Abstract
Influenza virus presents a significant and persistent threat to public health worldwide and current vaccines provide immunity to viral isolates similar to the vaccine strain. High affinity antibodies against a conserved epitope could provide immunity to the diverse influenza subtypes and protection against future pandemic viruses. Co-crystal structures were determined at 2.2 and 2.7 Å resolutions for broadly neutralizing human antibody CR6261 Fab in complexes with the major surface antigen (hemagglutinin, HA) from viruses responsible for the 1918 H1N1 influenza pandemic and a recent lethal case of H5N1 avian influenza. In contrast to all other structurally characterized influenza antibodies, CR6261 recognizes a highly conserved helical region in the membrane-proximal stem of HA1/HA2. The antibody neutralizes the virus by blocking conformational rearrangements associated with membrane fusion. The CR6261 epitope identified here should accelerate the design and implementation of improved vaccines that can elicit CR6261-like antibodies, as well as antibody-based therapies for the treatment of influenza.
Over the past century, three human influenza A pandemics (1918 H1N1 Spanish, 1957 H2N2 Asian, and 1968 H3N2 Hong Kong) have killed ∼50–100 million people worldwide. Each pandemic virus was derived, at least in part, from an avian influenza virus by direct interspecies transmission or exchange of genetic material between avian and human viruses (1–4). In each case, a novel hemagglutinin (HA) envelope glycoprotein was acquired that was antigenically distinct from the HAs of the human viruses in circulation at that time. HA is the primary target of neutralizing antibodies and rapidly and continuously accumulates mutations to escape recognition by the immune system. In pandemic years, HAs are shuffled from the vast reservoir of 16 HA subtypes in avian viruses into a circulating human virus to evade prevailing immunity in the human population. Thus, while many factors likely contribute to virulence and transmissibility, immune evasion is critical for the rapid spread of pandemic and epidemic viruses.
Several small molecules are in use for treatment of influenza. Most notable are neuraminidase (NA) inhibitors, oseltamivir (Tamiflu) and zanamivir (Relenza), that prevent release of nascent virions, and amantadine (5) that interferes with the M2 channel proton conducting activity. However, excessive use leads to resistant viruses (6–8) that often show surprisingly little attenuation from the escape mutations, thereby contributing to rapid spread worldwide (6). Recently, a binding pocket was characterized on the HA for the fusion inhibitor tert-butyl hydroquinone (TBHQ) (9), which shows great promise, but is still in the early stages of development. Consequently, vaccination remains the most effective countermeasure against influenza virus.
Current trivalent influenza vaccines elicit a potent neutralizing antibody response to the vaccine strains and closely related isolates, but rarely extend to more diverged strains within a subtype or to other subtypes (10). Selection of the appropriate vaccine strains presents many challenges and frequently results in sub-optimal protection (11, 12). Furthermore, predicting the subtype and clade of the next pandemic virus, including when and where it will arise, is currently impossible. A vaccine that stimulates production of antibodies capable of neutralizing multiple influenza A subtypes would eliminate much of the guesswork associated with strain selection and impede emerging pandemic viruses. While a few rare antibodies with broad, heterosubtypic patterns of neutralization have been reported, their epitopes remain obscure and have hampered attempts at rational vaccine design (13–15).
Antibody CR6261 was isolated from the immune repertoire of a healthy, vaccinated individual using phage display selection on recombinant H5 HA (15). Despite no known exposure to H5 viruses, several clones capable of neutralizing H5 viruses were obtained. Human IgG1 CR6261 neutralizes multiple influenza subtypes, including H1, H2, H5, H6, H8, and H9, and protects mice from lethal challenge with H1N1 and H5N1 viruses when administered up to five days post-infection (15). To characterize the CR6261 epitope on the HA and the mechanism of neutralization, we determined crystal structures of CR6261 Fab in complex with HAs from the human 1918 H1N1 pandemic virus (A/South Carolina/1/1918; SC1918/H1) and from a highly pathogenic avian H5N1 virus (A/Vietnam/1203/2004; Viet04/H5).
The SC1918/H1 and Viet04/H5 HA ectodomains were expressed in baculovirus and Fab CR6261 in mammalian cells (16). Co-crystal structures were determined at 2.2 Å and 2.7 A resolution by molecular replacement (see Table S1 and Supplementary Online Material), and revealed three antibodies bound per HA trimer (Fig. 1). Both HAs are very similar to their unliganded structures (Fig. S1) (17–19). Each HA polypeptide is proteolytically cleaved during viral maturation to two disulfide-linked chains, HA1 and HA2. HA1 consists primarily of the membrane-distal receptor binding and vestigial esterase domains, but its N- and C- terminal regions extend towards the viral membrane and are intertwined with the exterior surface of HA2. HA2 constitutes the core fusion machinery in the stalk region and is dominated by the long, central CD-helix (residues 75–126) that forms a trimeric coiled-coil and the shorter A-helix (residues 38–58) that packs against the central helical bundle (20). Exposure to low pH leads to major structural rearrangements in HA2 that facilitate membrane fusion (21, 22).
