Abstract
A method based on microchip electrophoresis (MCE) with chemiluminescence (CL) detection was developed for the determination of ascorbic acid (AA) and amino acids including tryptophan (Trp), glycine (Gly) and alanine (Ala) present in single cells. Cell injection, loading, lysing, electrophoretic separation, and CL detection were integrated onto a simple cross microfluidic chip. A single cell was loaded in the cross intersection by electrophoretic means through applying a set of potentials at the reservoirs. The docked cell was lysed rapidly under a direct electric field. The intracellular contents were MCE separated within 130 s. CL detection was based on the enhancing effects of AA and amino acids on the CL reaction of luminol with K3[Fe(CN)6]. Rat hepatocytes were prepared and analyzed as the test cellular model. The average intracellular contents of AA, Trp, Gly and Ala in single rat hepatocytes were found to be 38.3 fmol, 5.15 fmol, 3.78 fmol and 3.84 fmol (n=12), respectively.
Keywords: Microchip electrophoresis, Chemiluminescence detection, Single cell analysis, Ascorbic acid, Tryptophan, Rat hepatocytes
1. Introduction
High throughput analysis of single cells is very important for molecular biology, physicochemical modelling of biological processes, and clinical diagnosis of disease. The reason for this significance is that seemingly identical cells are often quite heterogeneous in their chemical composition and biological activity, and in the timing and magnitude of their responses to external stimuli [1–4]. Therefore, quantification of intracellular constituents is essential for a better understanding of basic cellular functions and intra- and intercellular communications [2, 5]. Such demand has promoted the development of analytical techniques for single cell analysis. The methods developed so far were based on flow cytometry [6–7], open tubular liquid chromatography [8], electrochemical method [9], fluorescence microscopy [10], mass spectrometry [11] and capillary electrophoresis (CE) [12–17]. Microchip electrophoresis (MCE), which is considered as a miniaturized version of classical CE, is one of the most successful applications of microfluidics in analytical chemistry [18]. High separation efficiency of MCE allows fast separations of many constituents present in single cells [19–22]. Ramsey and co-workers used a glass MCE device to achieve high throughput analysis of Jurkat cells [23]. Klepárník and Horký reported an MCE device on a plastic disk for the detection of DNA fragmentation in a single apoptotic cardiomyocyte [24]. Fang group developed a MCE system for the analysis of single cells with functional integration of cell sampling, single cell loading, docking, lysing, separation and laser induced fluorescence (LIF) detection. Using this system, glutathione (GSH) and several reactive oxygen species (ROS) in individual human erythrocytes were determined [25–27]. Zare and co-workers reported the determination of amino acids in single Jurkat cells using an integrated MCE with LIF detection [28]. Reviews on MCE for single cell analysis have been given [29, 30]. In most of these MCE applications, LIF was employed as the detection scheme. Although LIF is the most widely used detection scheme due to its high sensitivity, a conventional LIF detector is sophisticated in instrumentation, expensive in cost, and difficult to be miniaturized. Diode laser-based LIF detectors can be small, but only work at a limited number of wavelengths. Furthermore, pre-column derivatization of the analytes with a fluorophore is often necessary. CL detection is considered one of the most sensitive detection schemes. Its convincing advantage over fluorescence detection is that it does not require a bulky light source. This is attractive, particularly when multiple functions are integrated into a small microfluidic chip. In addition, since CL background signal is usually very low the detector can be operated at its maximum sensitivity. However, as far as we know few MCE-CL methods have been reported so far for single cell analysis. This is likely because CL detection integrated onto MCE chips was not sensitive enough. Recently, we developed MCE-CL assays of glutathione present in single human red blood cells [31] and of biogenic amines in human physiological fluids [32]. In both methods, samples were pre-column derivatized with a CL tagging reagent to achieve high assay sensitivity. In the present study, we extended our previous work by demonstrating analysis of underivatized ascorbic acid and amino acids in single cells using CL detection following electrophoretic separation on chips.
Ascorbic acid (AA) and amino acids are important molecules in biological systems. AA, as one of the most important cellular antioxidants [33] and a valuable biomarker of oxidative stress [34, 35], is an analyte of great importance. At the cellular level, AA mitigates reactive oxygen specie production triggered by lipopolys accharide and, therefore, prevents the induction of nitric oxide synthase and excessive production of nitric oxide that worsens oxidative stress in hepatocytes [36, 37]. Tryptophan (Trp) is an essential amino acid. It exists in liver cells and participates in the synthesis of many other important molecules such as niacin and serotonin [38]. Glycine (Gly) and alanine (Ala) are amino acid neurotransmitters. Therefore, quantification of these compounds in individual hepatocytes is significant for studying various biological processes in the liver.
The aim of this work was to develop an MCE method with CL detection for simultaneous determination of AA, Trp, Gly and Ala present in individual rat hepatocytes. Integration of cell loading and lysing, electrophoretic separation, and CL detection onto a microfluidic chip was investigated. Conditions for MCE separation and CL detection of the targeted compounds were studied. Finally, MCE-CL quantification of intracellular contents of AA, Trp, Gly and Ala in individual rat hepatocytes was demonstrated for the first time in this work.
