Abstract
Neuronal depolarization and CaM kinase IV signaling alter the splicing of multiple exons in transcripts for ion channels, neurotransmitter receptors, and other synaptic proteins. These splicing changes are mediated in part by special CaM kinase-responsive RNA elements, within or adjacent to exons that are repressed in the initial phase of chronic depolarization. The splicing of many neuronal transcripts is also regulated by members of the Fox (Feminizing gene on X) protein family, and these Fox targets are also often proteins affecting synaptic activity. We show that Fox-1/Ataxin 2-Binding Protein 1 (A2BP1), a protein implicated in a variety of neurological diseases, can counteract the effects of chronic depolarization on splicing. We find that exon 19 of Fox-1 is itself repressed by depolarization. Fox-1 transcripts missing exon 19 encode a nuclear isoform of Fox-1 that progressively replaces the cytoplasmic Fox-1 isoform as cells are maintained depolarizing media. The resulting increase in nuclear Fox-1 leads to the reactivation of many Fox-1 target exons, including exon 5 of the NMDA receptor 1, that were initially repressed by the high-KCl medium. These results reveal a novel mechanism for the slow modulation of splicing as cells adapt to chronic stimuli: The subcellular localization of a splicing regulator is controlled through its own alternative splicing.
Keywords: Depolarization, alternative splicing, A2bp1/Fox-1, neuronal exons
Alternative pre-mRNA splicing is an important mechanism for controlling gene expression in metazoan organisms. Changes in splice site or exon usage frequently determine the function of a gene product or eliminate its expression (Black 2003; Matlin et al. 2005; Blencowe 2006). The best understood systems of splicing regulation are those modulated by cell type, where stable differences in pre-mRNA-binding protein expression determine a splicing choice. However, splicing is also dynamically regulated by external stimuli and growth conditions. Most cellular signaling pathways that alter gene transcription also alter groups of alternative exons (Shin and Manley 2004; Blaustein et al. 2007; Stamm 2008). In some cases, these responses have been traced to particular regulatory elements in the pre-mRNA, to post-translational modifications of splicing regulators, or to changes in transcriptional control. However, the mechanisms that allow the splicing machinery to respond to dynamic stimuli are not well understood.
A particularly interesting regulatory mechanism in excitable cells is their response to membrane depolarization. Depolarization-induced changes in neuronal transcription are well studied and involve the activation of CaM kinase and other signaling pathways to induce transcription of both rapid primary response genes and more slowly developing secondary responses (Flavell and Greenberg 2008; Greer and Greenberg 2008). These transcriptional programs play important roles in neuronal plasticity and modulating synaptic activation. Ion channel mRNAs and other transcripts affecting membrane activity also often contain exons whose splicing responds to chronic depolarizing stimuli (Li et al. 2007; McKee et al. 2007). Two examples of depolarization and CaM kinase IV (CAMKIV)-dependent splicing are exons 5 and 21 in the NMDA receptor 1 transcript (NR1), a glutamate receptor that modulates processes of synaptic plasticity (Zukin and Bennett 1995; Cull-Candy et al. 2001; Llansola et al. 2005). Alternative exon 5 encodes the N1 cassette in the extracellular domain that regulates agonist binding and other pharmacological properties of the receptor (Traynelis et al. 1995; Prybylowski et al. 2000; Rumbaugh et al. 2000). Alternative exon 21 encodes the C1 peptide cassette in the intracellular domain that regulates the membrane delivery and protein–protein interactions of the receptor, as well as its ability to signal to the nucleus following stimulation (Tingley et al. 1993; Ehlers et al. 1998; Standley et al. 2000; Scott et al. 2001; Bradley et al. 2006).
Several elements were identified in the two NR1 exons that mediate their inducible splicing repression. A type 1 CaMKIV-responsive RNA element (CaRRE1) is present in the 3′ splice site of exon 5, and type 1 and type 2 CaRREs are both found within exon 21 itself (Xie and Black 2001; Xie et al. 2005; Lee et al. 2007). Other depolarization-sensitive motifs in NR1 exon 21 are binding sites (UAGG) for the splicing repressor hnRNP A1 (An and Grabowski 2007). The RNA-binding protein hnRNP L binds to CaRRE1 elements, and may in part mediate their effect (Yu et al. 2009). Proteins that bind to CaRRE2 elements are not identified, and how the activity of any of these proteins is altered by depolarization to affect splicing is not known.
One protein that controls many neuronally regulated exons is Feminizing gene 1 on X (Fox-1; also called Ataxin 2-Binding Protein 1 [A2BP1]), a mammalian homolog of the Caenorhabditis elegans Feminizing gene 1 on X (Jin et al. 2003; Nakahata and Kawamoto 2005; Underwood et al. 2005; Kuroyanagi et al. 2006; Li et al. 2007; Zhou et al. 2007). Fox-1 (A2BP1) also has two homologs in mammalian genomes Fox-2 (RBM9) and Fox-3 (hrnbp3). Fox-1 is specifically expressed in neurons, heart, and muscle. Fox-2 is also expressed in these tissues, but is also found in other cell types including stem cells and hematopoetic cells (Ponthier et al. 2006; Yeo et al. 2007). Fox-3 appears to be expressed exclusively in neurons (McKee et al. 2005). In addition to the three genes, the mammalian Fox protein family is further diversified through the use of multiple promoters and extensive alternative splicing to produce a wide variety of isoforms. These proteins each contain a single highly conserved RNA-binding domain that specifically recognizes the RNA hexanucleotide UGCAUG (Jin et al. 2003; Nakahata and Kawamoto 2005; Underwood et al. 2005; Auweter et al. 2006; Ponthier et al. 2006). This Fox-binding element acts as a splicing enhancer in the intron downstream from many exons, but can induce exon skipping when present in the upstream intron (Black 1992; Huh and Hynes 1994; Modafferi and Black 1997; Jin et al. 2003; Underwood et al. 2005; Zhang et al. 2008; Yeo et al. 2009).
Exons controlled by Fox proteins are found in many genes important for neuronal development and mature function, including NR1 exon 5 (Brudno et al. 2001; Minovitsky et al. 2005; Nakahata and Kawamoto 2005; Underwood et al. 2005; Zhang et al. 2008; Yeo et al. 2009). The significance of this regulation is underscored by the identification of human mutations in the Fox-1 gene that lead to severe neurodevelopmental phenotypes exhibiting mental retardation, epilepsy, and autism spectrum disorder (Bhalla et al. 2004; Barnby et al. 2005; Martin et al. 2007; Sebat et al. 2007). Moreover, Fox-1 (A2BP1) was first identified through its interaction with Ataxin 2, the protein mutated in Spinal Cerebellar Ataxia type 2 (SCA2), and Fox-2 was identified as interacting with Ataxin 1, which is mutated in SCA1 patients (Shibata et al. 2000; Lim et al. 2006). Thus, there is great interest in how Fox-mediated splicing regulation might contribute to human neurologic disease.
