Abstract
A common dichotomy exists in inhibitor design: should the compounds be designed to block the enzymes of animals in the preclinical studies or to inhibit the human enzyme? We report that a single mutation of Leu-337 in rat neuronal nitric oxide synthase (nNOS) to His makes the enzyme resemble human nNOS more than rat nNOS. We expect that the approach used in this study can unite the dichotomy and speed up the process of inhibitor design and development.
Introduction
Nitric oxide (NO), a pleiotropic signaling molecule, has a range of biological functions, including neurotransmission, regulation of blood-vessel tone, and the immune response.1 There are at least three known nitric oxide synthases (NOS) responsible for endogenous NO production in all mammals.2 Among them, neuronal nitric oxide synthase (nNOS), which is predominantly expressed in neurons, is a potential target for the treatment of neurodegeneration since NO overproduction has been demonstrated to be a biomarker for a variety of neurodegenerative diseases.3, 4, 5, 6 Therefore, specific inhibition of nNOS without detriment to the essential function of endothelial nitric oxide synthase (eNOS) and inducible nitric oxide synthase (iNOS) is a promising approach for the design of novel drugs to treat those diseases.7, 8 Based on this goal, numerous selective nNOS inhibitors over eNOS and iNOS have been developed.7, 9
Rat nNOS, sharing more than 90% sequence identity with human nNOS, is the most thoroughly investigated neuronal nitric oxide synthase. It was initially isolated by Bredt and Snyder10 and subsequently Bredt et al 11 cloned and expressed it in mammalian cells. It was not until 1995 that rat nNOS was successfully purified from overexpressed E. coli having full activity.12, 13 Because of the very high sequence identity with human nNOS and because of the reliable method to prepare it, rat nNOS has been widely used to screen selective nNOS inhibitors14, 15, 16, 17, 18 and for structural studies.14, 19, 20, 21
A series of potent rat nNOS inhibitors with high selectivity over eNOS and iNOS have been developed in our lab.9 When comparing the inhibition efficiency of our highly selective inhibitors, which target the substrate L-arginine binding site (oxygenase domain),22 with human nNOS and rat nNOS , we found rat nNOS was more sensitive to those inhibitors than human nNOS. With the aid of sequence alignment (Supporting Information Figure 1), we found that there was only a one amino acid difference in their substrate binding sites, that is Leu-337 in rat nNOS corresponds to His-342 in human nNOS. These amino acids are at the entrance of the substrate access channel and form one of the few key “hot spots” (that is, the regions of a protein surface that are major contributors to the binding free energy) for ligand binding, so they potentially could be important in inhibitor design.9 Site-directed mutagenesis was employed in this study to change Leu-337 to His in rat nNOS to determine if that one amino acid mutation was sufficient to produce a mutant rat nNOS with properties similar to those of human nNOS.
Results
Purification of rat nNOS, human nNOS, and L337H rat nNOS
Purification of each of the enzymes was carried out by chromatography on 2’,5’-ADP-sepharose, CaM-sepharose, and by Sephacryl S-300 gel filtration (see Supporting Information Figure 2). There is no significant impurity in the enzymes after gel filtration. Generally, ∼10 mg enzyme was obtained from 1 liter of cultured cells.
Comparison of Km and Vmax values for L-arginine with the three enzymes
The reaction rates for all three enzymes reached saturation when the L-arginine concentration was 5 µM or higher (Supporting Information Figure 3A). The Km and Vmax values for L-arginine were obtained by plotting 1/v versus 1/[S] (Lineweaver-Burk method, Supporting Information Figure 3B; Table 1). These Km values for all three enzymes are quite similar, however, the Vmax for human nNOS is nearly twice those of rat nNOS and L337H rat nNOS.
Table 1.