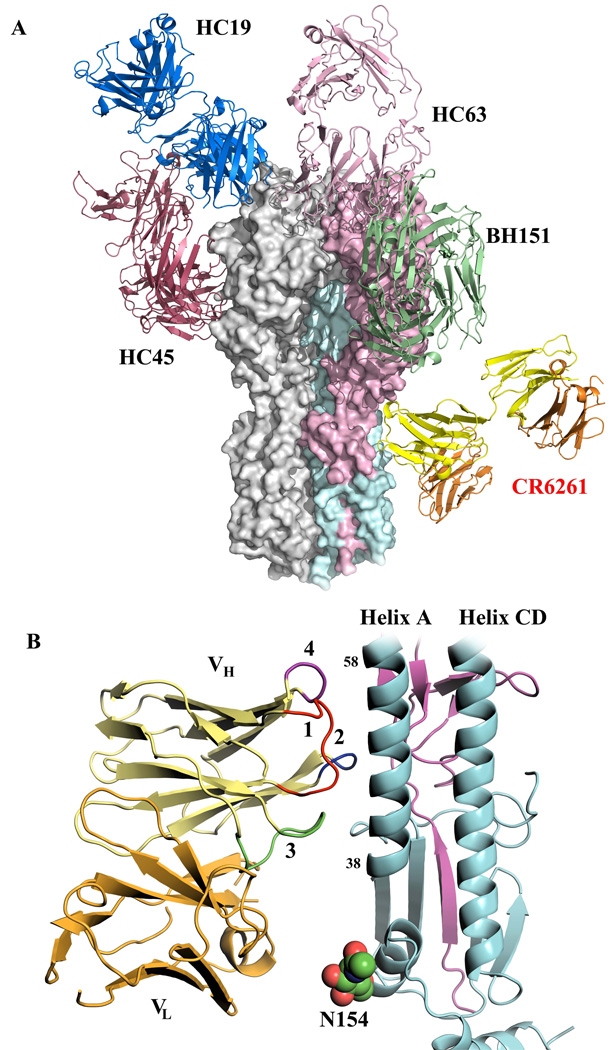
Figure 1. Crystal structures of broadly neutralizing antibody CRF6261 in complex with SC1918/H1 and Viet04/H5 HAs.
A) The trimeric CRF6261-SC1918/H1 complex. One HA/Fab protomer is colored with HA1 in purple, HA2 in cyan, Fab heavy chain in yellow, and Fab light chain in orange; the other two HA monomers and Fabs in the trimer are colored in grey for clarity. Glycans are depicted as their component atoms (carbon in green, oxygen in red and nitrogen in blue) with their van der Waals’ radii. B) The trimeric CRF6261-Viet04 complex, colored as in A. The HA1 globular heads are located at the top of the figure and the membrane-proximal stem is at the bottom where the Fab is bound.
Interpretable density is observed for HA1 11–325 and HA2 1–172 (SC1918/H1) and HA1 11–324 and HA2 1–177 (Viet04/H5) [H3 numbering (21, 23)]. The N-terminal fusion peptide in HA2 is embedded in the hydrophobic pocket between adjacent HA2 subunits (Fig. S2), indicating both HAs are in their pre-fusion states. Unlike other structurally characterized, neutralizing antibodies, CR6261 binds at the membrane proximal end of each HA, approximately parallel to the plane of the viral envelope (Fig. 2A) (24–27), within ∼25 Å of the bilayer. CR6261 interacts primarily with the HA2 A-helix, but also contacts HA1 residues in the stem region. The epitope and the Fab variable domains are well ordered in both complexes (Fig. S3), but the Fab constant domains in the SC1918/H1 complex are partially disordered (28). Unexpectedly, the Fab interaction is mediated exclusively by the heavy chain (Fig. 2B) (29). Furthermore, the relatively modest buried surface area [680 Å2 on HA (70% HA2), 624 Å2 on the Fab, 116 van der Waals’ contacts] presents fewer possibilities for escape mutations, particularly for highly conserved epitopes that are functionally important, and may also reduce the ability of masking glycans to interfere with antibody binding. Consistent with this notion, a universally conserved glycan on HA2 Asn154 lies directly between CR6261 VL and the HA (Fig. 2B and Fig. S4) (30). The selection of heavy-chain dominant antibodies may also be enhanced as a consequence of the large, diverse repertoire that can be accessed from the combinatorial pairings of heavy and light chains in the construction of phage libraries (31, 32).
Figure 2. Broadly neutralizing CR6261 binds in the HA stem distant from other strain-specific antibodies using only its heavy chain.
