Abstract
Central carbon metabolism is a basic and exhaustively analyzed pathway. However, the intrinsic robustness of the pathway might still conceal uncharacterized reactions. To test this hypothesis, we constructed systematic multiple-knockout mutants involved in central carbon catabolism in Escherichia coli and tested their growth under 12 different nutrient conditions. Differences between in silico predictions and experimental growth indicated that unreported reactions existed within this extensively analyzed metabolic network. These putative reactions were then confirmed by metabolome analysis and in vitro enzymatic assays. Novel reactions regarding the breakdown of sedoheptulose-7-phosphate to erythrose-4-phosphate and dihydroxyacetone phosphate were observed in transaldolase-deficient mutants, without any noticeable changes in gene expression. These reactions, triggered by an accumulation of sedoheptulose-7-phosphate, were catalyzed by the universally conserved glycolytic enzymes ATP-dependent phosphofructokinase and aldolase. The emergence of an alternative pathway not requiring any changes in gene expression, but rather relying on the accumulation of an intermediate metabolite may be a novel mechanism mediating the robustness of these metabolic networks.
Keywords: genetic interaction, metabolic flux, metabolomics, system biology, transaldolase
Introduction
Systematic phenome analysis of gene-deletion mutants combined with in silico predictions from genome-scale metabolic network models has been used to identify new genetic interactions and previously unknown gene functions in model microorganisms (Duarte et al, 2004; Joyce et al, 2006; Reed et al, 2006; Ohara et al, 2007). As this approach depends on a predicted or observed phenotype, genetic robustness, the phenomenon by which a majority of genes do not show a detectable phenotype when deleted (Giaever et al, 2002; Baba et al, 2006; Kato and Hashimoto, 2007), limits the availability of gene candidates. Such robustness could, in part, originate from redundancy such as the presence of an isozyme or other pathway with a duplicate function (Gu et al, 2003; Papp et al, 2004). In many cases, more than two redundant systems are present (Deutscher et al, 2006). Furthermore, the specialized functions of many genes for specific growth conditions, such as the availability of different carbon sources, could contribute to overall robustness. Systematic deletion of two or more genes, and fitness tests of the mutants under many conditions, would be powerful systems for the discovery of novel gene functions (Boone et al, 2007).
The central carbon metabolism of Escherichia coli is a model example of a very robust system. Despite the essential functions of both catabolism and anabolism, only four genes (out of >70 genes in the pathways shown in Figure 1) are essential for growth using glucose as the sole carbon source. Through the use of a novel method to systematically create multiple-gene knockouts, we tested for the presence of unknown metabolic reactions in the extensively examined network of one of the most thoroughly analyzed model organisms. By combining experimental and computational phenome analyses of systematic double and triple knockouts, grown on various carbon sources, we demonstrated the emergence of an unreported pathway formed by previously unknown activities of well-characterized glycolytic enzymes that allow transaldolase-deficient mutants to unexpectedly grow on some carbon sources.
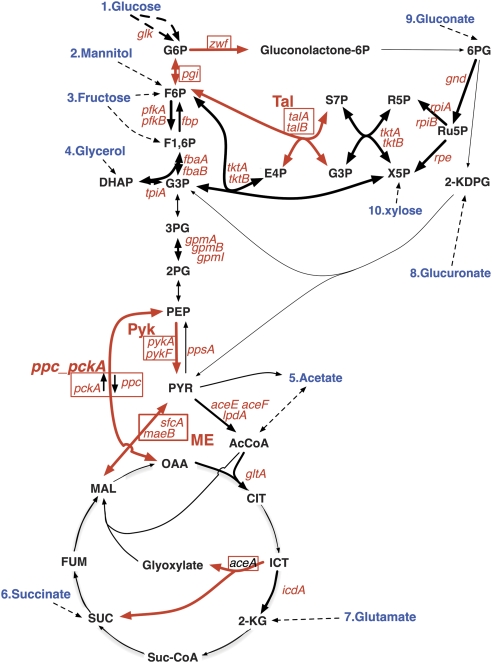
Figure 1.
Central carbon metabolism pathways examined in this study. Genes used for the first deletion are shown in red. Genes for the second deletion are boxed and abbreviations used in the text for the second deletion are indicated in boldface red if different from the deleted gene name. The reactions deleted by second deletion are shown with red arrows. For pckA and ppc, direction of the reaction catalyzing is shown by small arrows. The utilized carbon sources (blue) and entry points into central carbon metabolism are shown. See Supplementary Table I for gene and product names and Supplementary Table III for the abbreviations of metabolites not defined in the text.
Results
Systematic construction of multiple-knockout mutants and phenotype analysis
To efficiently analyze the phenotypes of multiple-knockout mutations in central carbon metabolism, seven key reactions were selected and a deletion of each reaction (second deletion) was combined with each of 31 single-gene deletions (first deletion). The names of the genes and their products are listed in Supplementary Table I, and the locations of the genes on the metabolic map are shown in Figure 1. Seven key reactions were selected to represent each of the following pathways: glycolysis (two reactions), the pentose phosphate pathway (two reactions), the anaplerotic pathway (two reactions), and the glyoxylate shunt (one reaction). As four of the selected reactions can be catalyzed by two isozymes, the deletion of two genes was required to attain the second deletion, and the resulting strain bore a triple deletion. To construct these multiple-deletion strains systematically, a derivative of P1 phage (P1dl) enabling multiple rounds of transduction in the liquid phase was constructed and used to transduce the second deletion into each single-gene-deletion strain. Two independently isolated single-deletion strains carrying the first deletion (31 metabolic-gene deletions and a control rrnH deletion) were used for duplicate analysis. We hereafter use the term ‘double' knockout even if the second deletion consists of two genes and the strain bears three-gene knockouts, and refer to the strain in which a first deletion X and a second deletion Y were combined as X–Y strain, for example, pgi-Tal or pgi-ppc_pckA.
The growth phenotypes of the resulting double-deletion cells were tested under various nutrient conditions, including rich medium, minimal media containing one of 10 different carbon sources, and a minimal medium with a combination of two carbon sources. The carbon sources were selected as they connect to central carbon metabolism at points distributed across most of the central pathways investigated (Figure 1). Cell growth was monitored by evaluating the OD600 at 24 and 48 h after inoculation (Supplementary Table II). Since we used an incubation method without forced aeration, the oxygenation level of the cultures is not known. However, growth of a wild-type strain on non-fermentative carbon sources, succinate and acetate (OD600 of 0.26 and 0.39 at 24 h, and 0.27 and 0.51 at 48 h, case numbers 5 and 6, respectively; Supplementary Table II), indicated that the oxygen supply was sufficient to support aerobic growth at least to this cell density.
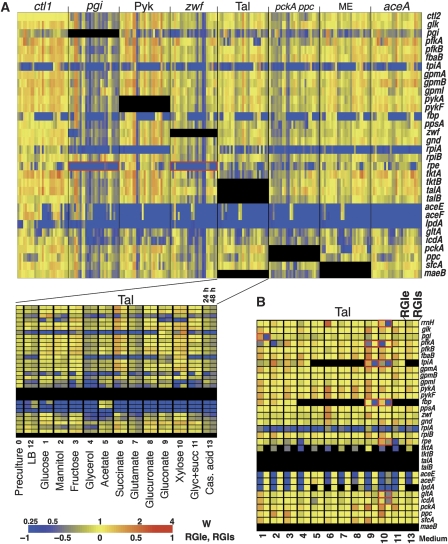
To eliminate the absolute growth difference on different media, relative growth (W) was calculated for each strain as the growth ratio of the test strain (single or double mutant) over the wild-type strain grown on the same medium (see Materials and methods for calculation of W). A heat map of the relative growth (W) is shown in Figure 2A.
Figure 2.
Heat maps showing global relative growth phenotype and comparison between measured and predicted values. (A) Growth rates of the multiple knockouts compared with wild type are shown in a heat map. Each row indicates the first gene deletion. Columns group indicates the second gene deletion, as well as the time point (24 or 48 h) and medium. The results for second mutations in talAB are enlarged in the lower panel. (B) RGIe and RGIs score are shown side by side for comparison. Tiles shown in black indicate conditions under which RGI scores were not calculated. The numbers at the bottom indicate the medium, as shown in panel A. The conditions mentioned in the text are highlighted by red boxes.
When compared with the parental single-deletion strains bearing only the first or second deletion, many double-knockout strains exhibited a slow-growth phenotype displayed as a shift of the relative growth distribution (the averages ±s.d. was −0.22±0.36; Supplementary Figure 1A and B). By contrast, when the comparison was made with the slower growing of the two parental strains, the distribution of the relative growth was centred around zero (average ±s.d. was −0.03±0.22; Supplementary Figure 1C), indicating that the single mutation that caused the larger growth defect generally determined the growth of the double mutants in most of the experiments.
However, some double mutants exhibited growth phenotypes that were very different from those of both corresponding single mutants. For example, rpe-pgi and rpe-zwf mutants exhibited very limited ability to utilize most carbon sources, whereas both of the parents could use a broad spectrum of carbon sources (Figure 2A, boxed).
