Abstract
Quantification of cellular responses to stimuli is challenging. Cells respond to changing external conditions through internal structural and compositional and functional modifications, thereby altering their transport and mechanical properties. By properly interpreting particle-tracking microrheology, we evaluate the response of live cells to cytoskeletal disruption mediated by the drug nocodazole. Prior to administering the drug, the particles exhibit an apparently diffusive behavior that is actually a combination of temporally heterogeneous ballistic and caged motion. Selectively depolymerizing microtubules with the drug causes actively crawling cells to halt, providing a means for assessing drug efficacy, and making the caged motion of the probes readily apparent.
I. INTRODUCTION
Mammalian cells, although composed mostly of water, are complex and highly organized biological systems that have extremely crowded interiors and are surrounded by outer phospholipid bilayer membranes. A dynamic cytoskeleton helps establish cell shape, elasticity, and mechanical strength. The cytoskeleton is a rapidly responding three-dimensional scaffold, composed of actin filaments, intermediate filaments, and microtubules bound together by cross-linking proteins that are essential for cell movement, for nuclear and cell division, and for organelle organization. The cytoskeleton adapts to facilitate cell survival, and microtubules function as molecular-motor highways for intracellular transport and as controllers of intracellular locomotion. However, real-time quantitative evaluation of the mechanical response of cells to changes in cytoskeletal structure, such as the depolymerization of microtubules (which is a frequent goal of anticancer drugs), is still poorly characterized. One technique that can elucidate dynamic changes in the internal structure and transport properties of live cells following disruption of the cytoskeleton is particle-tracking microrheology.1,2
Cytoskeletal disruption, by selective removal of specific molecules through genetic or biochemical approaches, has been used to control cell function and fate. Several drugs induce microtubule depolymerization, which inhibits cell division and causes arrest in the G2/M phase of the cell cycle.3 In addition, microtubule removal has been shown to affect filopodia and inhibit cell motility.4,5 Prolonged growth arrest typically results in cell death by apoptosis, or programmed cell death,6 making microtubule-dissociating drugs a front-line choice for combating cancer. Given the medical importance of microtubule dissociation, our study evaluates the adaptive response of cells to nocodazole, an anticancer drug that targets microtubule depolymerization.
Induced changes in cell micromechanics provide a quantifiable measure of the cellular response to microtubule depolymerization. Measurements made on the membrane using atomic force microscopy (AFM) have shown a marked decrease in membrane stiffness after incubation with nocodazole that reflects a dependence on tyrosinated, or labile, microtubules.7 However, membrane AFM studies cannot quantify changes in structural and transport properties within live cells, requiring a different approach. For this purpose, we used particle-tracking microrheology, which facilitates probing local regions deep within cells for rapid changes in rheology, without significantly perturbing cellular homeostasis.
Local rheological properties of a fluid, such as the heterogeneous active complex fluid comprising the cellular interior, are partially responsible for determining the response, or motion, of thermally driven small probes embedded within the fluid.1 In microrheology, the transport and motion of probes, typically fluorescent spheres, particles, or molecules is tracked and recorded over time, and local mechanical properties in the vicinity of the probes are deduced, depending upon the driving mechanisms. Through particle tracking, captured trajectories provide a measure of the type of motion the particle is undergoing, such as subdiffusive, diffusive, or a combination of diffusive and convective (sometimes known as superdiffusive). If the motion of the probes is governed solely by thermal fluctuations, then the local rheological properties of the material, such as the creep compliance or viscoelastic modulus, can be determined from probe trajectories through the fluctuation-dissipation theorem (FDT) and generalized Stokes-Einstein relation (GSER).1 The GSER is most applicable for inactive biofluids at equilibrium, including biopolymer solutions or ATP-depleted cells,8 and breaks down in active biofluids and cells that consume adenosine triphosphate (ATP) to respond to and generate force by reorganizing the cytoskeleton.9,10 Reviews of the theory and technical aspects of thermal diffusion microrheology in complex liquids11,12 and in cells13 are available. In this study, we determine and interpret the effect of nocodazole-induced microtubule depolymerization on the trajectories of fluorescent intracellular particles as local probes.
