Abstract
To further understand the roles of protein glycosylation in eukaryotes, we globally identified glycan-containing proteins in yeast. A fluorescent lectin binding assay was developed and used to screen protein microarrays containing over 5000 proteins purified from yeast. A total of 534 yeast proteins were identified that bound either Concanavalin A (ConA) or Wheat-Germ Agglutinin (WGA); 406 of them were novel. Among the novel glycoproteins, 45 were validated by mobility shift upon treatment with EndoH and PNGase F, thereby extending the number of validated yeast glycoproteins to 350. In addition to many components of the secretory pathway, we identified other types of proteins, such as transcription factors and mitochondrial proteins. To further explore the role of glycosylation in mitochondrial function, the localization of four mitochondrial proteins was examined in the presence and absence of tunicamycin, an inhibitor of N-linked protein glycosylation. For two proteins, localization to the mitochondria is diminished upon tunicamycin treatment, indicating that protein glycosylation is important for protein function. Overall, our studies greatly extend our understanding of protein glycosylation in eukaryotes through the cataloguing of glycoproteins, and describe a novel role for protein glycosylation in mitochondrial protein function and localization.
Keywords: glycosylation, lectin, localization, mitochondria, protein microarray
Introduction
Protein glycosylation is ubiquitous in eukaryotes and has been implicated in a wide variety of biochemical and cellular processes, including protein folding, maintenance of cell structure, receptor–ligand interactions and cell signalling, cell–cell recognition, and defence (Helenius and Aebi, 2004; Dube and Bertozzi, 2005). In spite of its importance, both the number and different types of proteins that are glycosylated are not known, and thus it is likely that the full range of biochemical and cellular functions is not understood.
Two major types of protein glycosylation have been identified thus far: N- and O-linked. N-linked glycans are attached to asparagine residues and O-linked glycans are connected to serine or threonine residues. The local motif required for attachment is known only for N-linked glycans: N-X-S/T, where the point of attachment is upstream of either a serine or threonine, and the intervening amino acid cannot be proline (Gavel and von Heijne, 1990). A variety of different glycans are often attached to the modified proteins by glycotransferases in a series of processing steps. The degree of processing can be heterogeneous, resulting in a complex mixture of isoforms for a particular protein.
N-linked glycosylation has been primarily associated with secretion, and most N-linked glycoproteins reside in compartments associated with this pathway. Glycan attachment is initiated in the endoplasmic reticulum (ER), where the nascent polypeptide chains are modified with an N-linked oligosaccharide at the appropriate site (Nilsson et al, 2003). The glycans on the glycoproteins are processed through the Golgi, and many proteins ultimately are shuttled to the plasma membrane or secreted. O-linked glycosylated proteins can be associated with membranes, in the cytosol, or within the nucleus (Cole and Hart, 1999; Wells and Hart, 2003).
Few studies have investigated the role of protein glycosylation in mitochondrial function. Glycosyltransferase activity has been found in both the inner and outer membranes of mouse liver mitochondrion, although modified peptide chains were not identified (Levrat et al, 1989, 1990). In Saccharomyces cerevisiae, a hypomorphic allele of ALG7, encoding an enzyme involved in the first N-linked glycan addition, caused mitochondrial dysfunction (Mendelsohn et al, 2005). A previous study demonstrated that a member of the ubiquinone complex in the mitochondrial inner membrane from rat liver tissue is mannosylated and is transported from the ER to the mitochondria (Chandra et al, 1998). To our knowledge, this is the only mammalian mitochondrial glycoprotein that has been identified.
To learn more about the roles of glycosylation, systematic studies would be useful. To date, a limited number of studies have been performed. Computational methods based on motif analysis are not sufficient to predict modified proteins. For example, a perusal of the S. cerevisiae proteome sequence indicates that the limited motif for N-linked glycosylation occurs ∼4.5 times per protein on average, yet most proteins are probably not glycosylated. Using covalent coupling to hydrazide resin, Zhang and Abersold purified glycoproteins from cell lysates, followed by mass spectrometry to identify hundreds of novel N-linked glycoproteins in mammalian cells (Kaji et al, 2003; Zhang et al, 2003; Tian et al, 2007). However, these studies do not identify low-abundance proteins.
Yeast is an ideal system for studying protein glycosylation (Kukuruzinska et al, 1987). Twenty to 50% percent of the yeast proteins are estimated to be glycosylated (Apweiler et al, 1999), and 175 N- and O-linked proteins (3% of total proteins) have been previously identified through studies of individual proteins (Costanzo et al, 2000; Csank et al, 2002). Two glycans are known to be present on yeast proteins; N-linked chains have N-acetylglucosamine (GlcNAc) and mannose, whereas O-linked chains have only mannose (Gemmill and Trimble, 1999). Many of the glycotransferases responsible for adding and trimming carbohydrates in yeast are known (Herscovics, 1999). Finally, proteomic and genomic tools are readily available for studying protein glycosylation. In particular, two versions of yeast proteome arrays (Kung and Snyder, 2006), which contain most yeast proteins with either N- or C-terminal tags spotted at high density onto microscope slides, are useful for surveying protein glycosylation. Gelperin et al (2005) performed a preliminary systematic analysis of protein glycosylation in yeast by probing a proteome chip containing 5573 C-terminally tagged fusion proteins purified from yeast with an antibody raised against the glycan backbone of yeast glycoproteins. This study increased the number of N-linked glycoproteins known in S. cerevisiae from 106 to 231, and the total number of known glycoproteins to 305. The sensitivity of that study was limited by the antibody and only N-linked proteins could be identified.
In this study, we have more comprehensively examined both the number and types of proteins that are glycosylated by developing and employing an assay using lectins to probe yeast proteome microarrays. Plant and fungal lectins have long been used as a tool to identify glycans on proteins (Chrispeels and Raikhel, 1991). Among the hundreds of known lectins, some (including Concanavalin A (ConA) and Wheat-Germ Agglutinin (WGA)) have been scrutinized for their glycan-binding spectra and affinities using glycan microarrays, thereby allowing specific detection of different glycoproteins (Hirabayashi, 2004). Using our approach, we greatly extended the number of known glycoproteins in yeast and identified several glycoproteins that localize to the mitochondrion. Treatment of cells with an inhibitor of protein glycosylation disrupted localization of two mitochondrial proteins (Gregan et al, 2001; Huh et al, 2003; Reinders et al, 2006). In addition to providing a more comprehensive understanding of protein glycosylation in general, this study thus defines new roles for protein glycosylation in mitochondrial protein function and localization.