Here, we refer to such specific slow- or fast-growth phenotypes that are specific to the double-knockout strain, but are not found in either of the parental single mutants as synthetic slow-growth phenotypes or synthetic fast-growth phenotypes, respectively. In general, the emergence of a synthetic slow- or fast-growth phenotype suggests functional interaction between the genes mutated or redundancy in their function. In the above case, the inability of the rpe-zwf mutant to grow on most carbon sources is due to its inability to produce the essential metabolite ribose-5-phosphate (R5P), as fluxes leading to this metabolite are blocked in both directions of the pentose phosphate pathway (see Supplementary Text 1 for details).
To evaluate the presence of such synthetic slow- or fast-growth phenotypes, we defined a Relative Growth Index from experiment (RGIe) score that weighs the growth of a mutant strain against that of its parental strain(s) (see Materials and methods).
Within a threshold of two standard deviations (s.d.s), we observed 229 cases (9.3%) that exhibited synthetic slow-growth phenotypes and 20 cases (0.8%) exhibiting synthetic fast-growth phenotypes out of the 2465 cases in which we could calculate the RGIe score. Similarly, using the RGIe score of single-knockout strains, we observed 30 cases (5.9%) with slow-growth and 6 (1.2%) with fast-growth phenotypes from a total of 493 experiments. Assuming normal distribution, the number of fast-growth phenotypes was within the range of experimental variation in both single- and double-knockout experiments; thus, most of them could result from experimental variation (see section Discussion), so we examined only synthetic slow-growth phenotypes. The rate of emergence of synthetic slow-growth phenotypes in double knockouts was slightly higher than the rate of emergence of slow-growth phenotypes by single deletions. However, controlling for the fact that four glycolysis genes (fbaA, gapA, pgk, and eno) could not be tested as first deletions because they are essential (Baba et al, 2006; Kato and Hashimoto, 2007), the rate of emergence of the slow-growth phenotype was not higher in the double mutants. This suggests that the first mutation did not cause drastic loss of the robustness of central carbon metabolism against the additional loss of the single gene.
Examining the 229 cases of synthetic slow-growth phenotypes, we found that most of them, except for five cases, were restricted to strains carrying a first deletion in rpe, pckA, aceE, aceF, lpdA, or rpiA. All of the rpe-related cases were rpe-pgi and rpe-zwf mutants, like those already mentioned.
All four pckA-related synthetic slow-growth phenotypes were observed in the pckA-ME mutant, which was deleted of all the reactions of the central carbon metabolism, connecting tricarboxylic acid (TCA) cycle to glycolysis. Thus, it is likely to result in a strain that cannot utilize gluconeogenic carbon sources entering metabolism from the TCA cycle intermediates. Three of the four synthetic slow-growth phenotypes were of such carbon sources: acetate, succinate, and glutamate. Additionally, pckA-ME mutants displayed synthetic slow-growth on minimal media supplemented with casamino acid, indicating that limited growth of many strains on casamino acid was dependent on some amino acid(s) feeding into the TCA cycle. In the cases of aceE, aceF, and lpdA, all of which encode subunits of the pyruvate dehydrogenase (PDH) complex, single mutants of any of these genes exhibited growth on most fermentative carbon sources, but growth on these carbon sources was abolished in many of the double mutants. This resulted in 53, 45, and 49 aceE aceF-, and lpdA-based synthetic slow-growth phenotypes, respectively. Similarly, while a single mutation in rpiA (encoding the major isozyme of ribose-5-phosphate isomerase (Rpi)) did not exhibited a drastic slow-growth phenotype, most of the secondary deletions affected its growth on many carbon sources, resulting in 52 cases of synthetic slow-growth phenotypes. The effect of secondary mutations of enzymes in the pentose phosphate pathway (zwf or Tal) was less pronounced compared with secondary mutations in other pathways.
Such extreme bias, as observed for the first deletion, was neither observed for the second mutations nor in the medium conditions employed.
Comparison of synthetic slow-growth phenotypes with those predicted by simulation
To determine the extent to which the observed phenotypes can be explained based on current knowledge of central carbon metabolism, and thereby possibly identifying missing elements of the current knowledge, we performed metabolic model-based predictions of growth rates and contrasted them with the experimental results. Several methods for growth rate prediction using genome-scale models have been proposed (Edwards and Palsson, 2000; Segre et al, 2002; Reed et al, 2003; Shlomi et al, 2005), but considering that one of our main objectives was to discover unknown reactions, we mainly used flux balance analysis (FBA), which attempts to use every non-constrained reaction in the model to obtain the best objective results. Additionally, prediction by Minimization of Metabolic Adjustment (MoMA) was also performed. This method was developed for more accurate predictions of growth phenotypes in non-evolved, knockout bacterial strains where the assumption of growth optimality used for FBA may not hold (Segre et al, 2002). The results of MoMA analysis were mentioned when differing from those made by FBA. Both predictions were made using the COBRA toolbox (Becker et al, 2007) to obtain the maximum rate of biomass production (μ) as the objective function. We used E. coli genome-scale metabolic reconstruction, iAF1260 (Feist et al, 2007), considering gene expression levels observed for aerobic growth on glucose as a base model, and incorporated several changes.
In the initial model iAF1260, 16 of the 31 genes for the first deletions were annotated with reactions harboring two or three isozymes (Table I). While deleting a gene associated with an isozyme from the in silico model does not affect the prediction, deleting the same gene in vivo could result in graded effects anywhere from no effective loss of the reaction to complete loss. Since all of the targeted reactions are well studied and the accepted major or minor roles of most isozymes are known (Sprenger, 1995; Romeo and Snoep, 2005; Keseler et al, 2009), we removed all minor isozymes from the model and included only the genes encoding the major isozymes. In this modified model, deleting the gene for any major isozyme (pfkA, fbp, gpmA, pykF, rpe, rpiA, tktA, or talB) resulted in complete loss of the corresponding reaction, whereas deleting the gene for a minor isozyme, which had already been removed, had no effect on the growth phenotype; thus, we could examine both extreme results.
Table 1.
Genes encoding enzyme having two or more isozyme, and used for the deletion study
| Reactiona | Majorb | Unknownc | Minord |
|---|---|---|---|
| PFK | pfkA | pfkB | |
| FBP | fbp | (glpx)e | |
| FBA | (fbaA)e | fbaB | |
| PGM | gpmA | gpmI | gpmB |
| PYK | pykF | pykA | |
| RPI | rpiA | rpiB | |
| RPE | rpe | (sgcE)e | |
| TKTf | tktA | tktB | |
| TALA | talB | talA |
aName of the reaction in the iAF1260 model.
bGene encoding the major isozyme of the reaction.
cGene encoding the isozyme of uncharacterized contribution.
dGene encoding the minor or lesser role of the reaction.
eNot selected for deletion.
fTKT1 and TKT2.
Since we used several different carbon sources, the necessary changes reflecting the expression of source-specific gene induction and flux direction constraints were considered. Comparison of the growth phenotype and the predicted growth of single-gene knockout strains was used to further tune the model by restricting the use of several reactions (see section Materials and methods).
Finally, several conditions (varying maximum oxygen consumption limits, and the presence or absence of anaerobic gene expression) were used for the growth rate prediction. To test consistency with the experimental data, correlation coefficients between the cell density at 24 h and the predicted growth (μ) were compared for each medium (Supplementary Figure 2A). For most carbon sources, predictions assuming a nearly unlimited supply of oxygen without anaerobic gene expression showed the best correlation with the experimental results, and therefore we selected these conditions for further comparisons. The prediction results obtained under these conditions are shown in Supplementary Table II.
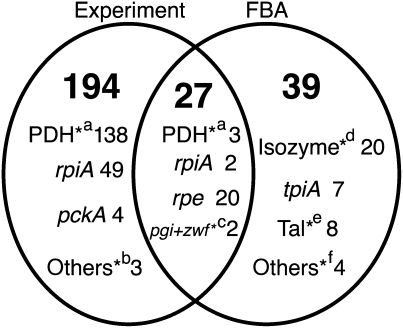
To evaluate the predicted growth of a mutant strain against its parental strain, we defined RGIs score similarly to the RGIe score used to evaluate the experimental data. Growth predicted by FBA was used instead of the OD600 for RGIe score calculation, and the RGIs scores were calculated for 2177 cases among 2465 for which we calculated the RGIe score of the double-knockout strains. The results obtained in complete medium (205) were not used for the prediction. In addition, the RGIs scores of the double mutants were not calculated when one of the parental single deletions was predicted to show no growth (83). Within the 2177 cases, 221 of the 229 cases that showed synthetic slow-growth phenotype in experiment were included, excluding the eight cases in complete medium. Using RGI scores and by applying a cut-off of ±2 s.d., 66 cases of slow-growth phenotypes were predicted whereas no cases of synthetic fast-growth phenotypes were predicted. Among the predicted 66 slow-growth phenotypes, 27 were experimentally observed (correct predictions; Figure 3).
Figure 3.
Experimental and FBA-simulated conditions that exhibited the synthetic slow-growth phenotype. Within the 2177 cases for which both RGIe and RGIs scores were obtained, the number of cases exhibiting the synthetic slow-growth phenotype in either of experiment, FBA, or both of experiment and FBA are shown. Each class was subdivided into categories and the number of cases in each category is also shown. Most of the categories were defined by the first deletion, and the first deletion is indicated. Other categories were marked by *. *a: Either of aceE, aceF, or lpdA in the first deletion; *b: one each in the case of pfkA-Tal-xylose, tpiA-Pyk-glucose, and icdA-pgi-acetate; *c: zwf-pgi and pgi-zwf on glucose; *d: cases in which genes of the first deletion encoded the major isozyme; *e: cases having Tal as second deletion and explained by the novel reactions (each case was shown in Table IIB); and *f: icdA-aceA on acetate and casamino acid, gnd-pgi-glucose, and rpe-pgi-xylose. Cases classified into each category are shown in Supplementary Table II.