Km and Vmax values of human nNOS, rat nNOS and L337H rat Nnos
| Human | Rat | L337H | |
|---|---|---|---|
| Km (µM) | 1.3 | 1.1 | 1.0 |
| Vmax (nmol NO min−1 mg−1) | 400 | 284 | 200 |
Inhibition of enzymes by inhibitors
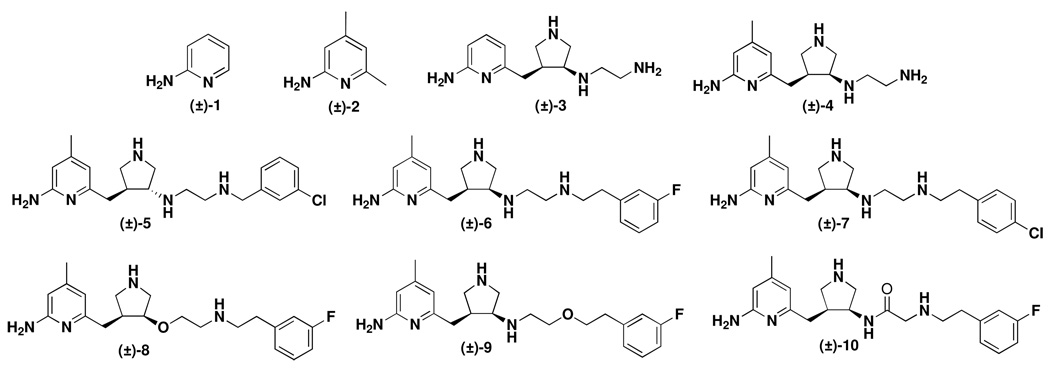
Several nNOS inhibitors (1–10 in Figure 1), mostly previously prepared in our lab,23,24,25,26 were tested. The IC50 values are summarized in Table 2. To confirm our results from the hemoglobin capture assay, we also performed a radioactivity assay for 8 (Supporting Information Figure 4B). The data from the radioactivity assay (data with a star in Table 2) are the same as those from the Hb assay.
Figure 1.
Chemical structures of nNOS inhibitors used in this study.
Table 2.
Inhibition of wild-type rat nNOS, human nNOS, and L337H rat nNOS§.
| Tested inhibitors | IC50(µM) | Selectivity | |||
|---|---|---|---|---|---|
| WT Rat nNOS | Human nNOS | L337H Rat nNOS | WT Rat/Human | WT / L337H Rat | |
| 1 | 8.7 ± 1.5 | 7.2 ± 0.6 | 9.3 ± 0.6 | 0.8 ± 0.08 | 1.1 ± 0.1 |
| 2 | 0.1 ± 0.00 | 0.1 ± 0.00 | 0.1 ± 0.01 | 1.0 ± 0.04 | 1.1 ± 0.1 |
| 3 | 6.3 ± 0.1 | 15.3 ± 2.1 | 16.0 ± 1.2 | 2.4 ± 0.3 | 2.5 ± 0.2 |
| 4 | 1.3 ± 0.3 | 4.0 ± 0.6 | 3.4 ± 0.3 | 3.1 ± 0.2 | 2.7 ± 0.4 |
| 5 | 3.3 ± 0.2 | 12.2 ± 0.6 | 13.0 ± 0.4 | 3.7 ± 0.05 | 3.9 ± 0.1 |
| 6 | 0.1± 0.01 | 0.7 ± 0.05 | 0.5 ± 0.03 | 7.0 ± 0.2 | 5.0 ± 0.2 |
| 7 | 1.1 ± 0.1 | 4.2 ± 0.2 | 5.0 ± 0.05 | 3.8 ± 0.2 | 4.6 ± 0.4 |
| 8 | 0.1 ± 0.01 | 0.6 ± 0.02 | 0.7 ± 0.05 | 6.0 ± 0.4 | 7.0 ± 0.2 |
| 0.1* | 0.7* | ||||
| 9 | 2.9 ± 0.6 | 6.6 ± 0.05 | 5.5 ± 0.4 | 2.4 ± 0.5 | 1.9 ± 0.3 |
| 10 | 0.7 ± 0.1 | 3.0 ± 0.5 | 4.0 ± 1.0 | 4.3 ± 0.6 | 5.7 ± 1.3 |
| L-NNA | 6.0 ± 0.8 | 4.4 ± 1.0 | 4.0 ± 1.5 | 0.7 ± 0.07 | 0.7 ± 0.07 |
Data are from duplicates and expressed as mean ± error.
Data are from radioassay; others are from Hb assay.
