Abstract
Peptide libraries have proven to be useful in applications such as substrate profiling, drug candidate screening and identifying protein-protein interaction partners. However, issues of fidelity, peptide length and purity have been encountered when peptide libraries are chemically synthesized. Biochemically produced libraries, on the other hand, circumvent many of these issues due to the fidelity of the protein synthesis machinery. Using thioredoxin as an expression partner, a stably folded peptide scaffold (avian pancreatic polypeptide) and a compatible cleavage site for human rhinovirus 3C protease, we report a method that allows robust expression of a genetically-encoded peptide library, which yields peptides of high purity. In addition, we report the use of methodological synchronization, an experimental design created for the production of a library, from initial cloning to peptide characterization, within a five-week period of time. Total peptide yields ranged from 0.8 to 16%, which corresponds to 2-70 milligrams of pure peptide. Additionally, no correlation was observed between the ability to be expressed or overall yield of peptide-fusions and the intrinsic chemical characteristics of the peptides, indicating that this system can be used for a wide variety of peptide sequences with a range of chemical characteristics.
Keywords: peptide library, fusion partner, avian pancreatic polypeptide, human rhinovirus 3C protease, methodological synchronization
Introduction
Peptide libraries are commonly used in a variety of endeavors including identifying peptide-DNA interactions [1], as surrogates for screening protein-protein interactions [2-5], and as a basis for finding potential peptidic drug molecules. Peptide libraries have also been used for profiling substrate specificities of proteases [6-11], phosphatases [12-14], kinases [15-18] and other drug targets [19-22]. Thus production of high quality peptide libraries is of wide interest for many applications.
Typically, peptides for libraries have been produced via solid-phase synthesis, which involves sequential coupling of amine-protected amino acids to resin-bound amino acids. The resulting libraries generally contain peptides no longer than 15-20 amino acids, which can prove limiting in applications requiring longer peptides. The length constraints for chemically synthesized peptides are the result of coupling and deprotection efficiencies at each step, such that an exponential decrease in sequence fidelity is observed as a function of length. In addition, enantiomerization of nineteen of the twenty naturally-occurring amino acids and the associated difficulties in purification of the D-isomer-containing peptides from the L-isomers set practical limits to peptide lengths in library synthesis [23, 24]. Sometimes peptide sequences are restricted due to steric clash of adjacent amino acids with bulky chemical catalysts or intermolecular aggregation which results in a low efficiency of the chemical coupling reaction [25-27]. Overall, chemically synthesized peptide libraries of longer lengths tend to have decreased sample quality, be costly and have increased production time.
Genetically-encoded peptide libraries offer several advantages in library design. The resulting libraries can contain longer peptide lengths and significantly increased yields while avoiding the more common limitations of chemical synthesis. Biological synthesis of peptides of longer lengths can be particularly important for production of isotopically-labeled peptides for use in heteronuclear Nuclear Magnetic Resonance (NMR) spectroscopy. The principle advantages of biological production result from the fidelity of protein machinery, particularly because the ribosome is not limited in sequence or length of synthesized peptides and has an error rate of only 0.01% error per amino acid added [28]. Genetically-encoded peptide libraries have not been widely used due to difficulties in the expression and purification of small peptides. This problem has been overcome by the use of protein-expression fusion partners such as glutathione-s-transferase [29], maltose binding protein [30] and thioredoxin [31] being the three most widely-used fusions. While fusion proteins often promote production, this method can be time consuming and requires post expression protease processing and purification of the peptide from the protease. However, the addition of fusion partners has resulted in the improvement of the overall yields by enhancing solubility of the peptide of choice.
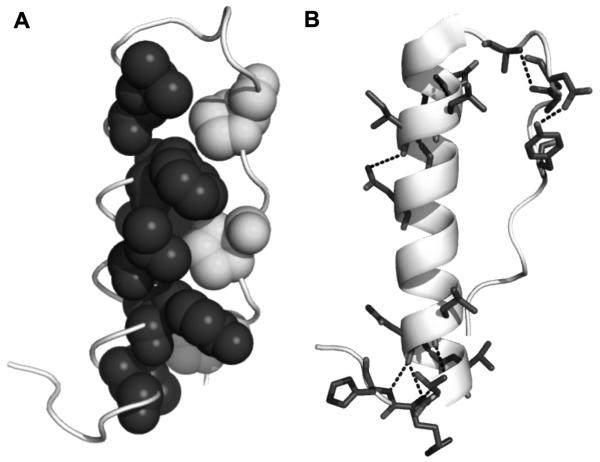
We report a method that allows robust expression of a genetically-encoded peptide library that addresses the issues of purity, yield, length of the purified peptide, batch-to-batch variability which we have observed with chemically synthesized peptides, in addition to cost and time of production. This genetically-encoded system features thioredoxin as a fusion tag for the stably-folding peptide scaffold avian pancreatic polypeptide (aPP). aPP is a 36 amino acid peptide that contains a hydrophobic core and hydrogen bond network between the α-helix and polyproline helix of the peptide [32, 33] (Fig. 1A and B). These properties are responsible for its fold and stability. Previous work has demonstrated that the presence of the hydrophobic core and hydrogen bond network render aPP insensitive to mutations over much of the amino acid sequence [34, 35] or to a c-terminal truncation [33, 36].
Figure 1. aPP structure.
(A) aPP backbone (lightest gray) has a hydrophobic core created by three prolines located on the poly-proline helix (medium gray) and various residues on the α-helix (dark gray). (B) aPP’s internal hydrogen bond network, highlighted in dashed lines, is responsible for the impressive structural stabilization of a peptide that is just 36 amino acids long.
A common difficulty with peptide expression is residual amino acid overhangs following the protease cleavage event. The native sequence of aPP allows inclusion of an N-terminal cleavage site for human rhinovirus 3C protease, a highly specific protease, without any unwanted amino acid additions to the peptides. Human rhinovirus 3C protease cleaves the sequence LEVLFQ-GP, generating a gly-pro overhang upon cleavage. In aPP the first two amino acids are gly-pro, so no residual amino acids are left upon cleavage. Combining this optimized expression system with methodological synchronization of site-directed mutagenesis, protein expression and purification, a twenty-member aPP variant library of peptides 27 amino acids in length was generated. This method allowed production of the desired library with overall improved yields, purity, and cost, all on a time frame that is comparable to synthetic peptide library methods on the same production scale.
