Abstract
Cellular defense mechanisms, including the unfolded protein response (UPR) and autophagy, attempt to resolve toxic protein aggregates, which are common denominators of neurodegenerative diseases. In this issue of Genes & Development, Hetz and colleagues (pp. 2294–2306) surprisingly show that inhibition of the UPR by knockout of XBP-1 causes a massive increase in autophagy, enhances clearance of superoxide dismutase 1 (SOD1) aggregates, and delays the development of amyotrophic lateral sclerosis. These findings suggest the existence of a homeostatic—if not hormetic—balance between distinct cellular defense mechanisms.
Keywords: Amyotrophic lateral sclerosis, unfolded protein response, endoplasmic reticulum stress, XBP-1, autophagy
In response to fluctuations of their microenvironment, cells must continuously activate a series of homeostatic mechanisms that aim at maintaining their bioenergetic, metabolic, ionic, and genomic integrity. The frontier between normal fluctuations and potentially pathogenic perturbations (“stress”) is not neat, and actually depends on the capacity of cells to adapt to distinct levels of stress. Hence, any genetically determined or acquired reduction in the capacity of cells to adapt to cellular stress is potentially pathogenic because it lowers the threshold at which endogenous or exogenous noxious agents cause irreversible cellular damage, culminating in the irreversible loss of cells by the activation of programmed cell death pathways. In this issue of Genes & Development, Hetz et al. (2009) report the intriguing and counterintuitive observation that inhibition of one particular stress response, the unfolded protein response (UPR), can improve the survival of cells responding to one peculiar stress; namely, the accumulation of a mutated, misfolded protein. How can this apparent paradox be resolved?
The UPR: a homeostatic mechanism
A plethora of distinct chemical agents and physical perturbations increase the rate at which unfolded or misfolded proteins accumulate in the endoplasmic reticulum (ER) (Scheper and Hoozemans 2009). In specific neurodegenerative diseases, mutations affecting the structure of neuron-specific proteins lead to the generation of constitutively misfolded proteins. This applies, for example, to α-synuclein in familial Parkinson's disease, to huntingtin (Htt) in Huntington's disease, and to superoxide dismutase-1 (SOD1) in amyotrophic lateral sclerosis (ALS) (Scheper and Hoozemans 2009). The best-characterized reaction to “ER stress” is the UPR (Ron and Walter 2007). In this pathway, accumulating unfolded or misfolded proteins stimulate a cascade of events leading to the activation of IRE1, a kinase with endoribonuclease activity that cleaves the mRNA of transcription factor XBP-1. XBP-1 then transactivates various chaperones and proteases that counteract protein aggregation and refold unfolded or misfolded proteins. In several neurodegenerative diseases, mutated proteins slowly accumulate in specific subpopulations of neurons and cause a progressive, age-dependent functional decline that is linked to pathological cell loss. Reportedly, the UPR, which partially depends on the IRE1/XBP-1 pathway, can dissolve potentially toxic protein aggregates. Accordingly, the UPR can protect against various neurodegenerative phenotypes, as this has been observed in multiple animal and cell culture models (Sado et al. 2009). Chronic activation of the UPR, however, can trigger cell death and, in one neurodegenerative condition, Pelizzaeus-Merzbacher disease, the ER stress response reportedly results in oligodendrocyte apoptosis (Forman et al. 2003). The severity of neurodegenerative diseases and the extent of the UPR could be correlated in animal models (Matus et al. 2008). In addition, post-mortem explorations of patients suffering from various neurodegenerative disorders revealed a marked activation of the UPR (Matus et al. 2008). This observation has been confirmed for familial and sporadic ALS, in which ER stress markers are up-regulated (Hetz et al. 2009). Thus, in recent years, the UPR has attracted increasing attention due to its potential neuroprotective function.
