Abstract
Traditionally, we understand that individual phenotypes result primarily from inherited genetic variants together with environmental exposures. However, many studies showed that a remarkable variety of factors including environmental agents, parental behaviors, maternal physiology, xenobiotics, nutritional supplements and others lead to epigenetic changes that can be transmitted to subsequent generations without continued exposure. Recent discoveries show transgenerational epistasis and transgenerational genetic effects where genetic factors in one generation affect phenotypes in subsequent generation without inheritance of the genetic variant in the parents. Together these discoveries implicate a key signaling pathway, chromatin remodeling, methylation, RNA editing and microRNA biology. This exceptional mode of inheritance complicates the search for disease genes and represents perhaps an adaptation to transmit useful gene expression profiles from one generation to the next. In this review, I present evidence for these transgenerational genetic effects, identify their common features, propose a heuristic model to guide the search for mechanisms, discuss the implications, and pose questions whose answers will begin to reveal the underlying mechanisms.
INTRODUCTION
A central tenet in genetics is that inherited variants together with life-style and environmental exposures account for phenotype and disease. By searching the genome of affected individuals and studying their life history, we hope to identify the factors that account for the associations between genotype, environment and phenotype, thereby linking cause and outcome. However, increasing evidence suggests an alternative mode of inheritance that is revolutionizing the ways that we understand the consequences of genetic factors and environmental conditions on phenotypic variation and disease susceptibility. In these examples, factors acting in the current generation affect gene expression in subsequent generations, in the absence of the inherited genetic variant or continued exposure to the environmental agent. If these transgenerational effects are found to affect common traits and diseases, the implications could be profound because breaking the bond between genotype, environment and phenotype complicates a fundamental premise of many disease studies. Recent reports bring this curious and potentially important mode of inheritance a step closer to the human condition where finding similar evidence is much harder. Several reviews emphasize genetic mechanisms in simpler organisms and environmental effects in higher organisms (1–6). Here, I focus on discoveries concerning transgenerational epistasis and parental genetic effects without transmission of the variant allele. In particular, I emphasize key features and general patterns, discuss possible mechanisms and consider the evolutionary and biomedical implications. A detailed examination of the evidence is provided given the complicated and unconventional nature of this mode of inheritance.
TRANSGENERATIONAL GENETIC EFFECTS
Three general classes of transgenerational genetic effects have been reported, one that involves interacting genes (epistasis) in different generations, another that involves parental effects without transmission of the genetic variant and a third that involves gene-diet interactions across generations.
Transgenerational gene interactions
Several studies highlight the evidence for interacting genes in different generations, which is perhaps the most provocative discovery to date in transgenerational research.
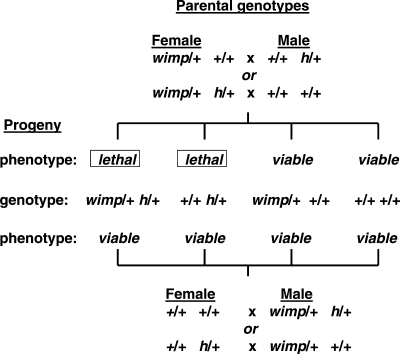
The first study involves the general problem of maternal RNAs and proteins that are deposited into oocytes to manage early embryogenesis until zygotic transcription begins after the ‘maternal zygotic transition’ (7,8). This study focused on the wimp mutation that interacts through the maternal lineage with many early-acting maternal effect and embryonic genes to compromise embryonic viability (9). wimp is a mutation in the gene that encodes the 140 kDa subunit of RNA polymerase II; the molecular nature of the mutation is not yet known. When wimp/+ heterozygous females are mated to males that are h/+ heterozygotes for the hairy (h) pair-rule gene, both h/+ wimp/+ and h/+ +/+ genotypic progeny classes show embryonic lethality, whereas progeny of the reciprocal cross of h/+ females to wimp/+ males are fully viable (Fig. 1). Thus, wimp acts premeiotically through the maternal but not the paternal lineage, affecting viability of h/+ offspring regardless of wimp inheritance. Remarkably, 23 of the 65 tested genetic variants interact in similar ways with wimp. In the cases that have been studied, wimp reduces transcript and protein levels of the interacting genes but not the genes that failed to interact, indicating specific rather than global effects on transcription and translation, and suggesting that organismal functions are sensitive to the level of the interacting proteins. As expected for loss-of-function mutations, the modified phenotype in heterozygous mutants that interact with wimp is reminiscent of the phenotype in mutant homozygotes, a phenomenon that is known as ‘pseudo-dominance’. wimp may be a special case of transgenerational effects, however, because its mRNA or protein is transferred from mother to offspring through oocytes, and lethality results if mutations in other genes sensitize early embryogenesis to reduced transcription of specific proteins. Although it is possible that these transcriptional changes have transgenerational effects, early embryonic lethality complicates direct tests to determine whether transmission to subsequent generations occurs in descendants of wimp heterozygotes.
Figure 1.
Transgenerational epistasis between wimp and interacting genes such as hairy (h) in Drosophila. Four parental crosses were made that each produced four identical genotypic classes among the offspring. Note that wimp acts in the parental generation and the interacting partners in the offspring and that maternal wimp leads to embryonic lethality of h/+ offspring regardless of wimp inheritance, but paternal wimp does not affect offspring viability. Similar results were found with many other maternal effect and early embryonic genes in interactions with wimp. See Parkhurst and Ish-Horowicz (9) for additional information.