Reversibility of the selective nNOS inhibitors
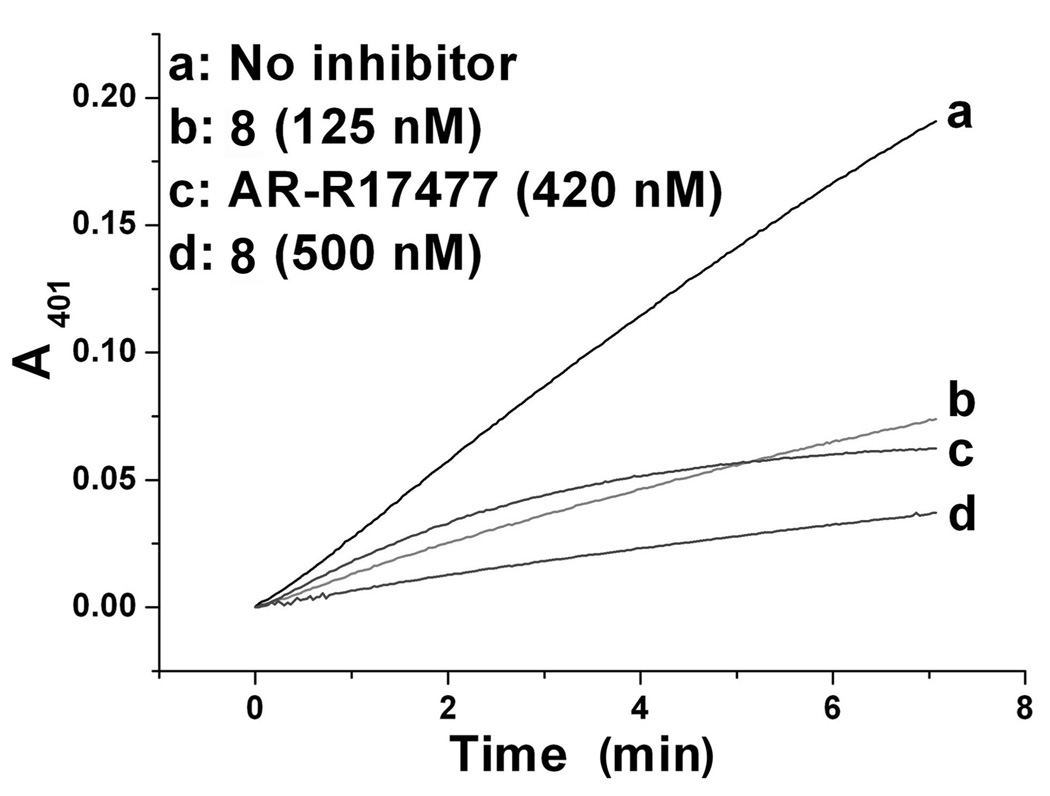
To determine the type of inhibition caused by our inhibitors, we first monitored the change in absorbance at 401 nm with time. As shown in Fig. 2, the rate of enzyme activity diminished but remained linear with time in the presence of inhibitor 8, while the rate of enzyme activity continually decreased with reaction time in the presence of a known irreversible rat nNOS inhibitor, AR-R17477 (Figure 3 and Supporting Information Figure 5).21 Reversibility of enzyme activity was observed with all of the inhibitors with human nNOS and L337H rat nNOS (only data for 8 is shown). To confirm the reversibility of the inhibitors and irreversibility of AR-R17477, we compared rat nNOS activity with 8 and AR-R17477 after removal of the inhibitor by dialysis. Again, we found the enzyme activity was rescued after removal of 8, but not with AR-R17477 (Supporting Information Fig. 5). Because of instability of the enzyme, about 15% of the control enzyme activity was lost after dialysis under the same conditions.
Figure 2.
Time dependence of rat nNOS inhibition. WT rat nNOS was incubated with or without 8, and the enzymatic reaction was monitored by the Hb assay for 7 min at room temperature. Only compound 8 with rat nNOS is illustrated; other inhibitors with the three enzymes had the same pattern as compound 8 shown here.
Figure 3.