Autophagy: a potent inhibitor of aging and neurodegeneration
While the UPR controls the degradation of smaller units of unfolded or misfolded proteins via transcription of chaperones, larger aggregates are detoxified via degradation in the lysosome, a process called autophagy. This perspective concentrates on macroautophagy (hereafter referred to as autophagy), because it is, besides the proteasome, probably the major cellular device responsible for the degradation of long-lived proteins (Rubinsztein 2006; Williams et al. 2006). During autophagy, cargos destined for degradation are sequestered into double-membrane-coated vesicles and targeted to the lysosome. Autophagy is strongly up-regulated by fasting or starvation and can also be triggered by protein aggregates and oxidative stress, as well as damage affecting distinct cytoplasmic organelles (Levine and Kroemer 2008). The induction of autophagy is strongly linked to that of the UPR. For example, the phosphorylation of the eukaryotic initiation factor 2α (eIF2α) is an integrated part of the UPR (and required for the translational arrest) (Hamanaka et al. 2005), yet is also stringently required for the induction of autophagy (Talloczy et al. 2002; Kouroku et al. 2007). Therefore, ER stress often stimulates autophagy.
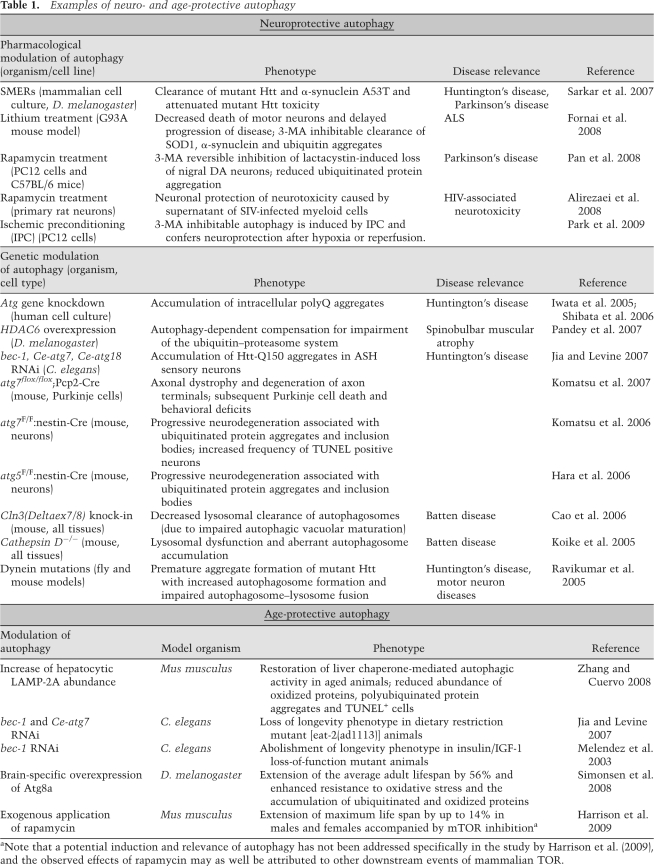
Many reports (some of which are summarized in Table 1) have addressed a potential neuro- and age- protective role of autophagy (Sarkar et al. 2009). Aging is the principal risk factor for neurodegenerative disorders (Thal et al. 2004). The only external regimen leading to life span extension in every organism tested to date is caloric restriction (CR), which is a reduction of the organism's caloric intake without malnutrition (Masoro 2005). New data suggest that autophagy, which is induced by CR, is the crucial determinant of life span extension. Thus, in the nematode Caenorhabditis elegans, autophagy is required for the life span prolongation mediated by CR (Jia and Levine 2007) or p53 inhibition (Tasdemir et al. 2008; Tavernarakis et al. 2008). Inhibition of the nutrient-responsive kinase target of rapamycin (TOR), which activates autophagy, increases life span in various model organisms (Wullschleger et al. 2006). In yeast, autophagy is required for rapamycin-mediated life span extension and delay of chronological aging (Alvers et al. 2009). The TOR inhibitor rapamycin can even prolong the healthy life of mice when the treatment is started at old age (Harrison et al. 2009).
Table 1.
Examples of neuro- and age-protective autophagy
aNote that a potential induction and relevance of autophagy has not been addressed specifically in the study by Harrison et al. (2009), and the observed effects of rapamycin may as well be attributed to other downstream events of mammalian TOR.