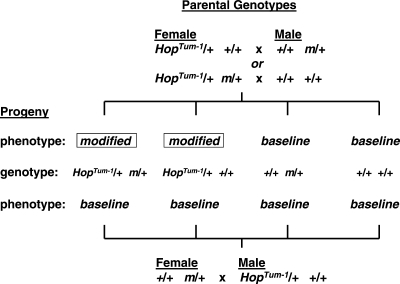
The second example of transgenerational gene interactions involves the hyperactive Janus kinase (JAK) HopTum-1 allele in Drosophila (10). (JAK kinase is member of a family of intracellular non-receptor kinases that mediate cytokine signaling through the JAK-STAT pathway.) Previous work showed that JAK signaling globally counteracts heterochromatin formation and JAK hyperactivity disrupts epigenetic reprogramming during development (11). In this study, HopTum-1/+ heterozygous females were mated to males that were heterozygous for variants in each of 37 different genes, 24 of which were shown to modify (enhance or suppress) hematopoietic tumorigenesis among HopTum-1/+ progeny, regardless of inheritance of the interacting partner (Fig. 2). Many of these modifiers modulate heterochromatin remodeling (see Table 1 in Ref. 10). This transgenerational effect requires HopTum-1 inheritance through the maternal germline, although the modifiers could be inherited through either germline. Remarkably, the effect of one of the modifiers (Kruppel, Kr) was transmitted through two subsequent generations without inheritance of the Kr mutation, but with progressively weaker effects in subsequent generations, an effect that the authors interpreted as segregation of many modified loci throughout the genome, although other explanations are possible. The authors then showed that HopTum-1 acted in a transgenerational manner to transmit established effects of Kr mutants on methylation and transcription of the fushi-tarazu pair-rule transcription factor gene. Thus hyperactivity of the JAK kinase pathway promotes transmission of heterochromatin changes to subsequent generations by disrupting the normal process of epigenetic reprogramming during early embryonic development, with adverse outcomes including enhanced tumorigenesis in subsequent generations.
Figure 2.
Transgenerational epistasis between JAK hyperactivity in HopTum-1 mutant and many partner genes (m) in flies. Three crosses were made, two with maternal HopTum-1 and one with paternal HopTum-1. All three crosses produced four identical genotypic classes among the offspring. Note that the interacting partners act in the parental generation and HopTum-1 in the offspring, and that only maternal HopTum-1 modified (enhanced or suppressed) hematopoietic tumorigenesis. See Xing et al. (10) for additional information.
The third example of interacting genes in different generations involves susceptibility to testicular germ cell tumors (TGCTs) in laboratory mice (12). Although susceptibility is highly heritable, its genetic control is unusually complex. Evidence for high heritability includes genetic effects accounting for ∼25% of TGCT susceptiblity in the Swedish Family Cancer Database of 9.6 million individuals (13), a relative risk that is higher than other cancers, at 10–13-fold for brothers of cases and 4-fold for sons of affected fathers (14–17), and finally a risk for a monozygotic twin of a TGCT case that is increased 75-fold (18). However, despite strong heritability, the genetic variants responsible for inherited susceptibility are remarkably elusive. In humans, the reported genetic variants together account for less than 10% of variation (19,20). Similar complexity is found in the laboratory mouse model where crosses between susceptible and resistant inbred strains resulted in a frequency of only 0.01% affected males (21). Difficulty finding the susceptibility genes for highly heritable traits is unusual.
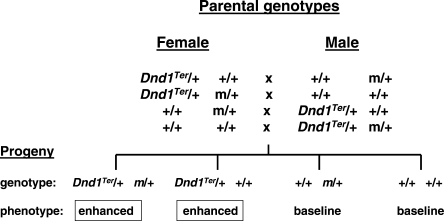
Given these challenges, focus shifted to single gene variants that modify susceptibility in genetically predisposed males from the 129 family of inbred strains (22). In a formal test for epistasis, six genetic TGCT modifiers (m) were unexpectedly found to interact in a transgenerational manner with the Ter mutation in the Deadend1 (Dnd1) gene (Fig. 3). Dnd1 shows sequence similarity with Apobec complementation factor (A1cf) (23), which is the RNA-binding subunit of the cytidine deaminase RNA editing complex (24). Dnd1 also controls access of specific microRNAs to their mRNA targets, several of which play key roles in primordial germ cells, which are the TGCT stem cells (25). The interaction between modifiers in parents and Dnd1Ter in sons is remarkably strong, with both Ter/+ m/+ and Ter/+ +/+ male offspring showing a 2–3-fold increased prevalence of affected males and a 6–7-fold increased prevalence of bilateral versus unilateral TGCT cases. The key evidence for transgenerational epistasis is the contrasting results for Ter/+ +/+ males, which show the expected baseline rate of affected males in control crosses that do not involve the modifiers, but increased risk when the modifier is present in one of the parents. In addition, the similar rates of affected Ter/+ m/+ and Ter/+ +/+ males suggests that m acts before meiosis in the parents to increase susceptibility in all Ter/+ sons. Together these results implicate RNA editing and miRNA control in transgenerational effects on germ cell biology, cancer susceptibility and early embryonic development.
Figure 3.
Transgenerational epistasis between the Deadend1 mutant Dnd1Ter and interacting partner genes (m) in laboratory mice. Each of the four crosses produced four identical genotypic offspring classes. Only Dnd1Ter sons, regardless of inheritance of the interacting partner gene showed enhanced tumorigenesis. Note that the modifier genes act in the parental generation and Dnd1Ter in the offspring. See Lam et al. (12) for additional information.
Transgenerational effects without epistasis
Several examples have been reported where genetic variants in the parental generation have phenotypic effects in their wild-type offspring, without inheritance of the predisposing genetic variant. In some cases, preliminary studies suggest that these effects do not depend on genetic background, but this point has not been tested rigorously, so an interacting gene in the progeny remains a possibility.
Rassoulzadegan et al. (26) showed that white-spotting of the feet and the tip of the tail associated with an engineered mutation at the Kit receptor gene in the mouse was transmitted to wild-type progeny who did not inherit the predisposing Kit variant, a phenomenon that the authors call ‘paramutation’ (see also Ref. 1). Binding of the Kit ligand to the Kit receptor tyrosine kinase leads to signaling through the P13K-Akt-mTOR effector pathway with impacts on transcription, proliferation, cell survival, invasiveness and angiogenesis (27). Mutants in the Kit ligand and its receptor typically affect melanogenesis, hematopoiesis and gametogenesis (28).
