Abstract
Mutations in superoxide dismutase-1 (SOD1) cause familial amyotrophic lateral sclerosis (fALS). Recent evidence implicates adaptive responses to endoplasmic reticulum (ER) stress in the disease process via a pathway known as the unfolded protein response (UPR). Here, we investigated the contribution to fALS of X-box-binding protein-1 (XBP-1), a key UPR transcription factor that regulates genes involved in protein folding and quality control. Despite expectations that XBP-1 deficiency would enhance the pathogenesis of mutant SOD1, we observed a dramatic decrease in its toxicity due to an enhanced clearance of mutant SOD1 aggregates by macroautophagy, a cellular pathway involved in lysosome-mediated protein degradation. To validate these observations in vivo, we generated mutant SOD1 transgenic mice with specific deletion of XBP-1 in the nervous system. XBP-1-deficient mice were more resistant to developing disease, correlating with increased levels of autophagy in motoneurons and reduced accumulation of mutant SOD1 aggregates in the spinal cord. Post-mortem spinal cord samples from patients with sporadic ALS and fALS displayed a marked activation of both the UPR and autophagy. Our results reveal a new function of XBP-1 in the control of autophagy and indicate critical cross-talk between these two signaling pathways that can provide protection against neurodegeneration.
Keywords: Amyotrophic lateral sclerosis, unfolded protein response, endoplasmic reticulum stress, XBP-1, autophagy
Most neurodegenerative disorders—such as amyotrophic lateral sclerosis (ALS), Alzheimer's, and Parkinson's and Huntington's diseases—share a common pathology characterized by abnormal protein inclusions containing specific misfolded proteins (Matus et al. 2008). ALS is a progressive and deadly adult-onset motor neuron disease characterized by muscle weakness, spasticity, atrophy, paralysis, and premature death. The pathological hallmark of ALS is the selective degeneration of motoneurons in the spinal ventral horn, most brainstem nuclei, and the cerebral cortex (Boillee et al. 2006; Pasinelli and Brown 2006). ALS is more frequent in males, and the disease lacks a defined genetic component in a majority of ALS patients, so-called sporadic ALS (sALS), while ∼10% of cases are familial (fALS). More than 100 mutations in the gene encoding superoxide dismutase-1 (SOD1) are linked to fALS and trigger its misfolding and abnormal aggregation, resulting in motoneuron dysfunction (Pasinelli and Brown 2006). Overexpression of human fALS-linked SOD1 mutations in transgenic mice recapitulates essential features of the human pathology, provoking age-dependent protein aggregation, paralysis, and motor neuron degeneration. Since sALS and fALS affect the same neurons with similar pathology, therapeutics effective in mutant SOD1 mouse models may translate to sALS.
The pathogenesis of mutant SOD1 toxicity remains unclear, and may include mitochondrial dysfunction, altered axonal transport, and nonneuronal inflammatory components (for review, see Boillee et al. 2006; Pasinelli and Brown 2006). Recent data suggest that stress signaling responses originating from the endoplasmic reticulum (ER) may contribute to ALS (Atkin et al. 2006; Kikuchi et al. 2006; Ilieva et al. 2007). A number of conditions interfere with oxidative protein folding processes in the ER, leading to the accumulation of intralumenal misfolded proteins, a cellular condition referred to as “ER stress” (Ron and Walter 2007). Adaptation to ER stress is mediated by engagement of the unfolded protein response (UPR), an integrated signal transduction pathway that transmits information about protein folding status in the ER lumen to the cytosol and nucleus to increase protein folding capacity. Conversely, cells undergo apoptosis if these mechanisms of adaptation are insufficient to handle ER stress. The most conserved UPR pathway is initiated by the stress sensor IRE1α (inositol-requiring transmembrane kinase/endonuclease) (Ron and Walter 2007). IRE1α is a Ser/Thr protein kinase and endoribonuclease that, upon activation, initiates the splicing of the mRNA encoding the transcriptional factor X-box-binding protein 1 (XBP-1), converting it into a potent activator (termed XBP-1s) that controls genes related to protein quality control, ER translocation, glycosylation, and ER/Golgi biogenesis (Ron and Walter 2007). XBP-1 is essential for the proper function of plasma B cells (Reimold et al. 2001), Paneth cells (Kaser et al. 2008), exocrine cells of the pancreas and salivary glands (Lee et al. 2005), and for liver lipogenesis (Lee et al. 2008), but its actual role in the nervous system remains speculative (Hetz et al. 2008; Matus et al. 2008).
Up-regulation of ER stress markers was described recently in human post-mortem samples from sALS patients (Ilieva et al. 2007; Atkin et al. 2008), and activation of IRE1α and XBP-1 mRNA splicing is observed in the spinal cord of transgenic rodents expressing different fALS-related SOD1 mutations (Atkin et al. 2006; Kikuchi et al. 2006; Nishitoh et al. 2008). A fraction of mutant SOD1 accumulates inside the ER and Golgi in vivo, where it forms insoluble high-molecular-weight species and physically interacts with ER chaperones (Atkin et al. 2006; Kikuchi et al. 2006; Urushitani et al. 2006). Importantly, a proteomic analysis revealed that two UPR target genes, PDI and Grp58, are among the most induced proteins in an ALS mouse model (Atkin et al. 2006). Finally, a recent study indicated that only vulnerable motoneurons in a fALS mouse model showed presymptomatic ER stress responses that occurred very early, prior to detectable axonal denervation (Saxena et al. 2009). Although a strong correlation between ER stress, motoneuron loss, and ALS progression has been established, the actual role of the UPR in the disease process has not been addressed directly. Genetic manipulation of the UPR is required to directly address this question.
Here, we investigated the role of the UPR in fALS using cellular and animal models of the disease. We developed a mutant SOD1 transgenic mouse with specific deletion of the transcription factor XBP-1 in the nervous system. Unexpectedly, despite predictions that XBP-1 deficiency would enhance the severity of experimental ALS, we observed that female mice were markedly more resistant to developing the disease. This phenotype was associated with the up-regulation of macroautophagy in motoneurons due to XBP-1 deficiency, a survival pathway involved in lysosome-mediated clearance of cytosolic components including damaged organelles and abnormal protein aggregates. Our results reveal a homeostatic connection between the UPR and macroautophagy in vivo, and identify XBP-1 as a possible therapeutic target for the treatment of neurological conditions linked to protein misfolding.