A) Comparison of the binding sites of Fab CR6261 (yellow and orange, as in Fig. 1)) that recognizes the HA stem and strain-specific antibodies that bind to the HA1 globular head domain The strain-specific antibody structures are depicted in green (BH151, PDB code 1EO8), copper (HC63, PDB code 1KEN), dark red (HC45, PDB code 1QFU), and blue (HC19, PDB code 2VIR). The HA trimer is shown as a surface representation, with the HA1 and HA2 from one protomer colored in purple and cyan, respectively. B) A close up view of the interaction of CR6261 with the A-helix. HA1 and HA2 are colored in purple and cyan, respectively. The HCDRs 1, 2, and 3 of the CR6261 VH domain are highlighted in red, blue, and green, respectively. In addition, the 72–73 loop from FR3 of VH, which is structurally analogous to the CDR HV4 of a TCR, is indicated in purple. CDR1 runs along the side of the A-helix, interacting with five consecutive helical turns. In contrast, the light chain makes no contacts with the HA and is separated from the nearest HA side chain by ∼8 Å.
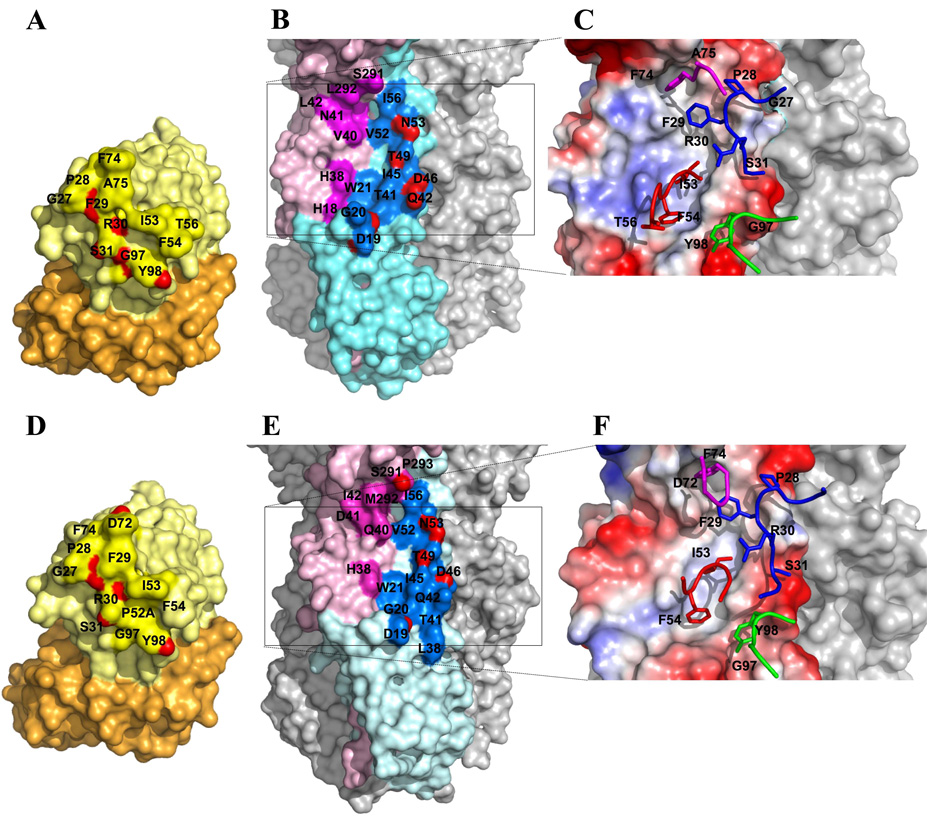
The CR6261-SC1918/H1 interaction is depicted in Fig. 3A–C, in which 8 hydrogen bonds are formed with HA2 (Table S2), five of which involve recognition of the A-helix by HCDR1, primarily by the main chain (33). In contrast, CR6261 makes mainly non-polar contacts with HA1 that can be grouped into two distinct clusters within the hydrophobic groove at the junction between the A-helix and HA1 (Fig. 3B–C, Table S3). The first region consists of HA1 Val40, Leu42, and Leu292, along with HA2 Thr49, Val52, and Ile56 from the membrane-distal end of the A-helix, which interact with HCDR1 Pro28 and Phe29. In addition, Phe74 in Framework Region 3 (FR3), reminiscent of HV4 in TCRs (34), interacts with this first hydrophobic patch, likely due to the binding interactions being focused exclusively on the heavy chain. The second cluster, close to the membrane-proximal end of the A-helix, includes His18 and His38 from HA1 and Trp21, Thr41, and Ile45 from HA2, that interact with the conserved hydrophobic tip of HCDR2 (Ile53 and Phe54), and Tyr98 from HCDR3.
Figure 3. Interaction of CR6261 with SC1918/H1 and Viet04/H5 HA illustrating the neutralizing epitope.
Surface representations of the interaction of CR6261 with SC1918/H1 (A–C) and Viet04/H5 (D–F). The coloring in A, B, D, and E corresponds to Fig. 1, with interacting epitope residues highlighted in a darker shade. Atoms making polar contacts are highlighted in red. C and F depict the electrostatic potential surface of the epitope on the HA (red – negative; blue-positive; grey-neutral) with HCDRs 1, 2, 3, and FR3 (CDR “HV4”) represented as sticks and colored blue, red, green, and purple, as in Fig. 2. Key interacting residues on the antibody and the HA are indicated on the top of the surface with labels.