II. EXPERIMENTAL PROCEDURE AND APPARATUS
A. Cell culture and nanoparticle introduction
Murine NIH3T3 fibroblasts are harvested at 60–80% confluence and transferred into CO2-independent media (Gibco, Invitrogen Life Technologies, Carlsbad, CA) with 10% fetal bovine serum, L-glutamine, sodium pyruvate, non-essential amino acids, and antibiotics. The CO2-independent medium maintains cell viability for prolonged periods (up to 72 h) with temperature control alone.7 Cells are plated at 2.5×104 cells/cm2 onto chambered coverglass wells (Nunc, Rochester, NY). Concurrent with the replating process, carboxylate-modified, yellow-green fluorescent polystyrene microspheres (505/515 excitation/emission), 100 nm in diameter (Molecular Probes, Invitrogen Life Technologies, Eugene, OR) are placed in the wells for uptake by the cells during their adherence to the coverglass. In this way, some particles became immobilized between the cells and the coverglass. These serve as markers for the cell-coverglass interface, establishing a fixed reference point for distinguishing cell crawling motion from a possible drift of the microscope stage. After 16 h in a controlled temperature incubator, cell media and any unincorporated particles are aspirated, cells are washed once with phosphate-buffered saline (Gibco, Invitrogen Life Technologies, Carlsbad, CA), and fresh CO2-independent media is added. Cells are then equilibrated for 6 h before conducting microrheology tests.
B. Microtubule depolymerization treatment
Microtubules are depolymerized with nocodazole (methyl[5-(2-thienyl-carbonyl)-1H-benzimidazol-2-yl]-carbamate, Sigma) in dimethyl sulfoxide (DMSO) added at a final concentration of 2 μM to each chamber well. NIH3T3 cells tolerate the DMSO carrier, used as a control in all experiments, and at least 10 μM nocodazole for up to 3 h with no increase in cell death by flow cytometry and no alterations in actin or intermediate filament cytoarchitecture by immunofluorescence microscopy.7 Drug washout was performed by washing the cells once with phosphate buffered saline followed by addition of fresh prewarmed DMSO-containing medium. The same cells were evaluated after washing; however, if cells were lost during washing, neighboring well-attached cells were examined.
C. Particle tracking in living cells
To identify probe locations and obtain trajectories, we image cells and particles with a Nikon TE2000U inverted fluorescent microscope equipped with a 100× oil immersion lens (NA 1.4) and mercury-lamp based fluorescence. Cells are maintained on the microscope for several hours at 37 °C, using a custom-made temperature-controlled stage and objective heater (Bioscience Tools, San Diego, CA). Particle motion within the cells is recorded directly onto a computer at 30 frames per second using a Sony X75 CCD camera. Following addition of nocodazole to the wells, 20-s videos of particle motion are collected every 2–3 min up to 30 min, to evaluate the immediate and short-term effects of the drug. We focus on specific cells and particles throughout each experiment to allow continuous evaluation of microrheological evolution. Photobleaching of the fluorescent particles leads to deterioration of the signal-to-noise ratio, but we are still able to measure useful trajectories over this limited time period. We obtain these trajectories by performing frame-by-frame localization of a video movie using specialized code in MATLAB 7.0 (Mathworks Inc., Natick, MA), based on established tracking protocols.14 Detailed analyses of experimentally obtained trajectories are performed in a separate MATLAB module.