Results
Knocking down XBP-1 and IRE1α decreases mutant SOD1 aggregation and toxicity
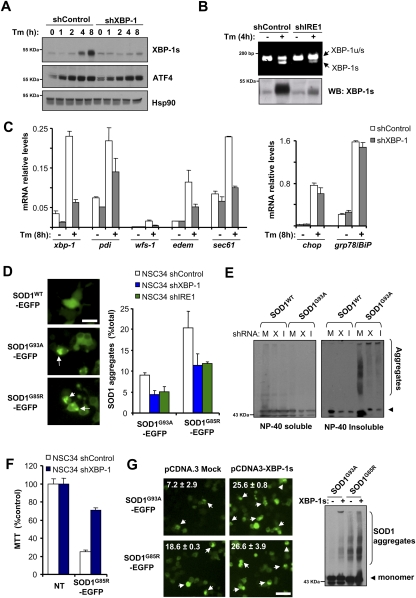
To define the role of the UPR in SOD1 pathogenesis, we reduced the expression levels of major UPR components in the NSC34 motoneuron cell line (Hetz et al. 2007) using lentiviral delivery of shRNAs. Targeted genes included IRE1α, XBP-1, activating transcription factor-6α (ATF6α) and ATF4. In addition to XBP-1, ATF6 and ATF4 constitute two parallel signaling branches of the UPR that regulate distinct transcriptional responses under ER stress (Ron and Walter 2007). To monitor SOD1 misfolding in these cells, we transiently expressed human SOD1WT or the mutants SOD1G93A and SOD1G85R as EGFP fusion proteins and examined the accumulation of intracellular SOD1 inclusions by fluorescent microscopy or SOD1 aggregation by Western blot analysis. In agreement with the known role of ATF4 and ATF6α in the transcriptional control of ER chaperones, knocking down these UPR components increased mutant SOD1 aggregation (Supplemental Fig. S1A–C). We verified functional knockdown of XBP-1 as evidenced by the significant reduction in expression of XBP-1s protein and many XBP1-target genes (Fig. 1A,C). Similarly, IRE1α knockdown reduced the levels of XBP-1 mRNA splicing and XBP-1s expression under ER stress conditions (Fig. 1B). Surprisingly, a drastic reduction in the generation of high-molecular-weight and detergent-insoluble SOD1 species was observed in XBP-1 and IRE1α knockdown NSC34 cells (termed shXBP-1 and shIRE1, respectively) (Fig. 1E; Supplemental Fig. S1D for total extracts). To complement these observations, we monitored the accumulation of SOD1 inclusions in these cells. A nearly 50% reduction in the number of cells harboring SOD1G93A and SOD1G85R intracellular inclusions was detected in shXBP-1 and shIRE1 (Fig. 1D). Similar results were obtained when IRE1α and XBP-1 knockdowns were performed in Neuro2a cells (data not shown). As control, to monitor the efficiency of protein expression/cell transfection, SOD1WT-EGFP was expressed in shXBP-1, shIRE1, and shControl cells, and then quantified by FACS (Supplemental Fig. S1E).
Figure 1.
XBP-1/IRE1α deficiency in motoneurons reduces abnormal mutant SOD1 aggregation and toxicity. (A) NSC34 motoneuron-like cells were stably transduced with lentiviral vectors expressing shRNA against XBP-1 or control luciferase mRNA (shXBP-1 and shControl, respectively), and expression of XBP-1s or ATF4 (negative control) were analyzed after treatment with tunicamycin (500 ng/mL, Tm) by Western blot. Levels of Hsp90 served as loading control. (B) In parallel, IRE1α mRNA was targeted with shRNA and analyzed as described in A. (Top panel) The levels of XBP-1 mRNA splicing were monitored by RT–PCR in the same samples. (C) Levels of XBP-1s-dependent (edem, sec61, pdi, and wfs1) and XBP-1s-independent (bip and chop) genes were evaluated by real-time PCR in cells treated or not with 500 ng/mL Tm for 8 h. Average and standard deviation is presented of results representative of three independent experiments performed in duplicate. (D) shXBP-1, shIRE1α, and shControl cells were transiently transfected with expression vectors for human SOD1WT-EGFP, SOD1G93A-EGFP, and SOD1G85R-EGFP fusion proteins. After 72 h, SOD1 intracellular inclusions were quantified by fluorescent microscopy (arrows). (Left panel) The number of cells displaying intracellular inclusions was quantified in a total of at least 300 cells per experiment. Results are representative of four independent experiments performed. Average and standard deviation are presented. Bar, 20 μm. (E) In parallel, detergent-insoluble SOD1 protein aggregates were measured in cell extracts prepared in NP-40 and centrifuged at high speed. NP40-insoluble and NP40-soluble SOD1 was analyzed by Western blot. Of note, high-molecular-weight SOD1 aggregates are observed. shRNA cells: (M) Mock; (X) XBP-1; (I) IRE1α. SOD1 monomers are indicated by an arrowhead. Results are representative of at least four independent experiments. (F) shXBP-1 or shControl cells were transiently transfected with expression vector for SOD1G93A. After 72 h, cell viability was monitored using the MTT assay and normalized to control nontreated (NT) cells. Data represent average and standard deviation of three determinations. (G) NSC34 cells were cotransfected with mutant SOD1-EGFP and an XBP-1s expression vector or empty pCDNA.3 vector. After 48 h, intracellular inclusions were observed by fluorescent microscopy (white arrowheads). (Insets) The percentage of cells harboring protein aggregates is indicated. Data represent average and standard deviation of three determinations. Bar, 50 μm. (Right panel) In parallel, SOD1 aggregation was determined by Western blot in total protein extracts. Data are representative of three independent experiments.
Consistent with a decrease in the levels of mutant SOD1 misfolding, shXBP-1 cells displayed increased survival after SOD1G85R expression as measured by monitoring mitochondrial activity with the MTT assay (Fig. 1F). We also investigated the effects of XBP-1s gain of function in mutant SOD1 aggregation. After cotransfection of an XBP-1s expression vector with SOD1G93A or SOD1G85R constructs, we observed increased aggregation of mutant SOD1 and augmented generation of intracellular inclusions (Fig. 1G). Taken together, these results revealed an unexpected role of the IRE1α/XBP-1 axis of the UPR on SOD1 pathogenesis.
Autophagy-mediated degradation of mutant SOD1 in XBP-1-deficient motoneurons
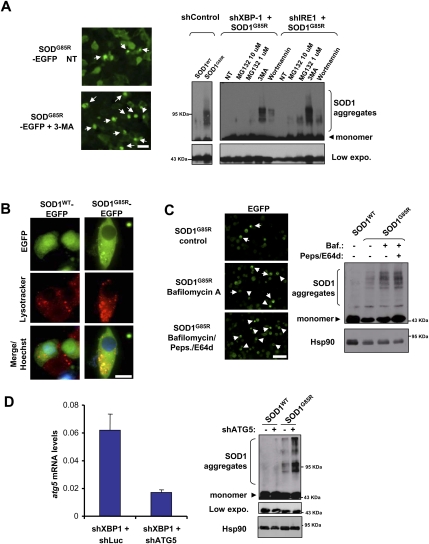
Diminished SOD1 aggregation in XBP-1 knockdown NSC34 cells might be explained by the up-regulation of protein degradation pathways involved in mutant SOD1 clearance. Both the proteasome and macroautophagy (referred to here as autophagy) (Rubinsztein 2006; Mizushima et al. 2008) pathways have been shown to mediate mutant SOD1 degradation in vitro (Kabuta et al. 2006). To define the contribution of these pathways to SOD1 clearance, we treated shRNA NSC34 cells with proteasome (MG-132) or phosphatidylinositol-3 (PI3) kinase inhibitors (3-methyladenine [3-MA] and Wortmannin), which block an early step controlling autophagosome formation (Levine and Kroemer 2008; Mizushima et al. 2008), and thus inhibit autophagy. Blocking PI3 kinases resulted in more SOD1 aggregation than did proteasome inhibition, with recovery of mutant SOD1 aggregation in knockdown cells (Fig. 2A). In agreement with these results, no changes in basal proteasomal activity were observed after knocking down XBP-1 (Supplemental Fig. S2A).
Figure 2.
