to solidify the agarose. This should make 8 - 10 plugs of 100 μl each.
Eject the solidified plugs into 1.2 mL of CPES (from Step 2) in a 1.5 mL microcentrifuge tube (making sure all plugs are submerged) and incubate at 37°C for 1 h 15 min, mixing periodically by inversion or by placing on a rotator.
Remove the CPES with a flame-polished Pasteur pipette carefully, so as not to tear the plugs. Add approximately 1.2 mL of Lysis Buffer with Proteinase K (1 mg/mL) until the plugs are completely submerged. Mix by inversion. Incubate plugs at 50°C overnight, inverting the tubes after 30 min to mix.
The next day, remove Lysis Buffer and add fresh Lysis Buffer with Proteinase K (1 mg/mL). Repeat the 50°C incubation overnight, inverting the tubes after 30 min to mix.
Remove the Lysis Buffer and add about 1.2 mL of TE with PMSF (1 mM) and incubate plugs at room temperature for 1 - 2 h, mixing periodically by inversion. The PMSF is needed to inactive any residual Proteinase K.
Remove the TE-PMSF and add TE until the plugs are covered completely. Mix on a rotator at room temperature for an hour. Repeat this step two more times.
After the final TE wash, store the plugs at 4°C for further use. The plugs and the DNA are stable for at least two years, though the freshest preparations are ideal.
Flow cytometry can be used to verify that replication, and therefore the initiation of meiosis, has occurred. Spin the cell samples stored in 70% ethanol from step 1 for 1 min at 13,000 rpm (16,000g), wash once in 1 mL of 50 mM trisodium citrate, spin again for 1 min, and resuspend the cells in 0.5 mL of 50 mM trisodium citrate. Add RNase A to 10 μg/mL and incubate at least 2 h at 37°C. Pipette cells into a 5 mL round bottom tube and add 0.5 mL of propidium iodide (4 μg/mL). Vortex and store at 4°C. Sonicate the cells at the lowest output setting with a microtip for 15 s to break any cell clumps just prior to flow cytometry.
3.3. Gel Electrophoresis of Recombination Intermediates
3.3.1. Pulsed-field Gel Electrophoresis for DSB Analysis
3.3.1.1. Whole Chromosomes
To perform DSB analysis on whole chromosomes, take an agarose plug with DNA from each time point of a meiotic induction and soak in 250 - 500 μl of 1 × TAE in a 1.5 mL microcentrifuge tube at room temperature for 1 h, mixing periodically to aid diffusion. Melt 0.8% Chromosomal Grade Agarose in 1 × TAE and cool to the touch (about 40°C). With the gel comb lying flat, place one plug on each comb tooth, in order from 0 - 6 h. S. pombe whole chromosome standards (BioRad #170-3633) can also be used if desired. Wick away any excess buffer from around the plugs, place the comb in the gel mold, and pour gel around comb and plugs, which should remain adhered to the comb. If a plug starts to pull away, use a spatula to replace it. Alternatively, the gel can be poured and solidified first, and the plugs then placed into the empty wells, making sure that the plug and gel are in complete contact.
Place the gel in a PFGE box in 2 L of cold 1 × TAE, and electrophorese for 48 h at 2V/cm, 100° angle, with a 30 min switch time (initial and final) at 14°C.
Stain the gel for 30 min with EtBr (0.5 μg/mL), destain in water for 15 min, and photograph under UV light (Fig. 1A).
3.3.1.2. 20 kb - 1 Mb DNA Fragments
The digestion of the chromosomes into DNA fragments allows one to visualize sites of DSBs. First, select a restriction enzyme that will give fragments of the desired size range. In order to prevent shearing of the DNA, the digestion is performed on agarose-embedded DNA. Use an agarose plug with DNA from each time point in the induction and soak in 250 μl of 1 × digestion buffer in a 1.5 mL microcentrifuge tube for 30 min at 4°C, mixing periodically to aid diffusion. Repeat this step once. Remove the buffer and add 250 μl of fresh 1 × buffer with 10 - 50 units of restriction enzyme (see Note 7). Incubate at 4°C for 4 - 6 h to allow the enzyme to diffuse into the plug (see Note 7), and then incubate at 37°C overnight.
The next day remove the buffer and soak the plugs in 0.5 × TBE at room temperature for 1 h. Melt 1.0% Chromosomal Grade Agarose in 0.5 × TBE and cool to the touch (about 40°C). Load the gel as above in Section 3.3.1.1, Step 1, and use appropriate sized PFGE markers.