In this context, it appears intriguing that enhancement of autophagy specifically in the brain of Drosophila melanogaster suffices to prolong life span by ∼50% (Simonsen et al. 2008), arguing in favor of aging as a program located within the brain. Alternatively, the brain may be the crucial vulnerable component of the organism that determines organismal aging. In several mouse models, brain-specific disruption of autophagy causes neurodegeneration, accompanied by accumulation of protein aggregates and a progressive decline of neuronal functions, connectivity, and survival (Table 1). These observations suggest that baseline autophagy is required for the avoidance of precocious neurodegeneration (Levine and Kroemer 2008; Winslow and Rubinsztein 2008).
Numerous studies performed in animal models suggest that pharmacological or genetic induction of autophagy delays the onset of age-related neurodegenerative diseases, such as Alzheimer's, Parkinson's, and Huntington's diseases and ALS (Table 1). In Drosophila, mutations affecting the regular function of the ubiquitin–proteasome system (UPS) led to a compensatory increase in autophagy that depended on the microtubule-associated deacetylase HDAC6 (Pandey et al. 2007). Strikingly, overexpression of HDAC6 in vivo was sufficient to rescue degeneration associated with UPS impairment in a Drosophila model of spinobulbar muscular atrophy, in an autophagy-dependent manner (Pandey et al. 2007). Rapamycin-induced autophagy can rescue cultured rat neurons from the toxic effects of the proteasome inhibitor lactacystin while it avoids the aggregation of ubiquitinated proteins. Loss of nigral dopaminergic neurons and protein aggregation caused by lactacystin treatment of C57BL/6 mice in vivo were similarly inhibited by post-treatment with rapamycin (Pan et al. 2008).
An initial screen in yeast for small molecule enhancers of rapamycin (SMERs) identified three compounds that were subsequently shown to modulate autophagy in mammalian cells independently of or downstream from Tor (Sarkar et al. 2007). Application of these SMERs induced the clearance of mutant Htt and mutant (A53T) α-synuclein in mammalian cell culture and attenuated the toxicity of mutant Htt in a Drosophila model of Huntington's disease (Sarkar et al. 2007). Furthermore, yet another autophagy inducer, lithium, which acts in a TOR-independent fashion, delayed the onset of ALS and augmented the life span of mice expressing a mutated (G93A) SOD1 protein, correlating with enhanced, presumably autophagy-mediated, clearance of SOD1 and ubiquitin aggregates (Fornai et al. 2008). Intriguingly, there is epidemiological evidence that patients with mood disorders that are treated with lithium exhibit a later onset of Alzheimer's disease than control populations (Yeh and Tsai 2008). Moreover, a clinical trial indicates that lithium treatment could be beneficial for ALS patients (Pradat et al. 2009). This suggests that the pharmacological induction of autophagy may constitute a valuable strategy for the prevention (and, perhaps, the treatment) of neurodegenerative diseases in humans.
Disturbance of UPR causes neuroprotection in ALS
Hetz et al. (2009) now provide provocative but compelling evidence that disturbance of the UPR paradoxically attenuates ALS in mice through the induction of autophagy. Human ALS occurs in either a familial (10%) or a sporadic (90%) form. Familial ALS is caused by mutations that affect the cytosolic SOD1 and that result in the intracellular accumulation and aggregation of misfolded SOD1, in particular in motorneurons (Pasinelli and Brown 2006). Hence, mice that express a transgenic, mutated (G93A) SOD1 provide an accurate model of ALS, showing characteristic hallmarks of the human disease. Hetz et al. (2009) deleted the three major mediators of the UPR—ATF4, ATF6, and IRE-1—from cells harboring mutated SOD1. Depletion of both the transcription factors ATF4 and ATF6 increased the amount of SOD1 aggregates, in line with the common idea that the UPR has been designed for this specific purpose—the removal of unfolded or misfolded proteins. However, knockdown of IRE-1 or that of its downstream target, XBP-1, the third transcription factor involved in the UPR (which regulates genes involved in protein folding and quality control), unexpectedly enhanced clearance of SOD1 aggregates and improved survival of cultured motorneurons. Hetz et al. (2009) subsequently found that deletion of IRE-1 or XBP-1 strongly stimulated autophagy, which then led to improved clearance of SOD1 aggregates. Thus, the low levels of aggregated SOD1 in XBP-1-depleted cells were increased to toxic levels when essential autophagy mediators such as ATG5 or Beclin1 were knocked down by RNAi. Hetz et al. (2009) then went on to demonstrate that XBP-1 deletion can also improve SOD1 clearance in vivo, in an established mouse model of familial ALS. Indeed, mice expressing a transgenic mutated SOD1 exhibited delayed ALS onset and increased life span when XBP-1 was deleted in the CNS. Again, this state of neuroprotection correlated with increased autophagy and reduced accumulation of SOD1 aggregates in the spinal chord. Unfortunately, no results are available that would demonstrate that CNS-specific inhibition of autophagy would counteract the beneficial effect of the XBP-1 knockout in vivo. Finally, in an attempt to translate their findings to human ALS, Hetz et al. (2009) analyzed post-mortem samples from sporadic and familial ALS brains and detected signs of both UPR and autophagy. This suggests that the findings obtained in mice can be applied to the human setting.