Similarly, we calculated the RGIe score of 1968 of 2465 cases using MoMA prediction (RGIe–MoMA). Within these, 105 of the 229 experimental cases exhibiting the synthetic slow-growth phenotype were included. This apparent difference with FBA predictions was due to the fact that parental single-deletion strains were predicted to show no growth and therefore RGIe–MoMA scores of derived strains (292) were not calculated. Among these were multiple cases of PDH-related double mutants that experimentally showed synthetic slow growth. Using RGIs-MoMA, 67 cases of slow-growth phenotype were predicted and 36 of them were experimentally observed (Supplementary Figure 3). All the differences in correct predictions (27 for FBA and 36 for MoMA) were of mutants related to PDH deletion.
Among the correctly predicted cases, 20 of these corresponded to the above mentioned rpe-based double knockouts. However, the experimentally observed slow-growth phenotype for all four cases of pckA-ME double mutants, and for many cases of the aceE, aceF, lpdA, or rpiA-based double knockout, were not correctly predicted (false positive). For pckA-ME mutants growing on a gluconeogenic carbon source, the presence of any single enzyme connecting the TCA cycle to glycolysis will result in such a false-positive prediction. In the model employed for the prediction, five such reactions were present and they could independently connect the TCA cycle to glycolysis (Supplemental Figure 4A–D). Deletion of all of these reactions from the model resulted in the prediction of complete loss of growth of all four cases of pckA-ME double knockouts while not affecting most other predictions (Supplementary Table II, compare raw RGIe and RGIe-2, cases 1513–1515, and 1520). The only exception was the prediction for ppsA-pck_ppc on gluconeogenic carbon sources. As for the pckA-ME, this double mutant lost all reactions connecting the TCA cycle to glycolysis upstream of PEP if all of the five additional constraints were applied in the model. However, the experimental results of the ppsA-pck_ppc mutant exhibited only a moderate slow-growth phenotype (Supplementary Table II, cases 2046–2048 and 2053), suggesting residual flux along the route from glyoxylate to 3PG (Supplementary Figure 4C) in this mutant.
Most of the synthetic slow-growth phenotypes of the aceE, aceF, and lpdA-based double mutants were not correctly predicted by FBA. Since the growth defect of the mutant in the PDH complex is due to the inability to supply acetyl-CoA (AcCoA) (Clark and Cronan, 2005; Keseler et al, 2009), the growth of strains with single knockouts of these genes on fermentative carbon sources were considered to be dependent on the supply of AcCoA, either from the main carbon source or from the degradation of the supplied casamino acid. We checked all the reactions that produce AcCoA in PDH mutants and found that the removal of five additional reactions related to AcCoA production (Supplementary Figure 4E) from the model completely abolished the predicted growth of PDH mutants, but had much less of an effect on the wild type. We had limited maximum possible flux of these reactions to mimic the growth of the PDH single mutant when making the simulation of double knockout. However, the failure to predict the synthetic slow-growth of the PDH-related double knockout indicated that the constraint used was not appropriate in these strains. In contrast, in the case of MoMA-based predictions, none of the bypass routes in Supplementary Figure 4E were utilized in any of the PDH-related mutants, because the flux was very different from that of wild type when one of the pathways was activated. In the real cell, however, the extent of utilization of these pathways could correspond to an intermediate between FBA and MoMA predictions. Since small differences in the flux of one of these pathways can cause this difference, more complete information about gene expression and/or regulation of these enzymatic activities in each case might be required for more accurate predictions.
Many synthetic slow-growth phenotypes of double-deletion strains carrying rpiA as a first deletion were also incorrectly predicted. In such cases, we found that the same amount of flux from R5P towards biomass passes from ribulose-5-phosphate (Ru5P) to lipidA biosynthesis, and then to the export of lipidA-related molecules (Supplementary Figure 5). Without Rpi (encoded by rpiA and rpiB), all reactions in the non-oxidative branch of the pentose phosphate pathway (catalyzed by Tkt and Tal enzymes) must operate in the reverse direction in order to produce the essential metabolite R5P. This results in the generation of xylulose-5-phosphate (Xu5P) through the Tkt-catalyzed reactions (Supplementary Figure 5A). However, the only known pathway that consumes Xu5P in Rpi-deficient cells is the one for biosynthesis of lipidA-related molecules (Supplementary Figure 5B), and the cellular requirements for R5P is larger than the requirements for these molecules. In the simulation, this problem was solved by allowing for extracellular export of lipidA-related molecules, thus ensuring production of the required amount of R5P. Although such a waste of a cellular component is unlikely to take place in the cell, export of lipidA-related molecules might occur at low levels depending on the strain. More likely, however, the minor isozyme of Rpi encoded by rpiB may carry sufficient flux to supply enough R5P and allow limited growth of some of the rpiA-related double mutants. Both hypotheses seemed consistent with the results observed for the second deletions in the pentose phosphate pathway, and thus limiting the flux of this pathway had less of an effect on the growth of rpiA-deletion mutants compared with second deletions in other pathways, although strong support from simulation was not obtained.
Excluding the 27 correct predictions, FBA incorrectly predicted slow-growth phenotypes for 39 of the 66 cases (false negatives). Since 20 of the false-negative predictions were for deletions in a major isozyme, the lack of a functional isozyme in the model can explain this result. In vivo, the flux carried by the minor isozyme is sufficient under these conditions. The remaining 19 cases, shown in Table IIA–C, are of particular interest because these unexpected growth phenotypes could be the result of reactions that were not present in the model, and thus might reveal as yet unknown isozymes or bypass reactions.
Table 2.
Conditions that showed unexpected growth phenotype
| Noa | Knockout | C source | Growth | RGI score | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1b | 2c | Exp | FBA | RGIe | RGIs | RGIs-2g | ||||
| Rawd | Relativee | Rawf | Relativee | |||||||
| A | ||||||||||
| 3135 | tpiA | pgi | Mannitol | 0.23 | 0.41 | 0.15 | 0.18 | 0.61 | −0.48 | −0.45 |
| 3136 | tpiA | pgi | Fructose | 0.20 | 0.53 | 0.14 | 0.17 | 0.00 | −0.36 | −0.31 |
| 3143 | tpiA | pgi | Xylose | 0.16 | 0.44 | 0.16 | 0.24 | −0.44 | −0.64 | −0.61 |
| 3147 | tpiA | zwf | Glucose | 0.03 | 0.08 | 0.14 | 0.17 | 0.10 | −0.33 | −0.34 |
| 3148 | tpiA | zwf | Mannitol | 0.14 | 0.25 | 0.15 | 0.18 | −0.02 | −0.48 | −0.45 |
| 3149 | tpiA | zwf | Fructose | 0.20 | 0.53 | 0.14 | 0.17 | 0.01 | −0.36 | −0.31 |
| 3156 | tpiA | zwf | Xylose | 0.21 | 0.58 | 0.16 | 0.24 | −0.39 | −0.64 | −0.61 |
| B | ||||||||||
| 503 | fbp | Tal | Gluconate | 0.34 | 0.78 | 0.00 | 0.00 | −0.06 | −1.00 | 0.00 |
| 504 | fbp | Tal | Xylose | 0.39 | 1.06 | 0.00 | 0.00 | 0.03 | −1.00 | 0.00 |
| 712 | gltA | Tal | Xylose | 0.35 | 0.95 | 0.05 | 0.07 | 0.01 | −0.48 | −0.01 |
| 1232 | icdA | Tal | Xylose | 0.35 | 0.94 | 0.05 | 0.07 | −0.09 | −0.48 | −0.01 |
| 1847 | pgi | Tal | Glucose | 0.41 | 1.08 | 0.11 | 0.14 | 0.14 | −0.86 | 0.00 |
| 2376 | rpe | Tal | Xylose | 0.04 | 0.12 | 0.06 | 0.09 | 0.09 | −0.38 | -0.01 |
| 3207 | tpiA | Tal | Gluconate | 0.43 | 0.98 | 0.00 | 0.00 | 0.10 | −1.00 | 0.00 |
| 3208 | tpiA | Tal | Xylose | 0.24 | 0.66 | 0.00 | 0.00 | −0.37 | −1.00 | −0.03 |
| C | ||||||||||
| 742 | gnd | pgi | Glucose | 0.20 | 0.53 | 0.06 | 0.08 | −0.49 | −0.92 | −0.03 |
| 1188 | icdA | aceA | Acetate | 0.14 | 0.53 | 0.06 | 0.21 | −0.33 | −0.74 | −0.76 |
| 1195 | icdA | aceA | CasaminoA | 0.14 | 0.70 | 0.03 | 0.54 | −0.20 | −0.41 | −0.42 |
| 2311 | rpe | pgi | Xylose | 0.04 | 0.11 | 0.00 | 0.00 | 0.04 | −1.00 | −1.00 |
| D | ||||||||||
| 88 | Ctl | Tal | Xylose | 0.38 | 1.04 | 0.09 | 0.14 | 0.04 | −0.86 | 0.00 |
| 1648 | pfkA | Tal | Xylose | 0.05 | 0.15 | 0.09 | 0.14 | −0.86 | 0.00 | −0.83 |
aCondition number shown in Supplementary Tables.
bFirst deletion used as recipient of transduction.
cSecond deletion used as donor of transduction (Tal, talA, and talB).
dOD600 at 24 h.
eThe raw growth relative to that of the wild type.
fMaximum rate of cell mass production (growth rate per hour) predicted.
gRGIs score when the novel reactions were taken into account.