This unconventional mode of inheritance of the white-spotting phenotype was found with two closely related genetically-engineered mice, but interestingly not with mice that have a classical mutation in the Kit gene. The primary focus of the Rassoulzadegan study involved a genetic variant where the β-galactosidase (lacZ) coding sequence was inserted down-stream of the Kit promoter and regulatory sequences. Similar paramutation effects were also found in mice in which the gene for green fluorescent protein (GFP) was inserted downstream of Kit transcriptional control elements. In contrast, mice with the Kitw-v mutation did not transmit the spotting phenotype to genetically wild-type mice. (This spontaneous mutation involves a C to T point mutation at nucleotide 2007 that causes a threonine to methionine substitution at amino acid 660.) To account for the contrasting effects among these allelic variants, it may be relevant that the two alleles that show transgenerational effects are transgene insertions that lead to loss-of-function, whereas the Kitw-v allele that does not show transgenerational effects is a missense mutation and does not lead to hemizygosity. But why contrasting allele types makes such a dramatic difference in outcome is unclear.
Inheritance of the anomalous white-spotting phenotype has interesting features. The effect was strongest in the second generation, was independent of the genetic backgrounds that were tested, and progressively disappeared over subsequent generations. As expected, the Kit transcript level in heterozygotes for the engineered mutant was reduced ∼50% from normal levels, but surprisingly was also reduced to similar levels in paramutated wild-type mice. In addition, Kit transcripts were found in stages of spermatogenesis as well as in epididymal sperm where expression is not normally found, and transcripts of heterogeneous sizes were found in testes of Kit mutant and paramutated mice. Finally, preliminary evidence showed that RNA from brain and sperm from engineered mutant or paramutated mice, when injected into wild-type one-cell embryos, resulted in the white-spotting phenotype in ∼50% of the resulting offspring. Similar effects were found after injection of two microRNAs (miR-221 and miR-222) that have Kit as their mRNA target. Analysis of mice with other mutant Kit alleles might be instructive about the molecular triggers for transgenerational effects. Finally, it was not evident from this report whether Kit paramutation is restricted to white-spotting (melanogenesis), or also affects other prototypic Kit phenotypes such as anemia (hematopoiesis) and sterility (gametogenesis).
Recently, the same group report paramutation effects of microRNA miR-1 on cyclin-dependent kinase 9 (Cdk9), resulting in cardiac hypertrophy (29). The authors injected mRNA fragments corresponding to coding sequence or the cognate miR-1 for Cdk9, a gene that plays a central role in heart development. Both RNAs led to a cardiac hypertrophy phenotype that was inherited epigenetically, with presence of miR-1 in the sperm nucleus. In contrast to the effects on the mRNA level for the Kit receptor (26), the introduced RNAs led to a substantial increase in the Cdk9 mRNA level. Molecular and histopathologic features were transmitted through both the female and male lineages for at least three generations after treatment. This transgenerational epigenetic effect might explain the variable occurrence and presentation in familial cases of cardiac hypertrophy in humans (30).
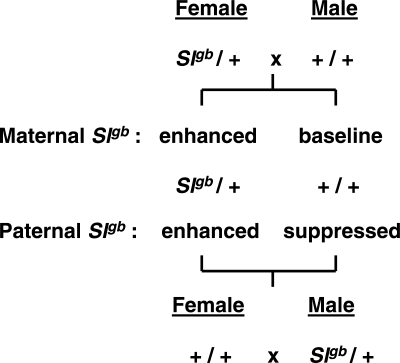
Transgenerational effects were reported for Kitl, the ligand for the Kit receptor (31). The authors showed that wild-type sons of males that are heterozygous for the steel grizzle-belly (Slgb) mutation in the Kitl gene are protected from spontaneous TGCTs, whereas wild-type sons of Slgb heterozygous females show the expected baseline rate of TGCTs that is characteristic of the host strain background on which the tests were done (Fig. 4). Slgb is a deletion mutation of the gene encoding the Kit ligand (32). In contrast to the paramutation white-spotting study (26), these wild-type sons of Slgb males did not show the grizzle-belly phenotype. It is also noteworthy that Kit receptor mutants do not affect susceptibility to TGCTs (31). The Rassoulzadegan study with the Kit receptor together with the Heaney study with the Kit ligand suggest that this signaling pathway plays a pivotal role in transmitting signals in a transgenerational manner, from parents to offspring, with effects on least two fundamental developmental pathways, namely melanogenesis and gametogenesis.
Figure 4.
Transgenerational epigenetic effects of the Kit ligand mutant Slgb on susceptibility to spontaneous testicular germ cell tumors in male offspring. Reciprocal cross between Slgb heterozygotes and wild-type mice on the same genetic background produced two identical genotypic classes among the offspring, but only wild-type sons of Slgb heterozygous male parents showed significantly reduced risk for TGCTs—to date affected males in this classes have not been found. See Heaney (31) for additional information.
Finally, to identify genes that control epigenetic reprogramming, Chong et al. (33) screened chemically-mutagenized mice for paternally-transmitted phenotypic modifications of a variegated expression pattern in GFP transgenic mice. The authors used a chemical mutagen N-ethylnitrosourea (ENU), which is a potent carcinogen, mutagen and alkylating agent that efficiently induces single base mutations. Two mutated genes were discovered, one in the Smarca5 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5) gene that is involved in chromatin remodeling, and the other in DNA methyltransferase 1 (Dnmt1) that is involved in the maintenance DNA cytosine methyltransferase activity. Smarca5 is expressed during spermatogenesis and in spermatids, and when inherited through the paternal lineage the induced mutant allele modifies the penetrance of the yellow coat color in agouti viable-yellow (Avy) mice. The phenotypic effects of the induced Dnmt1 mutation were also inherited through the paternal lineage and affected the range of coat colors found in Avy mice. The authors propose that transgenerational effects result from changes in dosage of chromatin proteins rather than from transmission of mRNas or miRNAs.