XBP-1 deficiency leads to autophagy-mediated degradation of mutant SOD1. (A, right panel) shXBP-1 and shControl cells were transfected with an expression vector for SOD1G85R and, after 48 h, cells were treated for 8 h with MG132 (10 and 1 μM) or for 16 h with 10 mM 3-MA or 10 μM wortmannin, and SOD1 aggregation was analyzed by Western blot. (Bottom panel) A shorter exposure of the same Western blot shows monomeric SOD1. As control, SOD1WT was expressed in shControl cells. (Left panel) Examples of SOD1 intracellular inclusions are presented visualized by fluorescent microscopy in cells treated with 3-MA or not. Arrows indicate intracellular SOD1-EGFP inclusions. Bar, 50 μm. (B) NSC34 shControl or shXBP-1 cells were transiently transfected with expression vectors for SOD1WT-EGFP and SOD1G85R-EGFP and stained with lysotracker and Hoechst, and the colocalization with SOD1 intracellular inclusions was determined by confocal microscopy. Bar, 10 μm. (C) SOD1WT and SOD1G85R were transiently expressed in shXBP-1 cells and, after 48 h, lysosomal activity was inhibited by treatment with 200 nM bafilomycin A1 (Baf.) alone or in combination with 10 μg/mL pepstatin (Peps.) and E64d. SOD1 oligomers were visualized by Western blot after 16 h of treatment. HSP90 levels served as loading control. (Left panel) Examples of mutant SOD1 inclusions after lysosome inhibition. Bar, 50 μm. (D) shXBP-1 cells were transiently transduced with a shRNA lentiviral constructs against ATG5 mRNA or control vector. After 7 d, cells were transfected with SOD1G85R-EGFP, and the levels of mutant SOD1 aggregation were evaluated by Western blot in total cell extracts. (Bottom panel) A shorter exposure (low expo.) of the same Western blot shows monomeric SOD1. HSP90 levels were monitored as loading control. (Left panel) Knockdown efficiency was monitored by real-time PCR and normalized with the levels of actin. Mean and standard deviation are presented. Data are representative of four determinations.
Autophagosomes fuse with lysosomes, forming autophagolysosomes where their content is degraded (Rubinsztein 2006; Mizushima et al. 2008). In order to study the role of the lysosomal compartment in the degradation of mutant SOD1, we first analyzed its possible localization at the lysosome. A clear colocalization between SOD1 mutant inclusions and acidic compartments was observed in NSC34 cells when compared with wild-type SOD1 (Fig. 2B; Supplemental Fig. S2B). To measure the functional degradation of mutant SOD1 by the lysosomal pathway, we treated shXBP-1 cells with a cocktail of lysosomal inhibitors (bafilomycin A1 and the protease inhibitors pepstatin and E64d). Using this approach, we observed an enhanced accumulation of SOD1 aggregates and inclusions in shXBP-1 cells after inhibiting lysosomal activity (Fig. 2C).
We extended our results by knocking down ATG5, a major autophagy regulator in the nervous system (Hara et al. 2006). We transduced shXBP-1 cells with shRNA lentiviruses against the atg5 mRNA, which reduced its mRNA levels by ∼70% (Fig. 2D). A significant increase in the levels of mutant SOD1 aggregation was observed in shXBP-1 cells when ATG5 expression was knocked down, reverting the phenotype of XBP-1 deficiency (Fig. 2D). Similar results were obtained when we targeted the expression of Beclin-1/ATG6, the first identified mammalian gene product shown to regulate autophagy (Liang et al. 1999; for review, see Mizushima et al. 2008), in shXBP-1 cells (Supplemental Fig. S2C). Thus, our results indicate that XBP-1 deficiency increases mutant SOD1 clearance due to autophagy-mediated degradation.
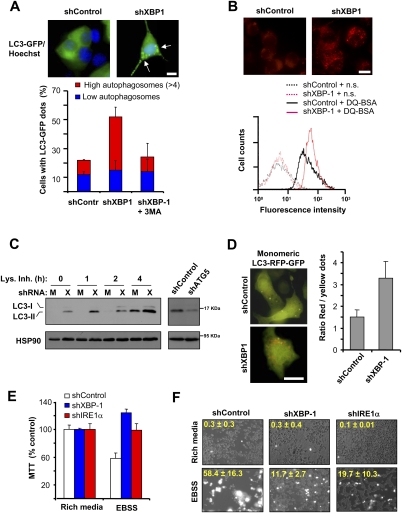
Targeting XBP-1 up-regulates basal autophagy activity
Based on the previous results, we then investigated the possible role of XBP-1 in the regulation of autophagy. LC3 (also known as ATG8 in yeast) is a commonly used marker of autophagy that localizes specifically to autophagosomes (Klionsky et al. 2008). Using LC3-EGFP fusion to determine autophagosome content, we observed a clear increase in the number of shXBP-1 cells containing autophagosomes compared with control cells (Fig. 3A). As control for the assay, shXBP-1 cells were treated with 3-MA, which drastically reduced the amount of LC3-positive vesicles to a similar level as shControl cells (Fig. 3A). We complemented these studies by measuring the activity of lysosomes using DQ-BSA, a dye that stains active proteolysis at the lysosome, and observed a significant increase in the content of active lysosomes in XBP-1 knockdown motoneurons (Fig. 3B). Similarly, increased lysosome content was detected after visualization of shXBP-1 cells with lysotraker, acridine orange staining, or electron microscopy (Supplemental Fig. S2D–F).
Figure 3.
Inhibition of XBP-1 expression increases the levels of basal autophagy. (A) Number of cells with autophagosomes was quantified by confocal microscopy in shControl or shXBP-1 NSC34 cells after expressing a LC3-EGFP expression vector. Cells were classified into two groups: those that contained low LC3-EGFP dots or those that contained more than four LC3-EGFP dots. As control, cells were treated for 16 h with 10 mM 3-MA. (Top panel) Representative images of LC3-EGFP-positive vacuoles (arrows). Bar, 10 μm. Average and standard deviation are presented of three determinations. (B) To monitor the proteolytic activity of lysosomes, shXBP-1 and shControl cells were loaded with DQ-BSA for 16 h and analyzed by FACS to monitor the dequenching of the dye associated with lysosomal degradation. (n.s.) Nonstained cells. (Top panel) Representative images of DQ-BSA staining are presented. Bar, 10 μm. (C) Basal levels of the endogenous lipidated LC3-II form were monitored by Western blot in shControl (M) or shXBP-1 (X) cells under resting conditions. To monitor the flux of LC3 through the autophagy pathway, experiments were performed in the presence or absence of a lysosome inhibitor cocktail (Lys. Inh.) containing 200 nM bafilomycin A1, 10 μg/mL pepstatin, and 10 μg/mL E64d for the indicated time points. (Right panel) As control, in shXBP-1 NSC34 cells, ATG5 was also knocked down with shRNAs and LC3-II levels were monitored by Western blot. (D) shControl and shXBP-1 cells were transiently transfected with a tandem monomeric LC3-RFP-GFP construct to monitor the active flux of LC3 though the autophagy pathway. After 48 h, LC3-positive dots were visualized by fluorescent microscopy in the red and green channels and the ratio between the number of red dots (autophagolysosomes, acidic compartment) versus colocalized yellow dots (representing autophagosomes) per cell was determined. Mean and standard deviation are presented. Representative overlapped fluorescent images are presented. Bar, 10 μm. (E,F) shControl, shXBP-1, and shIRE1α cells were maintained in rich culture medium or incubated in EBSS buffer for 2 h, and mitochondrial metabolism was determined using the MTT assay (E) or cell viability was monitored by propidium iodide staining (F). Overlapping phase contrast and propidium iodide staining images are shown. Average and standard deviation are presented of three to five determinations.
To determine whether the presence of LC3-positive vacuoles is related to augmented autophagy activity rather than to decreased lysosomal fusion/degradative activity, we monitored the flow of LC3 through the autophagy pathway. The level of endogenous LC3-II (the active phosphatidylethanolamine-conjugated form) was measured under resting conditions in shXBP-1 and control cells in the presence or absence of lysosomal inhibitors. We observed a clear increase in the expression of LC3-II in shXBP-1 cells when compared with control cells that was reverted by knocking down ATG5 (Fig. 3C). Interestingly, LC3-II levels were further augmented by blocking lysosomal activity (Fig. 3C), indicating elevated autophagy activity and LC3 flux in shXBP-1 cells. These data were also corroborated by expressing a tandem monomeric RFP-GFP-tagged LC3, where the LC3 flux into the lysosomal acidic compartment can be followed in living cells in the absence of drug treatment (Fig. 3D; Klionsky et al. 2008).