These 19 cases included three tpiA-pgi and four tpiA-zwf mutants on fermentative carbon sources (Table IIA). Since tpiA single mutants exhibited slow-growth phenotypes on many carbon sources, several constraints had to be incorporated to mimic these phenotypes in FBA (see Materials and methods). Since assigning different flux values to these reactions will result in different prediction results, these cases were not considered likely to result from unknown reactions, but can rather be explained by changes in gene or protein expression, and were not explored further.
Of the remaining 12 false-negative predictions, eight related cases possessed Tal second deletions (Table IIB), mainly when xylose or gluconate was the carbon source. Since no reason for this group of false-negative predictions was readily recognizable, we next focused on solving this discrepancy in the following sections. Of the remaining four cases (Table IIC), gnd-pgi on glucose can be explained by the expression of the Entner–Doudoroff (ED) pathway (Jiao et al, 2003), and the other three cases were not further examined in this study.
Essentially similar results were obtained for predictions by MoMA. A total of 31 false negatives could be categorized into 17 cases due to isozymes, two cases due to tpiA, five cases due to Tal, and seven other cases (Supplementary Figure 3).
Identification of uncharacterized reactions in talA talB mutants
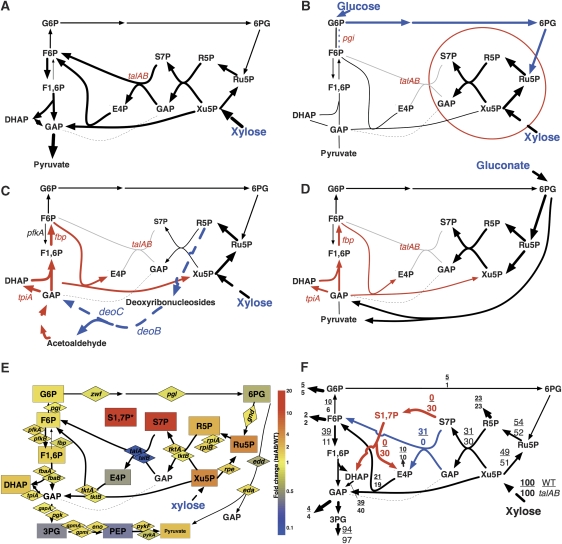
Slow growth on xylose was predicted for the multiple enzymatic knockouts as well as for a control deletion strain (ctl-Tal deletion; Table IID), which should have defects only in the transaldolase. However, the doubling time of the purified talA talB (talAB)-knockout strain on minimal medium containing xylose as the sole carbon source was confirmed to be 110 min, which is only slightly slower than the doubling time of the wild-type strain (100 min). According to the currently known metabolic pathways in E. coli, xylose flows into the central carbon metabolism as Xu5P and is further metabolized through the non-oxidative branch of the pentose phosphate pathway (Figure 4A). In the absence of transaldolase, xylose metabolism can be expected to stop after formation of sedoheptulose-7-phosphate (S7P) and D-glyceraldehyde-3-phosphate (GAP) by transketolase (Figure 4B). The prediction of the synthetic slow-growth of pgi-Tal strain on glucose occurred for the same reason, since most of the glucose is metabolized by the pgi mutant through the pentose phosphate pathway (Fraenkel and Levisohn, 1967). This strain did not exhibit a slow-growth phenotype on glucose (Table IIB).
Figure 4.
Pathways surrounding transaldolase. (A) Metabolic map of wild type during growth on xylose. (B) Currently accepted metabolism of transaldolase-deficient mutant (Tal deletion) on xylose and pgi-Tal deletion on glucose. Only the metabolites within the red circle could be produced in Tal deletion on xylose. Reactions indicated by blue arrows are additionally possible in the pgi-Tal knockout on glucose. (C) A map indicating a possible bypass pathway through deoxyribonucleosides (reactions shown in blue). The directions of the reactions shown in red are fixed to the reverse of the wild-type direction when the bypass is the sole source of carbon flux from xylose. (D) Possible metabolism of the Tal-deletion strain on gluconate, using currently known reactions. The reactions shown in red are required if ED pathway and/or the bypass shown in panel C is the sole source of carbon flux. (E) Relative amounts (talAB/wild type) of metabolite (box) and mRNA (diamond) are shown on the metabolic map. (F) Flux distribution of the wild type and talAB mutant growing on xylose. The novel reactions and the transaldolase reaction are shown in red and blue, respectively. The relative flux of each reaction (with incorporation of xylose set to 100) is shown at the respective position (upper, wild type; lower, talAB). See Supplementary Table IV B for detailed results.
The presence of an additional pathway beginning with R5P from deoxyribonucleosides and ending with the production of GAP and acetaldehyde (Figure 4C) can explain these phenotypes. Although, the additional pathway was included in the model for prediction, it was constrained to allow only limited flux that was not sufficient for normal growth, and to be consistent with the initial analysis of the tpiA single-knockout strain. Several additional results indicated that this pathway was not an important bypass allowing the Tal knockout to grow on xylose. First, growth on xylose was not affected by the additional deletion of deoC or deoB (data not shown), although each should be essential if this pathway is responsible for near-normal growth. Second, an additional mutation in either fbp or tpiA did not affect the growth of Tal mutants on xylose (Table IIB). Since F6P and E4P are essential metabolites that must be produced by gluconeogenesis from GAP or acetaldehyde, fbp and tpiA should be essential for Tal mutants on xylose if this alternative pathway is the main source of carbon flux from R5P (Figure 4C). Similar to growth on xylose, fbp-Tal and tpiA-Tal mutants were predicted to display synthetic slow growth on gluconate, but both strains showed normal growth (Table IIB). Although gluconate could be metabolized through the ED pathway (Peekhaus and Conway, 1998), tpiA and fbp were also required to produce F6P and E4P from the end products of the ED pathway (Figure 4D). Thus, the route from R5P to GAP and acetoaldehyde alone cannot explain the unexpected growth of fbp-Tal and tpiA-Tal mutants on gluconate.
Interestingly, the pfkA-Tal strain did not grow on xylose, although phosphofructokinase (pfkA gene product) activity would not be required even if the gluconeogenesis route (Figure 4C) is necessary in the talAB strain on xylose (Table IID). This surprising result strongly suggests the existence of an unknown pathway that involves pfkA and that connects S7P or its precursor to glycolytic intermediates in talAB-knockout cells.
The transcriptome and metabolome analyses of the talAB-knockout strain
To characterize the mechanism and metabolic reaction(s) that enable growth of the talAB knockout on xylose, we compared transcriptome and metabolome of this strain with those of wild-type strains during growth on minimal medium containing xylose as the sole carbon source (Supplementary Table III). Gene expression analysis performed with DNA microarrays revealed no notable changes in the mRNA levels of genes involved in related metabolic pathways (Figure 4E), and did not suggest interesting candidates for the novel pathway. On the other hand, metabolome analysis by Capillary Electrophoresis-Mass Spectrometry (CE-MS) (Soga et al, 2003, 2006), revealed a greater than 40-fold accumulation of S7P in the talAB-knockout cells and several-fold increase in pentose phosphates levels. Other metabolites showed only limited changes (less than twofold) in related pathways (Figure 4E). This accumulation of S7P is consistent with the genotype-based prediction that transaldolase activity was absent in this strain. Another interesting finding was the accumulation of an unidentified metabolite (m/z 369.0) that was comparable in magnitude to the increase in S7P. The m/z value of this compound matched the value of sedoheptulose-1,7-bisphosphate (S1,7P), a known metabolite, but not in the EcoCyc database (Karp et al, 2007). Although a standard sample of S1,7P was not available for direct confirmation of its identity, the migration time of the unknown compound by CE was consistent with the estimated time for S1,7P, which can be extrapolated from the difference in migration time of related sugar mono- and biphosphate pairs such as fructose-6-phosphate (F6P) and fructose-1,6-bisphosphate (F1,6P) or Ru5P and ribulose-1,5-bisphosphate.
Similar amount of S7P and the compound of m/z 369.0 were also found to accumulate in talAB cells when grown in minimal medium containing gluconate as the sole carbon source (Supplementary Table IIIC).
A bypass for S7P metabolism in the talAB mutant
The above mentioned finding suggests that S1,7P is produced in talAB mutant growing on either xylose or gluconate, although no enzyme has been reported for this activity in E. coli. The growth defect of the pfkA talAB knockout on xylose suggests that 6-phosphofructokinase I (the pfkA product that converts F6P to F1,6P) might act as a 7-phosphosedohepturokinase, converting S7P to S1,7P. The next metabolic step would be aldose cleavage of S1,7P to E4P and DHAP by an aldolase, such as the fbaA gene product that converts F1,6P to GAP and DHAP. Since glycolytic aldolase of some photosynthetic organisms is known to use both F1,6P and S1,7P as substrates (Flechner et al, 1999), homologous aldolase in E. coli (FbaA) might have the same activity. Such reactions could produce E4P and DHAP (reactions shown in red in Figure 4F) and provide a reasonable explanation for the unexpected growth of talAB-related mutants on xylose or gluconate, as shown by the FBA results considering the reactions (Table IIB and D, column RGIs-2).