Resolving the paradox
As outlined above, in many mouse models of neurodegeneration, as well as in post-mortem analyses of brains from patients with neurodegenerative diseases, the activation of the UPR correlates with the amount of aggregated proteins (Matus et al. 2008). Moreover, the UPR has generally been viewed as a homeostatic mechanism designed to avoid the accumulation of toxic protein aggregates. The finding by Hetz et al. (2009) that disturbance of the UPR by deletion of XBP-1 can prevent neurodegeneration is counterintuitive and, at this state of knowledge, may be explained in several nonexclusive ways (illustrated in Fig. 1).
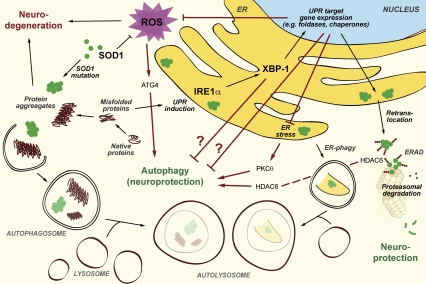
Figure 1.
Schematic view of UPR and autophagy interactions with relevance to neuroprotection. Unfolded or misfolded protein aggregates accumulate during neurodegenerative diseases within the cytosol or the ER of neurons. Several cellular defense mechanisms can be activated in order to resolve protein aggregates, thus serving as neuroprotective processes. This applies to the UPR, the ERAD pathway, and the lysosomal degradation pathway or autophagy. Upon depletion of IRE1α or XBP-1, a homeostatic activation of autophagy ensures efficient removal of SOD1 protein aggregates and is responsible for neuroprotection. However, the exact mechanism of how autophagy is activated when the UPR is impaired remains to be investigated and might involve several nonexclusive ways of interaction, illustrated by red colored arrows (see the text for details). Question marks indicate hypothetical interrelations.
First, beyond a certain threshold, intense and prolonged UPR can stimulate apoptosis, which hence can be initiated at the level of the ER (Haynes et al. 2004; Szegezdi et al. 2006). Thus, although the UPR stimulates the adaptive up-regulation of chaperones and hence mediates cytoprotection, prolonged UPR activation can lead to Bax-mediated apoptosis, presumably via activation of c-Jun N-terminal kinase (JNK) (Davis 2000). Therefore, under prolonged oxidative and protein folding stress, which is expected in ALS due to impairment of SOD1 function, knockdown of XBP-1 could rescue neurons due to inhibition of the apoptotic program in vivo, and not necessarily or solely via induction of autophagy. Indeed, deletion of the BH3-only protein Puma protects motoneurons from ER stress-induced apoptosis and delays motoneuron loss in mice expressing a mutant SOD1 transgene, yet does not prolong their life span (Kieran et al. 2007). In addition, ablation of CHOP, another key transcription factor of the UPR, reportedly restores motor function in a mouse model of Charcot Marie Tooth 1B neuropathy, possibly via inhibition of ER stress-induced apoptosis (Pennuto et al. 2008).