Materials and Methods
Cloning of Recombinant Peptide-Fusion Variants
In order to create the parent vector, pET32-Peptide, for the peptide-fusion DNA variants, the aPP gene was amplified by PCR from the pJC20[35] vector (from Alana Schepartz and Doug Daniels). Using the forward primer 5′GTACAACCATGGCTGGAAGTGCTGTTTCAGGGTCCGTCCCAGCCGACCTACCC3′ that included the human rhinovirus 3C cleavage sequence (bold) and NcoI endonuclease restriction site (italics) and the reverse primer 5′TCGAGCCTCGAGCTAGTAACGGTGACGGGTAACAACGTTCAGG3′ which encodes the XhoI restriction site (italics) and stop codon (bold), the desired gene was produced. This gene product was then ligated into the thioredoxin fusion tag, 6xHis purification tag containing vector, pET32b (Novagen) via restriction sites NcoI and XhoI. Insertion was confirmed by sequence analysis (Genewiz, Inc.).
The newly constructed vector, pET32-Peptide, was used for PCR-based site directed mutagenesis via QuikChange (Stratagene) to create the variable peptide sequences. Initial base constructs, B3 and B3II were created using two sets of primers. Mutational sites include Q4, T6, L17, I18, Y21, D23, Q25, L24 and Y27. Secondary base constructs B3DQ and B3NQ were produced from the B3 parent sequence by mutating sites D10, D11, and D16 while B3IQ and 3INQ were produced from the B3II sequence using two sets of primers. Remaining peptide sequences were produced from single step mutagenesis at position D11 utilizing one set of primers. The nucleotide sequences were optimized for expression in E. coli and all constructs were verified via sequence analysis.
Expression and Purification of Recombinant Peptide-Fusion Variants
DNA sequences encoding desired peptide sequences were transformed into E. coli strain BL21(DE3) competent cells (Novagen) for expression. These cells were inoculated into 2xYT media containing 100 μg/mL ampicillin (Sigma). Cells were grown at 37°C until an OD600 of 0.6 was reached. Isopropyl β-D-1-thiogalactopyranoside (IPTG), (Anatrace) was then added to a final concentration of 1mM. Cells were allowed to grow at 37°C for an additional 3 hrs and then harvested by centrifugation at 5000 rpm (Sorvall SLC-4000 rotor) for 10 minutes.
Cells were resuspended in lysis buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 2mM imidazole) and lysed using a microfluidizer system (Microfludics). The resulting solution was then centrifuged at 15,000 rpm (Sorvall SS-34 rotor) for one hour at 4°C to remove cellular debris. Lysates were passed over HiTrap™ Chelating HP columns (GE Healthcare) charged with nickel. aPP, B3, B3DQ, B3DE, B3DK, B3DR, B3NE, B3NQ, B3NK, and B3II bound proteins were washed and eluted using a step gradient that consisted of a wash step using 50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 10 mM imidazole and an elution step using 50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 250 mM imidazole. Peptide fusions B3, B3DK, B3DR, B3NQ, B3NE, B3NK, B3II eluted proteins were subjected to ion exchange chromatography to obtain additional purity. The previously eluted proteins were diluted 1:5 using Buffer A (20 mM Tris pH 8.0, 2mM dithiothreitol (DTT)) and bound to Macro-Prep HighQ cartridge (Biorad) at 5% Buffer B (Buffer B: 20 mM Tris pH 8.0, 2 mM DTT, 1M NaCl). Peptide-fusion proteins were eluted via a linear gradient that ranged from 50 mM to 350 mM NaCl over 240 minutes. Peptide fusions B3NR, B3IQ, B3IK, B3IE, B3IR, B3IL, 3INQ, 3INE, 3INR and 3INK were purified on a HiTrap™ Chelating HP columns (GE Healthcare) charged with nickel ions, using a gradient from 0 mM to 50 mM imidazole over 300 minutes. All peptide-fusions were analyzed by 16% SDS-PAGE gels that were stained with coomassie brilliant blue (Sigma). Peptide fusions were stored at -20°C until required for cleavage and separation.
Expression and purification of Human Rhinovirus 3C Protease
The human rhinovirus 3C gene in the pGEX vector (GE Healthcare) was transformed into E. coli strain BL21(DE3) competent cells (Novagen) for expression. These cells were inoculated into 2xYT media containing 100 μg/mL ampicillin (Sigma). Cells were grown at 37°C until an OD600 of 0.6 was reached. IPTG was then added to a final concentration of 1mM. Cells were allowed to grow at 20°C for 18 hours and then harvested by centrifugation at 5000 rpm (Sorvall SLC-4000 rotor) for 10 minutes.
Cells were resuspended in binding buffer (20 mM NaH2PO4 pH 7.4, 150 mM NaCl, 2mM DTT) and lysed using a microfluidizer system (Microfludics). The resulting solution was then centrifuged at 15,000 rpm (Sorvall SS-34 rotor) for one hour at 4°C to remove cellular debris. Lysates were passed over GSTrap™ FF column (GE Healthcare) and washed with 50 mM Tris HCl, 5mM reduced glutathione, 2mM DTT to remove loosely binding contaminants. The majority of the protease was eluted using 50 mM Tris HCl, 15mM reduced glutathione, 5mM DTT followed by a second elution step of 50 mM Tris HCl, 35mM reduced glutathione, 5mM DTT to release remaining protease and recharge the column.
The elution was diluted 1:5 using Buffer A (20 mM Tris pH 8.0, 2mM DTT) and bound to Biorad Macro-Prep HighQ cartridge at 2% Buffer B (Buffer B: 20 mM Tris pH 8.0, 2mM DTT, 1M NaCl). Protease was eluted via a linear gradient from 20 mM to 100 mM NaCl over 50 minutes.