Results
Development of a glycan-dependent lectin glycoprotein screen using proteome chips
We set out to globally identify glycoproteins in S. cerevisiae by probing the yeast proteome chips with lectins that recognize GlcNAc or mannose. Two lectins were used, ConA, which recognizes mannose residues, and WGA, which recognizes GlcNAc. As shown by the glycan microarray experiments from the Consortium for Functional Glycomics, the selectivity of these lectins for their preferred glycans is much higher than for the other related glycans (http://www.functionalglycomics.org/glycomics/publicdata/primaryscreen.jsp).
The lectins were used to screen two types of chips: (1) a chip containing ∼5800 proteins in which the proteins were fused at their N-termini to GST; (Zhu et al, 2001) and (2) an array containing ∼5600 proteins in which the proteins were fused at their C-termini to the IgG-binding domain of Protein A (Gelperin et al, 2005). In each case, the proteins were overexpressed in yeast and purified using the respective affinity tags. The proteome chips with the C-terminal protein fusions are ideal for screening type-I and the great majority of type-II and type-III membrane proteins that are glycosylated through the secretory pathway; these proteins have proper signal and translocation sequences and will be modified with N-linked glycans during or after translation. In contrast, proteome chips with the N-terminal protein fusions are ideal for the identification of proteins anchored to the membranes by a C-terminal signal sequence or transmembrane domain (e.g. cytochrome b5 and the SNARE proteins), as well as a portion of type-II proteins. Finally, both types of the yeast proteome chips are expected to detect O-linked glycoproteins, which do not have signal sequences for membrane transport.
Fluorescently labelled lectins were incubated with the chips, and the slides were then washed to remove unbound lectins. The slides were scanned and images were processed using an algorithm adapted from ProCAT (Zhu et al, 2006). One problem with lectin probing is that the affinity of lectin–carbohydrate interactions is rather weak; the dissociation constants for lectin–glycan interactions in solution are approximately ∼300 μM for ConA–mannose and ∼2 mM for WGA–GlcNAc (Clegg et al, 1981; Mackiewicz and Mackiewicz 1986; Bains et al, 1992). In addition, some lectins often exhibit non-specific binding, either to the slide itself or to the proteins on the chip. Furthermore, lectins tend to be sticky proteins due to their biological function; they recognize glycans that differ little except in the conformations of small side chains.
To circumvent the non-specific binding of ConA and WGA to the surfaces and ‘sticky' protein spots while simultaneously maintaining strong signals, we developed a glycan competition assay that allowed for sensitive detection of specific glycans. For each lectin, two proteome slides were probed in parallel, one in the presence of the glycan and the other in its absence (Figure 1). For ConA and WGA, 10 mM α-methyl-mannoside and 50 mM chitin hydrolysate, respectively, were used as inhibitors. These concentrations are well above the dissociation constants for the lectin–glycan associations and the concentrations of lectins used in the probings, and thus should completely prevent glycan-specific binding to the surface. By comparing the lectin binding in the presence and absence of the glycan inhibitors, the glycan-dependent interactions can be identified. All experiments were performed in duplicate on both the N- and C-terminal fusion libraries.
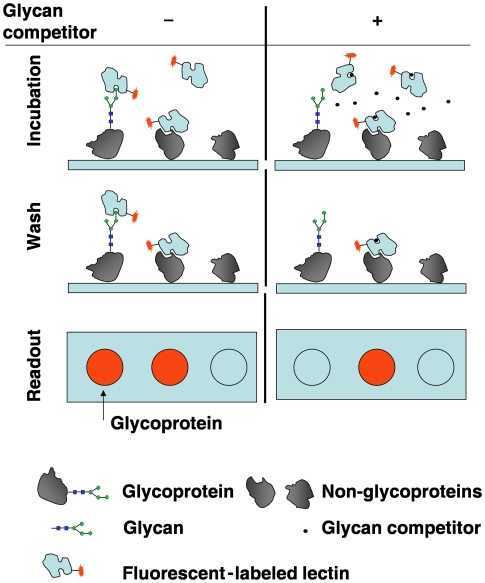
Figure 1.
A schematic representation of glycan competition assays. Proteome arrays fabricated with proteins of various types are probed with fluorescently labelled lectins. (Left track) A proteome chip is incubated with lectins, followed by a washing step to remove free lectins and some weak, non-specific interactions; however, stronger non-specific interactions, presumably caused by protein–protein interactions, still remain, leaving behind glycan-independent binding signals. (Right track) In parallel, a proteome chip is incubated with lectins in the presence of excess amount of glycan competitors, which should block glycan-dependent interactions. The comparison of signal intensities between the two tracks reveals proteins that show glycan-dependent binding activities.
Examples of the probings are shown in Figure 2. Although many proteins bound both in the presence and absence of the competing glycan inhibitors, a large number of proteins were identified that bound only in the absence of the competitors. All proteins recognized by the two lectins were identified and the glycoprotein candidates were determined as follows: set ORFx_Net Signal = ORFx_Signal (without sugar competitor)−ORFx_Signal (with sugar competitor), then the cut off is ORFx_Net Signal⩾3*Stdev (ORFx_Net Signal: ORFn_Net Signal), n equal the number of ORFs on the proteome chip. It was interesting to note that more binding signals were observed on the C-terminal proteome chips, which might reflect the differences in the tag positions of the fusion proteins and/or surface chemistries of the two types of proteome chips.
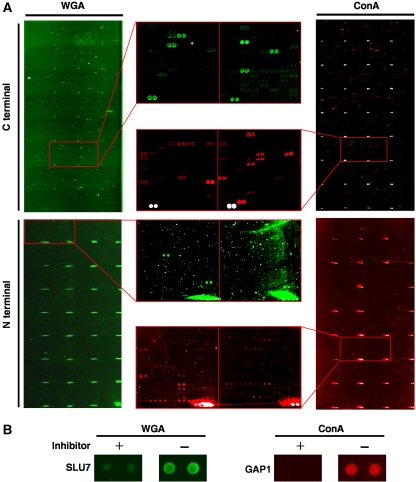
Figure 2.