Place the gel in a PFGE box in 2 L of cold 0.5 × TBE and electrophorese at 6 V/cm, 120° angle at 14°C. The length of run and the switch interval depends on the size of the restriction fragments; the longer each parameter is, the larger the fragments that can be resolved (consult BioRad with questions). If using a BioRad CHEF Mapper, use the Auto Algorithm function and enter the upper and lower limits of the desired separation.
Stain the gel for 30 min with EtBr (0.5 μg/mL), destain in water for 15 min, and photograph under UV light (Figs. 1B and 1C). Proceed to the Southern blotting procedure (Section 3.4) below.
3.3.2. Electrophoresis of Crossover DNA Products
In order to observe the physical (DNA) products of meiotic recombination, a proper arrangement of markers around a very intense DNA hotspot, one with approximately 10% DSBs or greater, is needed. Heterozygous changes that create or destroy restriction enzyme cleavage sites (see Note 8) have to be made in the DNA flanking the hotspot on each side. Additional homozygous restriction sites, one to each side, are also required. A diploid containing the heterozygous restriction sites is induced (see Note 1). Crossover DNA begins to appear at about 4 h and accumulates to its maximum at 6 h. The construct using the meiotic break site mbs1 is shown in Fig. 2A, which requires three restriction sites for analysis; use enzymes appropriate for the construct being analyzed. The restriction sites should be approximately, but not exactly, equally spaced and span about 2 - 15 kb; this will make the two parental fragments, and consequently the two intersister JMs, have different masses and hence allow them to separate upon gel electrophoresis.
Using plugs from an induction of a diploid described above, soak each plug in 250 μl of 1 × digestion buffer for 30 min at room temperature, mixing periodically to aid diffusion. Repeat this step. Remove buffer and melt the plugs at 65°C for 1 - 2 min, until the agarose is completely liquefied. Add 1 unit of β-agarase and 10 - 20 units of each of the restriction enzymes and digest at 37°C for 2 h. Concentrate the DNA to about 50 μl in a vacuum oven at 40° - 50°C (about 1 h).
Melt 0.8% agarose in 1 × TAE and pour a gel with wells 1 cm wide (see Note 9) and let it solidify. Add loading buffer directly to the DNA samples and load the gel with appropriate markers, radioactively labeled when possible. Run the gel at 1 V/cm at room temperature until desired separation is achieved (48 h for a 22 cm gel). Buffer circulation is highly recommended.
Stain the gel for 30 min with EtBr (0.5 μg/mL), destain in water for 15 min, and photograph under UV light. Proceed to the Southern blotting procedure (Section 3.4) below (see Fig. 2B for an example of a finished blot).
3.3.3. 2D Electrophoresis of Branched DNA Intermediates
3.3.3.1. Meiotic Time Course for JM Formation and Resolution
Select one or two restriction enzymes that generate a DNA fragment containing the DSB hotspot. The DNA fragment can be 5 - 15 kb long and should have the DSBs sufficiently far from the end of the fragment so that branch migration, and consequent loss, of the junction is not an issue. Using plugs from a meiotic induction (see Note 10), soak a plug from each time point in 250 μl of 1 × digestion buffer in a 1.5 mL microcentrifuge tube for 30 min at 4°C, mixing periodically to aid diffusion. Repeat this step. Remove the buffer and add 250 μl of fresh 1 × buffer with 10 - 50 units of enzyme (see Note 7). Incubate at 4°C for 4 - 6 h to allow the enzyme to diffuse into the plug (see Note 7), and then incubate at 37°C overnight.
The next day, remove the buffer and heat the plugs at 65°C for 1 - 2 min, until the agarose is completely liquefied. Add 1 unit of β-agarase and incubate at 37°C for 1 h. Add one-tenth volume of 3.0 M sodium acetate pH 5.2, put samples on ice for 15 min, and spin at 13,000 rpm (16000g) for 15 min to pellet any debris. From the supernatant, precipitate the DNA with ethanol. Resuspend the DNA pellet in 30 μl of TE and immediately load (see Note 11) into a 0.4% agarose gel (10 × 8.5 cm works well) in 1 × TAE with wells 0.5 cm wide, and electrophorese in 1 × TAE at 1 V/cm for 22 h at room temperature.
Stain the gel for 30 min with EtBr (0.5 μg/mL) and UV photograph. Carefully cut out each lane from the gel and place each along the top of a new gel mold, rotating the lane 90° counterclockwise so the DNA will run perpendicular from the gel slice during the second dimension of electrophoresis. Pour a 1.0% agarose gel in 1 × TAE with EtBr (0.5 μg/mL) around the gel slice and electrophorese in 1 × TAE at 4 V/cm for 14.5 h at 4°C (see Note 12).