Peptide-Fusion Cleavage by Human Rhinovirus 3C and Separation from Fusion Partners
Peptide-fusion and protease were incubated at 4°C for a 5-hour digestion period in a 1:25 protease to protein ratio. Reverse phase HPLC was carried out on the cleaved peptide-fusion samples to separate the peptide from the protein components. Cleaved peptide-fusion samples were filtered through 0.22 μm syringe filter and loaded onto a Waters Sunfire Prep C18 column (10×50 mm, 5μm, and 300Å). Separation of the peptide from fusion protein and contaminants was carried out by reverse phase HPLC on a Shimadzu HPLC system equipped with a SPD-20AV Prominence UV/Vis detector and LC-20AT Prominence liquid chromatograph. The mobile phase was 0.1% trifluoroacetic acid in water (Buffer A), with an elutant of 0.1% trifluoroacetic acid in acetonitrile (Buffer B). The column was developed with a three phase gradient consisting of a steep change in organic (5-30% Buffer B over 2.5 minutes) for buffer component elution, a shallow step (30-45% Buffer B over 15 minutes) for peptide elution, followed by a steep gradient (45-100% Buffer B over 4 minutes) to regenerate the column and elute larger proteins such as thioredoxin and human rhinovirus 3C. Absorbance was monitored at 214nm and 280 nm.
Mass spectrometry
To determine peptide molecular weight and degree of purity, peptide samples were analyzed using electrospray mass spectrometry. Lyophilized peptide samples were resuspended in water to a peptide concentration of 50 μM. 5 μl of sample was analyzed using an Esquire-LC electrospray ion trap mass spectrometer (Bruker Daltonics, Inc) set up with an ESI source and positive ion polarity. This system was equipped with an HP1100 HPLC system (Hewlett Packard). Scanning was carried out between 200 m/z and 2000 m/z, and the final spectra obtained were an average of 10 individual spectra. Equipment used was located at the Mass Spectrometry Center of the University of Massachusetts at Amherst.
Results
Construction of Peptide-Fusion Expression Vector and aPP variants
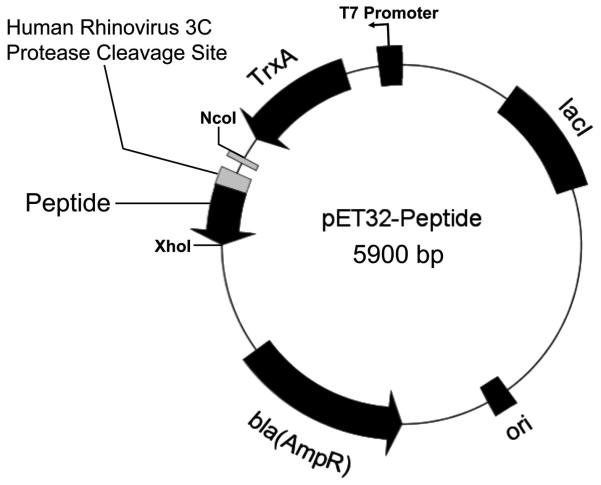
A peptide-expression system for the library of aPP-based peptides was generated that expressed thioredoxin fusion proteins. A goal of this project was to streamline all steps in the process of cloning, expression and purification. In order to limit subsequent subcloning events, the parent vector (Fig. 2) was created through a single PCR reaction that resulted in the gene for a human rhinovirus 3C protease cleavage site (LEVLFQ) and the truncated aPP sequence (GPSQPTYPGDDAPVEDLIRFYNDLQQY). This gene product was then ligated into the thioredoxin fusion gene and 6xHis purification tag containing vector, pET32b via restriction sites NcoI and XhoI. The resultant thioredoxin-peptide fusion construct then served as the first base sequence for further mutagenesis.
Figure 2. Schematic representation of expression vector, pET32-Peptide.
The T7 promoter, a fusion partner thioredoxin (TrxA), 6xHis tag, human rhinovirus 3C cleavable linker, gene for peptide, and ampicillin resistance are shown.
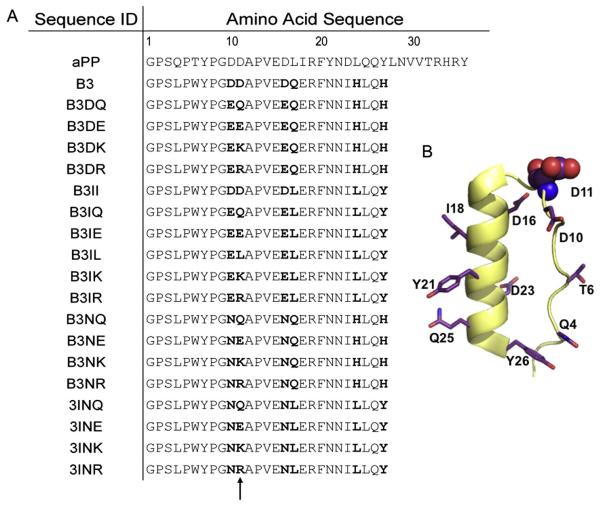
aPP variant sequences were created by mutating codons for six to twelve of the amino acids in the truncated 27 amino acid peptide scaffold (Fig. 3A) via a QuikChange (Stratagene) mutagenesis strategy. This limited library was designed as aPP mimics of natural caspase-inhibiting peptides. Mutations included the desired variations in the original peptide sequences and took into consideration amino acids that are known to be structurally important for scaffold stability. Sites of mutagenesis are throughout the truncated aPP sequence and are spread across the aPP structural elements (Fig. 3B). Mutational sites include Q4, T6, D10 and D11 on the polyproline helix and D16, D17, I18, Y21, D23, L24 Q25, and Y26 on the α-helix. In addition D11, was selected as a site to be extensively interrogated. By focusing our investigation in one region, we were able to produce many variants in a limited number of mutagenic steps. Moreover, all mutational oligonucleotides were designed to limit cost and allow rapid production of the new peptide encoding DNA constructs. These designs focused on producing an array of peptide sequences using the minimum number of rounds of mutagenesis. If mutagenesis was performed not by sequential means, 123 rounds of mutagenesis would have been performed to complete the library. Mutations were quickly assessed by growing the transformed E. coli cells for eight hours and plasmid prepping the DNA in time for same day sequencing by an outsourced company (Genewiz Inc). Upon sequence analysis, one out of two or more clones tested obtained the desired mutations. The success rate was directly related to the thoroughness of the DpnI digestion step during the QuikChange protocol (data not shown).