Glycoprotein identification using two types of proteome chips. (A) Lectin probing of the proteome chips. (Upper panel) C-terminal proteome chips probed with WGA–TAMRA (left) and ConA–Alexa647 (right). Many binding signals can be observed for both WGA and ConA in the absence of the sugar inhibitors; enlarged areas of the chips from both assays are illustrated in the middle. However, by comparing with the signals in the presence of sugar inhibitors (data not shown), 236 candidate glycoproteins were identified with ConA and 142 with WGA. (Lower panel) N-terminal proteome chips probed with WGA–TAMRA (left) and ConA–Alexa647 (right). Because of the N-terminal tags, less lectin binding signals were observed as expected. Using the same approach, ConA and WGA were found to identify 124 and 174 of positive proteins, respectively. (B) Examples of the sugar competition experiments. Representative glycoprotein candidates were shown. The inhibitors for WGA and ConA are chitin hydrolysate and α-methyl mannoside, respectively.
Chip probing results and identification of glycoproteins
Of the 5573 proteins on the proteome chips with C-terminal fusion proteins, 236 candidate glycoproteins were identified with ConA and 142 with WGA (Supplementary Table I). Of the 305 previously identified S. cerevisiae glycoproteins, 270 were present on the proteome chips, 105 were found by ConA, and 79 by WGA. Examples include DFG5 and YPS6. The glycoproteins that were not detected tended to be present at low levels on the arrays as evidenced by probing with antibodies to the tags. Nonetheless, these numbers represent very high ratio over the number expected by random selection of candidate proteins (9.56- and 12.05-fold, respectively) over that expected by chance. In addition, enrichment of proteins with predicted signal peptides is quite high (6.7- and 8.6-fold, respectively; Nielsen et al, 1997; Bendtsen et al, 2004). Thus, the candidate lists are highly enriched for glycoproteins and those with signal peptides.
Because all N-linked glycoproteins are modified with both GlcNAc and mannose, it was expected that many of the WGA-identified candidates would also appear in the ConA probing. A total of 112 (79%) of WGA-binding protein also bound ConA. The failure to of ConA to detect all WGA-reactive proteins may arise from the stringent cut-offs used in the determination of the candidate lists, the accessibility of the different sugars to the lectins, or their suitable presentation on the protein array surface.
Using the same protocol, ConA and WGA were used to probe the proteome chips printed with the N-terminal fusion proteins. Of the 5800 proteins, ConA and WGA identified 124 and 174 of positive proteins, respectively (Supplementary Table I). Of the 305 previously identified S. cerevisiae glycoproteins, 284 were on the N-terminal proteome chips and 15 were found by ConA. The enrichment ratio is about 2.5 over the number expected by random selection of candidate proteins. However, there is no enrichment for WGA; this is presumably because the GST tag located at the N-termini of the fusion proteins masks the N-terminal signal peptides in the type-I and the great majority of type-II and type-III membrane proteins.
Gene ontology analysis of candidate glycoproteins
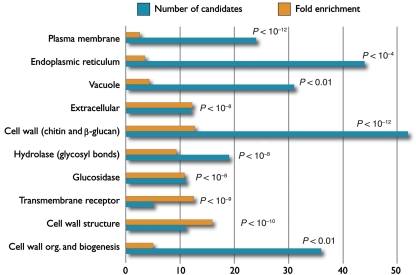
The candidate glycoproteins were analyzed using gene ontology (GO) annotations to identify common and enriched properties of these proteins compared with the rest of the S. cerevisiae proteome. The combined ConA and WGA list of 266 unique candidate glycoproteins identified using the C-terminal fusion protein chips was examined for enrichment in cellular component, biological process, and molecular function compared with the GO annotations of the proteins in the Saccharomyces Genome Database (Fisk et al, 2006). For cellular component, the candidate list was substantially (P<10−8) enriched for chitin- and β-glucan-containing cell wall components, proteins in the ER and vacuole, as well as secreted proteins (Figure 3). There was also enrichment (P<0.01) for membrane proteins. Of the 113 known chitin- and β-glucan-containing cell wall proteins, 52 (46%) were found by the lectin screening, representing 12.6-fold enrichment. For the other enriched components, about 10–15% of the known proteins were identified, representing 2.5- to 4.5-fold enrichment.
Figure 3.
GO analysis of candidate glycoproteins. Chart of enriched GO annotations in the list of candidate glycoproteins when compared with the entire S. cerevisiae proteome. Each category listed is significantly enriched (with P-value <0.01 as shown), and both the number of candidates belonging to the annotation (blue) and the fold enrichment over the proteome (yellow) are shown.
For biological process, the candidate list was substantially (P<10−12) enriched in cell wall organization and biogenesis, and more specifically (P<0.01) for chitin- and β-glucan-containing cell wall organization and biogenesis. The 36 candidates, involved in chitin- and β-glucan cell wall organization and biogenesis, represent 18% of the 199 proteins annotated to the category involved in the cell wall organization and biogenesis process, nearly fivefold enrichment.
For molecular function, the candidate list was substantially (P<10−4) enriched in hydrolase activity (used in the hydrolysis of glycosyl bonds in O-glycosyl compounds), as well as cell wall structure and glucosidase activity (P<0.001), and (membrane) receptor activity (P<0.01).
The large enrichment of cell wall components, processes, and function for C-terminal fusion proteins identified by lectin screening is expected, given the extensive glycosylation of these proteins. Enrichments were not observed for proteins identified from the N-terminal collection and might also be expected. These results validate that the lectin screen is indeed finding proteins, which are modified with glycans or interact with glycan chains.
Validation of glycoprotein candidates by secondary gel-shift assay
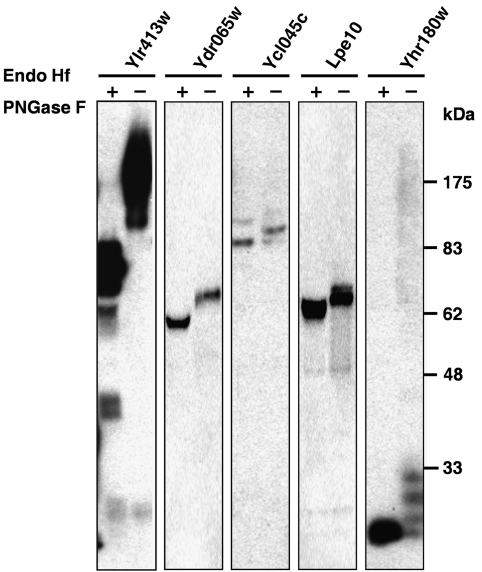
Candidate glycoproteins were further tested for glycosylation by determining if they exhibit a gel mobility shift after treatment with enzymes that remove glycan modifications (Figure 4 and Supplementary Figure 1). Purified candidate glycoproteins were subjected to digestion/treatment with two enzymes, which remove N-linked glycan chains, endoglycosidase Hf, and PNGase F. The digested proteins were compared with untreated samples by SDS–PAGE. Candidates showing a mobility difference between the two were scored as validated N-linked glycoproteins. The gel-shift assay had previously been shown to be quite robust, with 84% of known N-linked glycoproteins exhibiting shifts upon enzyme treatment and none of 19 negative controls showing shifts (Gelperin et al, 2005).