In thermal fluctuation microrheology, the two-dimensional (2D) time-dependent mean-square displacement (MSD), 〈Δr2(t)〉, can be used to accurately determine local mechanical properties of passive materials. However, probe particles moving in heterogeneous active viscoelastic media can be transported in a variety of ways, so naively applying thermal fluctuation microrheology is usually inappropriate and can lead to a misinterpretation of the local mechanical properties.8,15-18 Over certain time scales, the MSD can sometimes be described by a time-dependent power law, 〈Δr2(t)〉∼tα, where the diffusive exponent,19 α, can be related to the type of motion a particle is undergoing over a range of correlation times, t. The modes of probe motion observed are as follows: local elastic trapping (α≈0), sub-diffusion (α<1), diffusion (α=1), a combination of diffusion and convection (α>1), and ballistic motion (also known as pure convection) (α≅2). A combination of diffusion and convection can occur, for example, in thermally diffusing particles within a crawling cell. If the convective motion due to crawling does not have a fixed speed and direction, but instead changes continuously during the observation period, the diffusive exponent calculated by averaging over the entire time range may have a value between 1 and 2. In some cases, this exponent could appear to be constant over a fairly wide dynamic range in time. However, in the simplest case when the convection has a constant velocity, then the diffusive exponent would cross over from 1 (diffusive) at short times to 2 (ballistic) at long times; the diffusive exponent would still be between 1 and 2, but it would not remain constant over a wide dynamic range in time. Empirically, we choose a transition criterion between subdiffusive and trapping regimes to be α<0.25. For elastically trapped particles, we define a diameter of the probed microdomain, or cage, surrounding a particle. We assume that the cage is spherical and can be approximated at twice the probe-particle diameter plus the square root of the plateau value of the MSD.20 In flexible materials, such as the cytoskeleton, this approximation provides an overestimation of the local cage diameter.15
III. RESULTS
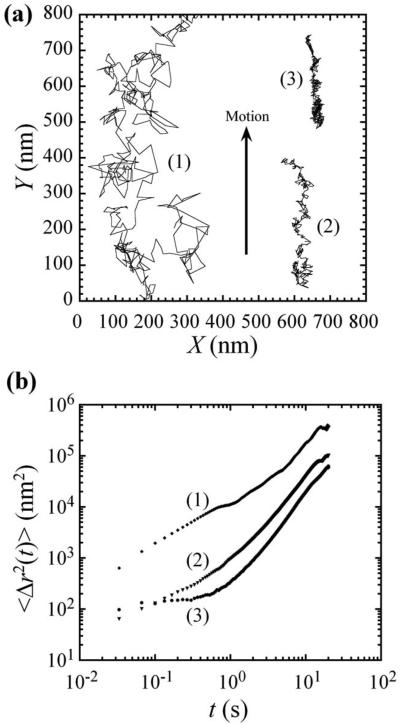
We evaluate embedded probe motion within equilibrated cells by picking one to three cells in a single field of view that adhere tightly to the coverglass, such that their filopodia are spread on the surface of the coverglass. Typically 20–50 particles per cell, distributed within the cytoplasm, are evaluated simultaneously. Due to their large size, probe particles remain outside the nucleus, as verified by confocal microscopy. Observations of particle transport in those cells revealed complex trajectories of superimposed ballistic, directional motion and a variety of thermally induced modes of motion, including intermittent subdiffusion and trapping. Ballistic motion (α→2) was detected in all cells examined and was from cells crawling on the substrate. This motion was in distinct directions at various regions within the same sample, excluding stage drift. The average crawling speed was 5–20 nm/s, as calculated from end-to-end distances of 20-s trajectories over 30 min. These speeds are two to three orders of magnitude lower than molecular motors,13 and become even lower when the glass substrate is coated with poly-L-lysine (data not shown). Therefore, this active element of probe motion did not result from molecular motor-induced transport.21 Prior studies have also shown that particles attached to the membranes of cells that have been prevented from crawling may exhibit a combination of convection and diffusion, or “superdiffusion,” with α≅1.6.10 These membrane-attached particles are only indirectly connected to the cytoskeleton. Since cells do not move at a constant speed or in a spatially coordinated manner, and a combination of convection and diffusion can also occur for motile cells, accounting for cell crawling in evaluating particle motion is complex, and convective trajectories may differ significantly in direction and magnitude even within a single cell.
In addition to cell crawling, during some limited intervals of time, we have observed diffusive motion, subdiffusive motion, and elastic trapping (Fig. 1). This spectrum of motions can reflect transport within crowded regions of the cell cytoplasm, where the cytoskeleton and other microstructures or organelles affect the motion of the particles.22 Probe particles very near to each other usually exhibited qualitatively similar transport behavior; however, in specific cases, significant variations in the trajectories have been observed for two nearby particles within a given cell, reflecting the heterogeneity of the intracellular microenvironments. For this reason, two-particle thermal microrheology,23-25 which relies on the relative diffusive motion of two closely located particles, when naively applied, joins single-particle thermal microrheology in its inability to accurately describe microscopic mechanical properties inside active living cells.