We show here that Fox-1 controls a secondary splicing response to cellular depolarization. Chronic treatment with high-KCl media leads to increased levels of nuclear Fox-1 isoforms through changes in the splicing of the Fox-1 transcript itself. This relocalization of Fox-1 to the nucleus increases the splicing of multiple Fox target exons, including NR1 exon 5.
Results
Fox-1 exon 19 (E19) is repressed after depolarization
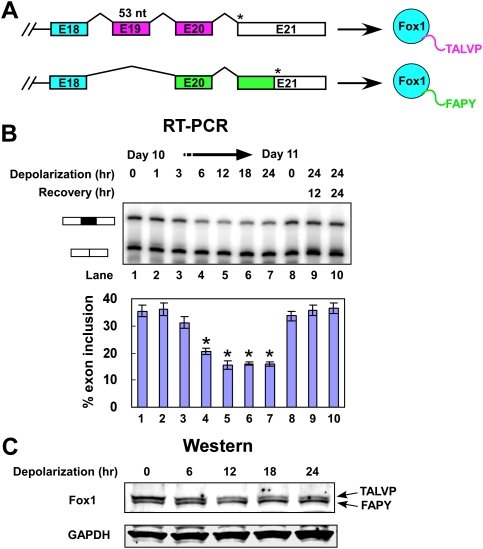
In a survey of exons that respond to depolarizing stimuli we found such an exon in the Fox-1/A2BP1 transcript. Fox-1 E19 is 53 nucleotides (nt) long, and its alternative use causes a frameshift in the 3′ portion of the mRNA to generate protein isoforms with different C termini (Fig. 1A; Nakahata and Kawamoto 2005; Underwood et al. 2005). The E19-included isoform has a C terminus ending with the residues TALVP (Fox-1-TALVP), whereas the E19-skipped isoform terminates with FAPY (Fox-1-FAPY). Sequences related to the FAPY sequence are observed in the Fox proteins of all species, including worms and flies (Nicoll et al. 1997; Jin et al. 2003; Kuroyanagi et al. 2006). The splicing that gives rise to an alternate Fox-1 TALVP terminus is conserved across vertebrate species including fish, and related alternative C termini are also observed in the Fox-2 and Fox-3 genes.
Figure 1.
Depolarization represses splicing of Fox-1 E19 in differentiated P19 cells. (A) Diagram of the alternative splicing within the 3′ region of the Fox-1 pre-mRNA. Alternative splicing of E19 alters the reading frame to generate two C-terminal peptide sequences, depicted as purple and green bars in the pre-mRNA. Asterisks (*) indicate the stop codons in the transcripts. The C-terminal peptides are indicated to the right. (B) Denaturing gel electrophoresis of Fox-1 RT–PCR products. The E19-included and E19-excluded bands are indicated. (Lanes 1–7) P19 cells were differentiated for 10 d and then depolarized for 0, 1, 3, 6, 12, 18, 24 h. Also shown are RNA from day 11 untreated cells (lane 8), and from cells depolarized for 24 h followed by recovery for 12 and 24 h (lanes 9,10). A graph of E19 inclusion is shown below the gel. Error bars represent SD; n = 3. Asterisks denote significant changes in splicing, indicated by a t-test P-value of <0.05. (C) Immunoblot analysis of Fox-1 collected from the same cells as in B. The identities of the two Fox-1 isoforms are indicated, and GAPDH was used as loading control for total protein. The protein samples were separated on a 4%–20% gradient gel.
The mouse Fox-1 gene is ∼2 MB long and is not expressed in most cell lines. To examine the effect of depolarization on Fox-1 splicing, we used mouse P19 embryonal carcinoma cells. Aggregation of these cells in the presence of retinoic acid and replating leads to efficient production of highly differentiated post-mitotic neuronal cells that express Fox-1 as well as most late differentiation markers for neurons, and that develop extensive fasciculated and highly connected axonal and dendritic processes (Finley et al. 1996; Lin et al. 1996; Ulrich and Majumder 2006). Unlike most primary neurons, these cells tolerate culture in high-KCl media and inhibition of CaMK pathways.
In the untreated P19 cells, Fox-1 E19 is included in 36% of the spliced mRNA. To examine the kinetics of E19 splicing repression, RT–PCR was performed on mRNA from cells depolarized in 50 mM KCl for 1–24 h. A decrease in E19 inclusion was observed within 6 h of treatment, and reached 16% inclusion between 12 and 24 h (Fig. 1B, lanes 1–7). To test the reversibility of this repression, KCl was washed out after 24 h of depolarization and the cells were cultured for an additional 12 or 24 h in normal media. As was observed previously for NR1 E21 splicing, Fox-1 E19 splicing was restored to its normal level after 12 h of recovery (Fig. 1B, lanes 9,10; Lee et al. 2007). An increase in cell death was not observed in the depolarizing media, and the recovery of the splicing after depolarization further indicated that the cells remain fully viable after these treatments. Thus, the repression of Fox-1 E19 is apparently a dynamic splicing response of cells adapting to the depolarizing media.
We found previously that CaMK signaling pathways are required for the depolarization-induced repression of NR1 E21 and other exons (Xie and Black 2001; Xie et al. 2005; Lee et al. 2007). Treatment of the cells with the CaMK inhibitor KN93 prior to depolarization also completely blocked Fox-1 E19 splicing repression (data not shown), indicating that repression of E19 is also mediated by CaMK pathways. Interestingly, E19 does not have apparent CaRRE1 or CaRRE2 elements, implying a different pathway of regulation from the NR1 and BK channel exons, where the CaRREs were identified (Xie and Black 2001; Xie et al. 2005; Lee et al. 2007). The predominant Fox-1 isoforms expressed in differentiated P19 cells migrate as a closely spaced doublet observed by immunoblot (Fig. 1C). The mobility difference of the two bands is equivalent to the difference seen for Flag-tagged Fox-1-TALVP and Fox-1-FAPY proteins separated by SDS-PAGE, with the FAPY form migrating faster, even though the Fox-1-TALVP protein has a slightly lower molecular weight (see Fig. 3A, below). Consistent with the RNA analysis, quantification of the protein bands over time confirmed that the Fox-1-TALVP form (Fig. 1C, top band) was more prevalent in untreated cells and gradually decreased after depolarization. The Fox-1-FAPY form (Fig. 1C, bottom band) increased during depolarization, becoming the more prevalent form by 18–24 h (Fig. 1C).
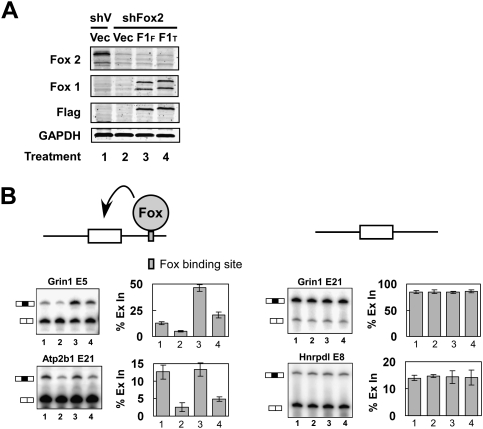
Figure 3.