2. Experimental Section
2.1. Chemicals and solutions
Luminol, AA, Trp, Gly and Ala were purchased from Sigma Chemicals (St. Louis, MO, USA). K3Fe(CN)6, Na2HPO4 and NaHCO3 were obtained from Guangzhou Second Chemical Reagent Factory (Guangzhou, China). Sylgard 184 (PDMS) silicone elastomer and curing agent were obtained from Dow Corning (Midland, MI, USA). All other chemicals used in this work were of analytical grade. Water was purified by employing a Milli-Q plus 185 from Millipore (Bedford, MA, USA) and used throughout the work. All solutions were filtered through a 0.22 µm membrane filter. AA and amino acid stock solutions (1.0 mM) were prepared in water. The electrophoresis buffer was 20 mM Na2HPO4 solution containing 2.5 mM luminol and 40 mM NaBr (pH10.0, adjusted with 1M NaOH solution). The CL reaction buffer was 50 mM NaHCO3 solution containing 0.8 mM K3Fe(CN)6 (pH12.5, adjusted with 1M NaOH solution). The physiological buffer (PBS) consisted of 0.135 M NaCl and 0.02 M NaH2PO4 -NaOH (pH7.4). The D-Hanks solution was prepared by dissolving 0.8 g of NaCl, 0.40 g of KCl, 0.06 g of Na2HPO4·H2O, 0.06 g of KH2PO4, and 0.35 g of NaHCO3 in 1 L of water with 0.02 g of phenol red (95.8%, Beijing Chemical Reagent Factory, Beijing, China) as a pH indicator. A 0.25% (w/v) trypsin (>2500 units/mg, Shanghai Chemical Reagents Co., Shanghai, China) solution was prepared by dissolving trypsin in D-Hanks solution at 4 °C. The pH of the solution was adjusted to 7.4 with NaHCO3 after filtering. The trypsin solution was then sealed and stored at −20 °C.
2.2. MCE-CL System
Analysis of single cells was carried out using a laboratory-built MCE-CL system as described previously [31]. The glass /PDMS microchip assembly was mounted on the X-Y translational stage of an inverted microscope (Olympus CKX41) that also served as a platform of CL detection. Use of the X-Y translational stage allowed viewing any point of the microchannel for introducing a cell. CL signal was collected by means of a microscope objective. After passing a dichroic mirror and a lens, CL emission with a wavelength maximum at 425 nm was detected by a photomultiplier (PMT, Hamamatsu R105). The PMT was mounted in an integrated detection module including HV power supply, voltage divider, and amplifier. The output signal of PMT was recorded and processed with a computer using a Chromatography Data System (Zhejiang University Star Information Technology, Hangzhou, China). A multi-terminal high voltage power supply, variable in the range of 0–8000 V (Shandong Normal University, Jinan, China), was used for cell loading, lysing, and MCE separation. A valuable practical aspect of the inverted microscope setup was the possibility of visually checking all field-controlled operations on the device through the eyepiece. The inverted microscope was placed in a black box.
2.3. Fabrication of microchip
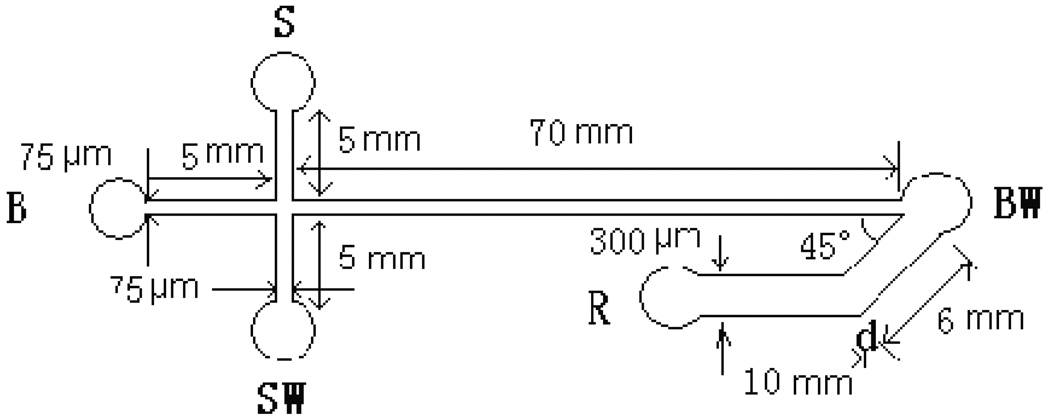
The glass layer with microchannels was fabricated by using a standard photolithography and wet chemical etching techniques. A 9.0 × 2.5 cm rectangle glass (Shaoguang Microelectronics Corp., Changsha, China) with predeposit 145 nm thick Cr layer and 570 nm thick AZ1518 photoresist was exposed beneath the designed mask under an UV lamp, and was then developed and etched using a wet chemical etching procedure [39]. A smooth PDMS surface for bonding to the etched glass slide was prepared by casting the PDMS over a flat As-Ga wafer. Sylgard 184 PDMS prepolymer was mixed thoroughly with its curing agent at 10:1, w/w, and then degassed by vacuum pump. The mixture was cured against the As–Ga wafer at 80 °C for 2 h. After the replica was peeled from the mold, holes were punched as reservoirs. The PDMS covering sheet was exposed under an UV lamp for 2 h, then the glass layer and PDMS covering sheet were bonded at room temperature for 24 h. According to a work reported previously, UV exposure strengthens the bonding between the glass chip and the PDMS cover [40]. The schematic of the microfluidic chip is shown in Figure 1. Microchannels measured 75 µm wide by 25 µm deep for cell introduction, separation and waste delivery, 300 µm wide by 25 µm deep for oxidizer introduction, respectively. The reservoirs S, B and SW were 3.5 mm in diameter and 1.5 mm deep. The reservoirs R and BW were 4.5 mm in diameter and 1.5 mm deep. The channel between reservoir S and SW was used for sampling, the channel between B and BW was used for the separation and the channel between R and BW was used for the oxidizer introduction. Join-point of the oxidizer introduction channel with the separation channel in BW was used for the collection of CL. A similar chip design was reported by Liu and Terabe et al. [41]. In our work, due to the rapid CL reaction and the limited size of detection spot afforded by the microscope objective excessive band broadening was effectively prevented. The MCE peaks observed were sharp as shown below.
Fig. 1.