Figure 3. Structure of peptide library.
(A) Amino acid sequences of the twenty-member peptide library. Highly interrogated residues, which vary relative to the starting B3 sequence, are bold. (B) Cartoon representation of truncated aPP peptide scaffold with sites of mutations shown in stick representation and highly interrogated site D11 represented in spheres.
Expression and Purification of Recombinant Peptide Library
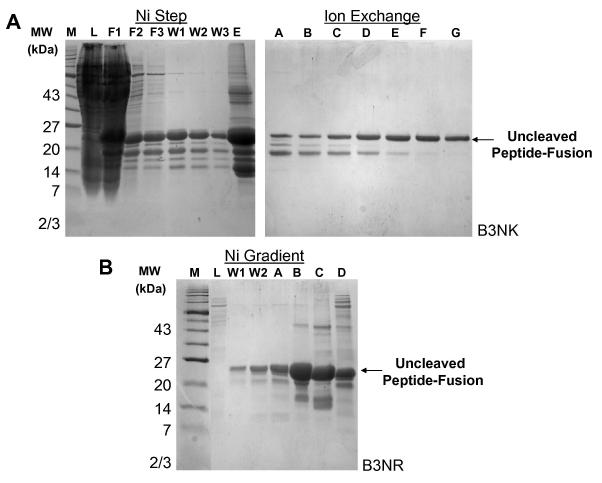
Once the desired genetic mutations were verified, the vectors were transformed into the BL21(DE3) strain of E. coli, expressed and harvested. The resulting cell pellets were lysed and prepared for purification. Various avenues were tested to optimize the purification of the peptide-fusion from other bacterial proteins. This was done in order to minimize the potential additive loss of protein during each purification step. Therefore, a nickel affinity step gradient, alone or in combination with ion exchange methods as well as nickel affinity linear gradients were explored in order to obtain a peptide-fusion sample of adequate purity without sacrificing yield. For example, peptide B3NK was purified on a nickel-affinity column using a step gradient and ion exchange methods while peptide B3NR was purified by a nickel-affinity column using a linear imidazole gradient only (Fig. 4). It is clear that a two-step purification yields protein of higher purity compared to nickel-affinity chromatography alone. However, since only a few contaminants are removed by the ion-exchange chromatography step in spite of a significant loss of sample, the single-step linear gradient using nickel-affinity chromatography became the preferred method of purification.
Figure 4. Peptide-fusion purifications.
(A) SDS-PAGE gels of peptide-fusion B3NK purified using nickel-affinity chromatography with a step gradient (Load-L, Flow through 1-F1, Flow through 2-F2, Flow through 3-F3, Wash 1-W1, Wash 2-W2, Wash3 W3, and Elution-E) and ion exchange (fractions A-G) are indicated. (B) SDS-PAGE gel of peptide-fusion B3NR purified using nickel-affinity chromatography and a linear imidazole elution gradient from 75 to 250 mM imidazole (Load-L, Wash1-W1, Wash 2-W2 and elution fractions A-C). Fraction D, which was eluted with 1M imidazole, contained a mixture of tightly bound B3NK and contaminants. Uncleaved peptide-fusion MW = 24 kDa.
Once the peptide-fusions were purified to a satisfactory degree, cleavage of the peptide from its fusion partner and linkers was accomplished by the highly specific human rhinovirus 3C protease (Fig. 5). Various protease to peptide fusion ratios were examined for optimal cleavage in minimal time. As a 5-hour digestion with a 1:25 protease to protein ratio was sufficient for full cleavage of peptide from the fusion tag, this method emerged as the time-optimized protocol. In order to separate the liberated peptide from the other components in solution, reverse phase HPLC was employed. By employing a three phase gradient which changes in organic phase from steep for buffer component elution, to shallow for peptide elution, to a second steep phase to regenerate the column and elute larger proteins such as thioredoxin and human rhinovirus 3C protease, the peptide was separated during a 21.5 minute gradient. For example, cleaved peptides B3DE, B3II and B3NR, all were successfully separated from the contaminating proteins and cleaved thioredoxin fusion partners via the three phase gradient (Fig. 6). Major peaks in the chromatogram correspond to buffer components (peak 1) and larger proteins such as the thioredoxin tag and protease (peak 3) as verified by SDS-Page analysis. To verify the peptide identity and purity from the expected peptide samples (peak 2) on the reverse phase HPLC chromatogram, electrospray ionization mass spectrometry (ESI-MS) was performed. The spectra indicate peptides are of the expected molecular weights (3,119.1 – 3,201.3 Da) (Fig. 7). In addition, the chromatogram for the liquid chromatography that is in line with the mass spectrometer showed only a single peak. Averaging of spectra from all regions of the peak resulted in uniform mass spectra indicating only one molecular species was present. These data suggest that the purified peptides are of extremely high purity.
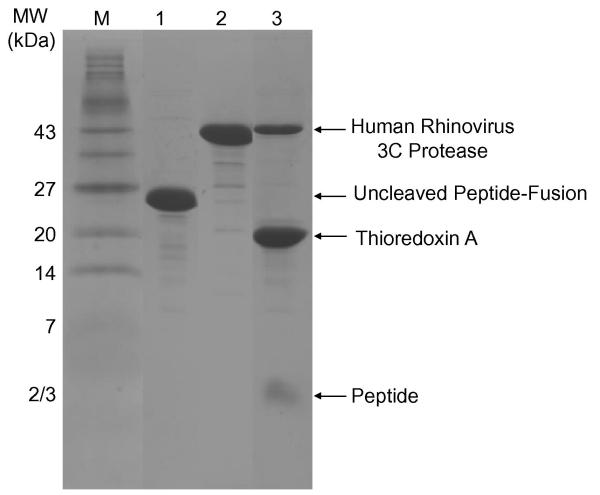
Figure 5. Peptide-thioredoxin fusion protein cleavage.