Observations and generalizations
Several features of the transgenerational gene interactions are noteworthy. First, although several of these examples show classic features of conventional epistasis, the interacting genes occur in different generations. Second, in all cases, to account for offspring phenotype, parental genotype must be known. Third, in the cases that have been studied (26,29), the phenotypic consequences of transgenerational effects persist beyond the first generation but with progressively weaker effects. Fourth, transgenerational effects tend to be sensitive to dosage rather than alternative functions of the interacting genes. Fifth, several of the parental effects must occur before the haploid phase so that all genetically predisposed progeny are affected regardless of inheritance of the parental gene. Sixth, some genetic variants are unitary parental effects with heterogeneous partners in progeny, e.g. wimp in the maternal lineage and 23 partners in the offspring (9), others are unitary in progeny with heterogeneous interacting partners in the parental generation, e.g. any of 37 genes in the parental genome and HopTum-1 offspring (10), and any of six variants in the parents and Dnd1Ter in the sons (12).
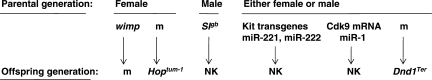
Two important generalizations emerge from these studies (Fig. 5). The first generalization involves the examples of paternal transmission that focus attention on mechanisms such as methylation and histone modifications as well as RNAs and proteins in sperm. For some traits, care must be taken to control for interactions between fathers and offspring that could lead to transgenerational effects on social, neurological, metabolic and perhaps other traits. In addition, similar molecular mechanisms probably operate in maternal transmission, but are confounded with effects resulting from the special physiological relationship between mother and her fetuses and pups.
Figure 5.
Summary of the parental gender effects together with parental and offspring genetics for the transgenerational genetic effects. See text for details.
The second generalization involves the remarkably diverse functions and pathways that mediate epigenetic transmission through the parental germlines, including Kit signaling [Kit ligand (31) and Kit receptor (26)], DNA methylation (Dnmt1, Chong 33), and RNA biology [transcription (9), mRNA (26,29) and miRNA biology (26,29)], whereas in the offspring generation, functions include JAK kinase and histone modifications (Hop) (10) as well as RNA editing (23) and miRNAs biology (25). Whether these are integrated or parallel pathways in the parental or in the offspring generations remains to be determined.
ENVIRONMENTAL EFFECTS AND TRANSGENERATIONAL GENE-DIET INTERACTIONS
Pioneering studies in animal models and epidemiological evidence in humans show that various environmental exposures have effects that in some cases are transmitted epigenetically for at least three generations (for review, see Refs. 3,4,34). These environmental factors include maternal metabolism and physiology (35–38), nutritional supplements (3,4), xenobiotics (39–43), maternal and paternal behaviors (44,45), irradiation and chemical mutagens (see below), and viruses (46,47) and perhaps symbiotic microbes. In some cases the strength of the effect persists across generations, although in other cases the effects eventually revert back to the original state. Some of exposures involve epigenetic modifications of the host genome, whereas others such as viruses are inherited independent of genetics and epigenetics.
These environmental factors sometimes involve metastable epialleles—loci that can be epigenetically modified in variable and reversible manners, resulting in different phenotypes despite genetic identity. The prototypic example is the agouti viable-yellow (Avy) allele at the agouti locus (48). This mutation results from a transposon (intracisternal A particle—IAP) insertion in the promoter region of the agouti gene. Stochastic effects modulate extent of methylation silencing of a cryptic promoter in the IAP, leading to a range in the degree of methylation and resulting in variation in the extent of yellow versus agouti coat color. When the diet of pregnant females is supplemented with folate and other methyl donors such as choline and betaine, IAP methylation patterns are modulated in ways that can be inherited in subsequent generations (48,49). Thus dietary factors during pregnancy interacting with specific kinds of genetic variants in fetuses control expression patterns and phenotypes in offspring—an example of transgenerational gene-diet interactions.
Studies with endocrine disruptors are particularly striking. Agents such as vinclozolin and methoxychlor have strong and diverse transgenerational effects (42,43). (Vinclozolin is a fungicide with antiandrogenic activity, and methoxychlor is a pesticide with estrogenic activity.) In particular, reduced fertility and spermatogenic abnormalities were transmitted for at least three generations after exposure of the pregnant females, formally excluding the possibility of exposure of primordial germ cells (the precursors of sperm and eggs) within the fetus of pregnant females (42,50). Vinclozolin also induces adult diseases of the prostate, kidney, immune system, testis, breast and lipid biochemistry (43) as well as anxiety and mate-choice (51). The many organs and tissues, systems and functions suggest diverse targets of transgenerational disease during embryogenesis, development and adulthood. Anway et al. (42,43) also provided evidence that these agents disrupt normal patterns of methylation and their transmission, suggesting that these epigenetic changes escape the normal reprogramming that occurs early in development to record the gender of the individual (52).
Mutagens
Mutagenesis studies with agents such as irradiation and chemical mutagens such as ENU (53–55) reveal a remarkable result. The conventional protocol is to induce deletions and nucleotide substitutions by exposing adult males to these various physical and chemical agents (56). These treatments result in temporary sterility because the later stages of spermatogenesis are disrupted, but fertility is eventually restored from surviving cells from earlier stages of spermatogenesis. Breeding these males converts mutagenized gametes to mutagenized mice that can then be tested for heritable induced phenotypic variants. These variant mice are then propagated for genetic and functional studies. It is commonly assumed that occurrence of new mutations returns to baseline rates in the first generation after exposure.
Several studies highlight a remarkable and unexpected transgenerational effect. In these studies, rates of ENU-induced instability of DNA repeats such as expanded simple-tandem repeats were measured in treated individuals as well as in succeeding generations. The authors found that the frequency of new genetic variants remains elevated, at rates comparable with the rate in treated individuals and at similar rates through females and males, for at least two generations after the original exposure, and leading to elevated rates of mortality and malformation among grand-offspring of exposed individuals (53–55). In addition, if mutagenized individuals are outcrossed to another inbred strain, new mutations are found both in DNA derived from the strain that was originally mutagenized as well as in DNA derived from the outcross strain, proving that new mutations arise at a high rate for several generations after the original exposure (54). Finally the authors showed that the high rate of induced mutations eventually returns to baseline suggesting either a dilution of the active agents, or more likely a response to environmental or even genetic stress that eventually returns homeostatically in the absence of persistent exposure. The most likely explanation is that repair of spontaneous processes is reduced or compromised, at least for several generations, in part perhaps as an epigenetic adaptation to stress conditions, allowing new mutations to persist at an elevated rate (57,58). It would then be interesting to determine whether other stress conditions lead to relaxed DNA repair and increased rates of new mutations, and whether these show transgenerational effects.