Layout and dimensions of the glass /PDMS microfluidic chip used in this work. S: Sample reservoir; B: buffer reservoir; SW: sample waste reservoir; BW: buffer waste reservoir; and R: oxidizer solution reservoir.
2.4. Preparation of rat hepatocytes
Rat hepatocytes were prepared following the procedure described by Jin et al [42]. A rat (~300 g) was sacrificed by cervical dislocation. Liver lobes were removed and washed with PBS solution, and then cut into pieces. They were washed twice with PBS solution and then transferred into a 5-mL centrifuge tube. A 0.25% (w/v) trypsin solution (~10-fold volume of the rat liver) was added to the centrifuge tube. After shaking gently, they were enzymolyzed for 20 min at 37 °C. The supernatant was discarded. The enzymolysis was stopped by addition of 3 mL of PBS solution, which was subsequently removed. This enzymolysis step was repeated twice. Then 3 mL of PBS solution was added to the centrifuge tube, and the cell mixture was disrupted by pipetting. The hepatocyte suspension was centrifuged for 10 min at 1000 rpm. The supernatant was discarded. The pellet was resuspended in 2 mL of PBS following by centrifugation at 1000 rpm for another 10 min. This was repeated several times until the supernatant was colorless and transparent. Before injection for single analysis, 15 µL the hepatocyte suspension was diluted to 1.5 mL with PBS as sample solution.
2.5. MCE-CL operations
The microchannels were rinsed with 1M NaOH for 30 min before the first use. Between two consecutive runs, they were rinsed sequentially with 0.1 M NaOH, water, and electrophoresis buffer for 10 min each. For calibration, reservoirs B, S, and SW were filled with the electrophoresis buffer, reservoirs R and BW were filled with CL reaction buffer and vacuum was applied to the reservoir BW in order to fill the separation channel with the electrophoresis buffer. Then, the electrophoresis buffer solution in reservoir S was replaced by AA and amino acids standard solution. For loading the standard solution, a set of electrical potentials were applied to five the reservoirs: reservoir S at 600 V, reservoir B at 150 V, reservoir BW at 200 V, reservoir SW at grounded, and reservoir R at 0 V. The standard solution was transported from reservoir S to SW in pinched mode. After 20 s, potentials were switched to reservoir B, S, SW and R at 2050, 1250, 1250 and 350 V, respectively, while reservoir BW was grounded for separation and detection.
2.6. Analysis of single cells
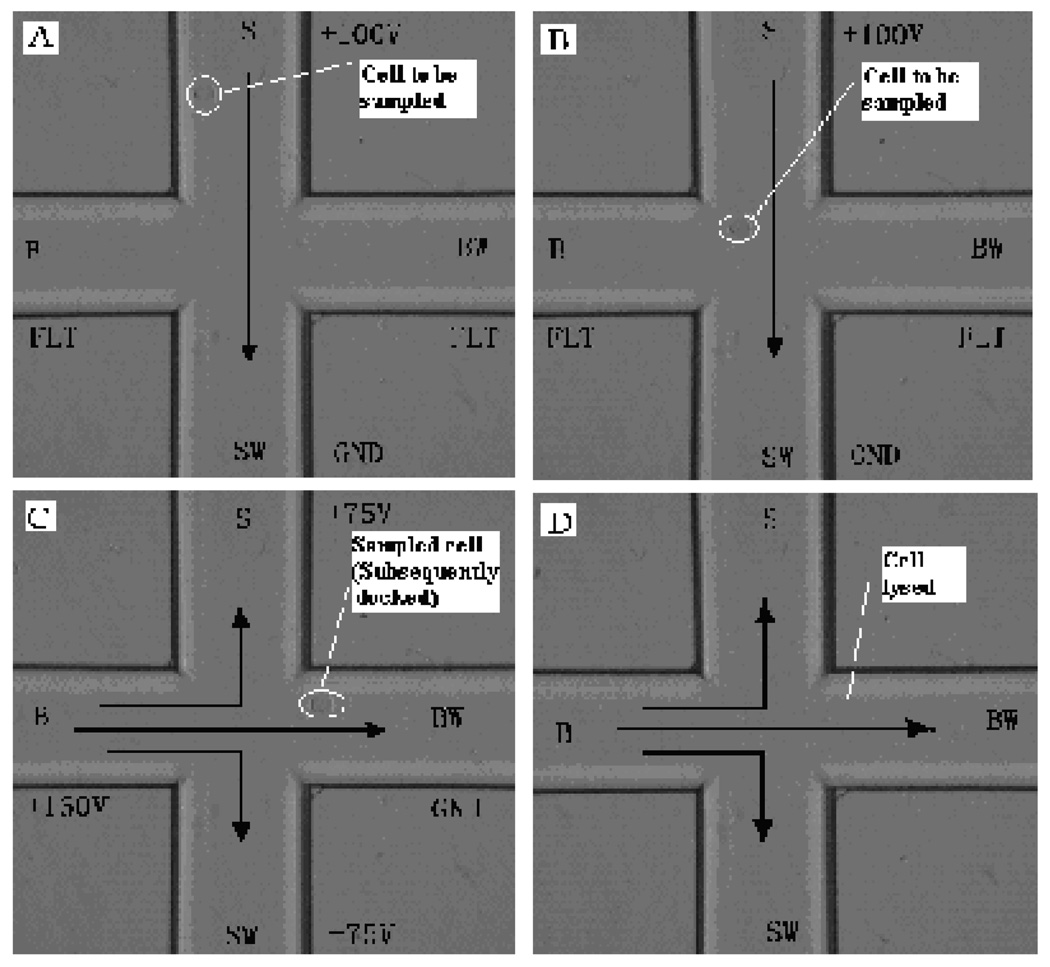
Electrokinetic loading and lysis of cells were used. Prior to MCE separation, reservoirs B, and SW were filled with the electrophoresis buffer, reservoir S was filled with PBS solution, reservoirs BW and R were filled with CL reaction buffer. Vacuum was applied to the reservoir BW in order to fill the separation channel with the electrophoresis buffer. The PBS solution in reservoir S was replaced by hepatocyte suspension. For loading cells, a voltage of 100 V was applied at reservoir S with the reservoir SW grounded and other reservoirs floating. Hepatocytes were driven into the sample channel as shown in Figure 2a. When a single cell moved within the cross intersection of the channels as being observed under the microscope (Figure 2b), potentials were switched to reservoir B, S, SW at 160, 75 and 75 V, respectively, reservoir BW was grounded and reservoir R floating to move the cell into separation channel. After the cell moved completely into the separation channel (Figure 2c), the potentials were turned off to let the sampled cell settle down and adhere to the channel wall. After the chip was shifted from the cross intersection viewing position to the detection point (i.e. the join-point of the oxidizer introduction channel with the separation channel in BW), the docked cell was lysed immediately by a set of electrical shocks with reservoir B set at 1800 V, reservoir BW grounded and other reservoirs floating 6 times intermittently, 0.5 s each time (Figure 2d). MCE separation was performed by applying a 2050 V voltage to reservoir B, 1250 V to reservoir S and SW with the reservoir BW held at ground. Separated AA and amino acids mixed with a K3Fe(CN)6 solution stream from in the reservoir R generated by EOF through applying a 350 V voltage to the reservoir R.