To confirm the existence of such a bypass by following the fate of xylose, we performed metabolic flux analysis (MFA) of wild-type and talAB-knockout strains. Cells were grown in minimal medium with 1-13C-xylose as the sole carbon source, and harvested during log phase. The mass isotopomer patterns of intermediate metabolites as detected by 13C incorporation were analyzed by CE-MS (Toya et al, 2007). The wild-type and talAB-knockout strains exhibited clearly distinct 13C isotopomer distributions for many metabolites (Supplementary Figure 6 and Supplementary Table IV A). The mass isotopic patterns of F6P, a product of transaldolase, observed for the talAB-knockout strain were particularly different from that of the wild type (Figure 5A). To determine whether these differences could be explained by the proposed novel reactions, we used the observed mass isotopic patterns to find the metabolic flux for each reaction, using the currently accepted pentose phosphate pathway model (‘current' model) or an updated model containing the reactions from S7P to S1,7P, and S1,7P to E4P and DHAP, but without transaldolase (‘novel' model). As expected, the results from the wild-type strain fit the ‘current' model, but the results from talAB did not (Supplementary Figure 7 and Supplementary Table IV B). Importantly, the characteristic mass isotopic patterns of F6P in the talAB mutant were not generated with the ‘current' model (Figure 5B). The results from the talAB cells fit better with the ‘novel' model than with the ‘current' model. Furthermore, the F6P pattern was reproduced in the ‘novel' model (Figure 5B). When both transaldolase and the new reactions were incorporated into the model (‘Both' model in Supplementary Table IV B), the most favorable flux distributions using the mass isotopic pattern from wild-type demonstrated no flux in the novel reactions, whereas the most favorable flux using the talAB mass isotopic pattern showed all flux from S7P was to the novel reactions and no flux in the transaldolase (Figure 4F).
Figure 5.
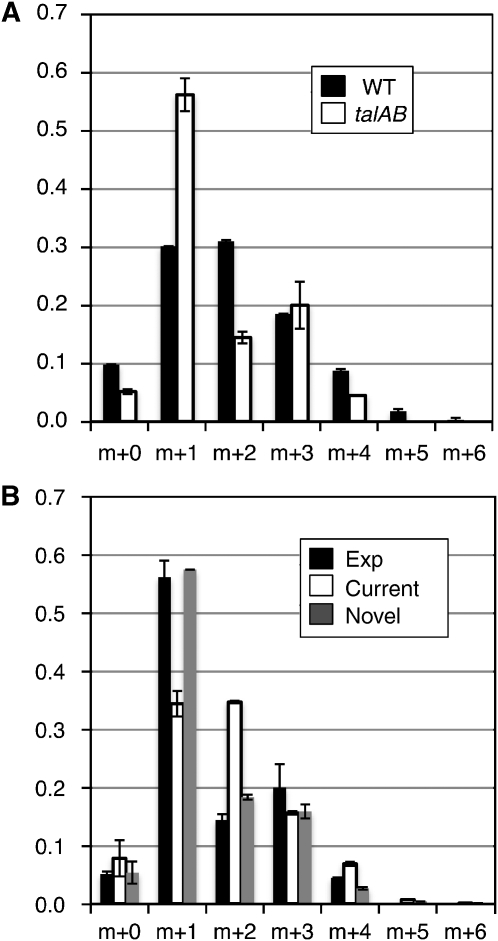
Mass isotopomer distribution of F6P. (A) Mass isotopomer distributions of F6P obtained from the wild type (WT) or the talAB mutant (talAB). m+n indicated the x-axis indicates the number of 13C incorporated, and on the y-axis indicates the ratio of each mass isotopomer. An average and s.d. value of results from two independent analyses are shown. (B) Math isotopomer distribution of F6P produced from the optimum fluxes result of MFA. The MFA results using the experimentally obtained isotopomer distributions from talAB were used to generate isotopomer distribution of F6P. The isotopomer distributions using the ‘current' pathway (current) or the ‘novel' pathway (novel), as well as the experimentally obtained one (exp.), are shown.
We also tested the fitness of the mass distribution from the talAB mutant with another proposed variant of the pentose phosphate pathway (the L-type pentose phosphate pathway; Williams et al, 1978; McIntyre et al, 1989; shown in Supplementary Figure 4B). Fitness using the L-type pathway was better than that obtained with the ‘current' pathway, but not as good a fit as with the ‘novel' pathway (Supplementary Figure 7 and Supplementary Table IV C online). In addition, the flux leading to the best fit resulted in an improbable direction of the transketolase reaction (F8 in Supplementary Figure 9B) towards Xu5P. These results suggest that the growth of the talAB knockout with xylose is most likely due to the presence of reactions that convert S7P to S1,7P and then to DHAP and E4P.
In vitro confirmation of the reactions that convert S7P to E4P and DHAP
To confirm whether PfkA and FbaA can directly convert S7P to E4P and DHAP, we purified recombinant PfkA and FbaA proteins and examined their activities in vitro. When S7P and ATP were incubated with PfkA, S7P and ATP were consumed in equivalent proportions, and a metabolite with an m/z ratio of 369.0 was produced (Table III). This metabolite co-migrated with the suspected S1,7P observed by metabolite profiling of talAB-knockout cells. Moreover, addition of FbaA to the reaction resulted in the production of E4P and DHAP, apparently from the metabolite (m/z 369.0). These results provide direct evidence that PfkA and FbaA have the capacity to catalyze a previously unreported transformation of S7P to E4P and DHAP through the intermediate, S1,7P. Since these in vitro reactions were performed with physiological enzyme levels near those found intracellularly (Ishii et al, 2007a), and metabolite concentrations equivalent to those observed in the talAB mutant, it is very likely that these reactions occur in vivo, at least in the talAB mutant, thus confirming the results from the MFA.
Table 3.
In vitro enzymatic assay for S7P kinase and S1,7P aldolase
| No enzyme | PfkA | PfkA+FbaA | FbaA | PfkA+FbaAa | No metabolite | |
|---|---|---|---|---|---|---|
| Metabolites, enzymes added | ||||||
| S7P | + | + | + | + | − | − |
| F6P | − | − | − | − | + | − |
| PfkA | − | + | + | − | + | + |
| FbaA | − | − | + | + | + | + |
| Metabolites detected (mM±s.d.) | ||||||
| S7P | 5.13±0.26 | 2.91±0.15 | 3.05±0.14 | 5.27±0.17 | ND | |
| S1,7P | ND | 1.15±0.13 | 0.79±0.11 | ND | ND | |
| DHAP | ND | ND | 0.23±0.02 | ND | 1.60±0.04 | ND |
| E4P | ND | ND | 0.39±0.03 | ND | ND | |
| F6P | 3.10±0.14 | |||||
| F1,6P | 0.51±0.09 | |||||
Blank: not determined; ND, not detected.
aF6P was used instead of S7P.
Improvement of the FBA model and results based on a deletion growth experiment
After considering the observed discrepancies between FBA and the experimental results, a second round of FBA analysis was performed using the model containing the novel reactions and with additional constraints deduced from the experimental results. Correlation coefficients between the FBA and experimental results in each medium are shown in Supplementary Figure 2B. The correlations showed clear improvement in most media, and the lower correlation coefficients obtained in the initial FBA results in gluconate and xylose notably improved.
Discussion
By generating systematic double mutants for the central carbon metabolism and testing their growth on many carbon sources, we observed significant number of cases exhibiting synthetic slow-growing strains, whereas the number of cases exhibiting the fast-growth phenotype was much less. While combinations of mutations are expected to display equivalent or more severe growth defects, they can also sometimes result in growth phenotype improvement (Typas et al, 2008). Such suppressive secondary mutations could result from overexpression of other protein due to the loss of an inhibitory factor (Sørensen and Hove-Jensen, 1996), or by blockade of the accumulation of toxic metabolite linked to the primary mutation, (Fuhrman et al, 1998). However, in this study, we could not readily assign a specific mechanism to the cases observed. Another possibility is that many fast-growing strains reached maximum cell density before the 24-h point. In that case, some of the cases exhibiting the fast-growth phenotype might not be due to an increased growth rate, but to increased biomass yield. The second hypothesis agrees with the result that half (13/20) of the synthetic fast-growth phenotypes were observed for strains whose second deletion was pgi or ppc_pckA as second deletion, which are known to show higher biomass yield, due to reduced secretion of metabolites like acetate, during aerobic growth conditions (Canonaco et al, 2001; Yang et al, 2003; Peng et al, 2004). However, overall the few improved growth phenotypes we observed appeared to be within experimental errors, and it was thus not possible to conclude about their physiological significance or the underlying mechanism.