Since autophagy was described originally as a survival pathway under limiting nutrient conditions, we tested the cellular consequence of increased basal autophagy in IRE1α- and XBP-1-deficient NSC34 cells by analyzing their susceptibility to nutrient starvation (assessment of preconditioning effect). Using two independent methods, we observed that shXBP-1 and shIRE1 cells were more resistant to starvation-induced cell death induced by incubating cells in Earle's balanced salt solution (EBSS) buffer (Fig. 3E,F). Together, these results indicate that functional autophagy is overactive in cells where IRE1α signaling is impaired.
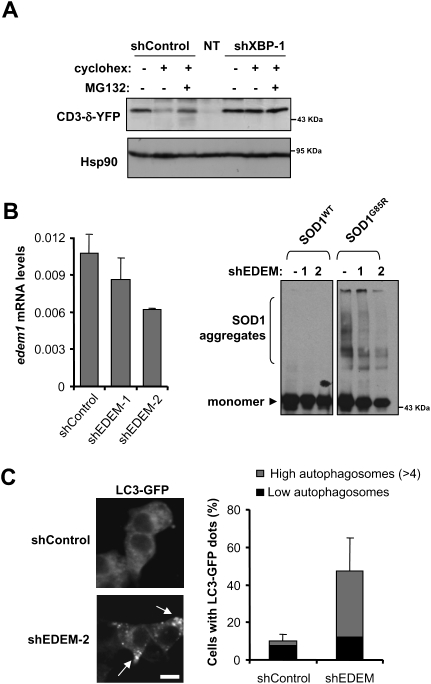
ER-associated degradation (ERAD) impairment increases basal autophagy
Through ERAD, misfolded proteins accumulated at the ER are retrotranslocated to the cytosol for degradation by the proteasome, and ERAD-related genes are a major target of XBP-1s (Lee et al. 2003). Autophagy has been suggested to act as a second ERAD pathway for degradation of ER-located misfolded proteins (Fujita et al. 2007). Because proteasome impairment has been shown to trigger autophagy (Ding et al. 2007; Pandey et al. 2007; Kaganovich et al. 2008), we hypothesized that XBP-1 deficiency may affect ERAD and other related processes, leading to increased basal levels of misfolded proteins at the ER, activating autophagy as a survival mechanism. To determine the effects of XBP-1 on ERAD activity in motoneurons, we first monitored the levels of a classical ERAD substrate, CD3-δ-YFP (Lerner et al. 2007), in shXBP-1 cells under resting conditions. To determine the flow of CD3-δ-YFP through the ERAD pathway for proteasome-mediated degradation, experiments were performed after inhibition of translation by cycloheximide treatment in the presence or absence of proteasome inhibitors. Western blot analysis of CD3-δ-YFP-expressing cells revealed a marked decrease in its degradation in shXBP-1 cells when compared with control cells (Fig. 4A). To test the possible involvement of ERAD-related XBP-1 target genes in the up-regulation of autophagy, we knocked down EDEM1, a key protein in ERAD (Molinari et al. 2003) that is specifically up-regulated by XBP-1 under ER stress conditions (Lee et al. 2003). Interestingly, decreasing EDEM1 levels in NSC34 cells recapitulated the phenotype of XBP-1 silencing, with reduced accumulation of SOD1G85R detergent-insoluble species (Fig. 4B). In addition, elevated numbers of LC3-labeled autophagosomes were observed in these cells (Fig. 4C). These results suggest that the protective effects of XBP-1 deficiency on SOD1 pathogenesis are related to its essential regulation of ERAD function.
Figure 4.
Reduced ERAD in XBP-1 knockdown cells and its connection with autophagy. (A) ERAD activity was measured in shXBP-1 and control cells by determining the degradation rate of the substrate CD3-δ-YFP by Western blot after 3 h of cycloheximide (cyclohex.) treatment to allow for the clearance of CD3-δ-YFP through the proteasome. As control, cells were cotreated with the proteasome inhibitor MG132. Nontransfected (NT) cells are also shown. (B) Mutant SOD1 aggregation was detected by Western blot analysis in NSC34 cells transduced with EDEM1 shRNA. Two different EDEM1 shRNA constructs (shEDEM) were used that reduced mRNA levels. Results are representative of three independent experiments. (Left panel) Knockdown levels were determined by real-time PCR. (C) In parallel, cells transduced with a shRNA construct against EDEM1 mRNA (construct 2 from B) or control vector were transiently transfected with an LC3-EGFP expression vector and the levels of autophagosomes were visualized and quantified after 48 h with a confocal microscope. Average and standard deviation are presented. Arrows indicate LC3-labeled autophagosomes. Bar, 10 μm.
XBP-1 deficiency prolongs the life span of a mutant SOD1 transgenic mouse
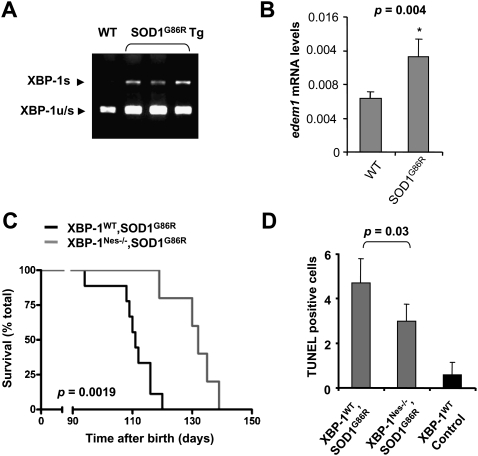
To establish the role of XBP-1 in fALS in vivo, we first monitored the levels of ER stress markers in the spinal cord of SOD1G93A or mouse SOD1G86R (the equivalent to human SOD1G85R mutation) transgenic mice. In agreement with previous reports, we observed the up-regulation of PDI, Grp78/BiP, and CHOP in symptomatic animals (Supplemental Fig. S3A). More importantly, XBP-1 mRNA splicing was observed in the SOD1G86R transgenic mice (Fig. 5A), which correlated with the up-regulation of its target genes edem (Fig. 5B), herp, erdj4, wfs-1, sec61, grp58, and other UPR-related genes (Supplemental Fig. S3B).
Figure 5.
XBP-1 deficiency in the nervous system prolongs life span of mutant SOD1 transgenic mice and decreases the levels of apoptosis. (A) The levels of XBP-1 mRNA splicing (XBP-1s) and nonspliced (XBP-1u/s) were determined in the spinal cord of three symptomatic SOD1G86R transgenic mice by RT–PCR. (B) The mRNA level of the XBP-1 target gene edem1 was analyzed by real-time PCR in total cDNA obtained from the spinal cord of five SOD1G86R or four littermate control mice. All samples were normalized to β-actin levels. Average and standard deviation are presented. P-value was calculated using Student's t-test. (C) XBP-1Nes−/− (N = 7) and control wild-type (N = 9) mice were bred onto SOD1G86R transgenic mice and survival was evaluated in female animals. P-value was calculated with Kaplan-Meier statistics. (D) TUNEL-positive cells in the right half of the ventral horn were quantified in a total of five animals per group in mice at the late stage of the disease. Average and standard deviation are presented. Indicated P-value was calculated using Student's t-test.