Fig. 2.
Photographs showing a single rat hepatocyte being transported, docked (adhering) and lysed in microfluidic channels. Arrows indicate the direction of liquid flow. (A) & (B): A cell was loaded; (C): the cell was injected from the sample channel into the separation channel, and then docked at an appropriate site on the channel walls; and (D): the docked cell was lysed in the MCE running buffer by means of electrical potentials and ready for MCE-CL separation.
3. Results and discussion
3.1. Cell loading and lysing
In single cell analysis by MCE, several approaches including syringe pump [23], hydrostatic pressure [25] and electric field [43] can be used to drive cells to the separation channel. In our preliminary studies, both hydrostatic pressure and the electric field methods were tested for cell loading. It was found that bringing cells to the cross intersection was not easy on a cross intersection chip by using the hydrostatic pressure approach. Therefore, the electric field method was employed for single cell sampling. After a cell suspension was transferred into the sample reservoir, an electric field at strength of 50–200 V/cm was applied between sample reservoir and sample waste reservoir. The traveling speed of cells (~ 0.1 mm /s) was dependent upon the electrical field strengths applied. A voltage of 100 V was used in this work. When a single cell moved close to the cross intersection, the electric field was turned off. In order to prevent the intracellular contents of a lysed cell from diffusing into the sampling channels, the cell loaded at the cross intersection was further pushed into the separation channel by applying an electric field to reservoir B, S, SW at 160, 75 and 75 V, respectively, with reservoir BW being grounded and reservoir R floated.
After docking a cell into the separation channel, the cell was lysed. The cell can be lysed easily by adding chemical reagents such as NaOH [44] and sodium dodecylsulfate (SDS) [45] in the running buffer. However, introducing additional chemical reagents into running buffer may interfere with MCE separation and CL detection. Therefore, lysing approach by means of a set of high electric potentials was used in this work. It was noticed that cells suspended in the electrophoresis buffer of relatively high pH values could be lysed readily at moderate electric field strength. In our experiments, cells were lysed effectively at 1800 V by setting the potential switch on and off for 6 times with an interval of 0.5 s between each switching. The lysing process was monitored under the inverted microscope.
3.2. MCE Separation
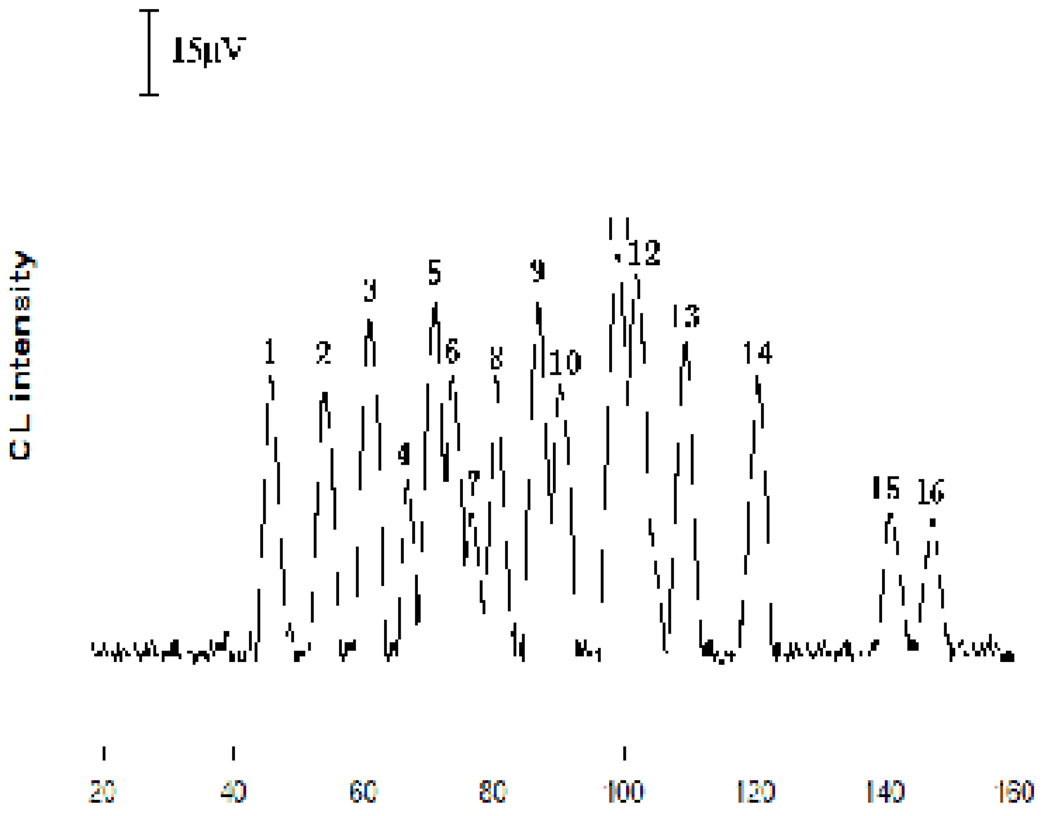
Luminol is a widely used CL reagent. Most amino acids catalyze the CL reaction of luminol with an oxidant such as hydrogen peroxide, producing peaks in an MCE-CL analysis. Therefore, MCE conditions such as running buffer concentration and pH were investigated to achieve an effective separation. In this work, Na2HPO4 buffer was chosen as the running buffer and the effect of Na2HPO4 concentration and the pH on the separation of AA and 20 protein amino acids was studied. Na2HPO4 concentration ranging from 10 to 35 mM was tested. Optimal resolutions were obtained with a 20 mM Na2HPO4 buffer. The effects of running buffer pH values from 9.0 to 10.5 were examined using the same background electrolyte composition. Best separation was oachieved at a pH value of 10.0. Figure 3 shows a typical electropherogram from the separation of a mixture of AA and amino acids under the selected MCE conditions. As can be seen, AA, Trp, Gly and Ala were separated not only from each other, but also from other amino acids.