MISSING HERITABILITY
Nearly 30 years ago the molecular genetic revolution began the systematic discovery of genes that are responsible for many simple (Mendelian) genetic traits (59,60). Until recently, however, more complex and common traits were beyond study. Now with largely complete genome sequences for human and other species, and with new high-throughput technologies that provide genotypes for very large numbers of genetic variants across the genome, remarkable progress is being made with the mapping and sometimes identification of genes and other functional DNA elements that contribute to phenotypic variation and disease risk (60,61). As hoped, these discoveries are providing exciting insights about sometimes unexpected protein functions and physiological pathways (62).
But these genetic studies have led to a remarkable and unexpected observation that has been termed ‘missing heritability’ (63). The heritable component of many traits is often substantial, with genetically complex conditions such as autism showing 90% heritability (61–63). [Heritability in its general (broad) sense is the fraction of the total phenotypic variance that is attributable to genetic factors, and measures the tendency for similar phenotypes across generations within versus between families.] However, the fraction of the phenotypic variation that is explained by the individual genetic variants that have been discovered, as well as the fraction that is explained by their cumulative action, is often disconcertingly small, usually less than 10% (61–63). The difference between the heritability of a trait and the explained variance implies that most genetic risk have eluded detected. As a result, a reasonably complete inventory of genes is not yet available for characterizing the genetic architecture of these traits, for modeling studies to prioritize targets for molecular interventions, and for identifying reliable diagnostic biomarkers. Several classes of factors could account for missing heritability including significant numbers of variants that are rare or that have weak effects (61–64), copy number variation (65), epistasis (66,67), polymorphic transposable elements and other functional repeat elements (49,68), and undetected genetic variants because of haplotype heterogeneity and recombination hotspots (69).
Transgenerational genetic effects could also contribute to missing heritability. With transgenerational effects, individuals show the trait not because they carry the responsible genetic variants, but instead because these variants were present in previous generations. Under these circumstances, genome surveys of affected individuals would fail to reveal the predisposing genetic factors. However, a recent analytical study showed that transgenerational epigenetic effects probably contribute significantly to average risk but not to recurrence risk or heritability unless the effects persist for at least tens of generations (70). Currently the literature is largely silent on the extent to which these effects persist. Transgenerational epistasis is also unlikely to significantly impact estimates of recurrence risk or heritability because familial effects are probably small.
Despite the limited impact on several standard measures of population genetic and evolutionary features, unconventional approaches might be pursued to assess the impact of transgenerational genetic effects in human biology and disease. First, transgenerational genetic effects could be tested by comparing genome-wide genotyping results in the parental generation with phenotypes and clinical information on the offspring generation for trios or multiplex families, using the same principles and practices of genome-wide association studies (GWAS), but testing instead whether genetic variants in parents account for phenotypes in offspring because of transgenerational genetic effects rather than direct genetic inheritance. Second, surveys for epigenetic marks in cases and controls might provide evidence for inherited risk. GWASs currently focus on DNA sequence polymorphisms or on DNA copy number variation. But if offspring phenotype can be impacted through epigenetic inheritance, then it should be possible to survey methylation and histone marks as well as parentally-derived RNAs and proteins to test for the molecular changes that account for traits and diseases in offspring. Third, a defining characteristic of epigenesis is change in gene expression without corresponding change in DNA sequence. It may be possible to design expression profile studies that could identify loci that are candidate targets of epigenetic modification (71). Thus although transgenerational genetic effects might not appreciably affect population measures of genetic variation, progress might still be made to find the transgenerational epigenetic marks for gene discovery, biological studies, risk estimation and disease diagnosis.
MECHANISMS AND MODELS
A simple heuristic model provides a framework for assessing the evidence for transgenerational genetic effects and for identifying mechanisms that remain to be identified (Fig. 6). This model has several features that warrant consideration.
Figure 6.
Principles of transgenerational genetic effects. See text for details.
Considerable evidence from many species shows that both genetic factors and environmental conditions can act as physiological stresses that lead to epigenetic changes. The various factors and conditions that initiate transgenerational epigenesis illustrate remarkable heterogeneity in molecular functions as well as chemical and physical properties. In addition, many of the genetic variants that act in the parental generation confer loss-of-function variants, suggesting that simple changes in gene (protein) dosage are sufficient to trigger epigenetic responses. However, the factors and conditions that trigger transgenerational effects in the parental generation probably do not directly affect DNA methylation and chromatin remodeling, RNAs or other epigenetic features. Instead these stresses are probably sensed by molecular mechanisms that in turn transduce the signal to epigenetic changes in DNAs or RNAs in somatic cells. These mechanisms are largely unknown.
The epigenetic signal must then be transferred from somatic cells to the germ line. This is a difficult issue, because exposure of the individual leads to stress signals and epigenetic changes in somatic cells, but gametes carry the epigenetic marks from one generation to the next. The Kit signaling pathway has features that illustrate how this might work, because the Kit ligand is expressed in somatic cells, adjacent to germ cells and the receptor is expressed in germ cells, and proper function of both is required for normal spermatogenesis (27,28). This pathway is implicated in several cases of transgenerational genetic effects (Fig. 5), but whether this is the key pathway, whether other pathways are involved, and how this pathway triggers epigenetic changes remain to be determined.
Next, the epigenetic marks must pass within gametes from one generation to the next. Recent studies document the nature of methylation and histone marks, although other studies characterize the RNAs that are present in human sperm. In contrast to the typical histone to protamine transitions that occur during spermatogenesis, some nucleosomes are retained at imprinted, miRNA, and developmental transcription factors and signaling molecules loci (72). In addition, considerable evidence shows that RNAs as well as DNAs are packaged in sperm (73). The factors that determine the location and nature of these epigenetic marks and RNAs, whether these are sensitive to genetic and environmental stresses, and whether anomalies lead to altered phenotypes in the next generation remain to be determined.