SDS-PAGE gel of the purified thioredoxin-peptide fusion (Lane 1). Purified human rhinovirus 3C protease (Lane 2) and cleavage reaction including human rhinovirus 3C protease, cleaved thioredoxin and free aPP peptide (Lane 3). Peptide MW = 3176 Da. Human rhinovirus 3C protease MW = 46,000 Da. Thioredoxin with tags MW = 27,912 Da.
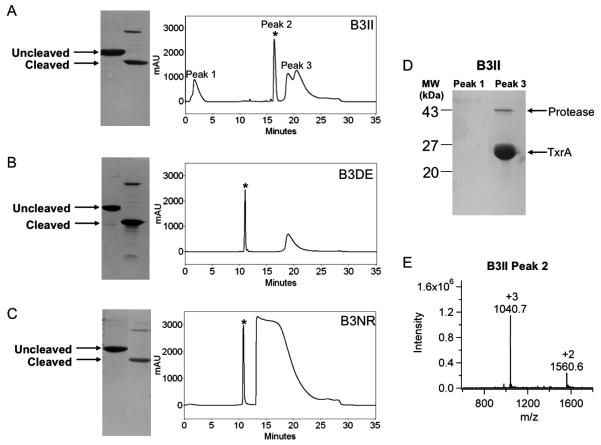
Figure 6. HPLC purification chromatograms of cleaved peptide-fusion samples.
(A-C) SDS-PAGE gel of uncleaved and cleaved peptide-fusion with corresponding HPLC separation chromatograms. Peak 1 consists of buffer components, peak 2 is eluted peptide indicated by * and peak 3 is cleaved thioredoxin and human rhinovirus 3C protease. (A) B3II (B) B3DE (C) B3NR (D) SDS-PAGE gel of peaks 1 and 3 from HPLC separation. (E) Mass spectra of HPLC peak 2, the purified peptide B3II, MW = 3119.1 Da.
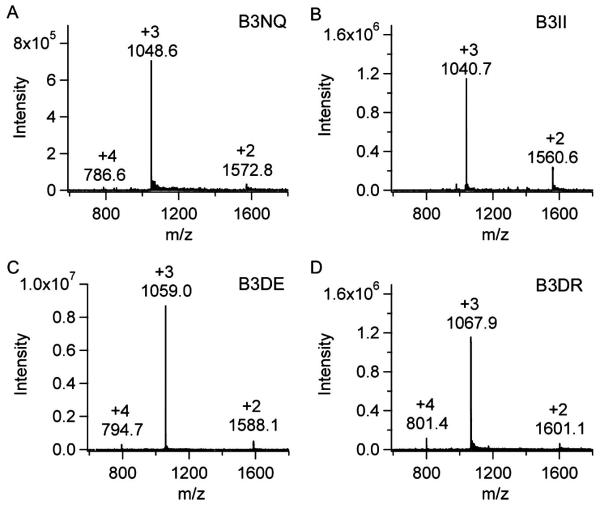
Figure 7. Mass spectra of peptides.
(A) B3NQ MW=3143.2 Da (B) B3II MW=3119.1 Da (C) B3DE MW=3174.2 Da (D) B3DR MW=3201.3 Spectra show double (+2) triply (+3), and quadruply (+4) charged ions.
In order to assess the overall utility of the peptide library construction system, the total amount of peptide fusion that was expressed in raw E. coli lysates is compared to the overall yield of each of the purification methods (Table 1). Total peptide yield ranged from 0.8 to 16% from the overall expression. A major contributor to low peptide yield is likely loss of protein at the step of column loading in the flow through during initial purification due to overloaded resin. In addition, other contributing losses are observed at the ionic exchange and reverse phase HPLC portions of the purification. The ability to be expressed and the overall yield after purification for each peptide was compared to different peptide features including charge, isoelectric point, and amino acid characteristics. Peptide charge ranged from -0.06 to -4.06 and isoelectric point from 3.77 to 6.92. The number of acidic residues in the peptide sequences ranged from two to five while one to three basic residues, six to twelve polar residues and seven to nine hydrophobic residues were present in the various sequences. Favorably, no correlation between the expressability of the peptide-fusion and intrinsic chemical characteristics of the peptides was found. Likewise, no correlation between peptidic character and overall yield of pure peptide was observed. This suggests that our system can be used for a wide variety of peptide sequences with a range of chemical characteristics.
Table 1. Peptide Expression, Yield and Chemical Characteristics.
Total peptide fusion expressed is compared to the overall yield of the peptide as a function of purification method used. Peptide characteristics such as charge, isoelectric point (IEP), the number of acidic, basic, polar, and non-polar amino acid residues are also tabulated.