To establish the contribution of XBP-1 to fALS in vivo, we generated an XBP-1 conditional knockout model in which xbp-1 was deleted in the nervous system using the Nestin-Cre system (XBP-1Nes−/−) (Hetz et al. 2008). These mice developed normally and did not show any overt spontaneous disease phenotype as we described recently (Hetz et al. 2008). We cross-bred XBP-1Nes−/− mice with SOD1G86R transgenic mice to evaluate the role of XBP-1 in animal survival. Consistent with our cellular studies, XBP-1 deficiency resulted in an average overall increase in life span of 10 d in SOD1G86R mice (Supplemental Fig. S4A). Interestingly, analysis by gender revealed that this was entirely due to the effect of XBP1 deficiency in female animals. XBP-1 deficiency in females resulted in a highly significant increase in life span of 22 d (P = 0.0019), with an average survival of 110 and 132 d for XBP-1WT–SOD1G86R and XBP-1Nes−/−–SOD1G86R mice, respectively (Fig. 5C). Analysis of male animals did not show a significant effect on life span (Supplemental Fig. S4B). Life span extension was associated with a delay in the disease onset in XBP-1Nes−/− mice, but the duration of the symptomatic phase of the disease was not affected by XBP-1 deficiency or gender when compared with control animals (Supplemental Fig. S4C,D). The increased life span in females correlated with a 30% decrease in apoptosis in the ventral horn of the spinal cord of XBP-1Nes−/−–SOD1G86R mice compared with control animals (P = 0.03) (Fig. 5D). Thus, despite our initial prediction that impairment of the IRE1α/XBP-1-dependent adaptive response would accelerate neuronal dysfunction and disease severity, therefore decreasing the life span of fALS mouse models, we observed significant protection against disease in female animals.
XBP-1 deficiency increases autophagy and SOD1 degradation in vivo
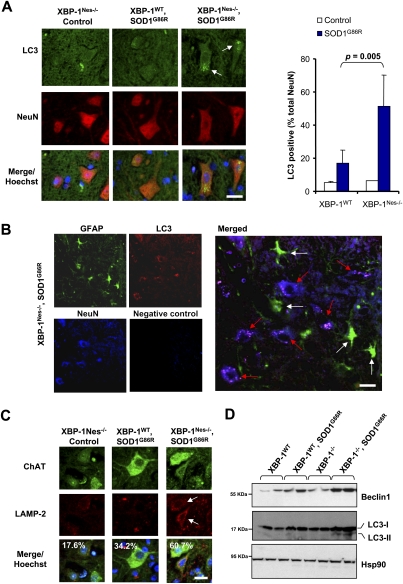
To test whether XBP-1 deficiency alters autophagy levels in fALS in vivo, we monitored the number of neurons containing autophagosomes in the ventral horn of the spinal cord. A marked increase of neurons containing LC3-labeled autophagosomes was observed in SOD1G86R/XBP-1Nes−/− mice when compared with control animals (Fig. 6A). Histological quantification of different animals revealed that ∼50% of neurons in the ventral horn contained LC3-positive autophagosomes (P = 0.005) (Fig. 6A). In contrast, SOD1G86R mice on a wild-type background showed only a slight increase in the number of autophagosomes in some animals when compared with nontransgenic animals, totaling, on average, ∼15% of neurons containing LC3-positive vesicles (Fig. 6A). Of note, the majority of LC3-positive cells were positive for NeuN staining (>90%) in the ventral horn (Supplemental Fig. S5A). In agreement with this observation, astrocytes were, in general, negative for LC3-positive dots (Fig. 6B), indicating a specific contribution of XBP-1 deficiency to the enhancement of autophagy in neurons. In addition, although SOD1G86R transgenic mice show clear signs of glial activation, this phenomenon was not drastically affected by XBP-1 deficiency (Supplemental Fig. S6).
Figure 6.
XBP-1 deficiency increases autophagy levels in neurons of SOD1G86R transgenic mice. (A) Autophagosomes were directly observed in the spinal cord of control of mSOD1G86R transgenic mice on an XBP-1WT or XBP-1Nes−/− background by inmunofluorescence using an anti-LC3 antibody (green). Neurons were costained with an anti-NeuN antibody (red) and with Hoechst (nucleus, blue). Images are representative of the analysis of five different animals per group of ∼125–130 d of age. (Right panel) Quantification of the percentage of NeuN-positive cells containing LC3-positive vacuoles. Values represent average and standard deviation. P-value was calculated using Student's t-test. Bar, 10 μm. (B) The colocalization between LC3-positive dots (red) with neurons (NeuN, blue) or astrocytes (GFAP, green) was analyzed in the spinal cord of a mSOD1G86R transgenic mice using coimmunofluorescence. A merged picture is presented where white arrows indicate astrocytes and red arrows indicate neurons. A negative control of staining without primary antibodies is presented. Bar, 20 μm. (C) Lysosomes were visualized in the samples in A after LAMP-2 staining (red). Colocalization with motoneurons was evaluated after ChAT (green) staining (white arrow). Total cells were stained with Hoechst (nucleus, blue). Quantification of LAMP-2-positive motoneurons is indicated in the inset. Bar, 20 μm. (D) Beclin-1, LC3, and Hsp90 expression were determined in spinal cord protein extracts from symptomatic mSOD1G86R transgenic or control mice by Western blot. LC3-II form is indicated. Two representative animals are shown per group.
Analysis of lysosomal content by LAMP-2 staining revealed a higher content in motoneurons of SOD1G86R–XBP-1Nes−/− mice when compared with control mice (Fig. 6C). Similar results were observed when autophagosomal structures were visualized by electron microscopy (see Supplemental Fig. S5B). In addition, we measured the expression of LC3-II in the spinal cord of symptomatic SOD1G86R mice bred onto a wild-type or XBP-1Nes−/− background. In agreement with the histological characterization, accumulation of LC3-II was only evident by Western blot analysis in XBP-1Nes−/−–SOD1G86R mice (Fig. 6D), which may be related to the low abundance of autophagosomes in SOD1G86R control mice as visualized by histology, and would be diluted in a total tissue extract. Interestingly, Western blot analysis revealed induction of the autophagy regulator Beclin-1 in the XBP-1-sufficient SOD1G86R strain that was further increased in XBP-1Nes−/−–SOD1G86R animals (Fig. 6D). Although alterations in Beclin-1 levels are not usually used as an indication of autophagy, changes in Beclin-1 expression have been reported to correlate well with autophagy levels in animal models of brain ischemia and injury (Diskin et al. 2005; Carloni et al. 2008; Rami et al. 2008), and autophagy-mediated clearance of aggregate-prone proteins in models of Huntington's and Alzheimer's disease (Shibata et al. 2006; Pickford et al. 2008). Overall, our results indicate that the slight increase in autophagy in SOD1 mutant spinal cords described previously (Morimoto et al. 2007) is significantly enhanced when XBP-1 is ablated in the nervous system.
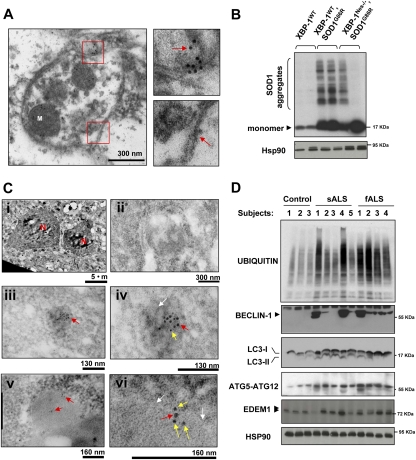
To monitor the active engulfment of SOD1 aggregates by autophagy in the spinal cord of XBP-1Nes−/− mice, we performed SOD1 immunogold staining and electron microscopy analysis. We were able to visualize SOD1 inside autophagosomes using this method (Fig. 7A). In addition, we detected a colocalization of SOD1 and LC3 in vesicular structures by double-immunogold staining of presymptomatic XBP-1Nes−/−–SOD1G86R mice (Fig. 7C). Consistent with increased autophagy levels in XBP-1Nes−/− mice and the colocalization of SOD1 with autophagosomal structures, analysis of female animals revealed almost complete elimination of mutant SOD1 aggregates in the spinal cord of some XBP-1Nes−/−–SOD1G86R animals when compared with littermate control female mice (Fig. 7B). Consistent with the animal survival data, the levels of SOD1 aggregation and numbers of LC3-positive cells were not significantly affected in male XBP-1Nes−/−–SOD1G86R animals compared with control mice (Supplemental Fig. S7A,B). Taken together, these data indicate that XBP-1 deficiency in female mice leads to increased autophagy, associated with augmented mutant SOD1 clearance and prolonged life span.