Proceed to the Southern blotting procedure (Section 3.4) below (see Fig. 3A for an example of a finished blot).
3.3.3.2. Identifying Intersister and Interhomolog JMs
Heterozygous restriction sites (Section 3.3.2. Step 1 and Fig. 2A) are needed to observe the presence of intersister (IS) and interhomolog (IH) JMs via 2D gel electrophoresis. The two IS species must have different masses; the IH species will have a mass intermediate between those of the IS JMs. DNA from 4.5 or 5 h usually has the maximum JMs; this is therefore the best time point to use for this analysis (see Note 10). Take two or three plugs from 0 h (as a control) and 4.5 h, and digest with restriction enzymes and β-agarase. Precipitate the DNA with ethanol as described above in Section 3.3.3.1 steps 1 and 2, pooling the DNA from each time point upon precipitation.
Dissolve the DNA in TE and immediately load the DNA (see Note 12) into a 0.35% agarose gel in 1 × TBE with wells 0.5 cm wide. Electrophorese at 0.75 V/cm for 44 - 48 h at room temperature. It is very important to get good DNA separation, so conditions are altered slightly (12) from those in Section 3.3.3.1 Steps 2 and 3.
Stain the gel for 30 min with EtBr (0.3 μg/mL) and UV photograph. Carefully cut out each lane from the gel and place each in another gel mold at the top, rotating the lane 90° counterclockwise. Pour a 1.0% agarose gel in 1 × TBE with EtBr (0.3 μg/mL) around the gel slice and then electrophorese in 1 × TBE with EtBr (0.3 μg/mL) at 2 V/cm for 44 - 48 h at 4°C (see Note 13).
Proceed to the Southern blotting procedure (Section 3.4) below (see Fig. 3B for an example of a finished blot).
3.4. Southern Blot Hybridization and Analysis of Intermediates
The gel can be prepared for Southern blot transfer by either UV nicking (in a Stratagene UV Stratalinker, for example) at 60 mJ/cm2 (for large DNA from PFGE), or by depurinating in 0.25 N HCl for 15 min at room temperature with gentle shaking.
Soak the gel in Alkaline Transfer Solution at room temperature for 15 min with shaking and transfer DNA to BioRad Zeta-probe membrane (or another positively charged nylon membrane) via the standard alkaline Southern blotting method (13).
After blotting for 24 h remove the membrane and neutralize by soaking in 0.40 M Tris-HCl pH 7.5 at room temperature for 5 min with shaking, followed by a brief rinse in 2 × SSC. The DNA becomes bound to positively charged nylon membranes during alkaline transfer; so, it is not necessary to bake the blot.
Preheat Church Buffer to 65°C. Place the membrane in a hybridization bottle and prehybridize (soak) in 10 mL of Church buffer for 30 min at 65°C with rotation in a hybridization oven.
DNA probes for hybridization are generated by PCR and gel purified. For DSB blots, 1 kb probes at either the right end or the left end of the restriction fragment to be probed are desirable for easier mapping. For crossover and JM blots, the 1 kb probe should be specific to the central region of the DNA fragments to detect all of the relevant DNA species. Label the probes by random primer synthesis with the incorporation of [α-32P] dCTP by taking 25 ng of DNA template, adding water to 30 μl, boiling for 5 min, and cooling for 5 min on ice. Add 10 μl of 5 × Labeling Buffer, 2 μl of 0.5 mM dNTPs (minus dCTP), 2 μl of 10 × BSA, 5 units of Klenow Fragment of DNA polymerase I, and 5 μl of 0.01 mCi/μl [α-32P] dCTP (3000 Ci/mmol). Incubate at room temperature for 1 h, and then purify the probe by spinning through a Microspin S-200 HR column at 3000 rpm (800g), and measure the specific radioactivity with a scintillation counter.
Remove the prehybridization buffer from the bottle (see step 4 above) and add 10 mL of Church Buffer and the labeled probe to 106 cpm/mL of buffer. The labeled probe should be placed in a boiling water bath for 2 min to denature it just prior to adding it to the membrane.
Incubate the membrane at 65°C for 16 - 22 h with rotation in a hybridization oven.