Chemical structure of AR-R17477
Molecular docking
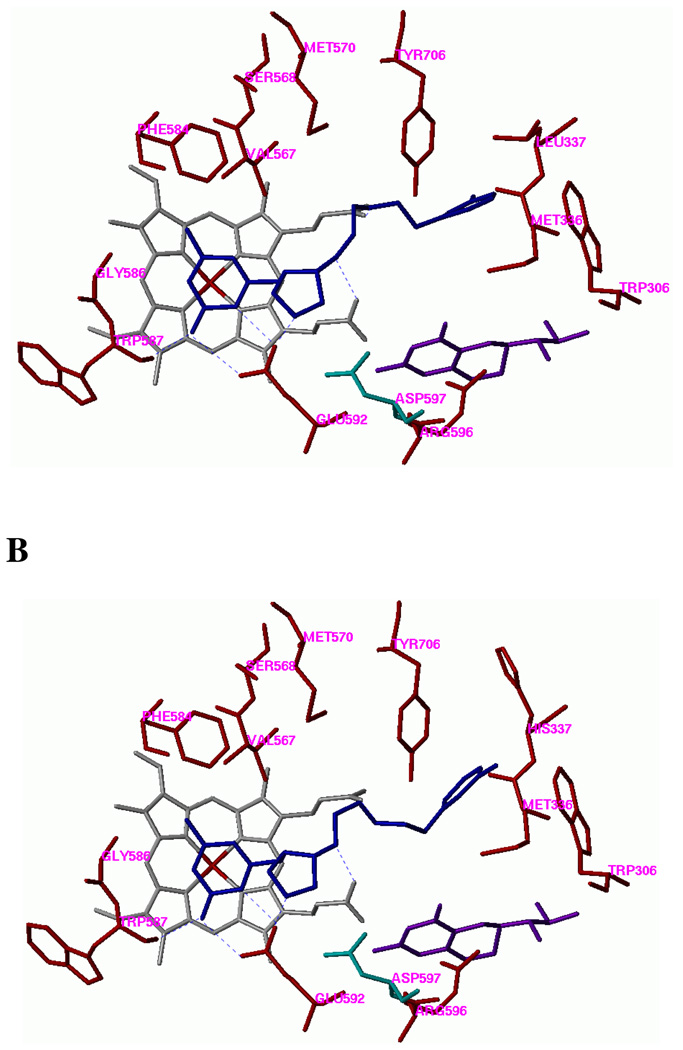
AutoDock 3.0.5 and AutoDock 4.1 were employed to perform the docking calculations.27 In AutoDock 3.0.5, the receptor structure is treated rigid, and the rotatable bonds of the ligand are allowed to rearrange. The docking mode of 6 in the WT rat nNOS is shown in Figure 4A. In AutoDock 4.1, not only the ligand but some of residues lining the active site of the receptor can be treated as flexible. In this study, the histidine side chain of L337H mutant rat nNOS was allowed to move freely. The docking mode of 6 in the L337H mutant enzyme is shown in Figure 4B.
Figure 4.
Docking conformations of 6 in (A) the active site of WT rat nNOS and (B) the L337H mutant rat nNOS. Inhibitor 6 is colored blue. Cofactors heme and H4B are shown in gray and purple, respectively. The potential H-bonds are indicated by dotted lines. Residue Asp597 of wild-type rat nNOS, which determines the selectivity between nNOS and eNOS, is colored green. The corresponding residue in bovine eNOS is Asn368.
Discussion
Whenever plans for translational research are considered, there is a common problem for the design of enzyme inhibitors or receptor antagonists: should the inhibitors or antagonists be designed to interfere with the target protein in a lower animal used in preclinical studies to accumulate data for clinical trials or should they be designed to interfere with the target protein in humans, in which case it may result in poor preclinical data? Because it is necessary to carry out preclinical studies prior to clinical studies, inhibitors must be identified that will have high activity in both systems. Therefore, it is important to determine what structural differences in target proteins of different species may be responsible for small molecule activity in those species. Although rat nNOS and human nNOS share more than 90% sequence identity, there are some fine differences. Based on an overlay of the crystal structures of rat and human nNOS, there appeared to be one amino acid different in the two species that would be significant in inhibitor design, namely, rat nNOS Leu-337 corresponds to human nNOS His-342.
Human nNOS and rat nNOS had almost the same affinity for their native substrate, L-arginine, because of the high sequence identity between them. The determined Km values for rat nNOS and human nNOS were 1.1 µM and 1.3 µM, respectively, which are quite consistent with previously reported data, i. e., 0.8 µM to 2.8 µM for rat nNOS and 1.6 µM for human nNOS.28, 29 Since the Km values for rat nNOS and human nNOS are indistinguishable, it is expected that the Km for the mutant enzyme, L337H rat nNOS, would have almost the same value, which it does (1.0 µM). Because L337 is at the entrance to the substrate access channel, not directly in the active site, its modification should not affect substrate binding. The reported Vmax values for rat nNOS ranged from 150 to 435 nmol NO min−1 mg−1.29, 30 The variability in Vmax values is probably because of the difficulty in acquiring an exact enzyme concentration. The similarity of the L337H rat nNOS Vmax value to that of WT rat nNOS is probably also the result of the mutation occurring at the entrance to the active site rather than directly in the active site.