Figure 7.
Decreased mutant SOD1 aggregation in XBP-1-deficient mice and enhanced autophagy in sALS and fALS patients. (A) Electron microscopy sections of SOD1G86R/XBP-1Nes−/− mice were analyzed by immunogold staining of SOD1. Autophagosomes were identified by the presence of double-membrane vesicles containing cytosol and organelles such as mitochondria (M). (Bottom panel) A higher magnification of the areas marked with red squares is shown to depict the double membrane of the vesicle and SOD1 immunogold staining (see arrows). Images are representative of three independent experiments. (B) SOD1 aggregation was determined by Western blot in XBP-1Nes−/− and control SOD1G86R presymptomatic female mice of the same litter (90 d of age). Two animals per group are presented. As loading control, Hsp90 levels were monitored. (C) Colocalization of SOD1 and LC3 was determined by double-immunogold staining. Gold spheres were 5 nm (SOD1) and 10 nm (LC3). (Panel i) Low-magnification picture to identify motoneurons by their size, morphology, and location in the spinal cord. (N) Nucleus. (Panel ii) Negative control for staining. (Panels iii,iv) Two magnifications showing the colocalization of LC3 and SOD1 inside a dense structure (white arrow). (Panels v,vi) Two magnifications showing the colocalization of LC3 and SOD1 in an autophagosomal structure. White arrows indicate double membranes. Arrows in panels iv and vi are SOD1 (yellow), 5-nm gold colloid; and LC3 (red), 10-nm gold colloid. Representative pictures of the analysis of three animals per group in two independent experiments are presented. (D) Increased levels of autophagy in post-mortem samples of sALS and fALS cases. The levels of BECLIN-1, ATG5–ATG12 complex, LC3, EDEM1, and polyubiquitinated proteins were investigated in total protein extracts derived from spinal cord post-mortem samples from sALS and fALS patients and three healthy control subjects. HSP90 levels were analyzed as loading control.
Up-regulation of autophagy in the spinal cord of sALS and fALS cases
Finally, to characterize the role of autophagy in sALS and fALS, we determined the levels of LC3-I, LC3-II, BECLIN-1, and the ATG5–ATG12 complex in human post-mortem spinal cord samples. A marked induction of autophagy markers was observed in the majority of samples analyzed when compared with age-matched, healthy control subjects (Fig. 7D). Interestingly, the relative levels of BECLIN-1 induction correlated well with increased amounts of polyubiquitinated proteins (Fig. 7D). Two recent reports (Ilieva et al. 2007; Atkin et al. 2008) indicated that signs of ER stress are observed in the spinal cord of sALS post-mortem samples. To determine whether XBP-1 and the UPR are active in sALS, we analyzed the expression levels of different UPR markers in post-mortem spinal cord samples from sALS patients. There was marked expression of XBP-1s and ATF4 in several sALS cases, up to a 4.5-fold increase, in addition to the ER chaperone Grp58 (Supplemental Fig. S8). Consistent with these results, increased expression of the XBP-1s target EDEM1 was observed in both sALS and fALS cases (Fig. 7D). These data corroborate previous findings suggesting that ER stress is observed in sALS patients, and suggest an active engagement of the UPR and autophagy in ALS.
Discussion
A common feature of many neurodegenerative diseases is the accumulation of misfolded proteins in the brain, affecting cognitive and motor functions. Increased expression of ER stress markers is observed in post-mortem brain tissues from patients affected with diseases such as Parkinson's disease, ALS, Alzheimer's disease, and Creutzfeldt-Jacob's disease, in addition to mouse models (for review, see Matus et al. 2008). The predicted functional significance of ER stress in the disease process is complex because it has two paradoxical interpretations: Activation of the UPR could result in protective responses to increase protein folding and quality control mechanisms (adaptive phase). Alternatively, ER stress may represent a deleterious signaling event during chronic stress due to the irreversible disturbance of ER homeostasis (proapoptotic phase). Here, we defined the role of XBP-1 in fALS models. To our surprise, silencing XBP-1 and IRE1α led to a drastic decrease in the misfolding of mutant SOD1, as established by a series of complementary methodologies. This phenotype was explained by the up-regulation of autophagy in XBP-1-deficient cells, uncovering an unexpected function of the IRE1α/XBP-1 axis of the UPR.
The occurrence of ER stress is associated with disease progression and motoneuron degeneration in ALS mouse models, and occurs during early presymptomatic stages (Saxena et al. 2009). Activation of the three major UPR stress pathways is observed in the spinal cord of mutant SOD1 transgenic mice (Atkin et al. 2006; Kikuchi et al. 2006). Interestingly, mutations in ALS8/VAPB, a gene recently discovered to be linked with fALS, may directly affect ATF6 and XBP-1 signaling (Gkogkas et al. 2008), increasing the susceptibility of cells to ER stress-mediated apoptosis (Suzuki et al. 2009). More importantly, analysis of post-mortem samples derived from sALS patients demonstrated up-regulation of UPR components (Ilieva et al. 2007; Atkin et al. 2008). Our results confirmed these observations and further demonstrated for the first time up-regulation of the UPR transcription factors ATF4 and XBP-1 in tissue from human sALS patients. Overall, these studies suggest that ER stress and protein misfolding is a common feature of sALS and different forms of fALS.
To address the function of XBP-1 in fALS in vivo, we studied the susceptibility of mice with selective deficiency of XBP-1 in the nervous system to experimental ALS. Targeting XBP-1 in the nervous system increased life span and motoneuron survival of mutant SOD1 transgenic mice, despite initial predictions that XBP-1 ablation would enhance disease severity based on its role as an ER stress survival factor. XBP-1 deficiency attenuated the rate of apoptosis in the ventral horn of SOD1G86R transgenic mice, associated with enhanced autophagy and diminished levels of mutant SOD1 aggregation. Since the Nestin-Cre transgene deletes in many cell types of the nervous system in addition to neurons, we cannot rule out a contribution of glial cells to the protective effects of XBP-1 ablation, as these cells are known to contribute to ALS pathogenesis (Pasinelli and Brown 2006). Of note, a recent study demonstrated that neuronal expression of mutant SOD1 in transgenic mice is sufficient to cause motoneuron degeneration and paralysis (Jaarsma et al. 2008). Conversely, we observed that most of the cells displaying LC3-labeled autophagosomes in the ventral horn of XBP-1Nes−/−/SOD1G86R mice were neurons and not astrocytes, suggesting a specific effect of XBP-1 deficiency on autophagy levels in neurons.
The effect of XBP-1 ablation was more evident in females, suggesting that the protective consequences of autophagy may be influenced by gender. A recent report also indicated that targeting BECLIN-1-dependent autophagy in the context of an Alzheimer's disease model had beneficial effects only in female and not in male animals (Pickford et al. 2008), suggesting that autophagy levels are influenced by gender. We obtained data supporting this hypothesis by monitoring the levels of LC3-II conversion in male and female mice using a classical model of autophagy in which nutrient starvation triggers the process in muscle tissue (Supplemental Fig. S7C; Mizushima et al. 2004). Interestingly, the incidence of human ALS is higher in males than females, and other reports have detected marked differences between genders in SOD1 transgenic mice (for examples, see Lepore et al. 2007; Stam et al. 2008), although the molecular basis of these gender differences is not understood. Along this line, we observed intrinsic differences in our SOD1G86R transgenic mice, where the accumulation of SOD1 aggregates was higher in males than females despite similar mRNA levels between genders (Supplemental Fig. S7D,E).