Second, given that autophagy induction is responsible for the cytoprotective effects observed by Hetz et al. (2009), at least in vitro, a major question emerges: How, exactly, is the homeostatic switch between UPR deficiency and autophagy induction achieved? Hetz et al. (2009) suggest that inhibition of the ER-associated degradation (ERAD) pathway secondary to the ablation of XBP-1 might account for the compensatory stimulation of autophagy. Alternatively, XBP-1 target genes may regulate autophagy directly by yet unknown mechanisms. Another (nonexclusive) possibility would be that a (transient?) accumulation of unfolded or misfolded proteins due to the absent UPR leads to cumulative ER stress, which in turn induces autophagy via activation of protein kinase C θ (PKCθ) (Sakaki et al. 2008). A few experiments could clarify this issue. First, a time course monitoring the generation of mutant SOD1 aggregates should be performed in the ALS cell culture model upon depletion of XBP-1. The prediction would be that there is a time window (prior to the earliest time point analyzed by Hetz et al. [2009], which is 48 h after application of XBP-1 shRNA), when depletion of XBP-1 leads to higher levels of SOD1 aggregates, preceding their homeostatic clearance by autophagy. Second, it should be clarified whether ablation of PKCθ would abolish the cytoprotective effects of XBP-1 depletion. Another possibility would be that HDAC6 would provide the link between the inhibited UPR and induction of autophagy. Indeed, activation of HDAC6 is responsible for the induction of autophagy when the UPS, yet another degradation pathway involved in clearance of misfolded proteins, is inhibited (Pandey et al. 2007).
Third, disturbance of the UPR reportedly triggers oxidative stress and depletion of glutathione (Xue et al. 2005), and there may be even more intimate connections between reactive oxygen species (ROS) generation and UPR. For example, tunicamycin, a prototypic inducer of ER stress, does not induce ROS generation in cultured cells per se, but leads to marked accumulation of ROS in cells lacking PERK or ATF4, two major mediators of the UPR (Harding et al. 2003). In addition, tumor necrosis factor-α induces ROS accumulation and cell death in a UPR-dependent fashion (Xue et al. 2005). Given that SOD1 mutations associated with familial ALS may themselves enhance the basal ROS level (Bogdanov et al. 1998), inhibition of the UPR might further exacerbate oxidative stress. At sublethal levels, ROS can act as signaling molecules and even prolong life span of C. elegans; for instance, in conditions of glucose restriction (Schulz et al. 2007). ROS can also stimulate autophagy by boosting the activity of ATG4, a cysteine protease that is a direct target for regulatory oxidation and activation (Scherz-Shouval et al. 2007). Thus, ROS can serve as signaling molecules for the induction of cytoprotection.
Hormesis: harming cells for longer life?
The term “hormesis” was coined by toxologists. It describes the, at first sight, paradoxical finding that noxious agents (e.g., chemical, thermal, or radiological) can have beneficial effects when they are applied at low, sublethal, and subtoxic doses (Calabrese et al. 1999). The teleological sense behind hormesis was first formulated by Friedrich Nietze in his proverb: “What does not kill him makes him stronger” (Nietzsche 2005; Gems and Partridge 2008). Indeed, low doses of cellular or organismal stressors (such as heat or mild doses of oxidative stress) have been reported to prolong life span in C. elegans (Cypser et al. 2006). This astonishing finding may be explained by a general enhancement of the anti-stress response leading to protection (possibly as an adaptive mechanism) against the ultimate physiological stress: aging.
The principle of hormesis is not restricted to heat, toxins, or oxidative stress, but can be observed and applied to numerous settings. For example, acquired immune responses constitute a protective system that has been shaped by sublethal infectious insults. Moreover, the induction of drug metabolizing enzymes upon xenobiotic stimulation can protect an organism against cancer (chemoprotection) (Talalay et al. 2003). Even more surprising is the strong and unexplained correlation between cigarette smoking and protection against Parkinson's disease (Quik 2004). The findings by Hetz et al. (2009) may hence be classified as a hormetic response in which the perturbation of the UPR stimulates a compensatory, cytoprotective response. Although this is not formally proven by in vivo experiments, it appears plausible that a large part of this neuroprotective response is mediated by autophagy. Irrespective of the yet-to-be-resolved molecular mechanisms, it is highly astonishing that already harmed cells can be rescued by disturbance of a cellular protection system. Hence, nature is more complex than we can imagine, and future treatments of neurodegenerative diseases may well be based on scientific surprises.