Figure 4.
Validation of glycosylation using gel-shift assays. Five representatives, for example, Ylr413wp, Ydr065wp, Ycl045cp, Lpe10p, and Yhr180wp, show the mobility of proteins digested with endoglycosidases (+) or without (−). The difference in mobility upon digestion indicates the cleavage of N-linked glycans from the protein, validating the candidate as a glycoprotein.
Of 118 candidate C-terminal glycoproteins and 24 candidate N-terminal glycoproteins screened, 51 (36%) were validated as N-linked glycoproteins with the gel-shift assay, six of them were known glycosylated protein, which were used as positive controls. Some of the candidates tested did not give conclusive results because of difficulties in purification or the known limitations of the gel-shift assay, such as detection difficulties due to smearing of heterogeneous glycan chains. Nonetheless, these studies validated 45 new glycoproteins (listed in Table I) and extended the total number of validated N-linked glycoproteins in yeast to 276, and the total number of the validated glycoproteins in S. cerevisiae to 350.
Table 1.
Validated novel glycoproteins
| Systematic name | Common name | Cellular component |
|---|---|---|
| YCR026C | NPP1 | Unknown |
| YDR434W | GPI17 | Cytoplasm/endomembrane system/ endoplasmic reticulum/membrane |
| YFR020W | YFR020W | Unknown |
| YLR413W | YLR413W | Unknown |
| YCL045C | YCL045C | Endoplasmic reticulum |
| YCR045C | YCR045C | Unknown |
| YDR065W | YDR065W | Mitochondrion |
| YGR279C | SCW4 | Cell wall |
| YHR039C | MSC7 | Endoplasmic reticulum |
| YIL039W | TED1 | Endoplasmic reticulum |
| YJL009W | YJL009W | Unknown |
| YLR414C | YLR414C | Cellular bud/cytoplasm |
| YMR215W | GAS3 | Membrane fraction/cell wall |
| YNL080C | EOS1 | Membrane/endoplasmic reticulum/ endomembrane system/cytoplasm |
| YNL283C | WSC2 | Site of polarized growth/membrane fraction/cellular bud/cytoplasm |
| YOR387C | YOR387C | Other |
| YBR298C-A | YBR298C-A | Unknown |
| YCR089W | FIG2 | Cell wall |
| YER158C | YER158C | Unknown |
| YER183C | FAU1 | Mitochondrion |
| YER185W | PUG1 | Membrane |
| YGL020C | GET1 | Endoplasmic reticulum/cytoplasm |
| YGR223C | HSV2 | Cytoplasm/vacuole |
| YHR202W | YHR202W | Cytoplasm/other/vacuole |
| YIL059C | YIL059C | Unknown |
| YJL038C | LOH1 | Unknown |
| YJL158C | CIS3 | Site of polarized growth/cell wall/ extracellular region/cellular bud/ plasma membrane/endoplasmic reticulum |
| YLR337C | VRP1 | Cellular bud/site of polarized growth/ other/cell cortex/cytoskeleton/cytoplasm |
| YMR238W | DFG5 | Membrane/plasma membrane |
| YNR015W | SMM1 | Nucleus/cytoplasm |
| YOL031C | SIL1 | Endoplasmic reticulum |
| YOR099W | KTR1 | Golgi apparatus |
| YOR214C | YOR214C | Cell wall |
| YOR246C | YOR246C | Cytoplasm |
| YOR292C | YOR292C | Cytoplasm/vacuole |
| YPL060W | LPE10 | Cytoplasm/membrane/mitochondrial envelope/mitochondrion |
| YPL277C | YPL277C | Membrane fraction |
| YGL258W | VEL1 | Other |
| YIR039C | YPS6 | Cell wall |
| YDR088C | SLU7 | Nucleus |
| YNL054W | VAC7 | Membrane/cytoplasm/vacuole |
| YNL094W | APP1 | Cell cortex/cytoplasm/cytoskeleton |
| YNL308C | KRI1 | Nucleolus |
| YOR027W | STI1 | Cytoplasm |
| YPL004C | LSP1 | Cytoplasm/cell cortex |
Several glycoproteins reside in the mitochondria
We found that a large number of glycosylated proteins resides in the nucleus (34) and mitochondria (30). Previous studies suggested that glycosylation might occur in mitochondria, but little effort has been devoted to this subject and the role of protein glycosylation in mitochondrial protein function has not been examined (Levrat et al, 1989, 1990; Chandra et al, 1998; Mendelsohn et al, 2005). Interestingly, we found 30 mitochondrial proteins that reacted with ConA and WGA. Two of them were identified by both the C-terminal and N-terminal proteome chips, and 14 were identified by the C-terminal chip only. Six of these proteins were validated as shown in Table II; three of them have also been validated by Gelperin et al (2005). The gel-shift results of two representative proteins, Ydr065wp and Lpe10p, are shown in Figure 4. Lpe10p, a mitochondrial inner-membrane magnesium transporter, is involved in the maintenance of magnesium concentrations inside mitochondria, and Ydr065wp is a protein of unknown function. For each of these proteins, most of the detectable protein is gel shifted upon enzyme treatment, suggesting that most, if not all, of the protein is modified.
Table 2.
Validated mitochondrial proteins
| Systematic name | Common name | Process | Function | Compartment |
|---|---|---|---|---|
| YMR306W | FKS3 | Biological_process unknown | 1,3-β-Glucan synthase activity | Mitochondrion |
| YGL068W | YGL068W | Biological_process unknown | Molecular_function unknown | Mitochondrion |
| YKL187C | YKL187C | Biological_process unknown | Molecular_function unknown | Mitochondrion |
| YER183C | FAU1 | Folic acid and derivative biosynthesis | 5-Formyltetrahydrofolate cyclo-ligase activity | Mitochondrion |
| YPL060W | LPE10 | Mitochondrial magnesium ion transport | Magnesium ion transporter activity | Mitochondrial inner membrane |
| YDR065W | YDR065W | Biological_process unknown | Molecular_function unknown | Mitochondrion |
The validated glycoproteins identified from the ConA and WGA probing, which are expected to localize to the mitochondrion. The columns show the systematic ORF name, the common name, and the expected process, function, and cellular compartment of each protein.