In contrast to our results, a recent report indicated that treatment of mutant SOD1 transgenic mice with salubrinal (Saxena et al. 2009), a small molecule that augments eIF2α phosphorylation with concomitant ATF4 up-regulation, significantly delays disease onset, increasing life span. These results are consistent with the effects of ATF4 knockdown in mutant SOD1 aggregation. Taken together with the current study, an intriguing scenario emerges in which the contribution of ER stress to protein misfolding disorders is more complex than previously anticipated, since each UPR signaling branch may have distinct effects on the cellular responses to mutant SOD1 protein misfolding.
In addition to its role as a survival pathway under conditions of nutrient starvation, autophagy is critical for the maintenance of neuronal homeostasis and contributes to basal elimination of misfolded proteins, and brain-specific ablation of essential autophagy-related genes results in spontaneous neurodegeneration (Hara et al. 2006; Komatsu et al. 2006). In addition, pharmacological activation of autophagy has proven beneficial in different models of neurodegeneration (for reviews, see Rubinsztein 2006; Matus et al. 2008). Our analysis of the XBP-1Nes−/− ALS model strain has uncovered a heretofore unappreciated cross-talk between autophagy and the UPR in the nervous system, where homeostatic balance related to XBP-1 deficiency lead to increased autophagy, possibly due to ERAD impairment. ERAD is an essential process controlled by XBP-1 that decreases the unfolded protein load at the ER. However, recent data indicate that disease-related protein aggregates linked to neurodegeneration are poorly degraded by the proteasome (Rubinsztein 2006), and in some cases can even block ERAD, triggering ER stress (Duennwald and Lindquist 2008; Nishitoh et al. 2008).
The functional connection between the UPR and autophagy may explain the lack of lethality or spontaneous disease in XBP-1Nes−/− mice (model in Supplemental Fig. S9). Because many aggregate-prone proteins, such as SOD1, cause disease by a gain-of-function mechanism, therapeutic strategies that reduce the levels of such proteins by increasing autophagy may be beneficial. In fact, we observed a clear induction of several autophagy markers in sALS and fALS spinal cord samples, indicating that autophagy is a relevant cellular response in ALS. Our results uncover a new function of XBP-1 in vivo, and suggest that small molecules such as IRE1α inhibitors and chemical chaperones in addition to autophagy activators may be a feasible strategy for the treatment of protein folding disorders in the nervous system.
Materials and methods
Knockdown of UPR and autophagy components in motoneurons
We generated stable motoneuron cell lines with reduced levels of XBP-1, IRE1α, Beclin-1, and EDEM1 using methods described previously (Hetz et al. 2007) by targeting the respective mRNA with shRNA using the lentiviral expression vector pLKO.1 and puromycin selection.
Assays for mutant SOD1 aggregation and detection of intracellular inclusions
We developed assays using the transient expression of human SOD1WT and the mutants SOD1G93A and SOD1G85R as EGFP fusion proteins. SOD1 oligomers were visualized in total cell extracts prepared in RIPA buffer and sonication, and then analyzed by Western blot. Alternatively, nuclear cell lysates were prepared in 1% NP-40 in PBS containing protease inhibitors. After solubilization for 30 min on ice, cell nuclei were precipitated by centrifugation at 3000 rpm for 5 min and cell extracts were centrifuged at 10,000g for 10 min to collect NP-40-soluble and -insoluble material. Pellets were resuspended in Western blot sampler buffer containing SDS.
Quantification of autophagy and cell viability
Different assays and control experiments were used to monitor autophagy-related processes following the recommendations and precautions described in Klionsky et al. (2008). Living cells were stained with 200 nM lysotraker or 600 nM Acridine orange for 45 min at 37°C and 5% CO2. Alternatively, cells were loaded with DQ-BSA to monitor lysosomal activity as described previously (Klionsky et al. 2008). Autophagy was monitored by analyzing LC3-positive dots or the levels of LC3-II by Western blot and its flux through the autophagosomal/lysosomal pathway by treating cells with a mix of 200 nM bafilomycin A1, 10 μg/mL pepstatin, and 10 μg/mL E64d. Alternatively, to monitor flux, we transiently expressed a tandem monomeric RFP-GFP-tagged LC3 (Klionsky et al. 2008). Cell viability was monitored using the MTT assay or propidium iodide staining (Lisbona et al. 2009). Proteasomal or ERAD activity was monitored after transient transfection of the fluorescent substrate GFPu or CD3-δ-YFP and FACS analysis.
Animal experimentation
XBP-1 conditional knockout mice were described previously (Hetz et al. 2008). We used as an ALS model the SOD1G86R transgenic strain (the equivalent of human SOD1G85R), which was generated in the FVB/N strain [strain FVB-Tg(Sod1-G86R)M1Jwg/J, The Jackson Laboratory]. All animal experiments were performed according to procedures approved by the Institutional Review Board's Animal Care and Use Committee of the Harvard School of Public Health (approved animal protocol 04137) and the Faculty of Medicine of the University of Chile (approved protocol CBA no. 0208 FMUCH). Disease progression was determined using methods described previously (Hetz et al. 2007, 2008).
Tissue analysis
To monitor SOD1 pathogenesis in vivo, animals were euthanized and tissue was collected for histology at different time points, depending on the analysis required. Spinal cord tissue was processed for immunohistochemistry using standard procedures as described (Hetz et al. 2007). Confocal microscopy was used to acquire images and then analysis was performed using the IP laboratory version 4.04 software (Beckon and Dickenson). Antibodies and dilutions used are described in the Supplemental Material.
Western blot analysis of spinal cord extracts
One centimeter of lumbar spinal cord tissue was collected and homogenized in RIPA buffer (20 mM Tris at pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% DOC, 0.5% Triton X-100) containing a protease inhibitor cocktail (Roche) by sonication. Antibodies and dilutions used are described in the Supplemental Material.
Electron microscopy studies and immunogold staining
Autophagosomes were also visualized by transmission electron microscopy as in Klionsky et al. (2008) and morphology was examined using standard methods (Eskelinen 2008). Immunogold electron microscopy staining was perfomed as we described previously (Court et al. 2008).
ALS human post-mortem spinal cord samples
Human post-mortem tissue from ALS and control subjects was obtained as frozen tissue from the Massachusetts General Hospital and then processed for biochemical analysis. The gender, genotype, age of death, and identity numbers are indicated in Supplemental Table 1.
Statistical analysis
Data were analyzed by Student's t-test, two-way ANOVA, or Kaplan-Meier statistics. The GraphPad Prism 5 software was used for statistical analysis. Complete methodological details are described in the Supplemental Material.
Acknowledgments
We thank Drs. Fabio Martinon, Sebastian Bernales, and Sergio Lavandero for helpful discussion, and Benjamin Caballero and Sylvia Flores for technical assistance. We thank Dianne Anne-Marie Wills (MD) for organizing tissue transfer from the ALS tissue bank, Patricia Boya and Maria Luisa Colombo for advice about autophagy assays, and Drs. Noburo Mizushima and Tamotsu Yoshimori for providing LC3-EGFP and LC3-RFP-EGFP constructs. We are grateful to Drs. Julie Atkin and Bradley Turner for kindly providing expression vectors for mutant and wild-type SOD1-EGFP, and Dr. Nir Hacohen and The Broad Institute for providing shRNA lentiviral constructs. We thank Dr. Ron Kopito for providing the GFPu expression vector, and Dr. Nico Dantuma for providing the CD3-YFP vector. This work was supported by the V. Harold and Leila Y Mathers Charitable Foundation and NIH grant AI32412 (to L.H.G.); Genzyme Sponsor Research Agreement, FONDECYT number 1070444, FONDAP grant number 15010006, Millennium Nucleus number P07-048-F, The Muscular Dystrophy Association, The Michael J. Fox Foundation for Parkinson's Research, National Parkinson Foundation, High Q Foundation, and ICGEB (to C.H.); FONDECYT number 3085017 and ALSA-The Milton Safenowitz Post-Doctoral Fellowship for ALS Research (to S.M.); CONICYT Ph.D. fellowship (to M.N. and G.M.); National Institute of Neurological Disorders and Stroke, the National Institute on Aging, the Al-Athel ALS Research Foundation, the ALS Association, the ALS Therapy Alliance, the Angel Fund, Project ALS, and the Pierre L de Bourgknecht ALS Research Foundation (to R.H.B.); FONDECYT number 1070377 (to (F.C.); and grant AG031782 (to A.M.C.).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1830709.