Fox-1-FAPY activates splicing of exons carrying downstream Fox-binding sites. (A) Immunoblot of Fox-2 and Fox-1 proteins in N2A cells. N2A cells were transfected with a plasmid expressing a shRNA targeting the Fox-2 3′ UTR (shFox2) or with a control vector (shV). Forty-eight hours after shRNA transfection, N2A cells were retransfected with Flag-Fox-1-FAPY (F1F), Flag-Fox-1-TALVP (F1T), or control (Vec) expression vectors; incubated for 24 h; and immunoblotted for Fox-2 and Fox-1 proteins. GAPDH served as a loading control. (B) RT–PCR analyses of exons carrying a Fox-binding site, UGCAUG (blue box) in the downstream intron (left panels), and control exons that lack Fox-binding sites (right panels). The exon IDs are shown above the gels. The cell conditions are as in A. Graphs of percent exon inclusion (% Ex In) are shown to the right of the gels. Error bars represent SD; n = 3.
Depolarization changes Fox-1 localization to increase its splicing activity
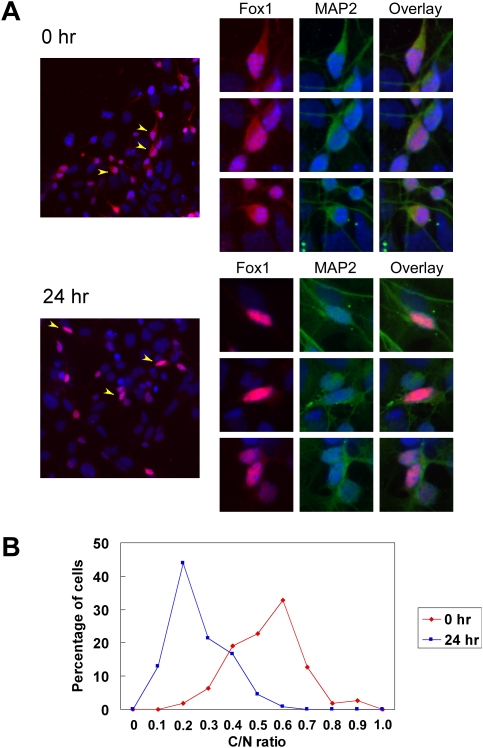
We next wanted to understand how the change in Fox-1 E19 splicing affected Fox-1 activity. It was shown previously that overexpressed Fox-1-FAPY protein is localized to both the nucleus and the cytoplasm, whereas the Fox-1-TALVP protein is mostly cytoplasmic when expressed in HeLa or Y79 cells (Nakahata and Kawamoto 2005). To confirm these observations, Flag-tagged Fox-1-FAPY and Fox-1-TALVP proteins were expressed in N2A and HEK cells, and were examined by immunostaining with anti-Flag antibody (Supplemental Fig. 1). In both cell types, Flag-Fox-1-FAPY was predominantly nuclear, and the Flag-Fox-1-TALVP was predominately cytoplasmic. Thus, the switch from Fox-1-TALVP to Fox-1-FAPY after depolarization should lead to an increase in the nuclear concentration of endogenous Fox-1 protein. To examine this, we compared untreated P19 cells and cells depolarized for 24 h by immunostaining with anti-Fox-1 antibody. Differentiated P19 cultures contain multiple cell types and, as expected, Fox-1 was seen only in the neuronal cells of the culture. In the untreated cells, endogenous Fox-1 protein is localized to both cytoplasm and nucleus, consistent with the splicing pattern of E19 (Fig. 2A). After incubation in depolarizing media, Fox-1 protein became predominantly nuclear with faint staining in the cytoplasm. The ratio of cytoplasmic to nuclear (C/N) staining was measured in 110 untreated and 132 depolarized cells, and was plotted in Figure 2B. The values for the C/N ratio under each condition were distributed normally, with a clear shift in these distributions upon depolarization. The mean C/N ratio shifted from 0.6 before treatment to 0.2 after depolarization. A two-sample t-test examining the likelihood that these distributions are the same yields a P-value that is very close to 0 (2.82 × 10−41). Thus, the endogenous Fox-1 protein is strongly relocalized to the nucleus upon depolarization.
Figure 2.
The subcellular distribution of Fox-1 protein is changed after depolarization. (A) Immunostaining of Fox-1 protein in differentiated P19 cells. Fox-1 (red) is found in the cytoplasm and nucleus in untreated cells (0 h, top left panel) and becomes mainly nuclear after depolarization (24 h of depolarization, bottom left panel). Three representative cells from each field (arrowheads) are shown at the right, with the individual channels of Fox-1 (red), cytoplasmic neuronal marker MAP2 (green), and the overlay indicated. DNA stain DAPI (blue) is shown in every panel. (B) The ratio of C/N Fox-1 was measured in 110 rested cells (0 h) and 132 depolarized cells (24 h). The C/N ratio significantly decreases after depolarization, with the peak ratio shifting from 0.6 in the rested cells to 0.2 in the depolarized cells (P-value = 2.82 × 10−41 from a two-sample t-test).
The different subcellular locations of the two Fox-1 isoforms should lead to differences in their ability to alter splicing. We examined this by expressing each isoform in cells and assaying Fox-dependent exon splicing. N2A cells express Fox-2 protein but not Fox-1 protein (Fig. 3A, lane 1). This Fox-2 expression was mostly eliminated by expression of a shRNA targeting the 3′ untranslated region (UTR) of Fox-2 transcripts (Fig. 3A, lane 2). These cells were then complemented with either Flag-Fox-1-FAPY or Flag-Fox-1-TALVP. Immunoblots with anti-Flag antibody indicated that the two isoforms were expressed nearly equally (Fig. 3A, lanes 3,4). Each isoform gave rise to two protein bands recognized by anti-Fox-1 antibody, where the lower band is presumably an N-terminal truncated protein as it does not bind the Flag antibody.
We next measured the splicing of Fox-dependent exons in the presence of these two Fox-1 isoforms. NR1 exon 5 (gene name Grin1) contains a UGCAUG element at position +8 of the downstream intron. This exon is 13% included in untreated N2A cells and reduced to 5% inclusion in the Fox-2-depleted cells. Complementation of these cells with mouse Fox-1-FAPY enhanced exon 5 inclusion to 46%, whereas the same amount of Fox-1-TALVP increased exon inclusion to only 20% (Fig. 3B). This lower activity is presumably due to the smaller portion Fox-1-TALVP protein that is nuclear-localized (Supplemental Fig. 1). Similar effects were observed with three other exons that contain a UGCAUG element in their downstream intron (Atp2b1, Adcyap1r1, and Nf2) (Fig. 3B; data not shown). In all cases, knocking down the Fox-2 protein reduced the exon inclusion and the Fox-1-FAPY isoform had a stronger activity in restoring this splicing than the Fox-1-TALVP isoform. Two control exons, NR1 E21 (Grin1) and Hhrpdl E8, were also tested. These exons are known to change in splicing with depolarization but do not carry Fox-binding sites. Neither of these exons was affected by the knockdown or re-expression of the Fox proteins (Fig. 3B).