Remove the hybridization solution and dispose of it according to the lab’s radioactive waste disposal policy. Briefly rinse the bottle with 2 × SSC, 0.1% SDS, and properly dispose. Wash the membrane as follows, each wash being for 15 min: twice in 2 × SSC, 0.1% SDS at 40°C; twice in 2 × SSC, 0.1% SDS at 52°C; and twice in 1 × SSC, 0.1% SDS at 65°C. A higher stringency wash of 0.1 × SSC, 0.1% SDS at 65°C can be done if needed.
Wrap the blot in plastic (Saran) wrap and place in a film cassette. Expose on BioMax MS film for 20 - 24 h at -80°C. Develop in a film processor (see Note 14).
A digital image can be made using a Typhoon Trio variable mode imager (or one similar). Expose the blot on a recently erased storage phosphor screen overnight at room temperature (see Note 15). Then place the screen in the imager and scan the appropriate area on the storage phosphor setting (under the “Acquisition Mode” when using the Typhoon) with with the desired pixel size. Quantitate the relative radioactive DNA signals from the gel image using Image Quant (or similar) software and then import the data points into an Excel spreadsheet.
The amount of DSBs at each site can be measured as ratio of the DSB signal to the total DNA signal for the entire lane, after an appropriate background has been subtracted for both. First, choose a time point at which DSBs have reached the maximum (3.5 or 4 h in rad50+; 5 or 6 h in rad50S) and a time point without DSBs (0 or 1 h, to establish the background) and subtract from all data points the lowest value present in either lane to establish the zero baseline. Next, sum the data points in each lane and adjust for different DNA contents (due to replication and differential recovery) by dividing the total counts in the early lane by the total counts in the late DSB lane to get a normalization ratio, and multiply every data point in the early lane by this ratio. Calculate the signal associated with the unbroken DNA fragment by subtracting the background established from the local minimum on each side of the peak signal. and then sum the values across the peak. For all data points below the unbroken fragment in the late DSB lane subtract the signal from the early lane to get the signal that is above background. Sum the values across each DSB peak [e.g. mbs1(*) in Fig. 1B and 1C] to calculate the associated signal. Divide the corrected signal from each individual band below the restriction fragment by the total DNA (the unbroken DNA plus all DSB signal above background) to calculate the DSB frequency at each site.
The amount of JMs for a time point (4.5 or 5 h is maximal) can be measured as the ratio of the JM signal (for each branched DNA species) to the total DNA (branched and linear), after subtracting the background locally for each DNA spot. Use an area that is the same size as the DNA spot but is in a “blank” region (no DNA) as the local background. Sum all distinct DNA species (Fig 3) for the total DNA, and divide each individual nonlinear JM species by this total to calculate JM frequency.
Acknowledgments
We are grateful to Sue Amundsen and Gareth Cromie for helpful comments on the manuscript. Our laboratory is supported by research grants GM031693 and GM032194 from the National Institutes of Health.
Footnotes
A completely prototrophic strain or a strain with only one auxotrophy is ideal. Strains with more than one auxotrophy can fail to induce completely or at all. Also, when inducing stable h+/h+ or h-/h- diploids selected and maintained by heterozygous auxotrophies, always grow in minimal medium to maintain diploidy. The complementing alleles ade6-M210 and ade6-M216 are useful for making such diploids.
This is for healthy strains. Mutant strains with growth defects (rad51, mus81, rad32, etc.) will need to grow an extra 4 - 6 h to reach the appropriate density. They will still, however, undergo a synchronous meiosis.
Whenever possible, maintain h+/h- diploids with ade6-M210/ade6-M216 complementation for easy selection and high stability. Mitotic recombination between these markers, which could produce a prototrophic haploid, is rare. Streak such a strain onto a YEA plate without added adenine and select a white colony (Ade+) to be sure that diploidy is maintained. Auxotrophic segregants make red colonies. It is also advantageous to keep any additional auxotrophic markers heterozygous to select for diploidy
This will vary depending on the growth of the strains. 1:100 is the appropriate dilution for a rad50S strain; for rad50+ strains, use 1:200.
The shift to 34°C is necessary only for strains with the rad50S mutation, since it is a temperature-sensitive mutation (14). It is not necessary for meiotic induction. Vigorous shaking of the cultures helps more of the cell population to proceed through meiosis. The use of baffled flasks to increase aeration is also recommended.