Supplemental material is available at http://www.genesdev.org.
References
- Atkin JD, Farg MA, Turner BJ, Tomas D, Lysaght JA, Nunan J, Rembach A, Nagley P, Beart PM, Cheema SS, et al. Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein-disulfide isomerase with superoxide dismutase 1. J Biol Chem. 2006;281:30152–30165. doi: 10.1074/jbc.M603393200. [DOI] [PubMed] [Google Scholar]
- Atkin JD, Farg MA, Walker AK, McLean C, Tomas D, Horne MK. Endoplasmic reticulum stress and induction of the unfolded protein response in human sporadic amyotrophic lateral sclerosis. Neurobiol Dis. 2008;30:400–407. doi: 10.1016/j.nbd.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Boillee S, Vande VC, Cleveland DW. ALS: A disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Carloni S, Buonocore G, Balduini W. Protective role of autophagy in neonatal hypoxia-ischemia induced brain injury. Neurobiol Dis. 2008;32:329–339. doi: 10.1016/j.nbd.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Court FA, Hendriks WT, Macgillavry HD, Alvarez J, van MJ. Schwann cell to axon transfer of ribosomes: Toward a novel understanding of the role of glia in the nervous system. J Neurosci. 2008;28:11024–11029. doi: 10.1523/JNEUROSCI.2429-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin T, Tal-Or P, Erlich S, Mizrachy L, Alexandrovich A, Shohami E, Pinkas-Kramarski R. Closed head injury induces upregulation of Beclin 1 at the cortical site of injury. J Neurotrauma. 2005;22:750–762. doi: 10.1089/neu.2005.22.750. [DOI] [PubMed] [Google Scholar]
- Duennwald ML, Lindquist S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes & Dev. 2008;22:3308–3319. doi: 10.1101/gad.1673408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen EL. Fine structure of the autophagosome. Methods Mol Biol. 2008;445:11–28. doi: 10.1007/978-1-59745-157-4_2. [DOI] [PubMed] [Google Scholar]
- Fujita E, Kouroku Y, Isoai A, Kumagai H, Misutani A, Matsuda C, Hayashi YK, Momoi T. Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: Ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II) Hum Mol Genet. 2007;16:618–629. doi: 10.1093/hmg/ddm002. [DOI] [PubMed] [Google Scholar]
- Gkogkas C, Middleton S, Kremer AM, Wardrope C, Hannah M, Gillingwater TH, Skehel P. VAPB interacts with and modulates the activity of ATF6. Hum Mol Genet. 2008;17:1517–1526. doi: 10.1093/hmg/ddn040. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hetz C, Thielen P, Fisher J, Pasinelli P, Brown RH, Korsmeyer S, Glimcher L. The proapoptotic BCL-2 family member BIM mediates motoneuron loss in a model of amyotrophic lateral sclerosis. Cell Death Differ. 2007;14:1386–1389. doi: 10.1038/sj.cdd.4402166. [DOI] [PubMed] [Google Scholar]
- Hetz C, Lee AH, Gonzalez-Romero D, Thielen P, Castilla J, Soto C, Glimcher LH. Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proc Natl Acad Sci. 2008;105:757–762. doi: 10.1073/pnas.0711094105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilieva EV, Ayala V, Jove M, Dalfo E, Cacabelos D, Povedano M, Bellmunt MJ, Ferrer I, Pamplona R, Portero-Otin M. Oxidative and endoplasmic reticulum stress interplay in sporadic amyotrophic lateral sclerosis. Brain. 2007;130:3111–3123. doi: 10.1093/brain/awm190. [DOI] [PubMed] [Google Scholar]
- Jaarsma D, Teuling E, Haasdijk ED, De Zeeuw CI, Hoogenraad CC. Neuron-specific expression of mutant superoxide dismutase is sufficient to induce amyotrophic lateral sclerosis in transgenic mice. J Neurosci. 2008;28:2075–2088. doi: 10.1523/JNEUROSCI.5258-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabuta T, Suzuki Y, Wada K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu,Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J Biol Chem. 2006;281:30524–30533. doi: 10.1074/jbc.M603337200. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi H, Almer G, Yamashita S, Guegan C, Nagai M, Xu Z, Sosunov AA, McKhann GM, Przedborski S. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc Natl Acad Sci. 2006;103:6025–6030. doi: 10.1073/pnas.0509227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepore AC, Haenggeli C, Gasmi M, Bishop KM, Bartus RT, Maragakis NJ, Rothstein JD. Intraparenchymal spinal cord delivery of adeno-associated virus IGF-1 is protective in the SOD1G93A model of ALS. Brain Res. 2007;1185:256–265. doi: 10.1016/j.brainres.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M, Corcoran M, Cepeda D, Nielsen ML, Zubarev R, Ponten F, Uhlen M, Hober S, Grander D, Sangfelt O. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol Biol Cell. 2007;18:1670–1682. doi: 10.1091/mbc.E06-03-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Lisbona F, Rojas-Rivera D, Thielen P, Zamorano S, Todd D, Martinon F, Glavic A, Kress C, Lin JH, Walter P, et al. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1α. Mol Cell. 2009;33:679–691. doi: 10.1016/j.molcel.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus S, Lisbona F, Torres M, Leon C, Thielen P, Hetz C. The stress rheostat: An interplay between the unfolded protein response (UPR) and autophagy in neurodegeneration. Curr Mol Med. 2008;8:157–172. doi: 10.2174/156652408784221324. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M, Calanca V, Galli C, Lucca P, Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- Morimoto N, Nagai M, Ohta Y, Miyazaki K, Kurata T, Morimoto M, Murakami T, Takehisa Y, Ikeda Y, Kamiya T, et al. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–117. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- Nishitoh H, Kadowaki H, Nagai A, Maruyama T, Yokota T, Fukutomi H, Noguchi T, Matsuzawa A, Takeda K, Ichijo H. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes & Dev. 2008;22:1451–1464. doi: 10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: Insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid β accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rami A, Langhagen A, Steiger S. Focal cerebral ischemia induces upregulation of Beclin 1 and autophagy-like cell death. Neurobiol Dis. 2008;29:132–141. doi: 10.1016/j.nbd.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- Stam NC, Nithianantharajah J, Howard ML, Atkin JD, Cheema SS, Hannan AJ. Sex-specific behavioural effects of environmental enrichment in a transgenic mouse model of amyotrophic lateral sclerosis. Eur J Neurosci. 2008;28:717–723. doi: 10.1111/j.1460-9568.2008.06374.x. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kanekura K, Levine TP, Kohno K, Olkkonen VM, Aiso S, Matsuoka M. ALS-linked P56S-VAPB, an aggregated loss-of-function mutant of VAPB, predisposes motor neurons to ER stress-related death by inducing aggregation of co-expressed wild-type VAPB. J Neurochem. 2009;108:973–985. doi: 10.1111/j.1471-4159.2008.05857.x. [DOI] [PubMed] [Google Scholar]
- Urushitani M, Sik A, Sakurai T, Nukina N, Takahashi R, Julien JP. Chromogranin-mediated secretion of mutant superoxide dismutase proteins linked to amyotrophic lateral sclerosis. Nat Neurosci. 2006;9:108–118. doi: 10.1038/nn1603. [DOI] [PubMed] [Google Scholar]