The relocalization of endogenous Fox-1 alters splicing
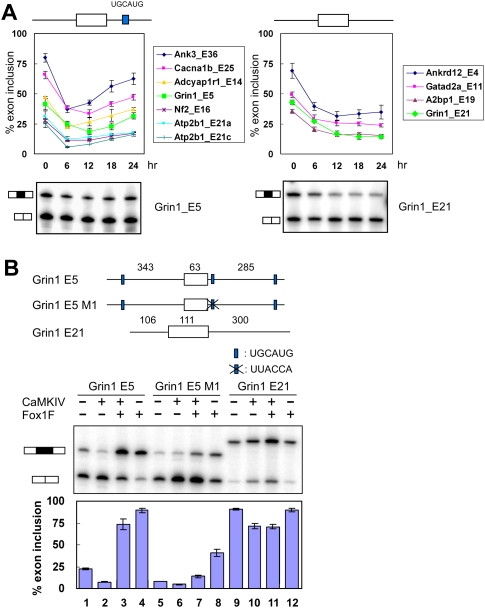
NR1 exon 5 is repressed by depolarization but carries a Fox site in its downstream intron that enhances splicing. From a group of depolarization-regulated exons, we found several others that contain Fox-binding sites in their downstream introns. For these exons, the change to the Fox-1-FAPY isoform should counteract the effect of depolarization. However, this restoration of splicing should occur after a delay during which Fox-1 E19 splicing is altered and the new nuclear Fox-1 protein is synthesized. To assess this, we analyzed a group of exons for the time course of their splicing change after depolarization, and compared exons carrying downstream Fox sites (within 200 nt) with exons that did not. These two groups showed different temporal profiles of splicing (Fig. 4A). The behavior of exons carrying downstream Fox-1 sites is typified by NR1/Grin1 exon 5, whereas exons without Fox-1-binding sites are typified by NR1/Grin1 E21. Nearly all exons showed a maximum repression within 6–8 h after the shift to depolarizing media. For exons without Fox sites, this repressed level of splicing was maintained as long as the cells remained in depolarizing media (Fig. 4A, right panel; Supplemental Fig. 2). In contrast, exons carrying downstream Fox-binding sites were initially repressed in depolarizing media, but then recovered. Their splicing begins to increase after 6 h, coincident with the nuclear accumulation of Fox-1-FAPY that occurs after this point (Fig. 4A, left panel; Supplemental Fig. 2). The magnitude of their recovery correlates with the level of their initial repression by depolarization. For these exons, the relocalization of Fox-1 has an apparent homeostatic effect that restores their splicing after depolarization-induced repression.
Figure 4.
Fox-1 antagonizes depolarization and CaMKIV-induced splicing repression. (A) The splicing repression of Fox-dependent exons is relieved during chronic depolarization. The inclusion levels of exons carrying a UGCAUG element within the first 200 nt of the downstream intron are shown in the left graph. The splicing of exons without Fox-binding sites are shown in the right graph. Exon inclusion was measured in untreated cells (0 h) and in cells treated with 50 mM KCl for 6, 12, 18, and 24 h. Error bars represent SD; n = 3. Representative RT–PCR gels for NR1 (Grin1) exon 5 and E21 are shown below the graph. Other gels are shown in Supplemental Figure 2. (B) Fox-1 can antagonize CaMKIV-induced repression. (Top panel) NR1 exon 5 and E21 were cloned into the pDup 4-1 splicing reporter. The lengths of exon and intron segments are indicated above the diagram. The three UGCAUG elements adjacent to exon 5 are indicated by blue boxes. The UGCAUG element proximal to the 5′ splice site was mutated to UUACCA in the exon 5 M1 construct. (Bottom panel) Primer extension assay of splicing reporters coexpressed with CaMKIV dCT (CaMKIV) and/or Fox-1-FAPY (Fox1F) expression constructs. The percent exon inclusion is shown below the gels. Error bars represent SD; n = 3.
Exons with upstream Fox-binding sites are often negatively regulated by Fox proteins and might be expected to be repressed by an increase in nuclear Fox-1 protein (Jin et al. 2003; Underwood et al. 2005; Zhang et al. 2008; Yeo et al. 2009). However, to see such an effect, splicing of the exon would need to increase rather than decrease during the initial phase of depolarization. We found one exon that shows this behavior. L-type calcium channel CaV1.2 exon 9* is negatively regulated by Fox proteins from binding sites both upstream and within the exon (Tang et al. 2009). This exon increased in splicing immediately after depolarization and then decreased as Fox-1 protein became more nuclear (data not shown).
We showed previously that these two NR1 exons respond to depolarization through the activation of calcium signaling pathways and specifically CaMKIV (Xie and Black 2001; Xie et al. 2005; Lee et al. 2007). To look at the ability of Fox-1 to counteract CaMKIV-mediated repression, we examined the splicing of an NR1 exon 5 minigene after cotransfection with CaMKIV and/or Fox-1-FAPY genes. This was compared with the splicing of a mutant exon missing one Fox site, and to NR1 E21, which does not have a Fox site. A mutation in the UGCAUG at position +8 downstream of exon 5 was generated, leaving additional Fox sites intact at position +262 in this intron and at position −319 in the upstream intron. The minigenes were transiently expressed in HEK 293T cells and assayed for exon 5 inclusion by primer extension. Without other expression plasmids, exon 5 is included in 22% of the minigene transcripts. Coexpression of constitutively active CaMKIV reduced this to 7% inclusion (Fig. 4B, lanes 1,2). This CaMKIV-induced repression is mediated by a CaRRE1 element in the 3′ splice site (Xie et al. 2005). Expression of Fox-1-FAPY reversed the induced repression by CaMKIV and strongly stimulated splicing (Fig. 4B, lane 3). Mutation of the binding site at position +8 significantly reduced the effect of Fox-1 on exon 5 splicing (Fig. 4B, lane 7). A control exon, NR1 E21, was repressed by CaMKIV, but this repression was not affected by the expression of Fox-1-FAPY (Fig. 4B, lane 11). Thus, Fox-1 protein can strongly counteract CaMKIV and depolarization-induced repression of splicing through its downstream binding site in NR1 exon 5.
Discussion
In previously described examples of splicing regulatory cascades, such as the Drosophila sex determination pathway, the regulatory module of the splicing factor acts as an on/off switch for its function (Black 2003). Here, the splicing alteration changes the nucleocytoplasmic balance of the splicing regulator, which in turn affects an ensemble of Fox target exons in transcripts important for neuronal function. The initial induced splicing change in Fox-1 leading to subsequent downstream changes in other spliced isoforms is analogous to a transcriptional primary response gene whose induction activates downstream target genes in a temporal program of transcriptional regulation. The time scale of this splicing regulatory cascade is slower than a typical transcriptional response, such as c-fos or CREB induction (West et al. 2001). Rather than a transient response to a brief stimulus, Fox-1 apparently mediates a slower cellular homeostatic mechanism through the modulation of its nucleocytoplasmic distribution.