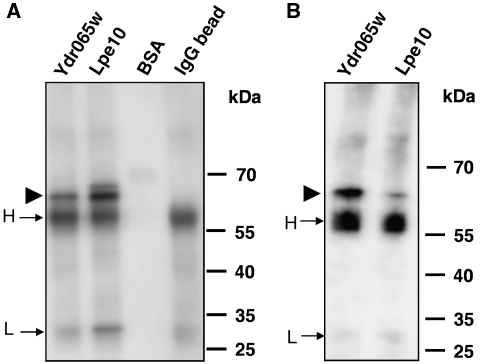
To further test whether these two proteins were glycosylated, lectin blotting analysis was employed using two types of cell extracts. First, Lpe10p and Ydr065wp were purified from unfractionated cell lysates of cells that overproduced the two proteins as C-terminal TAP-tagged fusions. Second, the mitochondrial fractions were prepared from cells that expressed the two TAP-tagged proteins close to endogenous level, followed by affinity purification of the two proteins. Purified proteins from unfractionated cell lysates or from the mitochondrial fractions were separated on a PAGE gel, blotted to a membrane, and subjected to lectin binding assays. As illustrated in Figure 5, ConA clearly recognizes protein bands close to the high-mobility sizes of Ydr065wp and Lpe10p, respectively, regardless whether they were overexpressed (Figure 5A) or close to the endogenous level (Figure 5B). As expected, both the heavy and light chains of IgG showed positive staining with ConA as IgG is known to be glycosylated. As a negative control, BSA did not show any signals. Thus, in addition to the evidence provided by the above gel-shift analysis (Figure 4), these ConA probing results demonstrate that Ydr065wp and Lpe10p from the mitochondrial fractions are glycosylated.
Figure 5.
Validation of glycosylation of two mitochondrial proteins using lectin blotting analysis. Two mitochondria proteins Ydr065wp and Lpe10p were purified from (A) unfractionated cell lysates of cells that overproduced the two proteins as C-terminal TAP-tagged fusions and (B) the mitochondrial fractions of cells that expressed the two TAP-tagged proteins close to endogenous level. The proteins were transferred to PVDF membrane and incubated with ConA. After a washing step, the membranes were incubated with HRP-conjugated anti-ConA antibodies and the binding signals were detected with an ECL kit. BSA and IgG were used as negative and positive controls, respectively. The two proteins, whose molecular weights are around 70 kDa, are indicated by an arrow. The heavy and light chains of IgG are marked as H and L, respectively.
Mitochondrial localization of candidate glycoproteins upon tunicamycin treatment
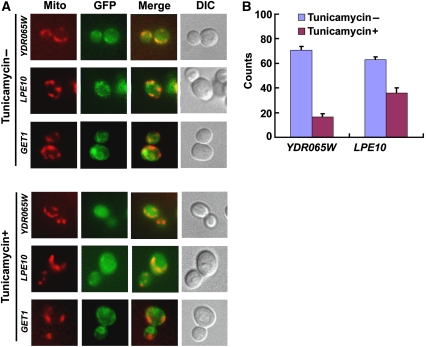
To further examine a potential role for glycosylation in the mitochondrial proteins identified in this study, we examined whether glycosylation is important for mitochondrial localization. Four candidates, Ydr065wp, Mnp1p, Pcp1p, and Lpe10p, were examined, along with Get1p, a known ER protein, as a control. Yeast strains that harbour chromosomally GFP-tagged candidate genes were collected at the mid-log phase, incubated in the presence and absence of tunicamycin, an inhibitor of N-linked glycosylation, and stained with MitoFluor Red 589 to visualize the mitochondria. In the untreated cells, the four candidate proteins showed clear mitochondrial localization, as described previously (Huh et al, 2003), and extensive overlapping staining with the MitoFlour (Figure 6A, upper panel). However, upon incubation with tunicamycin for1 h, the mitochondrial localization was almost absent and increased cytosolic localization of Ydr065wp and Lpe10p was observed (Figure 6A, lower panel). The effect of tunicamycin is strongest on Ydr065wp in which approximately 80% of cells lack mitochondrial staining as compared with 30% for untreated cells (Figure 6B). No effect of tunicamycin on mitochondrial localization was observed on Mnp1p and Pcp1p, or on the ER localization of Get1p. To quantify the effect of tunicamycin on Ydr065wp and Lpe10p, we compared their protein levels in the mitochondria fractionation with and without tunicamycin treatment. The results showed that treatment of tunicamycin reduced the amount of both Ydr065wp and Lpe10p in the mitochondria fractions by ∼8 and >50%, respectively (data not shown). Although the effect of tunicamcyin may be indirect, we note that the mitochondria appear intact using MitoFluor and only two of four miotchondrial proteins are affected, suggesting that a growth disruption of mitochondrial function has not occurred upon tunicamycin treatment. These results indicate that glycosylation is important for the localization of Ydr065wp and Lpe10p to the yeast mitochondria.
Figure 6.
Effect of glycosylation on mitochondrial protein localization. (A) Localization of mitochondrial proteins, Ydr065wp and Lpe10p, is regulated by glycosylation. Yeast strains that harbour chromosomally GFP-tagged candidate genes were collected at the mid-log phase, incubated in the presence and absence of tunicamycin, and stained with MitoFluor Red 589 to visualize the mitochondria. In the untreated cells (Tunicamycin−), Ydr065wp and Lpe10p showed clear mitochondrial localization (GFP) and extensive overlapping staining with the MitoFlour (Mito and Merged). However, upon incubation with tunicamycin (Tunicamycin+), mitochondrial localization of both proteins is reduced and increased cytosolic localization is observed. (B) Quantitative measurement of mitochondrial localization with and without tunicamycin treatment. Cell counting was performed three times independently and 100 cells were counted for each strain in the presence and absence of tunicamycin treatment. The error bars represent the standard deviation.