Acknowledgments
G.K. is supported by grants from Agence Nationale pour la Recherche and European Union (active p53, Apo-Sys, Apop-Train). We are grateful to the European Union for grant Apo-Sys (FP7) to F.M. and T.E., and to the Austrian Science Fund (FWF) for grant S-9304-B05 and Lipotoxicity to F.M.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1858009.
References
- Alirezaei M, Kiosses WB, Flynn CT, Brady NR, Fox HS. Disruption of neuronal autophagy by infected microglia results in neurodegeneration. PLoS One. 2008;3:e2906. doi: 10.1371/journal.pone.0002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvers AL, Wood MS, Hu D, Kaywell AC, Dunn WA, Jr, Aris JP. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy. 2009;5:847–849. doi: 10.4161/auto.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov MB, Ramos LE, Xu Z, Beal MF. Elevated “hydroxyl radical” generation in vivo in an animal model of amyotrophic lateral sclerosis. J Neurochem. 1998;71:1321–1324. doi: 10.1046/j.1471-4159.1998.71031321.x. [DOI] [PubMed] [Google Scholar]
- Calabrese EJ, Baldwin LA, Holland CD. Hormesis: A highly generalizable and reproducible phenomenon with important implications for risk assessment. Risk Anal. 1999;19:261–281. doi: 10.1023/a:1006977728215. [DOI] [PubMed] [Google Scholar]
- Cao Y, Espinola JA, Fossale E, Massey AC, Cuervo AM, MacDonald ME, Cotman SL. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem. 2006;281:20483–20493. doi: 10.1074/jbc.M602180200. [DOI] [PubMed] [Google Scholar]
- Cypser JR, Tedesco P, Johnson TE. Hormesis and aging in Caenorhabditis elegans. Exp Gerontol. 2006;41:935–939. doi: 10.1016/j.exger.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Forman MS, Lee VM, Trojanowski JQ. ‘Unfolding’ pathways in neurodegenerative disease. Trends Neurosci. 2003;26:407–410. doi: 10.1016/S0166-2236(03)00197-8. [DOI] [PubMed] [Google Scholar]
- Fornai F, Longone P, Cafaro L, Kastsiuchenka O, Ferrucci M, Manca ML, Lazzeri G, Spalloni A, Bellio N, Lenzi P, et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc Natl Acad Sci. 2008;105:2052–2057. doi: 10.1073/pnas.0708022105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Partridge L. Stress-response hormesis and aging: ‘That which does not kill us makes us stronger.’. Cell Metab. 2008;7:200–203. doi: 10.1016/j.cmet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, Bennett BS, Cullinan SB, Diehl JA. PERK and GCN2 contribute to eIF2α phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Mol Biol Cell. 2005;16:5493–5501. doi: 10.1091/mbc.E05-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Titus EA, Cooper AA. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol Cell. 2004;15:767–776. doi: 10.1016/j.molcel.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Hetz C, Thielen P, Matus S, Nassif M, Court F, Cuervo AM, Martinez G, Kiffin R, Brown RH, Glimcher LH. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes & Dev. 2009 doi: 10.1101/gad.1830709. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, Kopito RR. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- Kieran D, Woods I, Villunger A, Strasser A, Prehn JH. Deletion of the BH3-only protein puma protects motoneurons from ER stress-induced apoptosis and delays motoneuron loss in ALS mice. Proc Natl Acad Sci. 2007;104:20606–20611. doi: 10.1073/pnas.0707906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, Gotow T, Peters C, von Figura K, Mizushima N, et al. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease) Am J Pathol. 2005;167:1713–1728. doi: 10.1016/S0002-9440(10)61253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr, Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoro EJ. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Matus S, Lisbona F, Torres M, Leon C, Thielen P, Hetz C. The stress rheostat: An interplay between the unfolded protein response (UPR) and autophagy in neurodegeneration. Curr Mol Med. 2008;8:157–172. doi: 10.2174/156652408784221324. [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Nietzsche F. Ecce Homo: Wie man wird, was man ist. DTV-Verlag C.H. Beck; München: 2005. [Google Scholar]