FIG. 1.
Particle tracking microrheology with 100 nm diameter fluorescent polymer spheres embedded within NIH3T3 fibroblast cells. (a) 20 s trajectories of typical transport in NIH3T3 cells. The approximate speeds of particles (2) and (3), calculated by end-to-end displacement, are 18 and 12 nm/s, respectively. Particle 1 shows a partial trajectory that continues past the top of the figure. (b) Time-dependent mean-square displacements (MSDs) of the particles in (a). All the particles exhibit directional motion correlated to cell crawling (arrow).
We examine individual probe-particles to determine the micromechanical response of cells after selective microtubule depolymerization. After obtaining baseline tracking values in unperturbed cells, nocodazole is added to the culture and the time-dependent evolution of cellular micromechanics is evaluated as a function of “waiting time” (i.e., incubation time), twait, after the addition of the drug. During treatment, the extent of cellular attachment to the surface of the coverglass did reveal some qualitative differences, varying from cell to cell, especially in filopodia extension. Most cells retract their filopodia during nocodazole incubation, and some cells eventually dislodge. Those that dislodge are no longer available for microrheological characterization. For those that remain attached, filopodia retraction occurs quickly, typically within 0.1–1 s, leading to difficulty in probe tracking. Hence, filopodia-localized probes are not included in our tracking analyses, during and following this retraction. Very few cells remain visually unperturbed throughout the duration of the experiment (30 min), indicating excellent treatment efficacy of the nocodazole for nearly all NIH3T3 cells, consistent with previous AFM studies using nocodazole.7
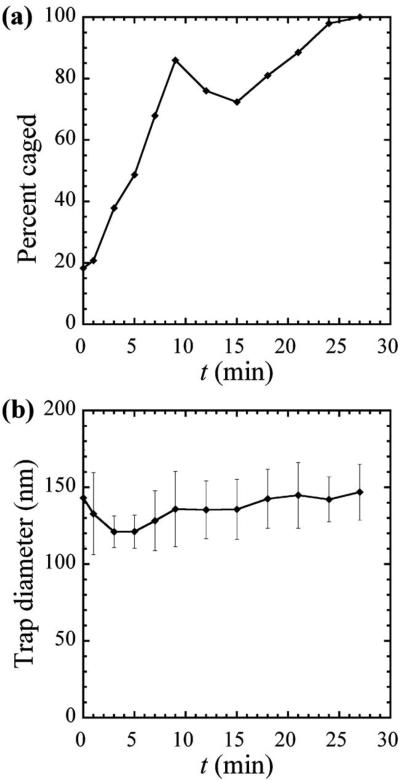
In our experiments, embedded particles are not actively transported and do not travel large distances within the cell, as the cell itself crawls on the substrate. Therefore, after accounting for cell crawling,23,26 each probe particle actually remains close to its starting point prior to nocodazole treatment. We do not ensemble-average individual traces to preserve the individuality of the trajectories and MSDs of specific particles in the heterogeneous cellular environments. However, on the whole, the time-dependent averages contain similar dynamical hallmarks. Up to twait=20 min, hundreds of particles can be considered, whereas for twait>20 min, due to photobleaching, only tens of particles could be reliably tracked. Cell viability is not affected during the examination period, as shown previously under similar conditions.7 Based on the average changes in the long-time behavior of the MSDs, where ballistic crawling motion of the locally caged particles plays an important role, we are able to show the efficacy of the drug over time after its introduction (Fig. 2). The depolymerization of the microtubules leads to a cessation of cell crawling, and most of the particles appear to be caged after about 5 min. Drug washout was then performed to determine the recovery of cell motion and mechanics. Although the effects from nocodazole were easily detected by 5–10 min, drug removal did not produce a discernible recovery of cell mechanics for up to 45 min of monitoring (data not shown).
FIG. 2.
Average probe transport characteristics as a function of incubation time, twait, in nocodazole, averaged over trajectories of particles in 15 cells. (a) Percent of the number of particles exhibiting locally trapped to subdiffusive motion in nocodazole, averaged over hundreds of particles. (b) Probed microdomain diameter in which particles become trapped, assuming a spherical cage, averaged over tens of particles.