Discussion
In this study, we have developed a lectin binding assay for identifying glycoproteins and used the assay to screen two types of the yeast proteome chips. These studies identified 534 yeast glycoproteins, 406 of them were novel and 45 of the novel glycoproteins were validated by gel shift. Combining our results with those of previous studies reveals a total of 350 validated glycoproteins, approximately 6% of the yeast proteome. This number likely is a minimum estimate since we only detected approximately 25% of known glycoproteins. Thus, we estimate that approximately 20–30% of yeast proteins are glycosylated, which is consistent with the previous estimation (Apweiler et al, 1999).
Our validation focused on N-linked glycosylation using EndoH and PNGase F, because dramatic changes can be observed in gel-shift assays. In principle, O-linked glycosylation can be followed using endo-β-N-acetylglucosaminidase F (Endo F), α-mannosidase, and O-glycanase. However, O-linked glycan chains are generally much shorter, and in yeast O-modified proteins possess only a linear chain of up to five mannose residues (Kukuruzinska et al, 1987); this makes validation with this approach more difficult.
The use of proteome chips is particularly well suited to the large-scale analysis of glycosylation for the following reasons: (1) the yeast proteins were purified from their original host that presumably preserves the native posttranslational modifications; (2) the native N- and C-termini ends of a given protein is likely to be preserved on the C- and N-terminal proteome chips, respectively; and (3) the use of the two types of chips likely increases the chances of detection; if a protein is failed to purify with a tag at one terminus, it has a second chance on the other proteome chip. Therefore, the use of two different types of chips should facilitate the identification of different types of glycosylations. This study used lectin probes for glycan detection rather than antibodies as was the case for a previous study (Gelperin et al, 2005). The number of lectins with highly specific glycan-binding activity is much more than the available anti-glycan antibodies, which are difficult to generate with high specificity and their characterization is often limited (this was the case in the previous study). Although some lectins are known to carry non-specific binding properties, the addition of glycan inhibitors can dramatically increase the fidelity of the binding results, as we demonstrated in this study. Thus, the types of glycans can usually be detected using lectin probes.
Yeast has only two glycan modifications and is simple compared with other eukaryotes. Nonetheless, the wide spectra of glycan recognition by a large number of lectins (∼100 are commercially available) should allow a similar strategy to be extended to higher eukaryotes, which have much more complicated glycan structures. Human protein chips containing over 8000 proteins are available (Invitrogen). Because of the larger numbers of possible modifications with different glycans (Gabius et al, 2004), a larger number of lectins may be needed to determine which modifications are present on each protein. Analysis of these modifications is expected to provide insight into the ‘glycan code,' for determining the functional role of each type of glycan modification.
In addition to identification of proteins that reside or traverse secretory compartments, a variety of other types of proteins were found, including nuclear and mitochondrial proteins. The finding of a nuclear protein is new in yeast but not surprising in mammalian systems. Hart and co-workers have identified O-linked glycosylation on nuclear and cytosolic proteins, including transcription factors, signalling components, and metabolic enzymes (Wells and Hart, 2003). Further studies by Hart and others demonstrated that O-linked glycosylation plays a role in modulating transcriptional activity, probably through control of protein stability, protein subcellular localization, and/or protein-O–GlcNAc interaction. We find that the nuclear proteins that are modified are involved in diverse cellular processes (e.g., Sox2p). We speculate that the modification of these proteins by glycans might provide a mechanism for tying transcription control to metabolic state; the concentration of glycans might affect the amount of modified transcription factor and thus regulate the transcriptional activity of the cell.
The finding that several mitochondrial proteins are glycosylated was surprising. To date, only one mammalian glycoprotein in the mitochondria has been described in a single study (Chandra et al, 1998). Additional mitochondrial proteins were found by Gelperin et al (2005), but the significance was not reported. Our finding of 30 mitochondrial glycoproteins, six of which were validated significantly, extends this list (Table II). Four of these proteins (Ykl187Cp, Fks3p, Ydr065wp, and Ygl068wp) react with both ConA and WGA, suggesting they have both mannose and GlcNAc.
The fact that the mitochondrial protein glycosylation is blocked by tunicamycin suggests that these proteins either transverse the normal secretory pathway to reach the mitochondria, or that tunicamycin-sensitive enzyme(s) reside in the mitochondria. Evidence provided for both mechanisms (Rizzuto et al, 1998; Bozidis et al, 2008) suggest that the ER makes close contact and can exchange membranes with mitochondria in virally infected cells, suggesting that direct contact can occur. Evidence for modification enzymes in the mitochondria comes from Louisot and co-workers who demonstrated N-linked glycoprotein synthesis occurs through dolichol intermediates in mammalian mitochondria, although they did not identify the protein substrates (Levrat et al, 1989, 1990). The use of lipid-linked oligosaccharides as donors in N-linked glycoprotein synthesis results in high-mannose structures that significantly differ from complex-type Asn-linked oligosaccharides in the plasma membrane. Perhaps, a similar, parallel N-linked glycosylation system exists in the mitochondria in the budding yeast that is also tunicamycin sensitive.
Our finding of 30 mitochondrial glycoproteins reveals candidate substrates for the glycotransferases and indicates that a number of mitochondrial proteins are glycoproteins. Although previous studies have shown glycotransferase activity and identified a glycosylated protein in mammalian mitochondria (Chandra et al, 1998), the biological relevance of such modifications remains elusive. Our study using tunicamycin inhibition demonstrates that glycosylation is important for the subcellular localization of at least two mitochondrial glycoproteins. Protein glycosylation has been shown to affect the distribution of proteins in the secretory pathway. Furthermore, Hart and co-workers showed that the balance between phosphorylation and glycosylation at Thr-58 regulated the distribution of c-Myc between the cytoplasm and the nucleus (Cole and Hart, 1999). Thus, protein glycosylation may be a general mechanism for regulation of protein localization in a variety of cellular compartments.
In summary, we developed a new protein microarray strategy to globally identify glycoproteins in yeast and reveal new roles for protein glycosylation. Further definition and characterization of the glycome of yeast and other eukaryotes is expected to reveal additional novel roles or protein glycosylation in eukaryotes.