The epigenetic marks in the germ line must then be transferred to appropriate somatic cell, and in cases where the marks persist to the next generation, the marks in one generation must be transferred to the next generation, either by maintaining methylation patterns or histone modifications in the germ line, or by maintaining RNAs in the germ line. Maintenance of methylation and histone profiles has been described. The ways in which RNAs are transmitted between generations is less obvious. But a recently discovered cytoplasmic element and the Deadend gene provides clues. Early embryos and germ cells, which are one of the first lineages of cells to be established during embryogenesis, have a cytoplasmic body called a germ cell (or polar) granule (74). These are found near the nuclear envelope and contain RNA editing enzymes, transposon-derived RNAs, Piwi proteins and Piwi-interacting RNAs, factors that control protein translation as well as key elements in RNA biology. These granules are thought to protect maternal (and paternal) transcripts until needed for development. In this context it may be relevant that a TGCT suppressor results from deletion of the Eif2s2 translation initiation factor (75) and that its locus shows transgenerational genetic effects (12,76, J.D. Heaney and J.H.N., unpubl.) Recently Deadend was shown to shuttle RNAs from the nucleus to these granule, based on its RNA recognition motif (77). Thus many of the key features of transgenerational genetic effects appear to be mediated through germ cell granules in the offspring generation.
At this point, several outcomes are possible, the next generation may return to the original phenotype in the absence or recurrent stress, or alternatively the genetics and phenotype of the offspring may represent a stress that reinitiates the cycles of transgenerational genetic effects.
NEXT QUESTIONS
Several obvious questions emerge:
Do specific or generic kinds of genetic and environmental factors trigger transgenerational genetic effects?
What is the molecular and physiological basis for sensing these genetic and environmental perturbations and transducing their signals to epigenetic changes?
What determines whether the transgenerational epigenetic mark is a change in methylation, a modification of histones, a difference in the number, kind or state of RNAs in the germline, or perhaps another class of molecular changes?
Do these molecular mechanisms target epigenetic changes at specific loci, or do they lead to epigenetic changes at variable loci in a stochastic manner across the genome?
What processes transfer the epigenetic mark from somatic cells to the germline in the parental generation, and then through meiosis to subsequent generations?
What is the rate of gain and loss of specific epigenetic marks across generations?
Do transgenerational genetic changes persist indefinitely, or do they require periodic reinforcement to avoid returning homeostatically to their original state?
What are the mechanisms for reversing transgenerational genetic changes, and under what conditions?
What determines the specificity of dysfunction and disease that result from epigenetic effects?
What is the normal function for transgenerational genetics?
What is the evolutionary impact of transgenerational genetics?
Ultimately these questions and the resulting investigations will identify the molecular marks and mechanisms that mediate transgenerational effects, they will reveal whether these are special cases or general effects, and they will establish the identity of epigenetic marks that can be used as diagnostic biomarkers for assessing transgenerational disease risk.
FUNDING
This work was supported in part by NIH (CA75056 and RR12305).
ACKNOWLEDGEMENTS
I thank Ron Conlon, Robert Elston, Jason Heaney, Sudha Iyengar, Vicki Nelson, Helen Salz, David Serre and Gunter Wagner for discussing these ideas and sharing references.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Chandler V.L. Paramutation: from maize to mice. Cell. 2007;128:641–645. doi: 10.1016/j.cell.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Galloway L.F., Etterson J.R. Transgenerational plasticity is adaptive in the wild. Science. 2007;318:1134–1136. doi: 10.1126/science.1148766. [DOI] [PubMed] [Google Scholar]
- 3.Gluckman P.D., Hanson M.A., Beedle A.S. Non-genomic transgenerational inheritance of disease risk. Bioessays. 2007;29:145–154. doi: 10.1002/bies.20522. [DOI] [PubMed] [Google Scholar]
- 4.Jirtle R.L., Skinner M.K. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleming J.L., Huang T.H., Toland A.E. The role of parental and grandparental epigenetic alterations in familial cancer risk. Cancer Res. 2008;68:9116–9121. doi: 10.1158/0008-5472.CAN-08-2184. [DOI] [PubMed] [Google Scholar]
- 6.Youngson N.A., Whitelaw E. Transgenerational epigenetic effects. Annu. Rev. Genomics Hum. Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- 7.Scheir A.F. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316:406–407. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- 8.Stitzel M.L., Seydoux G. Regulation of the oocytes-to-zygote transition. Science. 2007;316:407–408. doi: 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
- 9.Parkhurst S.M., Ish-Horowicz D. wimp, a dominant maternal-effect mutation, reduces transcription of a specific subset of segmentation genes in Drosophila. Genes Dev. 1991;5:341–357. doi: 10.1101/gad.5.3.341. [DOI] [PubMed] [Google Scholar]
- 10.Xing Y., Shi S., Le L., Lee C.A., Silver-More L., Li W.X. Evidence for transgenerational transmission of epigenetic tumor susceptibility in Drosophila. PLoS Genet. 2007;3:e151. doi: 10.1371/journal.pgen.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi S., Calhoun H.C., Xia F., Li J., Le L., Li W.X. JAK signaling globally counteracts heterochromatic gene silencing. Nat. Genet. 2006;38:1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam M.-Y.J., Heaney J.D., Youngren K.K., Kawasoe J.H., Nadeau J.H. Trans-generational epistasis between Dnd1Ter and other modifier genes controls susceptibility to testicular germ cell tumors. Hum. Mol. Genet. 2007;16:2233–2240. doi: 10.1093/hmg/ddm175. [DOI] [PubMed] [Google Scholar]