Fig. 3.
Electropherogram obtained from the separation of a standard mixture containing ascorbic acid and amino acids. MCE conditions: running buffer was a 20 mM Na2HPO4 solution containing 2.5 mM luminol and 40 mM NaBr (pH10.0); CL reaction buffer was a 50 mM NaHCO3 solution containing 0.8 mM K3Fe(CN)6 (pH12.5). The pinched sample injection and separation voltage are shown in the experimental section. Each analyte was at a concentration of 2.0 × 10−5 M. Peak identification: 1. Arg + Lys; 2. Ala; 3. Gly; 4. Val + Pro; 5. Asn + Met; 6. Gln; 7. Ser; 8. Cys; 9. Phe + Leu; 10. Ile + His; 11. Thr; 12. Tyr; 13. Trp; 14. AA; 15. Glu; and 16. Asp.
3.3. MCE-CL assay of AA and amino acids
To achieve a high sensitivity of CL detection, experimental conditions including the concentrations of luminol in MCE running buffer and of K3Fe(CN)6 in the reaction buffer were studied. Luminol concentration varied from 1.0 to 4.0 mM and K3Fe(CN)6 concentration from 0.5 to 1.0 mM. Maxima CL signal was obtained with luminol at 2.5 mM and K3Fe(CN)6 at 0.8 mM. The effects of pH values of CL reaction buffer was also investigated in a pH range of 10.5 – 13.5. The results indicate that the CL intensity increases with the increase of pH. However, the baseline noise was very high after the pH value became higher than 12.5. Therefore, the post-column introduced CL reaction solution was buffered at 12.5. Moreover, it was found that the addition of NaBr into running buffer enhanced the CL emission. It was not clear why CL signal increased in the presence of NaBr. However, it’s worth noting that the presence of 40 mM NaBr in the MCE running buffer increased the CL intensity by about 30%. Therefore, an MCE running buffer containing 40 mM NaBr was used.
Based on the experimental results described above, the MCE-CL conditions for assaying AA, Trp, Gly and Ala were selected as following: the MCE running buffer was a 20 mM Na2HPO4 buffer solution (pH10.0) containing 2.5 mM luminol and 40 mM NaBr; and CL reaction buffer was a 50 mM NaHCO3 buffer solution (pH12.5) containing 0.8 mM K3Fe(CN)6. Under the selected conditions, the number of theoretical plates for each analyte was studied by separating a mixture standard solution (1.0×10−5 M each analyte). The number of theoretical plates for AA, Trp, Gly and Ala were 23209, 31226, 29093 and 24362/m, respectively.
3.4. Analytical figures of merit
The present MCE-CL method was evaluated in terms of the response linearity, limit of detection, and reproducibility. Seven-point calibration curves were prepared by analyzing a series of analyte standard solutions at different concentrations. Peak areas were used for quantification. The results are summarized in Table 1. Since volumes of single cells vary from cell to cell, absolute amounts, instead of concentrations, are often determined for intracellular compounds in single cell analysis. Therefore, the concentration results shown in Table 1 were converted into absolute amounts using the calculated injection volume of 26 pL in the MCE-CL assays (Table 2). As can be seen, the limits of detection (LODs) for the targeted analytes ranged from 1.3 µM (or 0.33 fmol) for AA to 2.1 µM (or 0.46 fmol) for Trp. In the studies previously reported, an LOD of 0.39 µM for dansyl-labeled Gly was obtained from MCE followed by peroxalate CL detection [41]. In an MCE method with UV laser induced fluorescence detection, 0.1 µM underivatized Trp could be detected [22]. The detection limit for AA from a method based on classical CE with electrochemical detection was 5 µM [47]. The reproducibility of analyte response (RSD%) in terms of both CL intensity (peak area) and migration time were studied by separating a standard solution (1.0×10−5 M each analyte) for 7 times. The RSDs of peak area and migration time for all analytes were found < 4.7% and 4.1%, respectively.
Table 1.