The change in Fox-1 subcellular localization is also different from a typical change in the karyopherin-mediated nuclear transport of an existing protein (Terry et al. 2007). Such phosphorylation-mediated changes in distribution have been seen in the splicing factors hnRNP A1, Tra2β, PTB, and Sam68 (van der Houven van Oordt et al. 2000; Daoud et al. 2002; Xie et al. 2003; Ben Fredj et al. 2004; Allemand et al. 2005). With the exception of Tra2β, the relocalization of these proteins has not been shown to affect the splicing of endogenous transcripts and may be more important for their effects on translational control. Here, the splicing of a defined group of target transcripts clearly changes after Fox-1 relocalization, and the mechanism controlling the nucleocytoplasmic distribution of Fox-1 is notably different. Instead of the existing Fox-1 protein being relocalized due to modification of a nuclear localization sequence (NLS), the Fox-1-FAPY protein that is newly synthesized after depolarization will be taken up by the nucleus more readily than the pre-existing Fox-1-TALVP protein, and the nucleocytoplasmic ratio will change only as the pre-existing protein turns over. This does not rule out more rapid changes in Fox-1 localization also occurring in response to typical NLS modification. It remains to be determined whether the FAPY terminus actually mediates nuclear uptake, or if the NLS is elsewhere in the protein and the TALVP form is actively retained in the cytoplasm. Since the increase in nuclear Fox-1-FAPY protein is accompanied by a decrease in the cytoplasmic TALVP form, it will also be interesting to explore the consequences of this loss of cytoplasmic Fox-1 for translational regulation.
The two alternative exons of NR1 studied here exhibit different temporal responses to depolarization, and illustrate overlapping but independent control pathways for exons within the same transcript. Exon 5 is initially repressed due to the presence of a CaRRE1 in its 3′ splice site. Its inclusion level then increases due to the increase in nuclear Fox-1 after depolarization (Fig. 5). In contrast, E21 is repressed by depolarization due to CaRRE types 1 and 2 and hnRNP A1 sites within the exon itself. This exon lacks a Fox-1 site and maintains its repressed state unless the cells are shifted back to low-KCl media. Exon 5 adds the N1 peptide cassette to the extracellular domain of the receptor and alters its responses to glycine, protons, and other ligands (Traynelis et al. 1995; Prybylowski et al. 2000; Rumbaugh et al. 2000). E21 encodes the C1 cassette within the intracellular domain that is required for NMDA receptor-induced gene expression, among other functions (Tingley et al. 1993; Ehlers et al. 1998; Standley et al. 2000; Cull-Candy et al. 2001; Scott et al. 2001; Bradley et al. 2006). In other studies, changes in 3′ splice site choice within exon 22 of NR1 have been proposed to modulate receptor trafficking to the plasma membrane in a model for homeostatic plasticity (Mu et al. 2003; Perez-Otano and Ehlers 2005). These Fox-1-dependent and Fox-1-independent changes in splicing will clearly affect NR1 function, and their consequences need further elucidation.
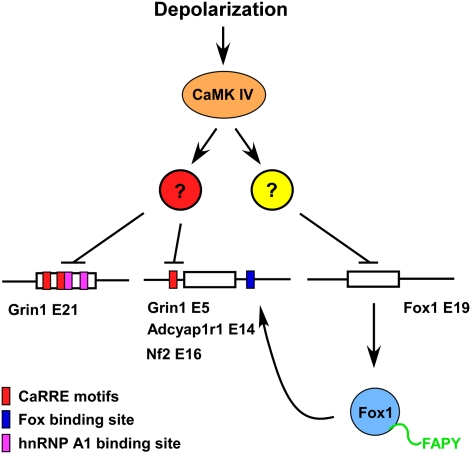
Figure 5.
Model for regulation of splicing recovery by Fox-1. The splicing of NR1 exon 5 and E21, Fox-1 E19, and many other exons is repressed by depolarization through a CaMKIV-dependent pathway and the effects of hnRNP A1. For Fox-1, this leads to increased expression of the nuclear Fox-1-FAPY isoform, which alleviates the repression of NR1 exon 5 and other exons carrying Fox-binding sites. The change in nuclear Fox-1 does not effect NR1 E21 or other exons lacking Fox-binding sites.
We also identified a number of exons in calcium and other ion channel proteins that are regulated by Fox1 and Fox2 (Tang et al. 2009; Z Tang and DL Black, unpub.). These ion channel targets point to the importance of examining the localization of Fox-1 and the splicing of Fox-1 target exons in settings that allow measurements of membrane physiology. Individual alternative protein isoforms of ion channels and neurotransmitter receptors have been shown to affect various types of long-term potentiation, and also to adapt physiological responses to specific cell types or conditions (Xie and McCobb 1998; Beffert et al. 2005; Huang et al. 2005; Lipscombe 2005; Li et al. 2007; Lipscombe and Raingo 2007; Raingo et al. 2007). However, changes in spliced isoform ratios depend on rates of mRNA decay, and are usually measured over the course of hours in response to chronic stimuli such as depolarizing media or drugs that block or stimulate synaptic activity. This slow time course of most splicing responses is compatible with longer-term changes in neuronal activity, such as circadian modulation of excitation, or circuit consolidation during development. Many splicing events, including NR1 exon 5, are seen to change during critical periods of circuit refinement in postnatal development, or in response to chronic stimuli such as kindling or other induced seizures in adult animals (Kamphuis et al. 1992; Laurie and Seeburg 1994; Kraus et al. 1996; Wang and Grabowski 1996; Rafiki et al. 1998; Ying et al. 1998; Daoud et al. 1999; Hoffmann et al. 2000; Musshoff et al. 2000; Prybylowski and Wolfe 2000; Yin et al. 2001; Bottai et al. 2002; Llansola et al. 2005; Jaekel et al. 2006). The role of Fox-mediated alternative splicing in modulating changes in membrane physiology during these processes thus invites further study.
How Fox-1-mediated splicing affects neuronal function is also a key issue for understanding its role in neurological disease. The interaction of Fox-1/A2BP1 with Ataxin 2 needs to be explored further. Ataxin 2 is found in the cytoplasm, and has been implicated in translational regulation (Kiehl et al. 2000; Ciosk et al. 2004; Nonhoff et al. 2007). Thus, the loss of Ataxin 2 in spinal cerebellar ataxia patients may affect cytoplasmic functions of Fox-1. Mutations in the Fox-1 gene itself have been observed in patients with severe epileptic, mental retardation, and autism spectrum disorders (Bhalla et al. 2004; Barnby et al. 2005; Martin et al. 2007; Sebat et al. 2007). The role of the Fox-1 splicing program in cellular adaptive responses provides an intriguing clue to the underlying physiology of these neurological disorders.