- 13.Czene K., Lichtenstein P., Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family Cancer Database. Int. J. Cancer. 2002;99:260–266. doi: 10.1002/ijc.10332. [DOI] [PubMed] [Google Scholar]
- 14.Sonneveld D.J., Sleijfer D.T., Schrafford Koops H., Sijmons R.H., van der Graaf W.T., Sluiter W.J., Hoekstra H.J. Familial testicular cancer ins a single-centre populations. Eur. J. Cancer. 1999;35:1368–1373. doi: 10.1016/s0959-8049(99)00140-9. [DOI] [PubMed] [Google Scholar]
- 15.Heimdal K., Olsson H., Tretli S., Flodgren P., Borresen A.L., Fossa S.D. Risk of cancer in relatives of testicular cancer patients. Br. J. Cancer. 1996;73:970–973. doi: 10.1038/bjc.1996.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forman D., Oliver R.T., Brett A.R., Marsh S.G., Moses J.H., Bodmer J.G., Chilvers C.E., Pike M.C. Familial testicular cancer: a report of the UK family register, estimation of risk and an HLA class 1 sib-pair analysis. Br. J. Cancer. 1992;65:255–262. doi: 10.1038/bjc.1992.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemminki K., Li X. Familial risk in testicular cancer as a clue to heritable and environmental etiology. Br. J. Cancer. 2004;90:1765–1770. doi: 10.1038/sj.bjc.6601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swerdlow A.J., De Stavola B., Swanwick M., Mangtani P., Maconochie N. Risk factors for testicular cancer: a case–control study in twins. Br. J. Cancer. 1999;80:1098–1102. doi: 10.1038/sj.bjc.6690470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanetsky P.A., Nandita M., Vardhanabhuti S., Li M., Vaughn D.J., Letrero R., Ciosek S.L., Doody D.R., Smith L.M., Weaver J. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat. Genet. 2009;41:811–815. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rapley E.A., Turnbull C., Al Olama A.A., Vermitzakis E.T., Linger R., Huddart R.A., Renwick A., Hughs D., Hines S., Seal S. A genome-wide association study of testicular germ cell tumor. Nat. Genet. 2009;41:807–810. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens L.C. Genetic influences on teratocarcinogenesis and parthenogenesis. Prog. Clin. Biol. Res. 1973;45:93–104. [PubMed] [Google Scholar]
- 22.Heaney J.D., Nadeau J.H. Testicular germ cell tumors in mice: new ways to study a genetically complex trait. Methods Mol. Biol. 2008;450:211–231. doi: 10.1007/978-1-60327-214-8_15. [DOI] [PubMed] [Google Scholar]
- 23.Youngren K.K., Coveney D., Peng X., Bhattacharya C., Schmidt L.S., Nickerson M.L., Lamb B.T., Deng J.M., Behringer R.R., Capel B., et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anant S., Davidson N.O. Molecular mechanisms of apolipoprotein B mRNA editing. Curr. Opin. Lipidol. 2001;12:159–165. doi: 10.1097/00041433-200104000-00009. [DOI] [PubMed] [Google Scholar]
- 25.Kedde M., Strasser M.J., Boldajipour B., Oude Vrielink J.A., Slanchev K., le Sage C., Nagel R., Voorhoeve P.M., van Duijse J., Ørom U.A., et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 26.Rassoulzadegan M., Grandjean V., Gounon V., Vincent S., Gillot I., Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 27.De Felici M., Farini D., Dolci S. In or out stemness: comparing growth factor signalling in mouse embryonic stem cells and primordial germ cells. Curr. Stem Cell Res. Ther. 2009;4:87–97. doi: 10.2174/157488809788167391. [DOI] [PubMed] [Google Scholar]
- 28.Russell E.S. Hereditary anemias of the mouse: a review for geneticists. Adv. Genet. 1979;20:357–459. [PubMed] [Google Scholar]
- 29.Wagner K.D., Wagner N., Ghanbarian H., Grandjean V., Gounon P., Cuzin F., Rassoulzadegan M. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev. Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Maron B.J. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 31.Heaney J.D., Lam M.-Y.J., Michelson M.V., Nadeau J.H. Loss of the transmembrane but not the soluble Kit ligand isoforms increases testicular germ cell tumor susceptibility in mice. Cancer Res. 2008;68:5193–5197. doi: 10.1158/0008-5472.CAN-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bedell M.A., Cleveland L.S., O'Sullivan T.N., Copeland N.G., Jenkins N.A. Deletion and interallelic complementation analysis of Steel mutant mice. Genetics. 1996;142:335–344. doi: 10.1093/genetics/142.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chong S., Vickaryous N., Ashe A., Zamudio N., Youngson N., Hemley S., Stopka T., Skoultchi A., Matthews J., Scott H.S., et al. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat. Genet. 2007;39:614–622. doi: 10.1038/ng2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nomura T. Transgenerational effects from exposure to environmental toxic substances. Mutat. Res. 2008;659:185–193. doi: 10.1016/j.mrrev.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Barker D.J.P., Osmond C., Winter P.D., Margetts B.M., Simmonds S.J. Weight in infancy and death from ischaemic heart disease. Lancet. 1989;2:577–580. doi: 10.1016/s0140-6736(89)90710-1. [DOI] [PubMed] [Google Scholar]
- 36.Barker D.J., Osmond C., Kajantie E., Eriksson J.G. Growth and chronic disease: findings in the Helsinki Birth Cohort. Ann. Hum. Biol. 2009:1–14. doi: 10.1080/03014460902980295. In press. [DOI] [PubMed] [Google Scholar]
- 37.Hasselquist D., Nilsson J.A. Maternal transfer of antibodies in vertebrates: trans-generational effects on offspring immunity. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2009;364:51–60. doi: 10.1098/rstb.2008.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stein A.D., Lumey L.H. The relationship between maternal and offspring birth weights after maternal prenatal famine exposure: the Dutch Famine Birth Cohort Study. Hum. Biol. 2000;72:641–654. [PubMed] [Google Scholar]
- 39.Blatt J., Van Le L., Weiner T., Sailer S. Ovarian carcinoma in an adolescent with transgenerational exposure to diethylstilbesterol. J. Pediatr. Hematol. Oncol. 2003;25:635–636. doi: 10.1097/00043426-200308000-00009. [DOI] [PubMed] [Google Scholar]
- 40.Brouwers M.M., Feitz W.F., Roelofs L.A., Kiemeny L.A., de Gier R.P., Roeleveld N. Hypospadias: a transgenerational effect of diethylstilbesterol? Hum. Reprod. 21:666–669. doi: 10.1093/humrep/dei398. [DOI] [PubMed] [Google Scholar]
- 41.Veurink M., Koster M., Berg L.T. The history of DES: lessons to be learned. Pharm. World Sci. 2005;27:139–143. doi: 10.1007/s11096-005-3663-z. [DOI] [PubMed] [Google Scholar]