- Pan T, Kondo S, Zhu W, Xie W, Jankovic J, Le W. Neuroprotection of rapamycin in lactacystin-induced neurodegeneration via autophagy enhancement. Neurobiol Dis. 2008;32:16–25. doi: 10.1016/j.nbd.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Park HK, Chu K, Jung KH, Lee ST, Bahn JJ, Kim M, Lee SK, Roh JK. Autophagy is involved in the ischemic preconditioning. Neurosci Lett. 2009;451:16–19. doi: 10.1016/j.neulet.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: Insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- Pennuto M, Tinelli E, Malaguti M, Del Carro U, D'Antonio M, Ron D, Quattrini A, Feltri ML, Wrabetz L. Ablation of the UPR-mediator CHOP restores motor function and reduces demyelination in Charcot-Marie-Tooth 1B mice. Neuron. 2008;57:393–405. doi: 10.1016/j.neuron.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradat PF, Camdessanche JP, Carluer L, Cintas P, Corcia P, Danel-Brunaud V, Echaniz-Laguna A, Gonzalez J, Nicolas G, Vandenberghe N, et al. Update on fundamental and clinical research in amyotrophic lateral sclerosis. Rev Neurol (Paris) 2009;165:532–541. doi: 10.1016/j.neurol.2009.01.038. [DOI] [PubMed] [Google Scholar]
- Quik M. Smoking, nicotine and Parkinson's disease. Trends Neurosci. 2004;27:561–568. doi: 10.1016/j.tins.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O'Kane CJ, Brown SD, Rubinsztein DC. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Sado M, Yamasaki Y, Iwanaga T, Onaka Y, Ibuki T, Nishihara S, Mizuguchi H, Momota H, Kishibuchi R, Hashimoto T, et al. Protective effect against Parkinson's disease-related insults through the activation of XBP1. Brain Res. 2009;1257:16–24. doi: 10.1016/j.brainres.2008.11.104. [DOI] [PubMed] [Google Scholar]
- Sakaki K, Wu J, Kaufman RJ. Protein kinase Cθ is required for autophagy in response to stress in the endoplasmic reticulum. J Biol Chem. 2008;283:15370–15380. doi: 10.1074/jbc.M710209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O'Kane CJ, Schreiber SL, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- Scheper W, Hoozemans JJ. Endoplasmic reticulum protein quality control in neurodegenerative disease: The good, the bad and the therapy. Curr Med Chem. 2009;16:615–626. doi: 10.2174/092986709787458506. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talalay P, Dinkova-Kostova AT, Holtzclaw WD. Importance of phase 2 gene regulation in protection against electrophile and reactive oxygen toxicity and carcinogenesis. Adv Enzyme Regul. 2003;43:121–134. doi: 10.1016/s0065-2571(02)00038-9. [DOI] [PubMed] [Google Scholar]
- Talloczy Z, Jiang W, Virgin HWt, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc Natl Acad Sci. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4:870–873. doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- Thal DR, Del Tredici K, Braak H. Neurodegeneration in normal brain aging and disease. Sci Aging Knowledge Environ. 2004;2004:pe26. doi: 10.1126/sageke.2004.23.pe26. [DOI] [PubMed] [Google Scholar]
- Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, Rubinsztein DC. Aggregate-prone proteins are cleared from the cytosol by autophagy: Therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- Winslow AR, Rubinsztein DC. Autophagy in neurodegeneration and development. Biochim Biophys Acta. 2008;280:723–729. doi: 10.1016/j.bbadis.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Xue X, Piao JH, Nakajima A, Sakon-Komazawa S, Kojima Y, Mori K, Yagita H, Okumura K, Harding H, Nakano H. Tumor necrosis factor α (TNFα) induces the unfolded protein response (UPR) in a reactive oxygen species (ROS)-dependent fashion, and the UPR counteracts ROS accumulation by TNFα. J Biol Chem. 2005;280:33917–33925. doi: 10.1074/jbc.M505818200. [DOI] [PubMed] [Google Scholar]
- Yeh HL, Tsai SJ. Lithium may be useful in the prevention of Alzheimer's disease in individuals at risk of presenile familial Alzheimer's disease. Med Hypotheses. 2008;71:948–951. doi: 10.1016/j.mehy.2008.03.049. [DOI] [PubMed] [Google Scholar]
- Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]