By contrasting the growth phenotypes of double mutants with those predicted by FBA, we obtained 194 false-positive predictions and 39 false-negative predictions. Most of the false-positive predictions and some of the false-negative predictions could be ‘corrected' by adjusting the constraints to some reactions connecting central carbon metabolism with peripheral metabolism. Well-known peripheral metabolism pathways, such as nucleotide and amino-acid synthesis and degradation, were examples of the possible bypass of central carbon metabolism. For accurate predictions of growth, information regarding the gene expression and maximum capabilities of these reactions will be very important. However, examining gene expression in all strains under all conditions of a large-scale phenotype analysis is a paradoxical effort, considering the benefit of high-throughput analysis and model-based simulation of large number of strains. Starting from the limited information of gene expression and enzymatic capability, and then refining the model based on the comparison with the experimental result might be also an important direction for analyzing large-scale metabolic network. Overall, the present study highlights how discrepancies between large-scale experimental results and predictions can be used to discover unsuspected or novel activities as well as bypasses in metabolic pathways. At the same time, these discrepancies drive the need to place additional constraints on metabolic networks in order to fine-tune the model according to experimental results, and thus improve its value in further applications such as metabolic engineering.
We have shown that previously unknown reactions in central carbon metabolism are functional in the talAB knockout, using xylose. Although the flux carried by these reactions in the wild-type strain seemed negligible, the reactions could carry enough flux for growth of the talAB-knockout cells at the rate comparable to the growth of wild-type cells.
The central carbon metabolism network in E. coli was shown to be robust and able to handle deletions in many single metabolic enzymes without significantly altering enzyme levels, by re-routing fluxes on existing pathways. These adjustments are made possible by changes in metabolite levels that are closely linked to the deleted enzyme (Ishii et al, 2007a). In other cases, E. coli induces an alternative pathway by gene regulation or mutation to cope with the lost pathway (Fong et al, 2006; Ishii et al, 2007a). The present study reveals yet another example of robustness in the metabolic network through a previously uncharacterized mechanism. With perturbation that is severe enough to abolish a normal pathway, de novo reactions could be produced with existing enzymes, and the reactions can support as much flux as central carbon metabolism. In this study, we observed the emergence of bypass reactions without any change in the gene expression of the enzymes associated with the reactions. Instead the bypass reactions are triggered by the accumulation of a metabolite (S7P). S7P is a less favorable substrate to E. coli PfkA and conversion of S7P to S1,7P is much slower than that of F6P to F1,6P in vitro (data not shown). Nevertheless, this activity was high enough to sustain growth of the talAB mutant at a wild-type-compatible rate following accumulation of S7P. Many enzymes are known to exhibit similar loose specificity (Kuznetsova et al, 2006), but our finding implicated that such ‘side' activity could potentially have important functions in vivo.
A phosphotransferase activity, which converts S7P to S1,7P, was reported for the pyrophosphate-dependent phosphofructokinase of parasitic amoeba (Susskind et al, 1982), but the finding that widespread ATP-dependent phosphofructokinase also has this activity suggests the presence of this pathway from S7P to E4P and DHAP in a wide range of organisms, spanning bacteria to higher eukaryotes. Recent genome analyses have shown that the gene for transaldolase is missing from the genomes of some microorganisms, whereas other genes in the non-oxidative branch of the pentose phosphate pathway are retained (Westberg et al, 2004; Makarova et al, 2006). Interestingly, one of these bacteria (Lactococcus lactis) was reported to use glycolysis and the pentose phosphate pathway to metabolize xylose when it was supplied at high concentrations (Erlandson et al, 2000; Ohara et al, 2007). The pfk gene from L. lactis, which encodes phosphofructokinase, complemented the growth of talAB pfkA-knockout E. coli on xylose (Supplementary Figure 8) and the gene product had 7-phosphosedohepturokinase activity in vitro (data not shown), indicating that L. lactis may naturally use the pathway through S1,7P.
Also, some mammalian tissues are known to lack transaldolase, and mutations in this enzyme are associated with liver cirrhosis (Grossman et al, 2004; Perl, 2007). An unusual pentose phosphate pathway (L-type pathway) that does not require transaldolase and involves S1,7P as an intermediate has been proposed to be functional in liver tissues (Williams et al, 1978), but its presence remains controversial. The L-type pathway involves three other intermediates that are not required in the ‘current' pentose phosphate pathway, as well as enzymatic activities that are not currently linked to any gene. Since universally expressed glycolytic enzymes are sufficient for our new pathway, it is necessary to examine the presence of this pathway, as well as the L-type pathway, when studying the metabolism of cells lacking transaldolase.
Systematic and exhaustive phenome analyses of multiple-knockout mutants, as reported in this study, can reveal the presence of novel pathways, even in a well-characterized network. Through similar analyses of more target genes using our P1dl system or a recently reported mating system (Butland et al, 2008; Typas et al, 2008), additional new reactions and pathways may be delineated that involve not only functionally uncharacterized genes, but also well-characterized genes.
Associating uncharacterized protein enzymes with a reaction using sequence homology has been the common approach used to fill in the holes in metabolic networks. Inferring new activities for well-characterized proteins based on the similarity of the reaction chemistry and/or the reactants' structure might be a more challenging task, but it will also be important for understanding the metabolic network, as highlighted in this study. Automated prediction of such new activities, combined with growth phenotype predictions using metabolic models, will be a useful tool to uncover more gene functions and approach a complete understanding of the metabolic networks.
Materials and methods
Media and culture methods
Construction and maintenance of the strains were carried out in LB medium supplemented with the required antibiotics. MOPS minimal medium (Wanner, 1994) was used for most of the experiments for phenotype analysis unless otherwise noted. Ninety-six-well, deep-well plates (3960; Coaning, USA), sealed with gas-permeable membranes (AB-0718; Nihon Genetics, Japan), were used for growing cells. The cells were incubated in an M-BR-11 (TAITEC, Japan) rotary shaker at 1200 r.p.m. For small numbers of cultures, 15-cm test tubes were used in a water bath shaker at 120 r.p.m. All cultures were performed at 37°C. Working concentrations of the antibiotics were 30 μg/ml for kanamycin (Km); 25 μg/ml for chloramphenicol (Cm), and; 25 μg/ml for tetracycline (Tet).
Bacterial strains and plasmids
All the deletion strains used for phenotypic assays were derivatives of BW25113 (Haldimann and Wanner, 2001), and differed only at the tested genes. Strains constructed as donors for P1 transduction are shown in Supplementary Table V. Genes targeted for second deletion were replaced with the Cm or the Tet resistance gene by Wanner's method (Datsenko and Wanner, 2000). pKD13Cm and pKD13tet plasmids were constructed as templates for PCR by replacing the XbaI fragment containing the Km-resistant gene of pKD13 (Datsenko and Wanner, 2000) with the fragment from pKD3 (Datsenko and Wanner, 2000) containing the Cm-resistant gene and the fragment from pAH162 (Haldimann and Wanner, 2001) containing the Tet-resistant gene. Nomenclature for protein, genes and gene symbols are shown in Supplementary Table I.
Construction of P1-derived phages
P1dl was made by removing the lpa gene, which is essential for lytic growth (Łobocka et al, 2004), from the P1kc phage. To delete the lpa gene from the P1kc genome, the lpa in a P1kc lysogenic strain, JE5530 (obtained from NBRP, NIG, Japan), was replaced with the Km-resistant gene from pKD13. Then, the resistant gene was removed by FRT-mediated homologous recombination to produce the P1dl lysogenic strain. Since P1dl cannot perform the lytic cycle, a helper plasmid, pTRC-lpa, which expresses the LPA protein under an inducible trc promoter, was constructed. The P1dl phage was obtained by introducing pTRC-lpa and inducing the phage by IPTG. The transducing lysate of P1dl was routinely obtained by the plate lysate method (Miller, 1992), with the exception that pTRC-lpa was introduced into the host and LPA was induced by 1 mM IPTG in soft agar.
Multiple parallel transductions by P1dl lysate
Two independent isolates of the bacteria with the targeted gene deletion, derived from the Keio collection (Baba et al, 2006), were purified by single-colony isolation, the gene deletion confirmed, and they were used as the recipients. As a control, a derivative of BW25113, with the entire rrnH operon replaced by the Km gene (rrnH:Km), was constructed. Each of the 64 recipients was placed in a 96-well microtitre plate, and most of the experiments were performed following this format. For transduction, the recipient strains were grown in 1 ml of LB broth supplemented with Km. A 100-μl volume of each of the overnight cultures was transferred into a new deep-well plate and 1 × 107 p.f.u. of P1dl-transducing lysate in 100 μl of SM were added. The mixture was kept at 37°C for 1 h, and 1 ml of LB supplemented with antibiotics (Km+Tet or Cm) was added. After overnight growth, the culture was maintained as a glycerol stock until needed for a growth check, in the case of a single round of transduction. In the case of a double round of transduction, to produce triple knockouts, 100 μl of the first round overnight culture was mixed with 5 × 107 p.f.u. of a second P1dl-transducing lysate. The mixture was selected overnight by growth in the presence of the three indicated antibiotics and stored as a glycerol stock.
Confirmation of genotype after gene deletion and transduction
Gene deletion and transduction were confirmed by checking the size of PCR fragments amplified by a set of primers located upstream and downstream of the targeted deletion(s). When the sizes of the deletion and non-deletion (wild type) alleles were similar, the fragment was digested by XbaI, which cuts at two sites within the deletion allele, and confirmed by electrophoresis. The primer sets (Up-primer and Down-primer) were designed with the Primer3 software for each tested gene (Rozen and Skaketsky, 2000). The Up-primer anneals at the 5′-end, 200–400 bp upstream of the coding region, and the Down-primer anneals on the complementary strand at the 3′-end, 100–250 bp downstream of the coding region.