Our observations of probe motion indicate that microtubule depolymerization within the cytoskeleton by nocodazole results in cessation of cell locomotion. About 15 min after the application of nocodazole, the directional motion of the probes observed before treatment has disappeared in nearly all cells, and the remaining motion resembles that of a particle in a dominantly elastic cage. The cessation of directional probe motion and appearance of caged diffusion correlates directly with the stoppage of intermittent cell locomotion. In addition, the intracellular microenvironment changes due to the depolymerization of microtubules. Figure 2 reveals the measured temporal dependence of the dynamics of particles in the cells on nocodazole. Specifically, we have examined the transition between a combination of ballistic motion plus subdiffusion or caging yielding α≈1 or α<1 at short time scales and α>1 at longer times [Fig. 1(b)], and simple caging with α≈0 at long times. These results are averaged from several experiments, including a total of 15 different cells containing several hundred probe particles. The proportion of caged particles, as apparent from the MSDs, increases over time after adding nocodazole. The transformation is sharp during the first 10 min, and then approaches a plateau corresponding to only elastically trapped particles near 100% toward 30 min [Fig. 2(a)]. The steep increase in Fig. 2(a) at small twait results from a reduction in the crawling of the cells due to the transport of nocodazole into them, leading to a nearly constant mean-square displacement at long times, indicative of caged diffusion. The peak around twait=10 min may be due to a combination of changes in the cells' internal structure and mechanics due to cell treatment and our empirical criteria for caging, but determining the exact origin of this feature requires further study. In membrane measurements with an AFM,7 we have observed similar time scales for changes in the Young modulus, an indicator of membrane stiffness, under similar conditions, including a sharp transition after 10 min of treatment. The correspondence between the time scales of microrheological and AFM measurements indicates a correlation between membrane and cytoplasm responses of the cells during microtubule depolymerization.
To better characterize the local interactions of the cytoplasm with the probe particles, we estimated the effective diameter of local microdomains where particles become caged. For short incubation times in nocodazole, the few particles that are locally trapped have been used for this calculation, whereas at longer times, most particles are trapped, providing improved statistics. We obtain a trap diameter, assuming a spherical trap,15,20 that is approximately constant at all measured times, within the statistical uncertainty [Fig. 2(b)]. The observed trap sizes, as we have defined them, depend on the probe's size, since the lower limit for detection is the probe diameter. Observed traps are typically between 110 and 250 nm in diameter, and a few traps are larger, around 300 nm. Particles exhibiting elastic trapping at time zero are lodged in microdomains close to their size during the pre-incubation period. In contrast, particles that initially had MSDs with α≈1 at long t and small twait subsequently evolved over the course of minutes to provide a signature of trapped behavior α≈0 due to the effect of the nocodazole.
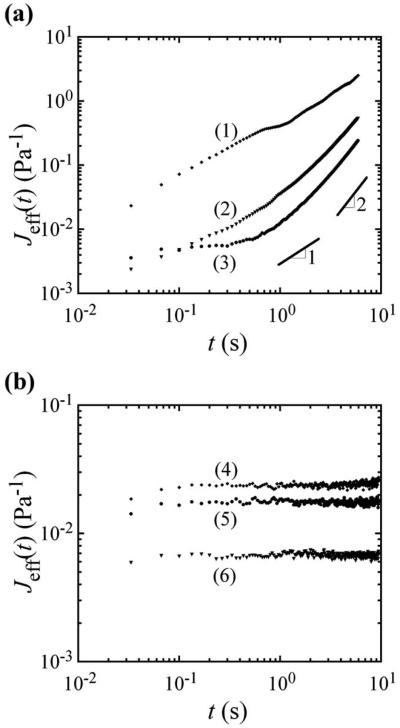
Because the cells can actively crawl, we report an “effective” creep compliance, Jeff, recognizing that, when ballistic motion contributes significantly to the MSD, the rheological meaning implied by the diffusion-based GSER should not be taken literally. This “effective” creep compliance is obtained directly from the MSD, Jeff(t)=πa〈Δr2(t)〉/kBT, where a is the particle's radius, kB is Boltzmann's constant, and T is the absolute temperature.27,28 The effective creep compliances for three representative trajectories both at small and large twait are presented in Figs. 3(a) and 3(b), respectively. At early times, t, subdiffusion is present even shortly after administering the drug [Fig. 3(a) and also at long twait, Fig. 3(b)]. However, right after administering the drug, at later times, Jeff(t), which is proportional to the MSD, appears to increase linearly. When the log-slope is 1, this could easily