To reveal the basis for cross-neutralization of the H5 subtype, we determined the structure of CR6261 with Viet04/H5 HA. Overall, the CR6261-Viet04/H5 interface is only slightly more restricted (Fig. 3D–F; Tables S2 and S3) with six H-bonds to HA2 (35). However, five of the six H-bonds to the A-helix are retained with Viet04/H5, highlighting the importance of the conserved A-helix recognition as the major determinant of specificity. An additional H-bond is formed between HA1 Ser291 and Asp72 (FR3). Thus, a somewhat peripheral contact with a homotypic antigen (SC1918/H1) is converted to a more direct and specific one with the heterotypic HA (Viet04/H5). Otherwise, the interactions with the two hydrophobic clusters at the junction of the A-helix with HA1 are qualitatively similar, with similar van der Waals’ contacts (104–122 for the two independent complexes) in the H5 interaction (total buried surface of 1220–1256 Å2; 640–656 Å2 for the HAs and 580–600 Å2 for Fabs).
It is interesting to note that many broadly neutralizing antibodies, including CR6261, have preferentially selected the Ig VH1–69 germline segment that codes for a conserved hydrophobic tip on HCDR2. CR6261 was isolated along with other independent VDJ recombinants, of which 12/13 broadly neutralizing clones were derived from VH1–69 (15). Similarly, many CD4-induced antibodies that target HIV-1 gp120 also use VH1–69 (36–38). Structures of several Ig VH1–69-derived Fabs with their antigens show their HCDR2 tips frequently interact with solvent-exposed hydrophobic clusters that may require electroneutral interactions (36, 37). Indeed, CR6261 HCDR2 is directed into a hydrophobic pocket adjacent to the A-helix in both structures (Fig. S5), which differs from that proposed from docking and mutational studies (15, 39). Furthermore, HCDR1, which makes the most critical contacts with the highly conserved A-helix, is also germline-encoded (40), although somatic mutations in the VH1–69 framework also make important contributions to binding (41). Perhaps more unusually, HCDR3 contributes relatively little, in contrast to its usual dominant role in antigen recognition (42). Thus, CR6261 binds with nanomolar affinity primarily using the germline-encoded CDR’s defined by Ig VH1–69 and complementary mutations in the framework regions with only a small contribution from HCDR3 and none from the light chain (15).
Most neutralizing antibodies against influenza HA recognize epitopes in the hypervariable regions that surround the receptor binding site and interfere with binding to host cells (Fig. 2A) (24, 26, 27, 43, 44). However, a few rare antibodies likely interfere with membrane fusion (13, 45). CR6261 does not inhibit agglutination of erythrocytes in vitro, suggesting that it might act by inhibiting membrane fusion (15). Thus, we assayed the ability of CR6261 to prevent conversion of SC1918/H1 and Viet04/H5 HAs to the post-fusion state upon exposure to low pH (46, 47). Both HAs are readily converted to their protease-susceptible post-fusion form at pH 4.9 or 5.3, but not at pH 8.0 (Fig 4A, lanes 7–9; Fig 4B, lanes 7–9). Significantly, CR6261 Fab prevents conversion of both HAs to their post-fusion conformations (Fig. 4A, lanes 10–12; Fig. 4B, lanes 10–12).
Figure 4. CR6261 recognizes a functionally conserved epitope in the stalk region and inhibits the pH-induced conformational changes in the SC1918/H1 and Viet04/H5 HAs.
CR6261 protects SC1918/H1 (A) and Viet04/H5 (B) HAs from the pH-induced protease sensitivity associated with membrane fusion. Exposure to low pH renders the SC1918-H1 and Viet04-H5 HAs sensitive to trypsin digestion (lanes 7–8 vs. 9), but CR6261 prevents conversion to the protease susceptible conformation (lanes 10–12). Note that the CR6261-SC1918-H1 crystals were grown at pH 5.3 that also indicates CR6261 blocks the extensive pH-induced conformational changes. C) Titration of SC1918-H1 trypsin-resistance versus varying pH-treatments (followed by neutralization to pH 8.0). The pH resulting in 50% conversion to protease sensitive conformation is 5.76 (95% confidence interval, 5.70–5.82). D) Superposition of the A-helix from SC1918/H1 (green) and an H3 HA (yellow, PDB code 2VIU) reveals that the CR6261 interacting surface is highly conserved among all subtypes. Helix positions are labeled according to the SC1918/H1 sequence, with the percent similarity across all subtypes (H1-16, from an analysis of 5261 sequences) indicated in parentheses. E–F) HA2 undergoes a dramatic and irreversible conformational change between the pre- and post-fusion states. This results in the translocation of the A-helix (red) from its initial position near the viral envelope (Fig. E) towards the target membrane at the opposite end of the HA trimer (Fig. F). The zipping up of coil H along the outside of the A and B-helices is thought to drive the fusion reaction. The orientation of helix C (yellow) is roughly identical in Figs. E and F. G) Polar residues on the CR6261-interacting surface of the A-helix form a network of interactions with coil H in the post-fusion state. These residues are well positioned to play a critical role in the late stages of membrane fusion, explaining the exceptional conservation the CR6261 epitope on the A-helix.