| Peptide | % Peptide in Lysate |
Purification Method |
Potential yield (mg) |
Yield (%) |
Charge | IEP | Acidic | Basic | Polar | Non- polar |
|---|---|---|---|---|---|---|---|---|---|---|
| aPP | 26.1 | Ni Step | 43 | 3.5 | -2.15 | 4.75 | 5 | 3 | 12 | 9 |
| B3 | 11.5 | Ni / IE | 2 | 1.0 | -4.06 | 4.42 | 5 | 1 | 6 | 7 |
| B3DQ | 44.5 | Ni Step | N/A | N/A | -4.06 | 4.49 | 5 | 1 | 6 | 7 |
| B3DE* | 45.0 | Ni Step | 50 | 5.0 | -4.05 | 4.62 | 5 | 1 | 6 | 7 |
| B3DK | 45.9 | Ni / IE | 14 | 0.8 | -2.06 | 5.40 | 4 | 2 | 6 | 7 |
| B3DR | 9.3 | Ni / IE | 5 | 1.4 | -2.06 | 5.40 | 4 | 2 | 6 | 7 |
| B3NQ | 25.4 | Ni / IE | 11 | 1.1 | -1.06 | 6.00 | 2 | 1 | 9 | 7 |
| B3NE | 20.9 | Ni / IE | 28 | 2.3 | -2.06 | 5.33 | 3 | 1 | 8 | 7 |
| B3NK | 11.5 | Ni / IE | 8 | 1.0 | -0.06 | 6.92 | 2 | 2 | 8 | 7 |
| B3NR | 24.5 | Ni Gradient | 20 | 1.7 | -0.06 | 6.92 | 2 | 2 | 8 | 7 |
| B3II* | 45.0 | Ni / IE | 70 | 5.8 | -4.23 | 3.77 | 5 | 1 | 6 | 9 |
| B3IQ | 32.2 | Ni Gradient | 31 | 3.9 | -3.23 | 4.10 | 4 | 1 | 7 | 9 |
| B3IE | 35.9 | Ni Gradient | 32 | 16.0 | -4.23 | 3.98 | 5 | 1 | 6 | 9 |
| B3IK | 36.1 | Ni Gradient | 25 | 2.5 | -2.23 | 4.49 | 4 | 2 | 6 | 9 |
| B3IR | 37.6 | Ni Gradient | 35 | 2.9 | -2.23 | 4.49 | 4 | 2 | 6 | 9 |
| B3IL | 30.8 | Ni Gradient | 40 | 4.0 | -3.23 | 4.10 | 4 | 1 | 6 | 10 |
| 3INQ | 27.0 | Ni Gradient | 30 | 3.0 | -1.24 | 4.53 | 2 | 1 | 9 | 9 |
| 3INE | 36.6 | Ni Gradient | 28 | 2.8 | -2.23 | 4.25 | 3 | 1 | 8 | 9 |
| 3INK | 19.2 | Ni Gradient | 25 | 2.5 | -0.24 | 6.14 | 2 | 2 | 8 | 9 |
| 3INR | 29.7 | Ni Gradient | 33 | 3.3 | -0.24 | 6.14 | 2 | 2 | 8 | 9 |
Percent peptide in raw E. coli lystates was calculated using GeneTools (SynGene).
Indicates % peptide calculated was estimated by inspection of the stained SDS-PAGE gels. Ni: Ni-Affinity chromatography. IE: ionic exchange chromatography. Acidic, Basic, Polar and Non-polar represent the number of amino acids in the 27-residue peptide that have the particular chemical characteristic.
Discussion
During chemical synthesis of peptides, sequence errors are a likely result due to the imperfect coupling efficiency at each step. For chemically synthesized peptides, the contaminants are extremely similar to the desired product in size and chemical characteristics and are therefore difficult to separate based on chromatography. Peptides produced by biochemical synthesis by the E. coli ribosome are expected to be very homogeneous due to the ribosome’s low error rate. In the system described here, any contaminants that remain after affinity purification and protease cleavage of the biochemically synthesized peptide fusions are unlikely to be chemically similar to the peptides. Our method for production of peptides uses reverse-phase HPLC purification as the final step. Because the non-peptide components are not chemically related to the peptides themselves, HPLC purification allows for simultaneous removal of the fusion partner, the protease, and any residual components of the buffer solution. This makes a simple, two-step purification possible. In addition, because of the high fidelity of biochemical synthesis, the homogeneity of our peptides is very impressive. This results in a library of extremely high purity.
All of the peptide-fusion proteins described here were purified using a six-histidine affinity tag. Following nickel-affinity chromatography using a step gradient purification the peptide fusions were approximately 50% pure. We probed the effect of more extensively purifying the peptide fusions to >95% purity by subsequent anion exchange chromatography or to 80% purity by using a linear gradient of imidazole on the nickel-affinity column. Employing reverse-phase HPLC, we were able to robustly purify the cleaved peptide to homogeneity after either of these protocols. This finding significantly shortened our production time by obviating the ion-exchange purification step.
An additional advantage of the method described is that it is unnecessary for the peptide and the protease to be tagged with the same affinity tag, as is the case in some commercial protein-fusion purification systems. The method described here is compatible with any fusion partner and any protease. Since both the fusion partner and the protease are likely to be much larger than peptide products, it will be possible to separate the product from the contaminants in the final reverse-phase HPLC purification step. This fact is of particular importance in production of peptide libraries where additional amino acids at either the N- or C-termini can have significant effects on the overall properties of the peptide. Since any protease can be used, the overhanging amino acids from the protease cleavage site can be matched with the desired peptide sequence, or a protease that does not leave any overhang can be selected [37]. The facts that this method is not dependent on use of any particular affinity tags or on utilizing matching affinity tags also limit sub-cloning steps that are necessary to use the library production method.
This method of peptide production using genetically encoded peptides offers clear advantages in the length and fidelity of peptides that can be produced and the resultant purity of the final samples. In the field of chemical peptide synthesis, the concept of “difficult” sequences (e.g. hydrophobic sequences) exists. Conversely, the genetically-encoded production method described here was insensitive to peptide sequence such that every sequence attempted worked the first time with no optimization of expression or purification procedure required. In order to make this method truly competitive with chemical synthesis, it is essential that libraries can be produced on the same general time scale as chemical synthesis. The scheduling strategy for the twenty-member library we present here lays the foundation for production of much larger libraries using the same processes. To this end, we have time-optimized our expression, purification and characterization protocols. Using the protocols we report and have routinely executed, we have constructed a map for methodological synchronization (Fig. 8). By coordinating mutagenesis cycles, sequencing, transformation, expression, purification and characterization, this peptide library can be produced rapidly. We generated this map reflecting the methods that were actually used, however we have also performed sequencing reactions after an eight-hour growth of cultures followed by mini-prepping of DNA for sequencing. If this were applied to each peptide construct, it would save one day from each segment of the mutagenesis portion of the plan. In addition, we have developed methodology so that QuikChange-mutagenized plasmids can be directly transformed into an expression strain of bacteria (such as BL21(DE3)) rather than into a cloning strain, which can save one step in the protocol. With these developments, the entire library could be generated even more rapidly. As presented, this methodological synchronization scheme allows construction of twenty peptide-expressing plasmids and purification of the resultant peptides to homogeneity in a five-week period using one chromatography system, one HPLC and one LC-MS.