be misinterpreted as diffusion of the probe in a viscous medium, and an effective viscosity could be extracted, but this would be incorrect and not rheologically meaningful. As shown in previous simulations of particle motion in heterogeneous active liquids,15 a combination of intermittent cell crawling and caged diffusion can also lead to a slope of the MSD of α≈1 for large t, even when the particle is not diffusing in a classical sense due to thermal fluctuations (see Fig. 5c in Ref. 15). In effect, our data support the results of the simulation, validating that what may look like passive diffusion of probe particles in cells is actually a result of more complex active dynamics. The crawling motion of the cell and caged diffusion are sufficient to yield this effect. Moreover, intermittent internal convection of particles by molecular motors inside the cell's crowded environment could also provide a similar signature, although this was not observed here. After incubation with nocodazole, most particles become elastically trapped with a log-slope close to zero at all time scales [Fig. 3(b)], corresponding to the disappearance of the active element resulting from motion of the whole cell on the glass substrate. As microtubules are depolymerized, the cell's crawling motion is greatly reduced or stopped, which also affects particle motion in the cells, leading to what appears to be more confinement in the MSD and Jeff(t). Even after microtubule dissociation, from the plateau MSDs in Fig. 3(b), we can estimate an approximate average scale for the cells' internal elastic modulus, G≈1/J≈100 Pa, which has a magnitude that is in the range corresponding to a cross-linked semiflexible biopolymer network of actin.
FIG. 3.
Effective creep compliance, Jeff, proportional to the measured MSD, as a function of correlation time, t, in nocodazole. (a) Typical Jeff for three particles at waiting times twait≈0. The represented particles are the same as in Fig. 1. At long times, t, the log-slope α≈1, but this cannot be interpreted as a viscosity because caged diffusion is modified by the ballistic motion of cell crawling. (b) Typical Jeff of three particles, not the same particles as in (a), after twait≈21 min in nocodazole. At long times, the log-slope α≈0, corresponding to caged diffusion and the disappearance of cell crawling. Corresponding trap diameters, calculated as for use in Fig. 2(b), are as follows from top to bottom: 202, 187, and 154 nm, respectively.
IV. CONCLUSIONS
Our measurements of changing particle-transport properties indicate an evolution of the internal structure of the cell after administering nocodazole, as expected during microtubule dissociation. The cells appear to reach a new metastable state after twait≈10 min, findings that also validate and confirm studies at the membrane using AFM,7 here using a distinct intracellular approach. After about twait≈20 min, nearly all cells have ceased their crawling, and particle trajectories more accurately reflect the locally caged dynamics within the crowded cytoplasm. The appearance of caged dynamics with α=0 is not absolute evidence that the elasticity inside the cell increases due to the depolymerization. By contrast, it clearly indicates that the ballistic component of the probe motion due to cell crawling is no longer present and dominating the MSDs at long times t. Cellular viability is not affected at the examined time scales. In addition, no stiffening is observed that may be attributed to rigor mortis. Moreover, local measurements on the cell membrane probing the region immediately beneath it have shown a decrease rather than an increase in stiffness.7 Such changes may be part of a compensatory response to reinforce the cell mechanical strength following microtubule depolymerization and are consistent with suggested tensegrity models of cellular mechanics.29,30 The observed response time depends most likely on diffusion of the drug into cells, internalization, and the cellular response. The cell is a highly dynamic, heterogeneous fluid-based system that adapts to changes and perturbations in its external and internal environments to maintain viability. Our study illustrates the dynamic response to changing the environment within cells and strengthens the need for approaches to quantifying mechanical cell responses to facilitate analyses of the involved molecular pathways.