We found that, in general, L337H rat nNOS behaved more like human nNOS than WT rat nNOS toward inibitor binding, provided the inhibitor was sufficiently large to reach the second sphere of amino acids, where L337 resides. The finding that our selective nNOS inhibitors can distinguish between human nNOS and rat nNOS suggests that there is a difference in binding of the inhibitors between the two enzymes. This could result from the increased size and polarity in going from leucine to histidine. As shown in Fig. 4A, in the WT rat nNOS, the 3-fluorophenyl group of 6 fits well into the hydrophobic pocket defined by Y706, M336, L337, and W306 (from the other monomer), and the 3-fluoro atom of the phenethyl group is located in an extra small pocket defined by L337 and Y706. Compared to leucine, the side chain of histidine is larger and more polar. These two properties of histidine shrink the hydrophobic cavity, making it difficult for the hydrophobic tail of the inhibitors to fit into that pocket based on an AutoDock 3.0.5 calculation when all of the residues of the L337H nNOS mutant enzyme are kept rigid. In an AutoDock 4.1 calculation, if the histidine side chain of the L337H mutant enzyme is allowed to rotate, the imidazole ring prefers to point outside of this pocket as shown in Fig. 4B; pi-pi stacking of the 3-fluorophenyl group of 6 with the imidazole of H337 is not favored because of the geometry of the inhibitor and this histidine in the active site. Although the 3-fluorophenyl side chain of 6 can still be in the same general pocket, the orientation of the 3-fluoro atom is different; it does not point into the small hydrophobic pocket as observed in the docking of 6 with WT rat nNOS. A similar report suggested that Leu-337 in rat nNOS may interact with the chlorophenyl group of AR-R17477, a potent selective nNOS inhibitor, via a hydrophobic interaction.21 It was suggested that this interaction allowed AR-R17477 to bind more tightly to nNOS compared with iNOS because Leu337 in rat nNOS corresponds to the polar amino acid Asp115 in iNOS. We found that our inhibitors bearing a halophenyl group (compounds 5–8, and 10) generally have higher selectivity than those without such a group (compounds 3 and 4). This observation suggests an important function of the halophenyl group in these inhibitors in binding to WT rat nNOS. As expected, Nω-nitro-L-arginine (L-NNA), 2-aminopyridine (1), and 2-amino-4,6-dimethylpyridine (2) have essentially no selectivity for the three enzymes (Table 2, and Supporting Information Fig. 6), consistent with the fact that they are too small to have any effect on the mutated site.
Conclusions
The major difference in amino acid sequence at or near the active site between rat and human nNOS is L337 in rat corresponds to H342 in human. Mutation of rat nNOS L337 to histidine makes the structure of the rat enzyme more similar to that of human nNOS. The three enzymes have indistinguishable Km values for L-arginine. A series of nNOS-selective inhibitors was tested for inhibitory properties against the three enzymes. Those inhibitors with a tail long enough to reach the mutation exhibited Ki values with L337H rat nNOS closer to those with human nNOS than with rat nNOS; those with no tails had exhibited no difference among the WT rat nNOS, the mutated rat nNOS, and human nNOS. This demonstrates that small changes in structure can have a significant effect on inhibitor design. By identifying the site responsible for inhibitor binding differences between the rat and human enzymes, more effective inhibitor design can be carried out to balance the problem associated with finding an appropriate animal model for preclinical compared to clinical drug development. In this case both rat and human enzymes are available. This approach should be even more significant when the enzyme for only the lower animal, not the human enzyme, is available. This site-directed mutation approach to mimic the human enzyme can be an excellent model to differentiate an enzyme in a lower animal from that in humans.
Experimental Materials and Methods
Materials
The synthesis of compounds 3–10 have been published elsewhere,23,24,25,26 and all compounds were used as their HCl salts. Compounds 1 and 2 were bought from Aldrich (Milwaukee, WI). All of the biochemicals and chemicals were purchased from Sigma (St. Louis, MO) except [14C]L-arginine (specific radioactivity 320 mCi/mmol), CaM-Sepharose, 2’,5’-ADP-Sepharose, and Sephacryl S-300 are products of Amersham. AR-R17477 is obtained from Proximagen. Complete protease inhibitors cocktail (EDTA-free) was bought from Roche.The site-directed mutagenesis kit and NOS activity radioassay kit were obtained from Stratagene. Tryptone and yeast extract were from BD Biosciences. Primers for site-directed mutagenesis were obtained from Sigma Genosys.