Acidification of endosomal compartments during endocytosis of influenza virus particles results in major structural rearrangements in HA2, leading to fusion of the viral envelope with internal, host cell membranes (22). Most notably, low pH exposure converts the connecting segment between the A- and CD-helices to an additional α-helical segment, extending the central HA2 trimeric coil towards the target endosomal membrane and dragging the A-helix and N-terminal fusion peptide along with it (Fig. 4E–F). Subsequent HA2 rearrangements are thought to bring the viral and target membranes into close proximity for fusion (22, 48). Surprisingly, the SC1918/H1 HA in complex with CR6261 is essentially identical to the unliganded, pre-fusion HA structure (Fig. S1), despite the crystals being grown at pH 5.3, well below the pH of membrane fusion. Exposure of SC1918/H1 HA to a range of pH conditions results in a dose-response curve with a pH of 5.76 for 50% conversion in 1h (95% confidence interval, 5.70–5.82; Fig. 4C). Since crystals did not appear for 2–3 days at pH 5.3, SC1918 HA would have been expected to adopt a post-fusion conformation in the crystals. Therefore, CR6261 appears to neutralize the virus by stabilizing the pre-fusion state and preventing the pH-dependent fusion of viral and cellular membranes.
In addition to its key role in receptor binding, HA1 is believed to act as a latch that locks HA2 in its pre-fusion conformation and prevents premature triggering of the fusion machinery. Around pH 7, the HA1 globular heads tightly pack atop HA2. However, pre- and post-fusion structures reveal that the HA1 heads must disassemble from the central 3-fold axis to initiate refolding of HA2 during membrane fusion (Fig. 4E–F). The electron density maps for CR6261-Viet04/H5 (crystals grown at pH 6.4) are consistent with a rigid molecule in a single conformation. However, although the SC1918/H1 maps (crystals grown at pH 5.3) are also excellent throughout the HA2/HA1 stem and Fab variable domains, they deteriorate towards the globular heads containing the receptor binding site at the membrane-distal end of the trimer (Fig. S6)(49). We, therefore, conclude the HA1 heads are flexible and adopt multiple conformations in our crystals, with a pivot axis running roughly through Val56 and Glu271 of HA1 in the base of the vestigial esterase domain (50). This pivot is suggestively located in the region where HA1 would be expected to peel away from HA2 during the fusion process, and may indicate that the low pH has loosened the HA1 interdomain associations. As CR6261 lies just below the hinge, it is well positioned to act as a molecular buttress to oppose the dissociation of the HA1 heads and inhibit membrane fusion. Consistent with this hypothesis, CR6261 Fab neutralizes virus as potently as intact IgG, suggesting that cross-linking adjacent HA protomers and interference by the Fc region are not critical for inhibition.
To understand why subtypes, such as H3 and H7, are not neutralized by CR6261, we conducted an extensive analysis of all non-redundant influenza A HA sequences in the NCBI FLU database (51, 52). Given its prominent role, we first examined A-helix conservation. Significantly, the CR6261 binding surface on the A-helix is almost invariant across all 16 subtypes, including H3 (Fig. 4D), whereas the opposite face of the helix, buried in the trimer interface, is more variable. A single polymorphism corresponding to Asn49 in H3 (Thr in most other subtypes) would still permit hydrogen bonding interactions with the backbone of HCDR1 Arg30 (53). Thus, to explain the low reactivity of CR6261 with H3 HA, we expanded our analysis to include the surrounding residues. Once again, most nearby residues were similar between SC1918/H1 and H3 HAs. However, whereas HA1 residues 38 and 40 of SC1918/H1 were His and Val, respectively, the same positions in H3 were Asn and Thr. Asn38 is, therefore, subject to N-linked glycosylation, as observed in H3 HA crystal structures (21, 27). As His38 lies near the center of the epitope and makes direct contact with the Fab heavy chain in the H1 and H5 complex structures, Asn 38 glycosylation would almost certainly abolish CR6261 binding to H3 HAs. Indeed, introduction of this glycosylation site in an H5 HA reduced CR6261 binding by ∼70% (15). The masking and unmasking of protein epitopes by carbohydrate to evade immune recognition play a major role in evolution of viral glycoproteins, such as HA and HIV-1 gp120 (21, 44, 54). Interestingly, Asn38/Thr40 in HA1 occurs only in the H3, H7, H10, and H15 subtypes (Fig. S7)(55) and correlates with lack of CR6261 binding or neutralization with H3, H5, and H10 isolates (15) (n.b. no data yet for H15). Thus, neutralization breadth of CR6261 is determined in large part by whether glycosylation is present at Asn38, although isolate-specific variation in other residues proximal to the epitope may also influence the interaction. Notwithstanding, CR6261 can likely neutralize isolates from 12 of the 16 influenza A subtypes: H1, H2, H4-H6, H8, H9, H11-H14, and H16.