ACKNOWLEDGMENTS
This work was partially supported by the Institute for Cell Mimetic Space Exploration—CMISE (NASA URETI Institute Grant No. NCC 2-1364) and NIH Grant Nos. CA90571, CA107300, and PN2EY018228, a NIH Roadmap for Medical Research. M.A.T. is a Scholar of the Leukemia and Lymphoma Society. T.G.M. thanks the American Chemical Society (Grant No. PRF 42858-AC7) for partial support.
Contributor Information
Daphne Weihs, Department of Pathology and Laboratory Medicine and Department of Chemistry and Biochemistry, University of California at Los Angeles, Los Angeles, California 90095, USA.
Thomas G. Mason, Department of Chemistry and Biochemistry, Department of Physics and Astronomy, and California NanoSystems Institute, University of California at Los Angeles, Los Angeles, California 90095, USA
Michael A. Teitell, Department of Pathology and Laboratory Medicine, California NanoSystems Institute, Institute for Cell Mimetic Studies, Broad Center of Regenerative Medicine and Stem Cell Research, and Jonsson Comprehensive Cancer Center, University of California at Los Angeles, Los Angeles, California 90095, USA.
References
- 1.Mason TG, Weitz DA. Optical measurements of frequency-dependent linear viscoelastic moduli of complex fluids. Phys. Rev. Lett. 1995;74:1250. doi: 10.1103/PhysRevLett.74.1250. [DOI] [PubMed] [Google Scholar]
- 2.Mason TG, Ganesan K, vanZanten JH, Wirtz D, Kuo SC. Particle tracking microrheology of complex fluids. Phys. Rev. Lett. 1997;79:3282. [Google Scholar]
- 3.Zieve GW, Turnbull D, Mullins JM, McIntosh JR. Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor Nocodazole: Nocodazole accumulated mitotic cells. Exp. Cell Res. 1980;126:397. doi: 10.1016/0014-4827(80)90279-7. [DOI] [PubMed] [Google Scholar]
- 4.Bershadsky AD, Vaisberg EA, Asiliev JM. Pseudopodia activity at the active edge of migrating fibroblasts is decreased after drug-induced microtubule depolymerization. Cell Motil. Cytoskeleton. 1991;19:152. doi: 10.1002/cm.970190303. [DOI] [PubMed] [Google Scholar]
- 5.Liao G, Nagasaki T, Gundersen GG. Low concentrations of nocodazole interfere with fibroblast locomotion without significantly affecting microtubule level: Implications for the role of dynamic microtubules in cell locomotion. J. Cell. Sci. 1995;108:3473. doi: 10.1242/jcs.108.11.3473. [DOI] [PubMed] [Google Scholar]
- 6.Mollinedo F, Gajate C. Microtubules, microtubule-interfering agents and apoptosis. Apoptosis. 2003;8:413. doi: 10.1023/a:1025513106330. [DOI] [PubMed] [Google Scholar]
- 7.Pelling AE, Dawson DW, Carreon DM, Christiansen JJ, Shen RR, Teitell MA, Gimzewski JK. Distinct contributions of microtubule subtypes to cell membrane shape and stability. Nanomedicine. 2007;3:43. doi: 10.1016/j.nano.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman BD, Massiera G, Van Citters KM, Crocker JC. The consensus mechanics of cultured mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10259. doi: 10.1073/pnas.0510348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizuno D, Tardin C, Schmidt CF, MacKintosh FC. Nonequilibrium mechanics of active cytoskeletal networks. Science. 2007;315:370. doi: 10.1126/science.1134404. [DOI] [PubMed] [Google Scholar]
- 10.Bursac P, Lenormand G, Fabry B, Oliver M, Weitz DA, Viasnoff V, Butler JP, Fredberg JJ. Cytoskeletal remodelling and slow dynamics in the living cell. Nat. Mater. 2005;4:557. doi: 10.1038/nmat1404. [DOI] [PubMed] [Google Scholar]
- 11.Gardel ML, Valentine MT, Weitz DA. Microdiagnostics. Springer-Verlag; Berlin: 2005. Microrheology. [Google Scholar]