Site-directed mutagenesis of rat nNOS
The L337H rat nNOS plasmid was constructed using rat nNOSpCW as a template. The following sets of forward and reverse primers were used to introduce mutations at Leu337 to His: Forward: ATG GGC TCG ATC ATG CAC CCT TCC CAG CAC ACG Reverse: CGT GTG CTG GGA AGG GTG CAT GAT CGA GCC CAT The mutant plasmid was constructed using the QuikChange® II XL kit from Stratagene. Mutagenesis was confirmed by automated nucleotide sequencing at the Feinberg School of Medicine sequencing facility, Northwestern University.
Overexpression and purification of rat nNOS, human nNOS, and L337H rat nNOS
The protocol used for overexpression of nNOS was essentially the same as that in the literature12, 13 , with some modifications as described in the Supporting Information.
Measurement of Km and Vmax values
Km and Vmax values were measured with L-arginine concentrations ranging from 0 to 30 µM in a final volume of 600 µL by the Hb assay.30 The amount of enzymes used was 1 µg, 0.4 µg, and 1.5 µg for rat nNOS, human nNOS, and L337H rat nNOS, respectively. The rate of NO production was detected by the Hb assay using ε401 = 60,000 M−1 cm−1 . Km and Vmax values were extrapolated from Lineweaver-Burk plots.
Inhibition of enzymes by Hb assay
The Hb assay mixture contained L-arginine (10 µM), NADPH (0.1 mM), tetrahydrobiopterin (10 µM), dithiothreitol (100 µM), Hb (0.1 mg/mL), CaM (10 µg/mL), CaCl2 (0.1 mM), and different amounts of inhibitors. The final volume was adjusted to 600 µL with 100 mM Hepes buffer, pH 7.4. The enzymatic reaction was initiated by addition of 3 µg of enzyme, and the rate of NO production was monitored by the change of absorbance at 401 nm in the initial 60 s on a Perkin-Elmer Lambda 10 UV/vis spectrophotometer. All assays were performed at room temperature.
Inhibition of enzymes by the radioassay
This assay method was adapted from the literature.30 The reaction mixture contained 250 µL of 2 x reaction buffer (supplied with the kit), 80 µL of 6.25 mM NADPH, 10 µl of [14C]-L-arginine, 50 µL of 6 mM CaCl2, and 10 µL of H2O. To 1.5-mL Eppendorf tubes, 40 µL of the above reaction mixture was added, and then 5 µL of CaM (supplied with the kit) was added to each tube. After equilibrating the mixture at room temperature for 10 min, 5 µL of 10 µg/mL rat nNOS was added. After incubation at room temperature for 15 min, the reaction was terminated by addition of 400 µL of stop buffer (supplied with the kit). [14C]-L-Citrulline was separated from the reaction mixture by the column supplied with the kit following the manufacturer’s procedure, and the radioactivity was quantified by liquid scintillation analysis.
Molecular docking
For the AutoDock 3.0.5 calculations, polar hydrogen atoms were added to the protein structure (PDB id: 1P6I), and Kollman united atom charges were assigned.32 Hydrogens were also added to the heme and H4B, and charges were calculated by the Gasteiger–Marsili method.32 The charge of the Fe atom bound to heme was assigned +3. The nonpolar hydrogen atoms of heme and H4B were removed manually, and their charges were united with the bonded carbon atoms. Atomic solvation parameters and fragmental volumes were assigned using the AddSol utility. The 3D structures of the ligands were built and partial atomic charges were also calculated using the Gasteiger–Marsili method. The rotatable bonds in the ligands were defined using AutoTors, which also unites the nonpolar hydrogens and partial atomic charges to the bonded carbon atoms. The grid maps were calculated using AutoGrid. The dimensions of the grid box was 27 × 26 × 31 Å, and the grid spacing was set to 0.375 Å. Docking was performed using the Lamarckian genetic algorithm (LGA), and the pseudo-Solis and Wets method were applied for the local search. The procedure in detail used was previously described.23, 33 The Leu-337 of rat nNOS (PDB id: 1P6I) was mutated into His and submitted to an AutoDock 4.1 calculation. The side chain of L337H was allowed to be rotatable. The Lamarckian genetic algorithm procedure was employed, and the docking runs were set to 100. The rest of the parameters were taken as default.