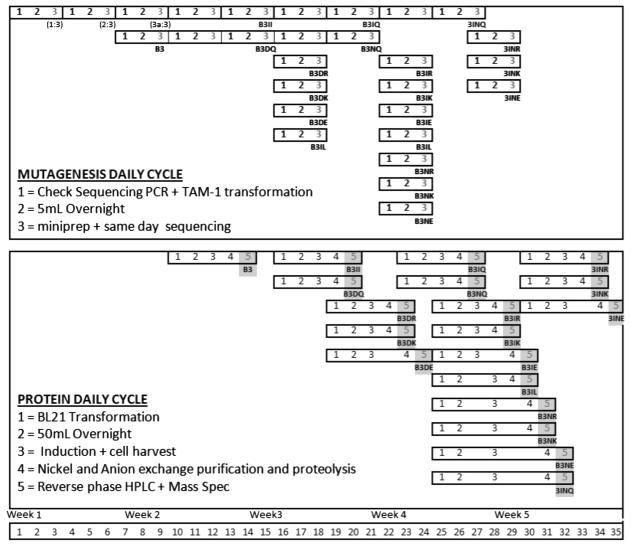
Figure 8. Methodological Synchronization.
An experimental map showing how synchronization of QuikChange-based mutagenesis and protein preparatory procedures allows the production of 20 peptides in a five-week time frame. Mutagenesis daily cycle (top) outlines the number of days in which cloning steps such as QuikChange mutagenesis, transformation, DNA purification and sequencing proceeds. 1:3, 2:3, and 3a:3 represents mutagenesis steps to create first peptide construct B3. Protein daily cycle (bottom) outlines the number of days in which expression, purification, and characterization for each peptide proceeds. The resultant DNA constructs are represented in light gray and purified peptides are represented in dark gray.
In this work, we have used no more than two liters of E. coli culture per peptide and have accepted losses at the affinity purification step. Nevertheless, the protocols discussed here could facilely be scaled up to produce even greater yields of pure peptide. For some of the peptide fusions we have constructed, the yield of the expressed fusion protein as a fraction of total E. coli proteins is high enough (45% of total E. coli protein) that we can nearly envision skipping purification of the fusion protein altogether. By co-expressing the protease and the fusion protein it may be possible to perform a one-step purification by HPLC to yield large quantities of pure peptide.
Acknowledgements
This work was supported by NSF Chemical Innovation Center for Fueling the Future CHE-0739227, National Research Service Award T32 GM08515 from the National Institute of Health (KLH), and NSF Award EEC-0649041 (KDO). We thank Alana Schepartz and Doug Daniels for kindly providing the synthetic gene of avian pancreatic polypeptide (Yale University New Haven, CT, USA) and Dr. Sumana Ghosh for helpful discussions.
Abbreviations
- LC-MS
liquid chromatography-mass spectrometry
- HPLC
high-performance liquid chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Winston RL, Gottesfeld JM. Rapid identification of key amino-acid—DNA contacts through combinatorial peptide synthesis. Chemistry & Biology. 2000;7:245–251. doi: 10.1016/s1074-5521(00)00099-5. [DOI] [PubMed] [Google Scholar]
- [2].Rodriguez M, Li SSC, Harper JW, Songyang Z. An Oriented Peptide Array Library (OPAL) Strategy to Study Protein-Protein Interactions. Journal of Biological Chemistry. 2004;279:8802. doi: 10.1074/jbc.M311886200. [DOI] [PubMed] [Google Scholar]
- [3].Sweeney MC, Wavreille AS, Park J, Butchar JP, Tridandapani S, Pei D. Decoding protein-protein interactions through combinatorial chemistry: sequence specificity of SHP-1, SHP-2, and SHIP SH2 domains. Biochemistry. 2005;44:14932–14947. doi: 10.1021/bi051408h. [DOI] [PubMed] [Google Scholar]
- [4].Baltrusch S, Lenzen S, Okar DA, Lange AJ, Tiedge M. Characterization of Glucokinase-binding Protein Epitopes by a Phage-displayed Peptide Library Identification of 6-Phosphofructo-2-kinase/fructose-2, 6-Bisphosphatase as a Novel Interaction Partner. Journal of Biological Chemistry. 2001;276:43915–43923. doi: 10.1074/jbc.M105470200. [DOI] [PubMed] [Google Scholar]
- [5].Newman JRS, Keating AE. Comprehensive Identification of Human bZIP Interactions with Coiled-Coil Arrays. American Association for the Advancement of Science. 2003:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- [6].Choe Y, Leonetti F, Greenbaum DC, Lecaille F, Bogyo M, Bromme D, Ellman JA, Craik CS. Substrate Profiling of Cysteine Proteases Using a Combinatorial Peptide Library Identifies Functionally Unique Specificities. Journal of Biological Chemistry. 2006;281:12824. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- [7].Gosalia DN, Salisbury CM, Ellman JA, Diamond SL. High Throughput Substrate Specificity Profiling of Serine and Cysteine Proteases Using Solution-phase Fluorogenic Peptide Microarrays*. Molecular & Cellular Proteomics. 2005;4:626–636. doi: 10.1074/mcp.M500004-MCP200. [DOI] [PubMed] [Google Scholar]
- [8].Gosalia DN, Salisbury CM, Maly DJ, Ellman JA, Diamond SL. Profiling serine protease substrate specificity with solution phase fluorogenic peptide microarrays. PROTEOMICS. 2005;5:1292–1298. doi: 10.1002/pmic.200401011. [DOI] [PubMed] [Google Scholar]
- [9].Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. National Acad Sciences. 2000:7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Turk BE, Huang LL, Piro ET, Cantley LC. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nature Biotechnology. 2001;19:661–667. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- [11].Cuerrier D, Moldoveanu T, Davies PL. Determination of Peptide Substrate Specificity for {micro}-Calpain by a Peptide Library-based Approach: The Importance of Primed Side Interactions. Journal of Biological Chemistry. 2005;280:40632. doi: 10.1074/jbc.M506870200. [DOI] [PubMed] [Google Scholar]
- [12].Garaud M, Pei D. Substrate profiling of protein tyrosine phosphatase PTP1B by screening a combinatorial peptide library. J. Am. Chem. Soc. 2007;129:5366–5367. doi: 10.1021/ja071275i. [DOI] [PubMed] [Google Scholar]