If psoralen cross-linking is needed for further manipulations, e.g., electron microscopy, do not wash the cells with 50 mM EDTA at this step. Instead, resuspend the cells in 1.5 mL of working psoralen solution (freshly prepared). Pipette the cell suspension into a plastic Petri dish and place on a long-wave UV box and irradiate with 360 nM UV light for 10 min (∼50 mJ/cm2), swirling the plate to mix the cells every few minutes. Pipette the cells back into the same tube and rinse the Petri dish once with 50 mM Tris-HCl pH 8.0, 50 mM EDTA pH 8.0 to collect the remainder of the cells (15). Centrifuge at 5000 rpm (3000g) at 4°C, wash once in 50 mM EDTA pH 8.0, decant and put on ice. Continue as above.
The amount of restriction enzyme needed for digestion of agarose-embedded DNA varies from enzyme to enzyme. Consult the manufacturer for this information. The 4°C preincubation keeps the enzyme stable during its diffusion into the agarose. A 4 - 6 h preincubation is sufficient for most enzymes.
Insertions and deletions have been used for similar constructions, but we prefer to use single bp mutations to minimize unexpected changes in DSB patterns and recombination. Such changes, which can be dramatic and affect events tens of kb from the DNA change, appear to result from unexplained changes in chromatin structure.
Wide gel wells prevent overcrowding of DNA samples and give sharper bands more readily quantitated after radioactive hybridization.
Analysis of HJs is greatly enhanced in a mus81 deletion strain, in which HJs accumulate. Without HJ accumulation it is more difficult (but not impossible) to recover enough DNA intermediates to get a strong signal after Southern blot hybridization, especially for IS and IH JMs.
Psoralen cross-linking is not necessary for preserving JMs as long as the cells and DNA remain embedded in agarose, in which branch migration of the junctions is inhibited. However, the agarose needs to be melted and treated with β-agarase for gel analysis; directly loading agarose plugs with DNA into a 2D gel gives poor resolution. After melting the agarose, Mg2+ ions inhibit branch migration of the junctions. Load the DNA as soon as it is dissolved in TE to limit the potential migration of the junctions in the absence of Mg2+.
If analysis by electron microscopy is to be performed, psoralen cross-linked DNA must be used. Digest the DNA and electrophorese in the first dimension as described. Keep the DNA ladder from this run. In the second dimension, use 1% low melting point agarose and electrophorese as described. Place the gel on a long-wave UV box and cut out an agarose slice, about 1 × 0.5 cm, starting just above the linear arc in the size range needed, using the first dimension DNA ladder as a guide. Extract the DNA by soaking the slice in 1 × β-agarase buffer, twice for 30 min at 4°C, and then remove the buffer and melt the agarose at 65°C. Precipitate the DNA with ethanol and resuspend in TE.
These separation conditions work best with a 22 cm gel. The electrophoresis can also be done in 1 × TAE buffer with higher voltage and shorter run times, but the separation will not be as good.
Exposure times will vary based on level of radioactivity. This film step can be skipped and a digital image can be created directly if preferred.
It is important not to expose the blot for too long. The signal can become saturated if overexposed (more than 105 counts per pixel), which will result in skewed quantitation data. 12 - 16 h is usually sufficient, depending on the level of radioactivity of the blot.
References
- 1.Bähler J, Wyler T, Loidl J, Kohli J. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J. Cell Biol. 1993;121:241–256. doi: 10.1083/jcb.121.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iino Y, Yamamoto M. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol. Gen. Genet. 1985;198:416–421. doi: 10.1007/BF00332932. [DOI] [PubMed] [Google Scholar]
- 3.Egel R, editor. The molecular biology of Schizosaccharomyces pombe. Springer; Berlin: 2004. [Google Scholar]
- 4.Cromie GA, Hyppa RW, Cam HP, Farah JA, Grewal SI, Smith GR. A discrete class of intergenic DNA dictates meiotic DSB hotspots in fission yeast. PLoS Genetics. 2007;3:e141. doi: 10.1371/journal.pgen.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beach D, Rodgers L, Gould J. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr. Genet. 1985;10:297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- 6.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 7.Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 8.Hyppa RW, Cromie GA, Smith GR. Indistinguishable landscapes of meiotic DNA breaks in rad50+ and rad50S strains of fission yeast revealed by a novel rad50+ recombination intermediate. PLoS Genetics. 2008;4:e10000267. doi: 10.1371/journal.pgen.1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- 10.Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
- 11.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krysan PJ, Calos MP. Replication initiates at multiple locations on an autonomously replicating plasmid in human cells. Mol. Cell. Biol. 1991;11:1464–1472. doi: 10.1128/mcb.11.3.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 14.Farah JA, Hartsuiker E, Mizuno K, Ohta K, Smith GR. A 160-bp palindrome is a Rad50-Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics. 2002;161:461–468. doi: 10.1093/genetics/161.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]