Concentration linear ranges and detection limits of AA, Trp, Gly and Ala
| Analyte | Linear regression equation | Linear ranges (µM) | R2 | LODc (µM) |
|---|---|---|---|---|
| AA | A=18.36C+13.87 | 3.0-600 | 0.9916 | 1.3 |
| Trp | A=19.15C+18.16 | 4.0-400 | 0.9913 | 2.1 |
| Gly | A=23.12C+12.39 | 3.5-400 | 0.9986 | 1.6 |
| Ala | A=20.19C+13.35 | 3.5-400 | 0.9994 | 1.8 |
R2: linear correlation coefficient; A: peak area (relative CL intensity); C: concentration of analytes (µM)
Table 2.
Linear ranges and detection limits expressed as in absolute amounts
| Analyte | Linear regression equation | Linear ranges (fmol) | R2 | LODm (fmol) |
|---|---|---|---|---|
| AA | A=71.62Q+13.87 | 0.7-153 | 0.9916 | 0.33 |
| Trp | A=74.69Q+18.55 | 1.0-102 | 0.9911 | 0.54 |
| Gly | A=90.18Q+12.57 | 0.9-102 | 0.9986 | 0.41 |
| Ala | A=78.85Q+13.21 | 0.9-102 | 0.9995 | 0.46 |
R2: linear correlation coefficient; A: peak area (relative CL intensity); Q: amount of analytes (fmol)
3.5. Quantification of AA, Trp, Gly and Ala in individual hepatocytes
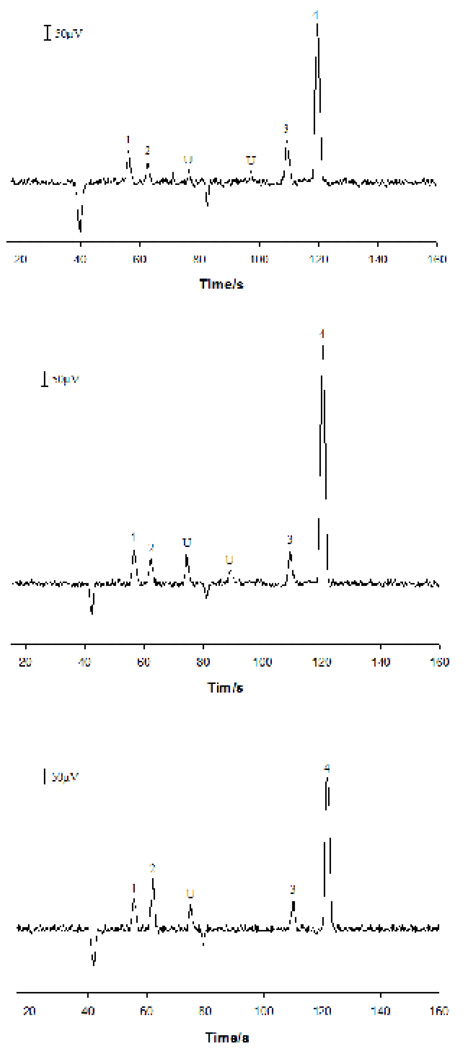
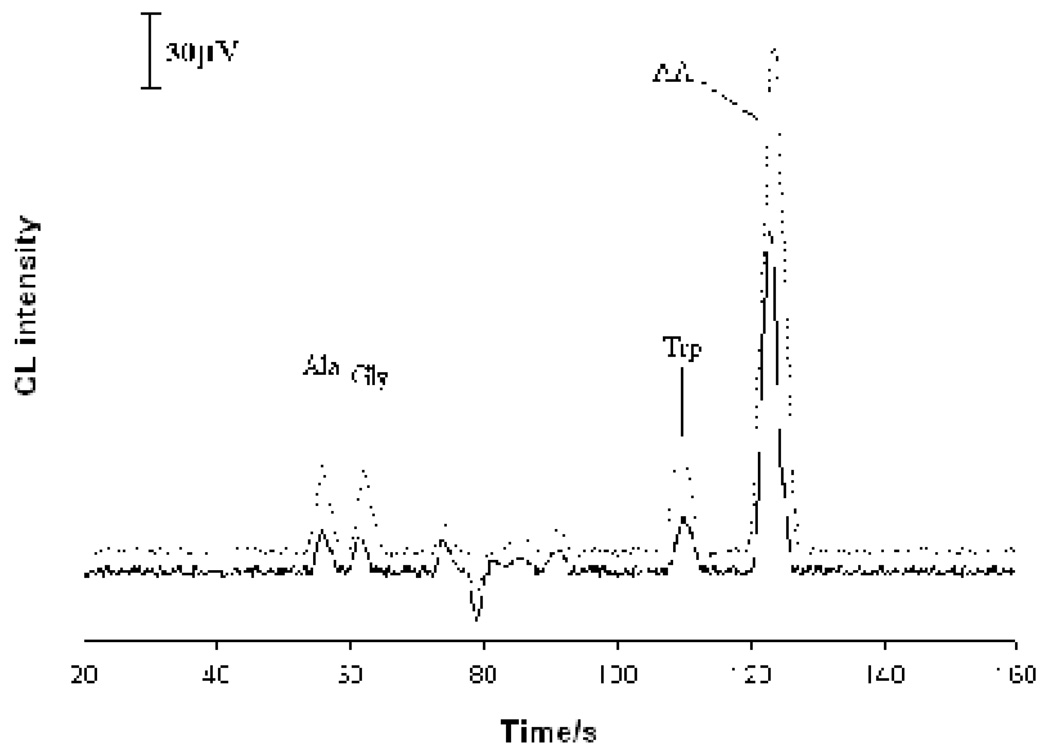
Electropherograms obtained from analyzing three individual hepatocytes are shown in Figure 4. Peaks at 55 s, 62 s, 112 s and 122 s were identified as AA, Trp, Gly and Ala, respectively, according to their migration times. Two unknown compounds were also detected. To verify the peak identification, an extract of rat hepatocytes was prepared. A portion of this extract sample was spiked with authentic AA (at 3.2×10−5 M), Trp, Gly and Ala (at 1.0×10−5 M each). Both samples were analyzed. Electropherograms obtained from the analysis are shown in Figure 5. As can be seen, the four peaks corresponding to Ala, Gly, Trp and AA were well identified. It should be pointed out that further work may be required to confirm the identity of the peaks detected since only migration time information was used in assigning peaks in this work. It is worth noting that the width at the peak half-height in single-cell analysis was narrower than that from analyzing standard solutions and these cell extract samples. This was because the sample plug from injecting a standard solution was longer than the diameter of individual cells. In addition, because of the difference in cell volumes, absolute amounts, instead of concentrations, of analytes were usually determined in single cell analysis [46]. Table 3 shows the amounts of AA, Trp, Gly and Ala measured for 12 individual hepatocytes. It was noticed that the amounts of AA, Trp, Gly and Ala in single hepatocytes differed from cell to cell. The average intracellular contents of AA, Trp, Gly and Ala were 38.3, 5.15, 3.78 and 3.84 fmol per cell, respectively. The Trp and AA contents in individual rat hepatocytes found in this work were very similar to those reported in literature (i.e. 5.0 fmol /cell for Trp and 37 fmol /cell for AA) [40, 47]. From the analysis, Trp, Gly, and Ala were the predominant amino acids in rat hepatocytes. Other amino acids didn’t occur or occurred at the levels below the limits of detection.