- [13].Huyer G, Kelly J, Moffat J, Zamboni R, Jia Z, Gresser MJ, Ramachandran C. Affinity Selection from Peptide Libraries to Determine Substrate Specificity of Protein Tyrosine Phosphatases. Analytical Biochemistry. 1998;258:19–30. doi: 10.1006/abio.1997.2541. [DOI] [PubMed] [Google Scholar]
- [14].Vetter SW, Zhang ZY. Probing the Phosphopeptide Specificities of Protein Tyrosine Phospha-tases, SH2 and PTB Domains with Combinatorial Library Methods. Current Protein and Peptide Science. 2002;3:365–397. doi: 10.2174/1389203023380594. [DOI] [PubMed] [Google Scholar]
- [15].Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. A rapid method for determining protein kinase phosphorylation specificity. Nature Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- [16].Mah AS, Elia AEH, Devgan G, Ptacek J, Schutkowski M, Snyder M, Yaffe MB, Deshaies RJ. Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening. BMC Biochemistry. 2005;6:22. doi: 10.1186/1471-2091-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Turk BE, Hutti JE, Cantley LC. Determining protein kinase substrate specificity by parallel solution-phase assay of large numbers of peptide substrates. [DOI] [PubMed]
- [18].Yaffe MB. Study of Substrate Specificity of MAPKs Using Oriented Peptide Libraries. Methods in Molecular Biology. 2004;250:237–250. doi: 10.1385/1-59259-671-1:237. [DOI] [PubMed] [Google Scholar]
- [19].Bartsevich VV, Juliano RL. Regulation of the MDR1 Gene by Transcriptional Repressors Selected Using Peptide Combinatorial Libraries. Molecular Pharmacology. 2000;58:1–10. doi: 10.1124/mol.58.1.1. [DOI] [PubMed] [Google Scholar]
- [20].El-Mousawi M, Tchistiakova L, Yurchenko L, Pietrzynski G, Moreno M, Stanimirovic D, Ahmad D, Alakhov V. A Vascular Endothelial Growth Factor High Affinity Receptor 1-specific Peptide with Antiangiogenic Activity Identified Using a Phage Display Peptide Library*. Journal of Biological Chemistry. 2003;278:46681–46691. doi: 10.1074/jbc.M308681200. [DOI] [PubMed] [Google Scholar]
- [21].Jahnke A, Wilmes T, Adebahr S, Pfeifer D, Berg T, Trepel M. Leukemia targeting ligands isolated from phage display peptide libraries. Leukemia. 2007;21:411. doi: 10.1038/sj.leu.2404548. [DOI] [PubMed] [Google Scholar]
- [22].Obata T, Yaffe MB, Leparc GG, Piro ET, Maegawa H, Kashiwagi A, Kikkawa R, Cantley LC. Peptide and Protein Library Screening Defines Optimal Substrate Motifs for AKT/PKB. Journal of Biological Chemistry. 2000;275:36108–36115. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- [23].Benoiton NL. Chemistry Of Peptide Synthesis. Taylor & Francis; 2006. [Google Scholar]
- [24].Scriba GKE. Peptide Diastereomers. Separation of.
- [25].Tam JP, Lu YA. Coupling Difficulty Associated with Interchain Clustering and Phase Transition in Solid Phase Peptide Synthesis. Journal of the American Chemical Society. 1995;117:12058–12063. [Google Scholar]
- [26].Kent SBH. Peptides:Structure and Function. In: CM Deber V.H.a.K.K., editor. Proceedings of the 9th Annual American Peptide Symposium; Rockford. 1985. pp. 407–414. [Google Scholar]
- [27].Tam JP. Peptides:Structure and Function. In: CM Deber V.H.a.K.K., editor. Proceedings of the 9th Annual American Peptide Symposium; Rockford. 1985. pp. 423–425. [Google Scholar]
- [28].Weaver RF. Molecular Biology. McGraw-Hill College; 2007. [Google Scholar]
- [29].Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- [30].Guan C, Li P, Riggs PD, Inouye H. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene. 1988;67:21–30. doi: 10.1016/0378-1119(88)90004-2. [DOI] [PubMed] [Google Scholar]
- [31].LaVallie ER, DiBlasio EA, Kovacic S, Grant KL, Schendel PF, McCoy JM. A Thioredoxin Gene Fusion Expression System That Circumvents Inclusion Body Formation in the E. coli Cytoplasm. Bio/Technology. 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]
- [32].Blundell TL, Pitts JE, Tickle IJ, Wood SP, Wu CW. X-Ray Analysis (1. 4-angstrom Resolution) of Avian Pancreatic Polypeptide: Small Globular Protein Hormone. Proceedings of the National Academy of Sciences. 1981;78:4175–4179. doi: 10.1073/pnas.78.7.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tonan K, Kawata Y, Hamaguchi K. Conformations of isolated fragments of pancreatic polypeptide. Biochemistry. 1990;29:4424–4429. doi: 10.1021/bi00470a023. [DOI] [PubMed] [Google Scholar]
- [34].Zondlo NJ, Schepartz A. Highly Specific DNA Recognition by a Designed Miniature Protein. Nature (London) 1993;363:38. [Google Scholar]
- [35].Chin JW, Grotzfeld RM, Fabian MA, Schepartz A. Methodology for optimizing functional miniature proteins based on avian pancreatic polypeptide using phage display. Bioorganic & Medicinal Chemistry Letters. 2001;11:1501–1505. doi: 10.1016/s0960-894x(01)00139-1. [DOI] [PubMed] [Google Scholar]
- [36].Shimba N, Nomura AM, Marnett AB, Craik CS. Herpesvirus Protease Inhibition by Dimer Disruption. Journal of Virology. 2004;78:6657–6665. doi: 10.1128/JVI.78.12.6657-6665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Feeney B, Soderblom EJ, Goshe MB, Clark AC. Novel protein purification system utilizing an N-terminal fusion protein and a caspase-3 cleavable linker. Protein Expression and Purification. 2006;47:311–318. doi: 10.1016/j.pep.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]