Determination of the optimal isolates to include in the annual flu vaccine flu season can be difficult and sometimes fails, resulting in dramatic increases in infection worldwide (11, 12). In light of the persistent threat posed by zoonotic influenza viruses, such as H5N1, H7N7 and H9N2 “bird flus”, and the three major pandemics in the last century, a vaccine that could provide heterosubtypic immunity would be a tremendous advance for public health. Broadly neutralizing antibodies against conserved epitopes on HA also have immediate clinical applications in the treatment of severe influenza infections, such as H5N1. However, given the extensive antigenic diversity between subtypes coupled with inherent high mutation rates, no clear strategies for development of a more broadly-based vaccine have emerged until now.
Furthermore, rational vaccine design, using potent and broadly neutralizing antibodies as a guide, is increasingly seen as the most promising solution to combat the immense diversity in certain pathogens, such as HIV-1 (56, 57). Immunization with HIV-1 and influenza leads to a robust response against hypervariable loops/regions of the envelope glycoproteins and elicitation of neutralizing antibodies. However, these hypervariable regions are generally not functionally important and virus can readily escape antibody binding. Occasionally, antibodies may be found that recognize highly conserved epitopes on the envelope proteins that are critical for viral entry. For HIV-1, three such classes of antibodies include b12 (which recognizes the gp120 CD4 binding site), 2G12 (conserved carbohydrate clusters on gp120), and 2F5 and 4E10 (membrane proximal regions of gp41 (57, 58), that are more akin to CR6261). For influenza, only a handful of such broadly neutralizing antibodies have been described. Most notably, a mouse monoclonal antibody C179 was reported to neutralize H1, H2, and H5 subtypes (13, 59) and, more recently, broadly neutralizing, human antibodies against H1 and H5 HA (14, 15), that may well map to the same or similar epitope as CR6261. Although CR6261 recognizes a conformational epitope between HA1 and HA2, it consists of two component parts: (i) the A-helix, which accounts for most of the interacting surface and most of the polar contacts, and (ii) the HA1 region adjacent to the A-helix, which makes primarily hydrophobic contacts with CR6261. Thus, it may only be necessary to mimic the A-helix as a linear peptide in any rationally designed antigen (60). Novel strategies may be required to improve its immunogenicity, as the epitope is not particularly exposed in intact virus, although it is certainly antigenic as it reacts with high affinity to CR6261.
The conservation of this epitope suggests a critical role in membrane fusion. In late stages of fusion, the HA2 C-terminus rearranges (Fig. 4E–F) and makes extensive contacts with the A-helix. In particular, the residues that mediate all of the hydrogen bonds between the A-helix and CR6261 form hydrogen bonds and a salt bridge to this HA2 C-terminal H segment in the post-fusion conformation (Fig. 4G), strongly suggesting that the CR6261 epitope is important in the membrane fusion process. Thus, identification of a highly conserved epitope in the HA provides a new lead for the design of the next generation of anti-virals and marks the first step towards the Holy Grail of influenza vaccinology: the development of a durable and cross-protective “universal” vaccine against influenza A.
Supplementary Material
References and Notes
- 1.Schafer JR, et al. Virology. 1993;194:781. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- 2.Reid AH, Fanning TG, Hultin JV, Taubenberger JK. Proc Natl Acad Sci U S A. 1999;96:1651. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawaoka Y, Krauss S, Webster RG. J Virol. 1989;63:4603. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bean WJ, et al. J Virol. 1992;66:1129. doi: 10.1128/jvi.66.2.1129-1138.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beigel J, Bray M. Antiviral Res. 2008;78:91. doi: 10.1016/j.antiviral.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowen AC, Palese P. Infect Disord Drug Targets. 2007;7:318. doi: 10.2174/187152607783018736. [DOI] [PubMed] [Google Scholar]
- 7.Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. JAMA. 2006;295:891. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 8.Kiso M, et al. Lancet. 2004;364:759. doi: 10.1016/S0140-6736(04)16934-1. [DOI] [PubMed] [Google Scholar]
- 9.Russell RJ, et al. Proc Natl Acad Sci U S A. 2008;105:17736. doi: 10.1073/pnas.0807142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The most common vaccine formulations contain influenza A H1N1, H3N2, and influenza B components.
- 11.Salzberg S. Nature. 2008;454:160. doi: 10.1038/454160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MMWR Morb Mortal Wkly Rep. 2008;57:393. [PubMed] [Google Scholar]
- 13.Okuno Y, Isegawa Y, Sasao F, Ueda S. J Virol. 1993;67:2552. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kashyap AK. Proc Natl Acad Sci U S A. 2008;105:5986. doi: 10.1073/pnas.0801367105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Throsby M, et al. PLoS ONE. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The complex was generated using a slight stoichiometric excess of antibody to HA (∼3:1) to saturate the potential binding sites on the HA trimer, and then purified by gel filtration.