Fig. 4.
Electropherograms obtained from assaying three individual rat hepatocytes. MCE conditions were as in Figure 3. Peak identification: 1. Ala; 2. Gly; 3. Trp; 4. AA; and U. unknown.
Fig. 5.
Electropherogram obtained from the separation of an extract of rat hepatocytes (solid line) and the extract spiked with Ala, Gly, Trp (at 1.0×10−5 M each) and AA at 3.5×10−5 M (dotted line). MCE conditions were as in Figure 3.
Table 3.
Contents of AA, Trp, Gly and Ala in individual hepatocytes (fmol per cell)
| Analyte | Cell |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | average | |
| AA | 12.9 | 49.4 | 51.6 | 34.0 | 24.5 | 14.6 | 39.9 | 63.2 | 48.7 | 41.3 | 53.4 | 26.5 | 38.3 |
| Trp | 5.45 | 4.21 | 7.08 | 4.93 | 6.99 | 4.46 | 2.92 | 5.14 | 6.1 | 4.51 | 6.31 | 3.73 | 5.15 |
| Gly | 2.02 | 2.84 | 4.15 | 3.27 | 2.53 | 5.0 | 5.69 | 3.40 | 3.01 | 4.87 | 6.21 | 2.41 | 3.78 |
| Ala | 3.10 | 2.60 | 1.96 | 4.32 | 5.03 | 3.9 | 4.35 | 5.33 | 6.07 | 4.11 | 3.05 | 2.21 | 3.84 |
4. Conclusion
An MCE-CL method has been developed for quantitative analysis of ascorbic acid and amino acids present in single cells. Compared with the MCE-LIF methods reported previously, the present MCE-CL method is simple in instrumental set-up and offers a comparable sensitivity. In addition, no pre-column derivatization of the sample is required. Analysis of individual rat hepatocytes was demonstrated in this work. Intracellular ascorbic acid and amino acids including Trp, Gly, and Ala were quantified. The average intracellular contents of ascorbic acid, Trp, Gly and Ala were found to be 38.3, 5.15, 3.78 and 3.84 fmol per cell, respectively.
Acknowledgment
Financial support from the National Natural Science Foundations of China (NSFC, Grant No. 20665002, 20875019 to SZ) and US National Institutes of Health (S06GM08047 to YML) is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 2.McClain MA, Culbertson CT, Jacobson SC, Allbritton NL, Sims CE, Ramsey JM. Anal. Chem. 2003;75:5646. doi: 10.1021/ac0346510. [DOI] [PubMed] [Google Scholar]
- 3.Greif D, Galla L, Ros A, Anselmetti D. J. Chromatogr. A. 2008;1206:83. doi: 10.1016/j.chroma.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Li XJ, Ling V, Li PCH. Anal. Chem. 2008;80:4095. doi: 10.1021/ac800231k. [DOI] [PubMed] [Google Scholar]
- 5.Dufva M. Methods Mol Biol. 2009;509:135. doi: 10.1007/978-1-59745-372-1_9. [DOI] [PubMed] [Google Scholar]
- 6.Kruth HS. Anal. Biochem. 1982;125:225. doi: 10.1016/0003-2697(82)90001-x. [DOI] [PubMed] [Google Scholar]
- 7.Naill MC, Roberts SC. Biotechnol. Prog. 2005;21:978. doi: 10.1021/bp049544l. [DOI] [PubMed] [Google Scholar]
- 8.Oates MD, Cooper BR, Jorgenson JW. Anal. Chem. 1990;62:1573. doi: 10.1021/ac00214a006. [DOI] [PubMed] [Google Scholar]
- 9.Troyer KP, Wightman RM. Anal. Chem. 2002;74:5370. doi: 10.1021/ac0203903. [DOI] [PubMed] [Google Scholar]
- 10.Ishijima A, Yanagida T. Trends Biochem. Sci. 2001;26:438. doi: 10.1016/s0968-0004(01)01860-6. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Golding RE, Whittal RM. J. Am. Chem. Soc. 1996;118:11662. [Google Scholar]
- 12.Hogan BL, Yeung ES. Anal. Chem. 1992;64:2841. doi: 10.1021/ac00046a031. [DOI] [PubMed] [Google Scholar]
- 13.Han FT, Lillard SJ. Anal. Biochem. 2002;302:136. doi: 10.1006/abio.2001.5519. [DOI] [PubMed] [Google Scholar]
- 14.Hu S, Zhang L, Cook LM, Dovichi NJ. Electrophoresis. 2001;22:3677. doi: 10.1002/1522-2683(200109)22:17<3677::AID-ELPS3677>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Jin WR. Electrophoresis. 2004;25:480. doi: 10.1002/elps.200305642. [DOI] [PubMed] [Google Scholar]
- 16.Fuller KM, Arriaga EA. Anal. Chem. 2003;75:2123. doi: 10.1021/ac026476d. [DOI] [PubMed] [Google Scholar]
- 17.Zhi Q, Xie C, Huang X, Ren J. Anal. Chim. Acta. 2007;583:217. doi: 10.1016/j.aca.2006.09.068. [DOI] [PubMed] [Google Scholar]