Growth measurements
The glycerol stocks of transductants were grown overnight in 1-ml aliquots of LB broth supplemented with the appropriate antibiotics and inoculated into 150 μl of LB broth or MOPS medium in a microtitre plate (167008; NUNC). The medium contained the carbon source indicated in Supplementary Table VI and was supplemented with 0.2% casamino acids (Difco Vitamin Assay; Becton Dickinson) and required antibiotics. The plates were sealed with gas-permeable membranes and incubated without shaking in a 37°C incubator in which humidity was kept above 80% by a water bath inside the incubator. After 24 h, precipitated cells were mixed and the OD600 of each well was measured using a SPECTRAMAX Plus 384 spectrophotometer (Molecular Devices, USA). The plates were re-sealed with new gas-permeable seals and incubated for another 24 h before a second measurement was taken. The growth of the single-knockout strains, parental strains used as recipient, was also examined. For later analysis, the average of OD600 values from two isogenic strains was used. If one of the two transductions was not successful, the result from the other transduction was used.
Model based prediction of the growth rate
Ec_iAF1260 (Feist et al, 2007), which contains 1260 ORF information of E. coli, was used as a base model. When a target reaction in this study involved two or more isozymes, only one gene considered to be the main isozyme was kept. Basically, thermodynamic constraint and gene expression information for aerobic growth on glucose reported in reference Feist et al (2007) were used as an initial constraint and additional changes depending on the carbon source and gene deletion were considered. Modifications to this default are listed in Supplementary Table VII. The maximum rate of the carbon source import (lower bound) was set to −6. The maximum rate for each amino-acid import was set to match the molar ratio calculated from the composition of casamino acids (supplied by Becton, Dickinson), and total amount was adjusted to match the observed growth of icdA and gltA single-knockout mutants, which required amino acids for growth. A solution that optimized the rate of biomass production in a given model condition was obtained by FBA or linearMoMA using COBRA toolbox (Becker et al, 2007). For prediction by MoMA, solution by the FBA using the same model, except that using the wild type pathway instead of the single or double deletion, was used for comparison. The maximum rate of biomass production was used as the simulated growth rate. The initial model included many reactions that could bypass known central carbon metabolism. However, many of these possible bypasses did not seem functional in the living cells, because most of the reactions cannot support the amount of flux required for central carbon metabolism. To validate these possible bypasses and refine the model, growth of the single-knockout strains, having first series of deletion, was compared with the prediction using the RGIe and RGIs value against the wild type. If difference between RGIe and RGIs was grater than 2, reactions that could result in false predictions were examined and maximum or minimum flux of the reaction was adjusted to match the prediction with the experimentally measured growth rate. The script used for the predictions using COBRA toolbox is shown in Supplementary text 2.
Calculation of RGIe or RGIs value for evaluating relative growth against parental single-knockout strains
1. At first relative growth of each strain against wild type at the same condition was calculated using the experimental growth result of double knockout w(xy), harboring x of the first deletion and y of the second deletion, and that of the wild type, w(wt).
2. For comparing the experimental growth result of double knockout [W(xy)] with the growth of parental single-knockout strains [W(x) or W(y), harboring x of the first deletion or y of the second deletion, respectively], the relative growth of the slower growing parental single-knockout strain was determined as follows:
3. The Relative Growth Index RGIe(xy) was determined as follows:
OD600 value at 24 h after inoculation was used as the experimental result. If WM(xy) was lower than 0.1, RGIe(xy) was not calculated because of the lower reliability of the control.
For evaluating the growth of single knockout of gene x, RGIe was calculated using wild type as a parental strain as follows:
For evaluation of FBA result, RGIs was calculated using predicted maximum rate of biomass production instead of the OD600 value used for RGIe.
Analysis of metabolite levels and MFA
Wild-type and talAB-knockout cells were grown in 50 ml of MOPS medium supplemented with 0.2% xylose or 0.22% sodium gluconate in 500 ml Erlenmeyer flasks until OD600 reached 1. Samples were collected and subjected to analysis of anions by CE-MS, as previously described (Soga et al, 2003; Ishii et al, 2007a). For analysis of metabolic flux by stable isotope labeling, 0.2% 1-13C-xylose was used in place of non-labelled xylose. The mass isotopic distribution of the anions was calculated, and optimization of the metabolic flux to fit the detected mass distribution was performed, as previously described (Toya et al, 2007). For evaluation of the fitness, the sum of [{(Xm–Xs)/0.06}2], where Xm is the relative amount of a mass isotopomer and Xs is a simulated amount of the corresponding mass isotopomer, was calculated for each metabolite, and the sum of the value for all employed metabolites was used. The pathway models are shown in Supplementary Figure 9 and Supplementary Table IV. Two biological replicates for each strain were performed from independent cultures. Optimization of flux was performed three times for each data set and model. The flux distribution with the best score for each data set and model is shown in Supplementary Table IV.
Transcriptome analysis
DNA microarrays were produced by spotting of the E. coli AROS V2.0 oligo-DNA set (Operon Biotechnologies, Huntsville, USA) on a GeneSlide (Toyo Kohan, Tokyo) according to the manufacturer's protocol. Cells were cultured and sampled under the same conditions as described for metabolome sampling. During sampling, RNA was stabilized by mixing with RNAprotect Bacteria reagent (Qiagen), and total RNA was isolated using the RNeasy Mini kit (Qiagen). Duplicate experiments employing dye swapping were performed. The protocol and conditions used for hybridization and washing were previously described (Oshima et al, 2002). Images were scanned by Affymetrix 428 in 10-μm resolution using Jaguar 2.0 software and processed by the Imagene 4.0 software (BioDiscovery, Segundo, USA) using the default settings. The raw numerical data files from Imagene were further processed using the GeneSpring 7.3 (Agilent) software package. A Lowess curve was fit to the log-intensity versus log-ratio plot. Twenty percent of the data was used to calculate the Lowess fit at each point. This curve was used to adjust the control value for each measurement.
The microarray data had been submitted to MIAMEexpress under accession no. E-MEXP-1841.
In vitro enzymatic assay of PfkA and FbaA proteins
The 6 × -histidine-tagged versions of the PfkA and FbaA proteins were purified from cells harboring the corresponding ASKA clone (Kitagawa et al, 2005), as previously described (Ishii et al, 2007b).
Briefly, 5 mM S7P (or F6P) was incubated with the indicated enzymes (2 μM) in 100 μl of 50 mM MOPS potassium buffer supplemented with 2 mM ATP at 37°C for1 h. All reactions, including an internal standard for analysis of metabolite recovery, were then diluted with ice-cold water. Protein was removed by ultra-filtration (cut-off of 5000 Da), and metabolites were quantified by CE-MS. Quantification of the metabolite with the m/z ratio of 367.0 (supposedly S1,7P) was performed by calculating the peak area/amount in relation to the F1,6P in the same CE-MS run. The average amount and s.d. of the metabolite level were calculated from five separate reaction samples and shown.
Supplementary Material
Supplementary Materials Table of Contents; Supplementary Text 1; Supplementary Figures S1-10; Supplementary Tables I, V-VII
Files for FBA
Growth and symulation datas
Transcript and metabolite levels
Mass isotopomer distribution, MFA models and results
Acknowledgments
We thank Kaori Igarashi, Chieko Kimura, Maki Oishi, and Yuko Narifusa for technical assistance, and Martin Robert for discussion and careful reading of the paper. We also thank the National BioResource Project (NIG, Japan): E. coli for providing JE5530 strain and pAH162 plasmid. This study was supported by funds from the Yamagata Prefectural Government and Tsuruoka City.
Footnotes
The authors declare that they have no conflict of interest.