Materials and methods
Culture conditions and protein purification
C-terminal fusion proteins were purified from 30-ml cultures of yeast strains as described previously (Gelperin et al, 2005). Briefly, the yeast strains were grown at 30°C in SC-ura/raffinose media. The cultures were grown overnight, induced at an OD600 of 0.6–0.8 by galactose at a final concentration of 2%, and harvested after 6 h. The cells were lysed by shaking with 300 μl acid-washed glass beads, and the supernatant were incubated with IgG–Sepharose (Amersham). The IgG beads were washed before elution of the proteins by cleavage with the 3C Protease in a final volume of 50 μl. N-terminal GST-fusion proteins were purified from 6-ml yeast culture as described previously (Zhu et al, 2001).
Protein chip production
Protein chips containing 5573 purified C-terminal fusion proteins from S. cerevisiae were produced as previously described (Gelperin et al, 2005). The purified fusion proteins were printed onto FAST slides (Whatman) using a 48-pin contact printer (BioRad ChipWriter Pro). Proteome chips with ∼5800 N-terminal GST-fusion proteins were fabricated on nickel-coated slides as described previously (Zhu et al, 2001). Each protein was spotted in duplicate. Control proteins, including Histone H3, Histone H4, BSA, and biotinylated BSA, were also spotted induplicate.
Lectins and glycan inhibitors
Fluorescently labelled lectins, Alexa 647-conjugated ConA and TAMRA-conjugated WGA (Invitrogen), were diluted into the TC buffer (1 mM Tris at pH 7.5 with 1 mM CaCl2, 1 mM MgCl2, 1 mM MnCl2, and 0.1% (v/v) Tween-20) to a final concentration of 20 μg/ml. The proteins were centrifuged at the maximum speed for 20 min and filtered through a 0.22-μm filter. Lectin solutions with inhibitory glycans were prepared in a similar manner. For ConA, 10 mM α-methyl-mannoside was used as the inhibitor and 50 mM chitin hydrolysate was used for WGA.
Lectin glycan competition assays
Protein chips were blocked with SuperBlock (Pierce) for 1 h and incubated with the lectin solution in the presence or absence of the glycan inhibitors for 15 min at room temperature. The chips were then subjected to three 2-min washes in TBST with gentle shaking, and dried by centrifugation at 1500 r.p.m. for 3 min. The slides were scanned with a GenePix 4200A slide scanner (Axon Instruments).
Data analysis and informatics
Signal intensity of each spot from each probing experiment was calculated and background normalized by subtraction of the median background of a local sliding window (22 by 22 spots). These intensities were used to calculate the standard deviation of each spot within another local sliding window (8 by 8 spots). The background subtraction and standard deviation calculations were performed using the ProCAT algorithm (Gelperin et al, 2005; Zhu et al, 2006). The standard deviations for each spot were then averaged over each slide of the same probing to calculate the score for each ORF. The differential of the scores of each ORF between slides without the inhibitor and slides with the inhibitor was used to generate a rank ordered list of candidate glycoproteins for each lectin. The cut-off of this list was set at three standard deviations of the score differential value.
Gel-shift assays for candidate validation
Candidate proteins were first denatured with denaturing buffer (0.5% SDS and 0.04 M DTT) and briefly boiled, followed by digestion with a mixture of endoglycosidase Hf and PNGase F (New England Biolabs) at 37°C for 3 h. The proteins were compared with the undigested proteins identified by SDS–PAGE and those with shifted bands were scored as positive.
Cell fractionation
Mitochondria fractionation was performed as described previously (Coelho et al, 2002; Fisk et al, 2006). Briefly, yeast strains with TAP-tagged candidate genes from the MORF collection were cultured in YAPD media to saturation and then inoculated in lactate media. The cells were harvested and converted to spheroplasts, homogenized, and separated into mitochondria fraction by centrifugation at 10 000 g for 10 min. The final mitochondrial pellets were resuspended in breaking buffer (0.6 M sorbitol, 20 mM HEPES–KOH, pH 7.4) with 1 mM PMSF, and stored at −80°C.
Lectin blotting
Lectin blotting was modified from Gravel P (Gavel and von Heijne, 1990). Proteins were transferred from SDS–PAGE gel to PVDF membrane by using the iBlot dry blotting system (Invitrogen). The membrane was subjected to three 5-min washes with water followed by blocking with 0.5% Tween-20 in PBS (pH 7.5) for 1 h at room temperature. The membrane was then incubated with 1 μg/μl ConA (EY Lab) at room temperature for 1 h. After six 10-min washes with 0.5% Tween-20 in PBS (pH 7.5), the membrane was incubated with HRP-conjugated anti-ConA antibodies (EY Lab) at 1000-fold dilution for 1 h. After the same wash step, the membrane was incubated with ECL substrate (GE healthcare). The signals were recorded using the LAS-3000 imager (Fujifilm).
Mitochondrial localization
Chromosomally tagged GFP strains (Gavel and von Heijne, 1990) were purchased from Invitrogen. Each strain was cultured to a mid-log phase in synthetic defined (SD) medium and two aliquots were prepared. One aliquot was treated with 10 μg/ml tunicamycin at 30°C on a roller drum for 1 h, whereas the other was used as an untreated control. MitoFluor Red 589 was added to both aliquots to stain the mitochondria. The cells were visualized immediately with Zeiss Axioskop2 mot plus multiple wavelength fluorescence and visible-light inverted microscope capable of digital imaging. An oil-immersion objective at × 100 magnification was used. Using the Zeiss AxioVision Rel. 4.4 software, fluorescence microscopy images for the signal from both the GFP and MitoFluor Red 589 channels were recorded, and compared with Nomarski/DIC (differential interference contrast) images. To quantify the localization results, the images were analyzed in two steps. First, individual cells were selected only if they showed both clear mitochondria staining and GFP signal from the target proteins. Second, the selected cells were counted only when mitochondrial staining and target protein GFP signal well overlapped. Cell counts were performed three times independently and 100 cells were counted for each strain in the presence or absence of tunicamycin treatment. The error bars represent the standard deviations.
Supplementary Material
Supplementary figure and table
Acknowledgments
We thank Dr Robert Jensen for help with yeast mitochondria staining. LK was supported by an NIH postdoctoral training fellowship. ST was supported by Shanghai Jiaotong University 985 grant. HZ was supported, in part, by NIH grants and the WW Smith Charitable Trust. MS was supported by NIH grants.
Footnotes
The authors declare that they have no conflict of interest.