Supplementary Material
Acknowledgments
We are grateful for financial support from the National Institutes of Health (Grants GM049725 to R.B.S. and GM52419 to Dr. Bettie Sue Masters, with whose laboratory L.J.R. is affiliated). B.S.S.M. also is grateful to the Welch Foundation for a Robert A. Welch Foundation Distinguished Professorship in Chemistry (AQ0012).
Abbreviations
- CaM
calmodulin
- Hb
hemoglobin
- NOS
nitric oxide synthase
- nNOS
neuronal nitric oxide synthase
- eNOS
endothelial nitric oxide synthase
- iNOS
inducible nitric oxide synthase
Footnotes
Supporting Information Available:
Supporting figures are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Calabrese V, Mancuso C, Calvani M, Rizzarelli E, Butterfield DA, Stella AM. Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity. Nat. Rev. Neurosci. 2007;8:766–775. doi: 10.1038/nrn2214. [DOI] [PubMed] [Google Scholar]
- 2.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function and inhibition. Biochem. J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson's disease. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 5.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-Nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 6.Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, Uehara T, Lipton SA. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erdal EP, Litzinger EA, Seo J, Zhu Y, Ji H, Silverman RB. Selective neuronal nitric oxide synthase inhibitors. Curr. Top. Med. Chem. 2005;5:603–624. doi: 10.2174/1568026054679317. [DOI] [PubMed] [Google Scholar]
- 8.Tafi A, Angeli L, Venturini G, Travagli M, Corelli F, Botta M;Computational studies of competitive inhibitors of nitric oxide synthase (NOS) enzymes: towards the development of powerful and isoform-selective inhibitors Curr. Med. Chem 2006131929–1946. [DOI] [PubMed] [Google Scholar]
- 9.Silverman RB. Design of selective neuronal nitric oxide synthase inhibitors for the prevention and treatment of neurodegenerative diseases. Acc. Chem. Res. 2009;42(3):439–451. doi: 10.1021/ar800201v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bredt DS, Snyder SH. Isolation of nitric oxide synthetase, a calmodulin-requiring enzyme. Proc. Natl. Acad. Sci. U. S. A. 1990;87:682–685. doi: 10.1073/pnas.87.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH;Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase Nature 1991351714–718. [DOI] [PubMed] [Google Scholar]
- 12.Gerber NC, Ortiz de Montellano PR. Neuronal nitric oxide synthase Expression in Escherichia coli, irreversible inhibition by phenyldiazene, and active site topology. J. Biol. Chem. 1995;270:17791–17796. doi: 10.1074/jbc.270.30.17791. [DOI] [PubMed] [Google Scholar]
- 13.Roman LJ, Sheta EA Martá, Martasek P, Gross SS, Liu Q, Masters BS. High-level expression of functional rat neuronal nitric oxide synthase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1995;92:8428–8432. doi: 10.1073/pnas.92.18.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossiter S, Smith CL, Malaki M, Nandi M, Gill H, Leiper JM, Vallance P, Selwood DL. Selective substrate-based inhibitors of mammalian dimethylarginine dimethylaminohydrolase. J. Med. Chem. 2005;48:4670–4678. doi: 10.1021/jm050187a. [DOI] [PubMed] [Google Scholar]
- 15.Boulouard M, Schumann-Bard P, Butt-Gueulle S, Lohou E, Stiebing S, Collot V, Rault S. 4-Substituted indazoles as new inhibitors of neuronal nitric oxide synthase. Bioorg. Med. Chem. Lett. 2007;17:3177–3180. doi: 10.1016/j.bmcl.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 16.Patman J, Bhardwaj N, Ramnauth J, Annedi SC, Renton P, Maddaford SP, Rakhit S, Andrews JS. Novel 2-aminobenzothiazoles as selective neuronal nitric oxide synthase inhibitors. Bioorg. Med. Chem. Lett. 2007;17:2540–2544. doi: 10.1016/j.bmcl.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Cottyn B, Acher F, Ramassamy B, Alvey L, Lepoivre M, Frapart Y, Stuehr D, Mansuy D, Boucher JL, Vichard D. Inhibitory effects of a series of 7-substituted-indazoles toward nitric oxide synthases: particular potency of 1H-indazole-7-carbonitrile. Bioorg. Med. Chem. 2008;16:5962–5973. doi: 10.1016/j.bmc.2008.04.056. [DOI] [PubMed] [Google Scholar]