References
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker SA, Feist AM, Mo ML, Hannum G, Palsson BØ, Herrgard MJ (2007) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc 2: 727–738 [DOI] [PubMed] [Google Scholar]
- Boone C, Bussey H, Andrews BJ (2007) Exploring genetic interactions and networks with yeast. Nat Rev Genet 8: 437–449 [DOI] [PubMed] [Google Scholar]
- Butland G, Babu M, Díaz-Mejía JJ, Bohdana F, Phanse S, Gold B, Yang W, Li J, Gagarinova AG, Pogoutse O, Mori H, Wanner BL, Lo H, Wasniewski J, Christopolous C, Ali M, Venn P, Safavi-Naini A, Sourour N, Caron S et al. (2008) eSGA: E. coli synthetic genetic array analysis. Nat Methods 5: 789–795 [DOI] [PubMed] [Google Scholar]
- Canonaco F, Hess TA, Heri S, Wang T, Szyperski T, Sauer U (2001) Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol Lett 204: 247–252 [DOI] [PubMed] [Google Scholar]
- Clark DP, Cronan JE (2005) Two-carbon compounds and fatty acids as carbon sources. In Escherichia coli and Salmonella: Cellular and Molecular Biology [Online], http://www.ecosal.org, Böck A, III RC, et al. (eds), Washington, DC: ASM Press [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher D, Meilijson I, Kupiec M, Ruppin E (2006) Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nat Genet 38: 993–998 [DOI] [PubMed] [Google Scholar]
- Duarte NC, Herrgard MJ, Palsson BØ (2004) Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res 14: 1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JS, Palsson BO (2000) Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions. BMC Bioinformatics 1: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandson KA, Park JH, Wissam, El Khal, Kao HH, Basaran P, Brydges S, Batt CA (2000) Dissolution of xylose metabolism in Lactococcus lactis. Appl Environ Microbiol 66: 3974–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BØ (2007) A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechner A, Gross W, Martin WF, Schnarrenberger C (1999) Chloroplast class I and class II aldolases are bifunctional for fructose-1,6-biphosphate and sedoheptulose-1,7-biphosphate cleavage in the Calvin cycle. FEBS Lett 447: 200–202 [DOI] [PubMed] [Google Scholar]
- Fong SS, Nanchen A, Palsson BO, Sauer U (2006) Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J Biol Chem 281: 8024–8033 [DOI] [PubMed] [Google Scholar]
- Fraenkel DG, Levisohn SR (1967) Glucose and gluconate metabolism in an Escherichia coli mutant lacking phosphoglucose isomerase. J Bacteriol 93: 1571–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman LK, Wanken A, Nickerson KW, Conway T (1998) Rapid accumulation of intracellular 2-keto-3-deoxy-6-phosphogluconate in an Entner–Doudoroff aldolase mutant results in bacteriostasis. FEMS Microbiol Lett 159: 261–266 [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Grossman CE, Qian Y, Banki K, Perl A (2004) ZNF143 mediates basal and tissue-specific expression of human transaldolase. J Biol Chem 279: 12190–12205 [DOI] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421: 63–66 [DOI] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL (2001) Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure–function studies of bacteria. J Bacteriol 183: 6384–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, Hirasawa T, Naba M, Hirai K, Hoque A, Ho PY, Kakazu Y, Sugawara K, Igarashi S, Harada S, Masuda T, Sugiyama N, Togashi T, Hasegawa M, Takai Y et al. (2007a) Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316: 593–597 [DOI] [PubMed] [Google Scholar]
- Ishii N, Suga Y, Hagiya A, Watanabe H, Mori H, Yoshino M, Tomita M (2007b) Dynamic simulation of an in vitro multi-enzyme system. FEBS Lett 581: 413–420 [DOI] [PubMed] [Google Scholar]
- Jiao Z, Baba T, Mori H, Shimizu K (2003) Analysis of metabolic and physiological responses to gnd knockout in Escherichia coli by using C-13 tracer experiment and enzyme activity measurement. FEMS Microbiol Lett 220: 295–301 [DOI] [PubMed] [Google Scholar]
- Joyce AR, Reed JL, White A, Edwards R, Osterman A, Baba T, Mori H, Lesely SA, Palsson BØ, Agarwalla S (2006) Experimental and computational assessment of conditionally essential genes in Escherichia coli. J Bacteriol 188: 8259–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Keseler IM, Shearer A, Latendresse M, Krummenacker M, Paley SM, Paulsen I, Collado-Vides J, Gama-Castro S, Peralta-Gil M, Santos-Zavaleta A, Peñaloza-Spínola MI, Bonavides-Martinez C, Ingraham J (2007) Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res 35: 7577–7590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J, Hashimoto M (2007) Construction of consecutive deletions of the Escherichia coli chromosome. Mol Syst Biol 3: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Bonavides-Martinez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, Peralta-Gil M, Santos-Zavaleta A, Shearer AG, Karp PD (2009) EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res 37: D464–D470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H (2005) Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 12: 291–299 [DOI] [PubMed] [Google Scholar]
- Kuznetsova E, Proudfoot M, Gonzalez CF, Brown G, Omelchenko MV, Borozan I, Carmel L, Wolf YI, Mori H, Savchenko AV, Arrowsmith CH, Koonin EV, Edwards AM, Yakunin AF (2006) Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem 281: 36149–36161 [DOI] [PubMed] [Google Scholar]
- Łobocka MB, Rose DJ, Plunkett G III, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR (2004) Genome of bacteriophage P1. J Bacteriol 186: 7032–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D et al. (2006) Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA 103: 15611–15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre LM, Thorburn DR, Bubb WA, Kuchel PW (1989) Comparison of computer simulations of the F-type and L-type non-oxidative hexose monophosphate shunts with 31P-NMR experimental data from human erythrocytes. Eur J Biochem 180: 399–420 [DOI] [PubMed] [Google Scholar]
- Miller JH (1992) A Short Course in Bacterial Genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press [Google Scholar]
- Ohara H, Owaki M, Sonomoto K (2007) Calculation of metabolic flow of xylose in Lactococcus lactis. J Biosci Bioeng 103: 92–94 [DOI] [PubMed] [Google Scholar]
- Oshima T, Wada C, Kawagoe Y, Ara T, Maeda M, Masuda Y, Hiraga S, Mori H (2002) Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol Microbiol 45: 673–695 [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD (2004) Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature 429: 661–664 [DOI] [PubMed] [Google Scholar]
- Peekhaus N, Conway T (1998) What's for dinner? Entner–Doudoroff metabolism in Escherichia coli. J Bacteriol 180: 3495–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Arauzo-Bravo MJ, Shimizu K (2004) Metabolic flux analysis for a ppc mutant Escherichia coli based on 13C-labelling experiments together with enzyme activity assays and intracellular metabolite measurements. FEMS Microbiol Lett 235: 17–23 [DOI] [PubMed] [Google Scholar]
- Perl A (2007) The pathogenesis of transaldolase deficiency. IUBMB Life 59: 365–373 [DOI] [PubMed] [Google Scholar]
- Reed JL, Patel TR, Chen KH, Joyce AR, Applebee MK, Herring CD, Bui OT, Knight EM, Fong SS, Palsson BO (2006) Systems approach to refining genome annotation. Proc Natl Acad Sci USA 103: 17480–17484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JL, Vo TD, Schilling CH, Palsson BO (2003) An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol 4: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo T, Snoep JL (2005) Glycolysis and Flux Control. In Escherichia coli and Salmonella: Cellular and Molecular Biology [Online], http://www.ecosal.org Böck A, III RC, et al. (eds). Washington, DC: ASM Press [Google Scholar]
- Rozen S, Skaketsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols, Krawetz S, Misener S (eds), pp 365–386. Totowa: Humana Press [DOI] [PubMed] [Google Scholar]
- Segre D, Vitkup D, Church GM (2002) Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci USA 99: 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Berkman O, Ruppin E (2005) Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proc Natl Acad Sci USA 102: 7695–7700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soga T, Baran R, Suematsu M, Ueno Y, Ikeda S, Sakurakawa T, Kakazu Y, Ishikawa T, Robert M, Nishioka T, Tomita M (2006) Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J Biol Chem 281: 16768–16776 [DOI] [PubMed] [Google Scholar]
- Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T (2003) Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res 2: 488–494 [DOI] [PubMed] [Google Scholar]
- Sørensen KI, Hove-Jensen B (1996) Ribose catabolism of Escherichia coli: characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression. J Bacteriol 178: 1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger GA (1995) Genetics of pentose-phosphate pathway enzymes of Escherichia coli K-12. Arch Microbiol 164: 324–330 [DOI] [PubMed] [Google Scholar]
- Susskind BM, Warren LG, Reeves RE (1982) A pathway for the interconversion of hexose and pentose in the parasitic amoeba Entamoeba histolytica. Biochem J 204: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toya Y, Ishii N, Hirasawa T, Naba M, Hirai K, Sugawara K, Igarashi S, Shimizu K, Tomita M, Soga T (2007) Direct measurement of isotopomer of intracellular metabolites using capillary electrophoresis time-of-flight mass spectrometry for efficient metabolic flux analysis. J Chromatogr A 1159: 134–141 [DOI] [PubMed] [Google Scholar]
- Typas A, Nichols RJ, Siegele DA, Shales M, Collins SR, Lim B, Braberg H, Yamamoto N, Takeuchi R, Wanner BL, Mori H, Weissman JS, Krogan NJ, Gross CA (2008) High-throughput, quantitative analyses of genetic interactions in E. coli. Nat Methods 5: 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B (1994) Gene expression in bacteria using TnphoA and TnphoA0 elements to make and switch phoA gene, lacZ (op), and lacZ (pr) fusions. In Methods in Molecular Genetics, Adolph K (ed), Vol. 3, pp 291–310. Orlando: Academic Press [Google Scholar]
- Westberg J, Persson A, Holmberg A, Goesmann A, Lundeberg J, Johansson KE, Pettersson B, Uhlén M (2004) The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1T, the causative agent of contagious bovine pleuropneumonia (CBPP). Genome Res 14: 221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JF, Blackmore PF, Clark MG (1978) New reaction sequences for the non-oxidative pentose phosphate pathway. Biochem J 176: 257–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Hua Q, Baba T, Mori H, Shimizu K (2003) Analysis of Escherichia coli anaplerotic metabolism and its regulation mechanisms from the metabolic responses to altered dilution rates and phosphoenolpyruvate carboxykinase knockout. Biotechnol Bioeng 84: 129–144 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials Table of Contents; Supplementary Text 1; Supplementary Figures S1-10; Supplementary Tables I, V-VII
Files for FBA
Growth and symulation datas
Transcript and metabolite levels
Mass isotopomer distribution, MFA models and results