- 17.Stevens J, et al. Science. 2006;312:404. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 18.Gamblin SJ, et al. Science. 2004;303:1838. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 19.Yamada S, et al. Nature. 2006;444:378. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 20.The helices are designated A and CD (see Figs. 4E–F) in accordance with the nomenclature established in ref. 22.
- 21.Wilson IA, Skehel JJ, Wiley DC. Nature. 1981;289:366. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 22.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Nature. 1994;371:37. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 23.While baculovirus-expressed HAs lacking a polybasic cleavage site are typically secreted in the uncleaved form (HAO), treatment with trypsin yields the fusion-competent HA1/HA2. In our crystals, both HAs have been cleaved to HA1 and HA2. CR6261 binds to both cleaved and uncleaved HA.
- 24.Fleury D, Daniels RS, Skehel JJ, Knossow M, Bizebard T. Proteins. 2000;40:572. [PubMed] [Google Scholar]
- 25.Barbey-Martin C, et al. Virology. 2002;294:70. doi: 10.1006/viro.2001.1320. [DOI] [PubMed] [Google Scholar]
- 26.Fleury D, et al. Nat Struct Biol. 1999;6:530. doi: 10.1038/9299. [DOI] [PubMed] [Google Scholar]
- 27.Fleury D, Wharton SA, Skehel JJ. Nat Struct Biol. 1998;5:119. doi: 10.1038/nsb0298-119. [DOI] [PubMed] [Google Scholar]
- 28.Presumably due to the well-established flexibility of the constant domains relative to the variable domains, the paucity of crystal contacts to the constant region, and the insertion of four extra residues in the light-chain elbow region. Indeed, we ultimately succeeded in building approximately 35 residues each of CH1 and CL in the region closest to the elbow. A comparison of the CR6261 Fabs from the Viet04/H5 structure with those from the SC1918/H1 structure revealed a ∼13°rotation of the constant domains with respect to the variable domains between the two structures.
- 29.The closest distance is approximately 8 Å To our knowledge, broadly neutralizing antibody b12, which targets HIV-1 gp120, is the only other example of a non-camelid/shark antibody that makes absolutely no light chain interactions with its protein antigen (38). Whether 15 heavy-chain only binding correlates with the potency and cross-reactivity of antibodies, such as CR6261 and b12, is provocative, but not yet proven (15, 36).
- 30.5241of 5261 HA protein sequences were found to have a glycosylation consensus motif (Asn-Xaa-Ser/Thr) at position 154 of HA2. While only the first one to two sugar residues are well ordered in our crystals, the entire glycan is presumably much larger, and would likely interfere with light-chain interactions with the HA protein surface in the orientation adopted by CR6261. Details of the sequence dataset used can be found in footnote 51 and in the Supporting Online Material.
- 31.The isolation of heavy chain dominant binders may be enhanced in phage display by the separation and random repairing of heavy and light chains during library preparation. This is expected to favor heavy chains that can bind antigen with little or no contribution from the light chain.
- 32.Lerner RA. Angew Chem Int Ed Engl. 2006;45:8106. doi: 10.1002/anie.200603381. [DOI] [PubMed] [Google Scholar]
- 33.Four of these five H-bonds are mediated by the HCDR1 backbone, with the hydroxyl of Ser31 making the only polar side-chain contact. A sixth H-bond to the A-helix is formed between the backbone amide of Tyr98 from HCDR3 and Gln42 (Table S2).
- 34.Garcia KC, et al. Science. 1996;274:209. [Google Scholar]
- 35.Asp19 is unchanged in the CR6261-Viet04/H5 complex and accepts a single H-bond via its backbone carbonyl group. However, in CR6261-SC1918/H1, the density maps indicate some flexibility in this region that would allow Asp 19 to rotate and reach over to H-bond with Thr56OG1, replacing the existing water-mediated H-bond with Ile18O.
- 36.Huang CC, et al. Proc Natl Acad Sci U S A. 2004;101:2706. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luftig MA, et al. Nat Struct Mol Biol. 2006;13:740. doi: 10.1038/nsmb1127. [DOI] [PubMed] [Google Scholar]
- 38.Zhou T, et al. Nature. 2007;445:732. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.In other previously characterized structures, VH1–69 restriction was proposed to result from the role of the unique protruding hydrophobic HCDR2 in anchoring HCDR3 interaction rather than in preserving precise molecular interactions (36).
- 40.definitions after CDR, Chothia C. J Mol Biol. 1992;227:799. doi: 10.1016/0022-2836(92)90224-8. [DOI] [PubMed] [Google Scholar]
- 41.Thr28Pro and Ser30Arg in the FR1 loop directly preceding HCDR1 and Ser74Phe mutation in FR3.