References
- Apweiler R, Hermjakob H, Sharon N (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta 1473: 4–8 [DOI] [PubMed] [Google Scholar]
- Bains G, Lee RT, Lee YC, Freire E (1992) Microcalorimetric study of wheat germ agglutinin binding to N-acetylglucosamine and its oligomers. Biochemistry 31: 12624–12628 [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Bozidis P, Williamson CD, Colberg-Poley AM (2008) Mitochondrial and secretory human cytomegalovirus UL37 proteins traffic into mitochondrion-associated membranes of human cells. J Virol 82: 2715–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra NC, Spiro MJ, Spiro RG (1998) Identification of a glycoprotein from rat liver mitochondrial inner membrane and demonstration of its origin in the endoplasmic reticulum. J Biol Chem 273: 19715–19721 [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ, Raikhel NV (1991) Lectins, lectin genes, and their role in plant defense. Plant Cell 3: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg RM, Loontiens FG, Van Landschoot A, Jovin TM (1981) Binding kinetics of methyl alpha-D-mannopyranoside to concanavalin A: temperature-jump relaxation study with 4-methylumbelliferyl alpha-D-mannopyranoside as a fluorescence indicator ligand. Biochemistry 20: 4687–4692 [DOI] [PubMed] [Google Scholar]
- Coelho PS, Bryan AC, Kumar A, Shadel GS, Snyder M (2002) A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA. Genes Dev 16: 2755–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RN, Hart GW (1999) Glycosylation sites flank phosphorylation sites on synapsin I: O-linked N-acetylglucosamine residues are localized within domains mediating synapsin I interactions. J Neurochem 73: 418–428 [DOI] [PubMed] [Google Scholar]
- Costanzo MC, Hogan JD, Cusick ME, Davis BP, Fancher AM, Hodges PE, Kondu P, Lengieza C, Lew-Smith JE, Lingner C, Roberg-Perez KJ, Tillberg M, Brooks JE, Garrels JI (2000) The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res 28: 73–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank C, Costanzo MC, Hirschman J, Hodges P, Kranz JE, Mangan M, O'Neill K, Robertson LS, Skrzypek MS, Brooks J, Garrels JI (2002) Three yeast proteome databases: YPD, PombePD, and CalPD (MycoPathPD). Methods Enzymol 350: 347–373 [DOI] [PubMed] [Google Scholar]
- Dube DH, Bertozzi CR (2005) Glycans in cancer and inflammation. Potential for therapeutics and diagnostics. Nat Rev Drug Discov 4: 477–488 [DOI] [PubMed] [Google Scholar]
- Fisk DG, Ball CA, Dolinski K, Engel SR, Hong EL, Issel-Tarver L, Schwartz K, Sethuraman A, Botstein D, Cherry JM (2006) Saccharomyces cerevisiae S288C genome annotation: a working hypothesis. Yeast 23: 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabius HJ, Siebert HC, Andre S, Jimenez-Barbero J, Rudiger H (2004) Chemical biology of the sugar code. ChemBioChem 5: 740–764 [DOI] [PubMed] [Google Scholar]
- Gavel Y, von Heijne G (1990) Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng 3: 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, Gerstein M, Dumont ME, Phizicky EM, Snyder M, Grayhack EJ (2005) Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev 19: 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmill TR, Trimble RB (1999) Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim Biophys Acta 1426: 227–237 [DOI] [PubMed] [Google Scholar]
- Gregan J, Bui DM, Pillich R, Fink M, Zsurka G, Schweyen RJ (2001) The mitochondrial inner membrane protein Lpe10p, a homologue of Mrs2p, is essential for magnesium homeostasis and group II intron splicing in yeast. Mol Gen Genet 264: 773–781 [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem 73: 1019–1049 [DOI] [PubMed] [Google Scholar]
- Herscovics A (1999) Processing glycosidases of Saccharomyces cerevisiae. Biochim Biophys Acta 1426: 275–285 [DOI] [PubMed] [Google Scholar]
- Hirabayashi J (2004) Lectin-based structural glycomics: glycoproteomics and glycan profiling. Glycoconj J 21: 35–40 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T (2003) Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol 21: 667–672 [DOI] [PubMed] [Google Scholar]
- Kukuruzinska MA, Bergh ML, Jackson BJ (1987) Protein glycosylation in yeast. Annu Rev Biochem 56: 915–944 [DOI] [PubMed] [Google Scholar]
- Kung LA, Snyder M (2006) Proteome chips for whole-organism assays. Nat Rev Mol Cell Biol 7: 617–622 [DOI] [PubMed] [Google Scholar]
- Levrat C, Ardail D, Louisot P (1990) Biosynthesis of glycoconjugates in mitochondrial outer membranes. Preliminary characterization of the oligosaccharide moiety of a N-glycoprotein. Biochem Int 20: 1–11 [PubMed] [Google Scholar]
- Levrat C, Ardail D, Morelis R, Louisot P (1989) Study of the N-glycoprotein biosynthesis through dolichol intermediates in the mitochondrial membranes. Int J Biochem 21: 265–278 [DOI] [PubMed] [Google Scholar]
- Mackiewicz A, Mackiewicz S (1986) Determination of lectin-sugar dissociation constants by agarose affinity electrophoresis. Anal Biochem 156: 481–488 [DOI] [PubMed] [Google Scholar]
- Mendelsohn RD, Helmerhorst EJ, Cipollo JF, Kukuruzinska MA (2005) A hypomorphic allele of the first N-glycosylation gene, ALG7, causes mitochondrial defects in yeast. Biochim Biophys Acta 1723: 33–44 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Nilsson I, Kelleher DJ, Miao Y, Shao Y, Kreibich G, Gilmore R, von Heijne G, Johnson AE (2003) Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J Cell Biol 161: 715–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A (2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5: 1543–1554 [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T (1998) Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280: 1763–1766 [DOI] [PubMed] [Google Scholar]
- Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H (2007) Solid-phase extraction of N-linked glycopeptides. Nat Protoc 2: 334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L, Hart GW (2003) O-GlcNAc turns twenty: functional implications for post-translational modification of nuclear and cytosolic proteins with a sugar. FEBS Lett 546: 154–158 [DOI] [PubMed] [Google Scholar]
- Zhang H, Li XJ, Martin DB, Aebersold R (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol 21: 660–666 [DOI] [PubMed] [Google Scholar]
- Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M (2001) Global analysis of protein activities using proteome chips. Science 293: 2101–2105 [DOI] [PubMed] [Google Scholar]
- Zhu X, Gerstein M, Snyder M (2006) ProCAT: a data analysis approach for protein microarrays. Genome Biol 7: R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure and table