- 42.Anway M.D., Cupp A.S., Uzumcu M., Skinner M.K. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anway M.D., Leathers C., Skinner M.K. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006;147:5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arai J.A., Li S., Hartley D.M., Feig L.A. Transgenerational rescue of a genetic defect in long-term potentiation and memory formation by juvenile enrichment. J. Neurosci. 2009;29:1946–1503. doi: 10.1523/JNEUROSCI.5057-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weaver I.C., Cervoni N., Chanpagne F.A., D'Alessio A.C., Sharma S., Secki J.R., Dymov S., Szyf M., Meaney M.J. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 46.Ferrari R., Berk A.J., Kurdistani S.K. Viral manipulation of the host epigenome for oncogenic transformation. Nat. Rev. Genet. 2009;10:290–294. doi: 10.1038/nrg2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferrari R., Pellegrini M., Horwitz G.A., Xie W., Berk A.J., Kurdistani S.K. Epigenetic reprogramming by adenovirus e1a. Science. 2008;321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf G.L., Kodell R.L., Moore S.R., Cooney C.A. Maternal epigenetics and methyl supplements affect agout expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 49.Waterland R.A., Jirtle R.L. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skinner M.K. What is an epigenetic transgenerational phenotype? F3 or F2. Reprod. Toxicol. 2008;25:2–6. doi: 10.1016/j.reprotox.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skinner M.K., Anway M.D., Svaenkova M.I., Gore A.C., Crews D. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS ONE. 2008;11:e3745. doi: 10.1371/journal.pone.0003745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 53.Dubrova Y.E., Plumb M., Gutierrez B., Boulton E., Jeffreys A.J. Transgenerational mutation by radiation. Nature. 2000;405:37. doi: 10.1038/35011135. [DOI] [PubMed] [Google Scholar]
- 54.Barber R., Plumb M.A., Boulton E., Roux I., Dubrova Y.E. Elevated mutation rates in the germ line of first- and second-generation offspring of irradiated male mice. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6877–6882. doi: 10.1073/pnas.102015399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dubrova Y.E., Hickenbotham P., Glen C.D., Monger K., Wong H.P., Barber R.C. Paternal exposure to ethylnitrosourea results in transgenerational genomic instability in mice. Environ. Mol. Mutagen. 2008;49:308–311. doi: 10.1002/em.20385. [DOI] [PubMed] [Google Scholar]
- 56.Justice M.J., Noveroske J.K., Weber J.S., Zheng B., Bradley A. Mouse ENU mutagenesis. Hum. Mol. Genet. 1999;8:1955–1963. doi: 10.1093/hmg/8.10.1955. [DOI] [PubMed] [Google Scholar]
- 57.Galhardo R.S., Hastings P.J., Rosenberg S.M. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hastings P.J., Lupski J.R., Rosenberg S.M., Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glazier A.M., Nadeau J.H., Aitman T.J. Finding genes that underlie complex traits. Science. 2002;292:2345–2349. doi: 10.1126/science.1076641. [DOI] [PubMed] [Google Scholar]
- 60.Manolio T.A., Brooks L.D., Collins F.S. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Altshuler D., Daly M.J., Lander E.S. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirschhorn J.N. Genomewide association studies—illuminating biologic pathways. N. Engl. J. Med. 2009;360:1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 63.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 64.McCarthy M.I., Hirschhorn J.N. Genome-wide association studies: potential next steps on a genetic journey. Hum. Mol. Genet. 1998;17:R156–R165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharp A.J., Cheng Z., Eichler E.E. Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:407–442. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- 66.Carlborg O., Jacobsson L., Ahgren P., Siegel P., Andersson L. Epistasis and the release of genetic variation during long-term selection. Nat. Genet. 2006;38:418–420. doi: 10.1038/ng1761. [DOI] [PubMed] [Google Scholar]
- 67.Shao H., Burrage L.C., Sinasac D.S., Hill A.E., Ernest S.R., O'Brien W., Courtland H.W., Jepsen K.J., Kulbokas E.J., Daly M.J., et al. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19910–19914. doi: 10.1073/pnas.0810388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waterland R.A., Jirtle R.L. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 69.Kruglyak L. The road to genome-wide association studies. Nat. Rev. Genet. 2008;9:314–318. doi: 10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- 70.Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182:845–850. doi: 10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johannes F., Porher E., Teixeira F.K., Saliba-Colombani V., Simon M., Agier N., Bulksi A., Albuisson J., Heredia F., Audigier P., et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009;5:e1000530. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hammoud S.S., Nix D.A., Zhang H., Purwar J., Carrell D.T., Cairns B.R. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krawetz S.A. Paternal contribution: new insights and future challenges. Nat. Rev. Genet. 2005;6:633–642. doi: 10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- 74.Anderson P., Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell. Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 75.Heaney J.D., Michelson M.V., Youngren K.K., Lam M.Y., Nadeau J.H. Deletion of eIF2beta suppresses testicular cancer incidence and causes recessive lethality in agouti-yellow mice. Hum. Mol. Genet. 2009;18:1395–1404. doi: 10.1093/hmg/ddp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lam M.Y., Youngren K.K., Nadeau J.H. Enhancers and suppressors of testicular cancer susceptibility in single- and double-mutant mice. Genetics. 2004;66:925–933. doi: 10.1534/genetics.166.2.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Slanchev K., Stebler J., Goudarzi M., Cojocaru V., Weidinger G., Raz E. Control of dead end localization and activity—implications for the function of the protein in antagonizing miRNA function. Mech. Dev. 2009;126:270–277. doi: 10.1016/j.mod.2008.10.006. [DOI] [PubMed] [Google Scholar]