- 18.Seo J, Igarashi J, Li H, Martasek P, Roman LJ, Poulos TL, Silverman RB. Structure-based design and synthesis of Nω-nitro-L-arginine-containing peptidomimetics as selective inhibitors of neuronal nitric oxide synthase. Displacement of the heme structural water. J. Med. Chem. 2007;50:2089–2099. doi: 10.1021/jm061305c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Martasek P, Paschke R, Shea T, Masters BSS, Kim J-JP. Crystal structure of the FAD/NADPH-binding domain of rat neuronal nitric-oxide synthase Comparison with NADPH-cytochrome P450 oxidoreductase. J. Biol. Chem. 2001;276:37506–37513. doi: 10.1074/jbc.M105503200. [DOI] [PubMed] [Google Scholar]
- 20.Garcin ED, Bruns CM, Lloyd SJ, Hosfield DJ, Tiso M, Gachhui R, Stuehr DJ, Tainer JA, Getzoff ED;Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase J. Biol. Chem 200427937918–37927. [DOI] [PubMed] [Google Scholar]
- 21.Fedorov R, Vasan R, Ghosh DK, Schlichting I. Structures of nitric oxide synthase isoforms complexed with the inhibitor AR- R17477 suggest a rational basis for specificity and inhibitor design. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5892–5897. doi: 10.1073/pnas.0306588101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proskuryakov SY, Konoplyannikov AG, Skvortsov VG, Mandrugin AA, Fedoseev VM. Structure and activity of NO synthase inhibitors specific to the L-arginine binding site. Biochemistry (Mosc) 2005;70:8–23. [PubMed] [Google Scholar]
- 23.Ji H, Stanton BZ, Igarashi J, Li H, Martasek P, Roman LJ, Poulos TL, Silverman RB. Minimal pharmacophoric elements and fragment hopping, an approach directed at molecular diversity and isozyme selectivity Design of selective neuronal nitric oxide synthase inhibitors. J Am Chem Soc. 2008;130:3900–3914. doi: 10.1021/ja0772041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji H, Li H, Martasek P, Roman LJ, Poulos TL, Silverman RB. Discovery of highly potent and selective inhibitors of neuronal nitric oxide synthase by fragment hopping. J Med Chem. 2009;52:779–797. doi: 10.1021/jm801220a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji H, Tan S, Igarashi J, Li H, Derrick M, Martásek P, Roman LJ, Vásquez-Vivar J, Poulos TL, Silverman RB. Selective Neuronal Nitric Oxide Synthase Inhibitors for Prevention of Cerebral Palsy, Ann. Neurol. 2009;65:209–217. doi: 10.1002/ana.21555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawton GR, Ranaivo HR, Chico LK, Ji H, Xue F, Martásek P, Roman LJ, Watterson DM, Silverman RB. Analogues of 2-aminopyridine-based selective inhibitors of neuronal nitric oxide synthase with increased bioavailability. Bioorg. Med Chem. 2009;17:2371–2380. doi: 10.1016/j.bmc.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comp. Chem. 1998;19:1639–1662. [Google Scholar]
- 28.Roman LJ, Sheta EA, Martásek P, Gross SS, Liu Q, Masters BSS. High-Level Expression of Functional-Rat Neuronal Nitric-Oxide Synthase in Escherichia-Coli. Proc. Natl. Acad. Sci. U.S.A. 1995;92:8428–8432. doi: 10.1073/pnas.92.18.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMillan K, Bredt DS, Hirsch DJ, Snyder SH, Clark JE, Masters BS. Cloned, expressed rat cerebellar nitric oxide synthase contains stoichiometric amounts of heme, which binds carbon monoxide. Proc. Natl. Acad. Sci. U. S. A. 1992;89:11141–11145. doi: 10.1073/pnas.89.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hevel JM, Marletta MA. Nitric-oxide synthase assays. Methods Enzymol. 1994;233:250–258. doi: 10.1016/s0076-6879(94)33028-x. [DOI] [PubMed] [Google Scholar]
- 31.Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Weiner P;A new force field for molecular mechanical simulation of nucleic acids and proteins J. Am. Chem. Soc 19841063765–784. [Google Scholar]
- 32.Gasteiger J, Marsili M. Iterative paratial equalization of orbital electronegativity –a rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 33.Ji H, Li H, Flinspach M, Poulos TL, Silverman RB. Computer modeling of selective regions in the active site of nitric oxide synthases: implication for the design of isoform-selective inhibitors. J Med Chem. 2003;46(26):5700–5711. doi: 10.1021/jm030301u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.