Materials and methods
Cell culture
P19 cells were differentiated into post-mitotic neuronal cells as described previously (Lee et al. 2007). Briefly, P19 cells were grown in α-MEM with 10% fetal bovine serum (FBS). Differentiation was induced by transfer to medium containing α-MEM, 5% FBS, and 0.5 μM all-trans retinoic acid (R-2625, Sigma Aldrich) in bacteriological Petri dishes to promote cell aggregation. After 2 d, cell aggregates were transferred to fresh induction medium and cultured for another 2 d. The cell aggregates were then trypsinized, and 107 cells were plated per 100-mm tissue culture plate in α-MEM plus 10% FBS. The tissue culture plates were coated previously overnight with 0.1 mg/mL poly-L-lysine in 1.25% boric acid/1.91% sodium tetraborate solution (1:1; pH 8.2), and washed three times with sterilized water. Two days after plating, the medium was changed to Neurobasal-A medium (GIBCO) with 1× B27 supplement (GIBCO), and maintained through the experiment. For depolarization, 10-d-old cultures of differentiated P19 cells were shifted to medium containing 50 mM KCl for 1–24 h. For recovery, 24 h after depolarization, the KCl-containing medium was replaced with fresh culture medium, and the cells were grown for another 12 or 24 h. HEK 293T cells and N2A cells were grown in DMEM with 10% FBS.
RT–PCR assay
One microgram of the total RNA was reverse-transcribed with random hexamers. One-tenth of this reaction was then amplified in 22–25 cycles of PCR with exon-specific primers, one of which was 32P-labeled. The PCR products were resolved on 8% polyacrylamide/7.5 M urea denaturing gels. The gel was dried, exposed, and scanned in a PhosphorImager (Fuji Medical Systems). PCR products were also isolated and confirmed by sequencing.
Western blotting
Immunoblotting was performed on cell lysates isolated in RIPA buffer containing benzonase (Sigma) and incubated for 30 min on ice to remove genomic DNA. Antibodies were used at the following dilutions: α-Fox-1 1D10, 1:1500; α-Fox-2, 1:2500; α-Flag, 1:5000 (Sigma); α-GAPDH, 1:400,000 (Fitzgerald); and ECL Plex C3-conjugated goat α-mouse and C5-conjugated goat α-rabbit secondary antibodies, 1:2500 (GE Healthcare).
Immunostaining and analysis
Cells were grown on coverslips coated overnight with 0.1 mg/mL ploy-L-lysine in 1.25% boric acid/1.91% sodium tetraborate solution (1:1; pH 8.2), and washed three times with sterilized water. All manipulations were performed at room temperature unless otherwise noted (Hall et al. 2004). Cells were fixed in freshly prepared 4% paraformaldehyde in 1× PBS (pH 7.4) for 20 min, and then washed once with 1× PBS and twice with 1× PBS with 0.1% Triton X-100. Cells were permeabilized in 1× PBS with 0.5% Triton X-100 for 10 min. Cells were then blocked in 1× PBS, 0.1% Triton X-100, 2% bovine serum albumin (Sigma-Aldrich), and 3% normal goat serum for 30 min. The cells were then incubated with the primary antibody in blocking buffer overnight using the following dilutions: α-Fox1 1D-10, 1:1000; and α-MAP2, 1:200 (Chemicon). Cells were washed three times with 1× PBS with 0.1% Triton X-100, and incubated with secondary antibody of either Alexa568 goat α-mouse IgG or Alexa488 goat α-rabbit IgG (Molecular Probes) at 1:1000 dilution in blocking buffer for 1 h. Cells were rinsed three times with blocking buffer and mounted with ProLong Gold anti-fade reagent with DAPI (Invitrogen). The intensities of the Fox-1 staining in the nucleus and cytoplasm were measured by ImageJ, and the similarity of the ratios of C/N Fox-1 before and after depolarization was examined by a two-sample t-test (Abramoff et al. 2004).
Short hairpin design and knockdown
The design of the shRNA was described previously (Boutz et al. 2007), and targets the 3′ UTR of mouse Fox-2/Rbm9. The oligos used were siFOX-2forward (GAGAGAAGACTAAAAAGAGGCCTGGCTATTGGAATAATAAGAGTTA) and siFOX-2reverse (CAGAGAAGACTAAAAAGAGGCCTGGCTATTGCAATAACTCTTATTA). A single nucleotide mismatch was introduced close to the 3′ end of the sense strand of the hairpin duplex to destabilize the end and ensure the selection of the antisense strand in the RISC complex. The short hairpin sequence was cloned behind the H1 promotor of plasmid pBlsH1 to generate pBlsH1Fox-2. N2A cells were trypsinized and resuspended in growth medium (DMEM 10% FBS) at 2.5 × 105 cells per milliliter. Four micrograms of EGFP N1 expression vector and 20 μg of either the control vector pBlsH1 or the pBlsH1Fox-2 were combined with 60 μL of Lipofectamine 2000 (Invitrogen) and were transfected into 2.5 × 106 N2A cells according to the manufacturer's protocol. Two days after shRNA transfection, the cells were retransfected with pcDNA3.1, pcDNA3.1-Flag-Fox-1FAPY, or pcDNA3.1-Flag-Fox-1TALVP using Lipofectamine 2000 and were cultured for one more day. The knockdown and complementation were tested by Western blot.
Plasmid construction and mutagenesis
The expression construct of pcDNA3.1-Fox-1TALVP was constructed by cloning the E19 sequence between the exon 18 and exon 20 sequences in pcDNA3.1-Fox-1FAPY using PCR with Pfu DNA polymerase. The pDup minigene reporters were constructed by cloning the regulated exons with partial flanking introns between the ApaI and BglII sites of pDup4-1 (Modafferi and Black 1997). The Grin1 E21 minigene was called NME21D300 previously (Lee et al. 2007). Mutations were made by PCR using Pfu DNA polymerase. The sequences and mutations were confirmed by DNA sequencing.
Transfection and primer extension assay of minigene splicing
HEK 293T cells were grown in six-well plates and transfected using 10 μL of Lipofectamine 2000 (Invtrogene) with 1.5 μg of the pDup minigene reporter and with or without 2 μg of the pcDNA3.1-CaMK IV-dCT expression construct and with or without 0.5 μg of pcDNA3.1-Fox-1FAPY. pcDAN3.1 was used to complement the DNA to 4 μg. RNA was purified 24 h after transfection using RNeasy kit (Qiagen) and reverse-transcribed using SuperScript II (Invitrogen) with 32P-labeled DNA oligo DUP3 (AACAGCATCAGGAGTGGACAGATCC). The primer extension products were resolved on 8% polyacrylamide/7.5 M urea denaturing gels. The gel was dried, exposed, and scanned in a PhosphorImager.
Acknowledgments
We thank Kelsey Martin and Toh Hean Chng for critical reading and discussion, and members of the Black laboratory for helpful discussions. This work was supported by NIH grant RO1 GM49662 to D.L.B. D.L.B. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1837009.