- 18.Janasek D, Franzke J, Manz A. Nature. 2006;442:374. doi: 10.1038/nature05059. [DOI] [PubMed] [Google Scholar]
- 19.Di DC, Lee LP. Anal. Chem. 2006;78:7918. doi: 10.1021/ac069490p. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y, Yin XF. J. Chromatogr. A. 2006;1117:228. [Google Scholar]
- 21.Sun Y, Lu M, Yin XF, Gong XG. J. Chromatogr. A. 2006;1135:109. doi: 10.1016/j.chroma.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 22.Hellmich W, Greif D, Pelargus C, Anselmetti D, Ros A. J. Chromatogr. A. 2006;1130:195. doi: 10.1016/j.chroma.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 23.McClain MA, Culbertson CT, Jacobson SC, Allbritton NL, Sims CE, Ramsey JM. Anal. Chem. 2003;75:5646. doi: 10.1021/ac0346510. [DOI] [PubMed] [Google Scholar]
- 24.Klepárník K, Horký M. Electrophoresis. 2003;24:3778. doi: 10.1002/elps.200305667. [DOI] [PubMed] [Google Scholar]
- 25.Gao J, Yin XF, Fang ZL. Lab. Chip. 2004;4:47. doi: 10.1039/b310552k. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y, Yin XF, Ling YY, Fang ZL. Anal. Bioanal. Chem. 2005;382:1472. doi: 10.1007/s00216-005-3352-8. [DOI] [PubMed] [Google Scholar]
- 27.Ling YY, Yin XF, Fang ZL. Electrophoresis. 2005;26:4759. doi: 10.1002/elps.200500232. [DOI] [PubMed] [Google Scholar]
- 28.Wu H, Wheeler A, Zare RN. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12809. doi: 10.1073/pnas.0405299101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sims CE, Allbritton NL. Lab. Chip. 2007;7:423. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 30.Huang WH, Ai F, Wang ZL, Cheng JK. J. Chromatogr. B. 2008;866:104. doi: 10.1016/j.jchromb.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 31.Zhao SL, Li XT, Liu YM. Anal. Chem. 2009;81:3873. doi: 10.1021/ac900391u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao SL, Huang Y, Shi M, Liu YM. J. Chromatogr. A. 2009;1216:5155. doi: 10.1016/j.chroma.2009.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frei B, England L, Ames BN. Proc. Natl. Acad. Sci. U.S.A. 1989;86:6377. doi: 10.1073/pnas.86.16.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lykkesfeldt J, Loft S, Poulsen HE. Anal. Biochem. 1995;229:329. doi: 10.1006/abio.1995.1421. [DOI] [PubMed] [Google Scholar]
- 35.Lykkesfeldt J, Prieme H, Loft S, Poulsen HE. Brit. Med. J. 1996;313:91. doi: 10.1136/bmj.313.7049.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De la Fuente M, Victor VM. Free Radic. Res. 2001;35:73. doi: 10.1080/10715760100300611. [DOI] [PubMed] [Google Scholar]
- 37.Kim JY, Lee SM. Life Sci. 2004;75:2015. doi: 10.1016/j.lfs.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Fukuwatari T, Morikawa Y, Sugimoto E, Shibata K. Biosci. Biotechnol. Biochem. 2002;66:1196. doi: 10.1271/bbb.66.1196. [DOI] [PubMed] [Google Scholar]
- 39.Kera Y, Aoyama H, Matsumura H, Hasegawa A, Nagasaki H, Yamada R. Biochim. Biophys. Acta. 1995;1243:283. [PubMed] [Google Scholar]
- 40.Chen L, Ren J, Bi R, Chen D. Electrophoresis. 2004;25:914. doi: 10.1002/elps.200305766. [DOI] [PubMed] [Google Scholar]
- 41.Liu B-F, Ozaki M, Utsumi Y, Hattori T, Terabe S. Anal. Chem. 2003;75:36. doi: 10.1021/ac026096s. [DOI] [PubMed] [Google Scholar]
- 42.Jin W, Li X, Gao N. Anal. Chem. 2003;75:3859. doi: 10.1021/ac0207022. [DOI] [PubMed] [Google Scholar]
- 43.Li PCH, Harrison DJ. Anal. Chem. 1997;69:1564. doi: 10.1021/ac9606564. [DOI] [PubMed] [Google Scholar]
- 44.Wand W, Xin H, Shao H, Jin W. J. Chromatogr. B. 2002;789:425. doi: 10.1016/s1570-0232(03)00105-3. [DOI] [PubMed] [Google Scholar]
- 45.Chen S, Lillard SJ. Anal. Chem. 2001;73:111. doi: 10.1021/ac0009088. [DOI] [PubMed] [Google Scholar]
- 46.Xia F, Jin W, Yin X, Fang Z. J. Chromatogr. A. 2005;1063:227. doi: 10.1016/j.chroma.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 47.Sun X, Niu Y, Bi S, Zhang S. J. Chromatogr. B. 2008;870:46. doi: 10.1016/j.jchromb.2008.05.035. [DOI] [PubMed] [Google Scholar]