- 12.Waigh TA. Microrheology of complex fluids. Rep. Prog. Phys. 2005;68:685. doi: 10.1088/0034-4885/79/7/074601. [DOI] [PubMed] [Google Scholar]
- 13.Weihs D, Mason TG, Teitell MA. Bio-microrheology: A frontier in microrheology. Biophys. J. 2006;91:4296. doi: 10.1529/biophysj.106.081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crocker JC, Grier DG. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 1996;179:298. [Google Scholar]
- 15.Weihs D, Teitell MA, Mason TG. Simulations of complex particle transport in heterogeneous active liquids. Microfluid. Nanofluid. 2007;3:227. [Google Scholar]
- 16.Bursac P, Fabry B, Trepat X, Lenormand G, Butler JP, Wang N, Fredberg JJ, An SS. Cytoskeleton dynamics: Fluctuations within the network. Biochem. Biophys. Res. Commun. 2007;355:324. doi: 10.1016/j.bbrc.2007.01.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trepat X, Deng L, An SS, Navajas D, Tschumperlin DJ, Gerthoffer WT, Butler JP, Fredberg JJ. Universal physical responses to stretch in the living cell. Nature. 2007;447:592. doi: 10.1038/nature05824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenormand G, Chopin J, Bursac P, Fredberg JJ, Butler JP. Directional memory and caged dynamics in cytoskeletal remodelling. Biochem. Biophys. Res. Commun. 2007;360:797. doi: 10.1016/j.bbrc.2007.05.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saxton MJ, Jacobson K. Single-particle tracking: Applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 1997;26:373. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 20.Tseng Y, Lee JSH, Kole TP, Jiang I, Wirtz D. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J. Cell. Sci. 2004;117:2159. doi: 10.1242/jcs.01073. [DOI] [PubMed] [Google Scholar]
- 21.Caspi A, Granek R, Elbaum M. Enhanced diffusion in active intracellular transport. Phys. Rev. Lett. 2000;85:5655. doi: 10.1103/PhysRevLett.85.5655. [DOI] [PubMed] [Google Scholar]
- 22.Weiss M, Elsner M, Kartberg F, Nilsson T. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys. J. 2004;87:3518. doi: 10.1529/biophysj.104.044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason TG, Dhople A, Wirtz D. Concentrated DNA rheology and microrheology. In: Wirtz D, Halsey TC, editors. MRS Proceedings on Statistical Mechanics in Physics and Biology. Vol. 463. Materials Research Society; Pittsburgh: 1997. pp. 153–158. [Google Scholar]
- 24.Crocker JC, Valentine MT, Weeks ER, Gisler T, Kaplan PD, Yodh AG, Weitz DA. Two-point microrheology of inhomogeneous soft materials. Phys. Rev. Lett. 2000;85:888. doi: 10.1103/PhysRevLett.85.888. [DOI] [PubMed] [Google Scholar]
- 25.Levine AJ, Lubensky TC. One- and two-particle microrheology. Phys. Rev. Lett. 2000;85:1774. doi: 10.1103/PhysRevLett.85.1774. [DOI] [PubMed] [Google Scholar]
- 26.Kuo SC, Sheetz MP. Force of single kinesin molecules measured with optical tweezers. Science. 1993;260:232. doi: 10.1126/science.8469975. [DOI] [PubMed] [Google Scholar]
- 27.Mason TG. Estimating the viscoelastic moduli of complex fluids using the generalized Stokes-Einstein equation. Rheol. Acta. 2000;39:371. [Google Scholar]
- 28.Xu JY, Viasnoff V, Wirtz D. Compliance of actin filament networks measured by particle-tracking microrheology and diffusing wave spectroscopy. Rheol. Acta. 1998;37:387. [Google Scholar]
- 29.Ingber DE. Tensegrity I. Cell structure and hierarchical systems biology. J. Cell. Sci. 2003;116:1157. doi: 10.1242/jcs.00359. [DOI] [PubMed] [Google Scholar]
- 30.Ingber DE. Tensegrity II. How structural networks influence cellular information processing networks. J. Cell. Sci. 2003;116:1397. doi: 10.1242/jcs.00360. [DOI] [PubMed] [Google Scholar]