- 42.Davis MM. Semin Immunol. 2004;16:239. doi: 10.1016/j.smim.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Gerhard W, Yewdell J, Frankel ME, Webster R. Nature. 1981;290:713. doi: 10.1038/290713a0. [DOI] [PubMed] [Google Scholar]
- 44.Wiley DC, Wilson IA, Skehel JJ. Nature. 1981;289:373. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 45.Barbey-Martin C, et al. Virology. 2002;294:70. doi: 10.1006/viro.2001.1320. [DOI] [PubMed] [Google Scholar]
- 46.Skehel JJ, et al. Proc Natl Acad Sci U S A. 1982;79:968. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The HA pre-fusion state is highly protease resistant, while the post-fusion state is much more susceptible to protease degradation (46).
- 48.Chen J, Skehel JJ, Wiley DC. Proc Natl Acad Sci U S A. 1999;96:8967. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Despite considerable efforts to reposition HA1, including rigid body refinement of various subdomains, simulated annealing, and manual rebuilding, no improvement of fit was achieved for the membrane-distal portion of HA. More details can be found in the Supporting Online Material.
- 50.The best fit of HA1 to the density results in a 2 Å radial displacement of the receptor binding site compared to the unliganded form.
- 51.Description of the analyzed sequence dataset: A total of 5261 clones comprising all full-length, non-redundant influenza A hemagglutinin protein sequences were analyzed. Only lab strains were excluded from the analysis, to avoid the effects of any mutations involved in adaption to culture in vitro. The dataset included 854 H1, 125 H2, 1582 H3, 144 H4, 1384 H5, 261 H6, 359 H7, 17 H8, 334 H9, 59 H10, 71 H11, 25 H12, 26 H13, 2 H14, 5 H15, and 13 H16 sequences. Details of the methods used for analysis are available in the Supporting Online Material. Sequences were retrieved from the URL listed below on January 14, 2009. http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html
- 52.Our analysis included a total of 5261 protein sequences, encompassing all 16 subtypes and isolates from all species, including humans.
- 53.Although we cannot rule out the possibility that the slightly larger Asn could result in unfavorable interactions between the Fab, sufficient space is available to accommodate the Asn side chain.
- 54.Kwong PD, et al. Nature. 1998;393:648. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.2004 of 2005 clones are predicted to be glycosylated based upon the presence of an Asn-Xaa-Ser/Thr motif at residues 38–40 (1581/1582 H3, 359/359 H7, 59/59 H10, and 5/5 H15 clones). Only a single H3 clone (A/New York/252/1999(H3N2)) lacked the consensus sequence. In contrast, not a single glycosylation motif was present in 3256 clones from the H1, H2, H4-H6, H8, H9, H11-H14, and H16 subtypes. See Fig. S7.
- 56.Walker BD, Burton DR. Science. 2008;320:760. doi: 10.1126/science.1152622. [DOI] [PubMed] [Google Scholar]
- 57.Burton DR, Stanfield RL, Wilson IA. Proc Natl Acad Sci U S A. 2005;102:14943. doi: 10.1073/pnas.0505126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zwick MB, Burton DR. Curr HIV Res. 2007;5:608. doi: 10.2174/157016207782418443. [DOI] [PubMed] [Google Scholar]
- 59.Smirnov YA, et al. Acta Virol. 1999;43:237. [PubMed] [Google Scholar]
- 60.Whereas polar interactions impose charge, spatial, and steric constraints on an interacting surface, hydrophobic interactions are less specific but provide binding energy. For example, formation of a hydrogen bond to a particular donor requires a suitable acceptor group (“charge”) to be placed on a particular position of the interaction surface and in a particular orientation (spatial), and the interacting residues and their neighbors must be able to physically accommodate the interacting residues (steric). In contrast, it is relatively likely that two or more aliphatic side chains will end up in close proximity and be able to exclude solvent. Thus, while the hydrophobic interactions with HA1 may help to stablize CR6261 binding to HA and increase the free energy of binding, the polar interactions with conserved residues in the A-helix are probably most critical for the specific recognition of muliple HA subtypes.
- 61.This work was supported in part by NIH grant AI-058113 (I.A.W.), a pre-doctoral fellowship from the ARCS Foundation (D.C.E.) and The Skaggs Institute. The JCSG is supported by NIH NIGMS (U54 GM074898). The GM/CA CAT 23-ID-B beamline has been funded in whole or in part with Federal funds from NCI (Y1-CO-1020) and NIGMS (Y1-GM-1104). Use of the Advanced Photon Source (APS) was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. The authors would also like to thank the Henry Tien of the Robotics Core at the JCSG for automated crystal screening; the staff of the APS GM/CA CAT 23-ID-B for beamline support; X. Dai, R. Stanfield, for excellent assistance with data collection, and processing and analyses, Max Crispin, Mark Wormald and Dennis Wolan for valuable comments, and Richard Lerner for insightful discussion. This is publication 19980 from The Scripps Research Institute. Coordinates and structure factors are deposited in the Protein Data Bank (XXXX and YYYY for CR6261-SC1918/H1 and CR6261-Viet04/H5, respectively).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.