Supplemental material is available at http://www.genesdev.org.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with image J. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Allemand E, Guil S, Myers M, Moscat J, Caceres JF, Krainer AR. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc Natl Acad Sci. 2005;102:3605–3610. doi: 10.1073/pnas.0409889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An P, Grabowski PJ. Exon silencing by UAGG motifs in response to neuronal excitation. PLoS Biol. 2007;5:e36. doi: 10.1371/journal.pbio.0050036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnby G, Abbott A, Sykes N, Morris A, Weeks DE, Mott R, Lamb J, Bailey AJ, Monaco AP. Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: Evidence of association at GRIN2A and ABAT. Am J Hum Genet. 2005;76:950–966. doi: 10.1086/430454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffert U, Weeber EJ, Durudas A, Qiu S, Masiulis I, Sweatt JD, Li WP, Adelmann G, Frotscher M, Hammer RE, et al. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron. 2005;47:567–579. doi: 10.1016/j.neuron.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Ben Fredj N, Grange J, Sadoul R, Richard S, Goldberg Y, Boyer V. Depolarization-induced translocation of the RNA-binding protein Sam68 to the dendrites of hippocampal neurons. J Cell Sci. 2004;117:1079–1090. doi: 10.1242/jcs.00927. [DOI] [PubMed] [Google Scholar]
- Bhalla K, Phillips HA, Crawford J, McKenzie OL, Mulley JC, Eyre H, Gardner AE, Kremmidiotis G, Callen DF. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet. 2004;49:308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- Black DL. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Blaustein M, Pelisch F, Srebrow A. Signals, pathways and splicing regulation. Int J Biochem Cell Biol. 2007;39:2031–2048. doi: 10.1016/j.biocel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Bottai D, Guzowski JF, Schwarz MK, Kang SH, Xiao B, Lanahan A, Worley PF, Seeburg PH. Synaptic activity-induced conversion of intronic to exonic sequence in Homer 1 immediate early gene expression. J Neurosci. 2002;22:167–175. doi: 10.1523/JNEUROSCI.22-01-00167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley J, Carter SR, Rao VR, Wang J, Finkbeiner S. Splice variants of the NR1 subunit differentially induce NMDA receptor-dependent gene expression. J Neurosci. 2006;26:1065–1076. doi: 10.1523/JNEUROSCI.3347-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M, Gelfand MS, Spengler S, Zorn M, Dubchak I, Conboy JG. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 2001;29:2338–2348. doi: 10.1093/nar/29.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, DePalma M, Priess JR. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development. 2004;131:4831–4841. doi: 10.1242/dev.01352. [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: Diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Daoud R, Da Penha Berzaghi M, Siedler F, Hubener M, Stamm S. Activity-dependent regulation of alternative splicing patterns in the rat brain. Eur J Neurosci. 1999;11:788–802. doi: 10.1046/j.1460-9568.1999.00486.x. [DOI] [PubMed] [Google Scholar]
- Daoud R, Mies G, Smialowska A, Olah L, Hossmann KA, Stamm S. Ischemia induces a translocation of the splicing factor tra2-β 1 and changes alternative splicing patterns in the brain. J Neurosci. 2002;22:5889–5899. doi: 10.1523/JNEUROSCI.22-14-05889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD, Fung ET, O'Brien RJ, Huganir RL. Splice variant-specific interaction of the NMDA receptor subunit NR1 with neuronal intermediate filaments. J Neurosci. 1998;18:720–730. doi: 10.1523/JNEUROSCI.18-02-00720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley MF, Kulkarni N, Huettner JE. Synapse formation and establishment of neuronal polarity by P19 embryonic carcinoma cells and embryonic stem cells. J Neurosci. 1996;16:1056–1065. doi: 10.1523/JNEUROSCI.16-03-01056.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Greenberg ME. From synapse to nucleus: Calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Hall MP, Huang S, Black DL. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol Biol Cell. 2004;15:774–786. doi: 10.1091/mbc.E03-09-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H, Gremme T, Hatt H, Gottmann K. Synaptic activity-dependent developmental regulation of NMDA receptor subunit expression in cultured neocortical neurons. J Neurochem. 2000;75:1590–1599. doi: 10.1046/j.1471-4159.2000.0751590.x. [DOI] [PubMed] [Google Scholar]
- Huang CS, Shi SH, Ule J, Ruggiu M, Barker LA, Darnell RB, Jan YN, Jan LY. Common molecular pathways mediate long-term potentiation of synaptic excitation and slow synaptic inhibition. Cell. 2005;123:105–118. doi: 10.1016/j.cell.2005.07.033. [DOI] [PubMed] [Google Scholar]
- Huh GS, Hynes RO. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes & Dev. 1994;8:1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- Jaekel B, Muhlberg K, Garcia de Arriba S, Reichenbach A, Verdaguer E, Pallas M, Camins A, Norenberg W, Allgaier C. Neuroprotection associated with alternative splicing of NMDA receptors in rat cortical neurons. Br J Pharmacol. 2006;147:622–633. doi: 10.1038/sj.bjp.0706471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis W, Monyer H, De Rijk TC, Lopes da Silva FH. Hippocampal kindling increases the expression of glutamate receptor-A Flip and -B Flip mRNA in dentate granule cells. Neurosci Lett. 1992;148:51–54. doi: 10.1016/0304-3940(92)90802-e. [DOI] [PubMed] [Google Scholar]
- Kiehl TR, Shibata H, Pulst SM. The ortholog of human ataxin-2 is essential for early embryonic patterning in C. elegans. J Mol Neurosci. 2000;15:231–241. doi: 10.1385/JMN:15:3:231. [DOI] [PubMed] [Google Scholar]
- Kraus JE, Nadler JV, McNamara JO. Regulation of alternative splicing of NMDAR1 in the kindling model. Brain Res Mol Brain Res. 1996;41:97–104. doi: 10.1016/0169-328x(96)00072-1. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat Methods. 2006;3:909–915. doi: 10.1038/nmeth944. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Seeburg PH. Regional and developmental heterogeneity in splicing of the rat brain NMDAR1 mRNA. J Neurosci. 1994;14:3180–3194. doi: 10.1523/JNEUROSCI.14-05-03180.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Xing Y, Nguyen D, Xie J, Lee CJ, Black DL. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 2007;5:e40. doi: 10.1371/journal.pbio.0060040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, et al. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Lin P, Kusano K, Zhang Q, Felder CC, Geiger PM, Mahan LC. GABAA receptors modulate early spontaneous excitatory activity in differentiating P19 neurons. J Neurochem. 1996;66:233–242. doi: 10.1046/j.1471-4159.1996.66010233.x. [DOI] [PubMed] [Google Scholar]
- Lipscombe D. Neuronal proteins custom designed by alternative splicing. Curr Opin Neurobiol. 2005;15:358–363. doi: 10.1016/j.conb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Lipscombe D, Raingo J. Alternative splicing matters: N-type calcium channels in nociceptors. Channels (Austin) 2007;1:225–227. doi: 10.4161/chan.4809. [DOI] [PubMed] [Google Scholar]
- Llansola M, Sanchez-Perez A, Cauli O, Felipo V. Modulation of NMDA receptors in the cerebellum. 1. Properties of the NMDA receptor that modulate its function. Cerebellum. 2005;4:154–161. doi: 10.1080/14734220510007996. [DOI] [PubMed] [Google Scholar]
- Martin CL, Duvall JA, Ilkin Y, Simon JS, Arreaza MG, Wilkes K, Alvarez-Retuerto A, Whichello A, Powell CM, Rao K, et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: Towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol. 2005;5:14. doi: 10.1186/1471-213x-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee AE, Neretti N, Carvalho LE, Meyer CA, Fox EA, Brodsky AS, Silver PA. Exon expression profiling reveals stimulus-mediated exon use in neural cells. Genome Biol. 2007;8:R159. doi: 10.1186/gb-2007-8-8-r159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 2005;33:714–724. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modafferi EF, Black DL. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y, Otsuka T, Horton AC, Scott DB, Ehlers MD. Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron. 2003;40:581–594. doi: 10.1016/s0896-6273(03)00676-7. [DOI] [PubMed] [Google Scholar]
- Musshoff U, Schunke U, Kohling R, Speckmann EJ. Alternative splicing of the NMDAR1 glutamate receptor subunit in human temporal lobe epilepsy. Brain Res Mol Brain Res. 2000;76:377–384. doi: 10.1016/s0169-328x(00)00030-9. [DOI] [PubMed] [Google Scholar]
- Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll M, Akerib CC, Meyer BJ. X-chromosome-counting mechanisms that determine nematode sex. Nature. 1997;388:200–204. doi: 10.1038/40669. [DOI] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, Krobitsch S. Ataxin-2 interacts with the DEAD/H-Box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Otano I, Ehlers MD. Homeostatic plasticity and NMDA receptor trafficking. Trends Neurosci. 2005;28:229–238. doi: 10.1016/j.tins.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Ponthier JL, Schluepen C, Chen W, Lersch RA, Gee SL, Hou VC, Lo AJ, Short SA, Chasis JA, Winkelmann JC, et al. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J Biol Chem. 2006;281:12468–12474. doi: 10.1074/jbc.M511556200. [DOI] [PubMed] [Google Scholar]
- Prybylowski KL, Wolfe BB. Developmental differences in alternative splicing of the NR1 protein in rat cortex and cerebellum. Brain Res Dev Brain Res. 2000;123:143–150. doi: 10.1016/s0165-3806(00)00079-1. [DOI] [PubMed] [Google Scholar]
- Prybylowski K, Rumbaugh G, Wolfe BB, Vicini S. Increased exon 5 expression alters extrasynaptic NMDA receptors in cerebellar neurons. J Neurochem. 2000;75:1140–1146. doi: 10.1046/j.1471-4159.2000.0751140.x. [DOI] [PubMed] [Google Scholar]
- Rafiki A, Ben-Ari Y, Khrestchatisky M, Represa A. Long-lasting enhanced expression in the rat hippocampus of NMDAR1 splice variants in a kainate model of epilepsy. Eur J Neurosci. 1998;10:497–507. doi: 10.1046/j.1460-9568.1998.00054.x. [DOI] [PubMed] [Google Scholar]
- Raingo J, Castiglioni AJ, Lipscombe D. Alternative splicing controls G protein-dependent inhibition of N-type calcium channels in nociceptors. Nat Neurosci. 2007;10:285–292. doi: 10.1038/nn1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh G, Prybylowski K, Wang JF, Vicini S. Exon 5 and spermine regulate deactivation of NMDA receptor subtypes. J Neurophysiol. 2000;83:1300–1306. doi: 10.1152/jn.2000.83.3.1300. [DOI] [PubMed] [Google Scholar]
- Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci. 2001;21:3063–3072. doi: 10.1523/JNEUROSCI.21-09-03063.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet. 2000;9:1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J Biol Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- Standley S, Roche KW, McCallum J, Sans N, Wenthold RJ. PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron. 2000;28:887–898. doi: 10.1016/s0896-6273(00)00161-6. [DOI] [PubMed] [Google Scholar]
- Tang ZZ, Zheng S, Nikolic J, Black DL. Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol Cell Biol. 2009;29:4757–4765. doi: 10.1128/MCB.00608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: Hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- Tingley WG, Roche KW, Thompson AK, Huganir RL. Regulation of NMDA receptor phosphorylation by alternative splicing of the C-terminal domain. Nature. 1993;364:70–73. doi: 10.1038/364070a0. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Hartley M, Heinemann SF. Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science. 1995;268:873–876. doi: 10.1126/science.7754371. [DOI] [PubMed] [Google Scholar]
- Ulrich H, Majumder P. Neurotransmitter receptor expression and activity during neuronal differentiation of embryonal carcinoma and stem cells: From basic research towards clinical applications. Cell Prolif. 2006;39:281–300. doi: 10.1111/j.1365-2184.2006.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Houven van Oordt W, Diaz-Meco MT, Lozano J, Krainer AR, Moscat J, Caceres JF. The MKK(3/6)–p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J Cell Biol. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Grabowski PJ. Cell- and stage-specific splicing events resolved in specialized neurons of the rat cerebellum. RNA. 1996;2:1241–1253. [PMC free article] [PubMed] [Google Scholar]
- West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, Greenberg ME. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Black DL. A CaMK IV responsive RNA element mediates depolarization-induced alternative splicing of ion channels. Nature. 2001;410:936–939. doi: 10.1038/35073593. [DOI] [PubMed] [Google Scholar]
- Xie J, McCobb DP. Control of alternative splicing of potassium channels by stress hormones. Science. 1998;280:443–446. doi: 10.1126/science.280.5362.443. [DOI] [PubMed] [Google Scholar]
- Xie J, Lee JA, Kress TL, Mowry KL, Black DL. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc Natl Acad Sci. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Jan C, Stoilov P, Park J, Black DL. A consensus CaMK IV-responsive RNA sequence mediates regulation of alternative exons in neurons. RNA. 2005;11:1825–1834. doi: 10.1261/rna.2171205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Xu X, Liang TY, Muotri AR, Carson CT, Coufal NG, Gage FH. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol. 2007;3:1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA–protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin ZQ, Deng ZM, Crewther SG, Crewther DP. Altered expression of alternatively spliced isoforms of the mRNA NMDAR1 receptor in the visual cortex of strabismic cats. Mol Vis. 2001;7:271–276. [PubMed] [Google Scholar]
- Ying Z, Babb TL, Comair YG, Bingaman W, Bushey M, Touhalisky K. Induced expression of NMDAR2 proteins and differential expression of NMDAR1 splice variants in dysplastic neurons of human epileptic neocortex. J Neuropathol Exp Neurol. 1998;57:47–62. doi: 10.1097/00005072-199801000-00007. [DOI] [PubMed] [Google Scholar]
- Yu J, Hai Y, Liu G, Fang T, Kung SK, Xie J. The heterogeneous nuclear ribonucleoprotein L is an essential component in the Ca2+/calmodulin-dependent protein kinase IV-regulated alternative splicing through cytidine-adenosine repeats. J Biol Chem. 2009;284:1505–1513. doi: 10.1074/jbc.M805113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes & Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol Cell Biol. 2007;27:830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukin RS, Bennett MV. Alternatively spliced isoforms of the NMDARI receptor subunit. Trends Neurosci. 1995;18:306–313. doi: 10.1016/0166-2236(95)93920-s. [DOI] [PubMed] [Google Scholar]