Abstract
The accurate control of cell proliferation and survival is critical for animal development. The Hippo tumor suppressor pathway regulates both of these parameters by controlling the nuclear availability of the transcriptional coactivator Yorkie (Yki), which regulates downstream target genes together with Scalloped (Sd), a DNA-binding protein. Here we provide evidence that Yki can also regulate target genes in conjunction with Homothorax (Hth) and Teashirt (Tsh), two DNA-binding transcription factors expressed in the uncommitted progenitor cells of the Drosophila eye imaginal disc. Clonal analyses demonstrate that Hth and Tsh promote cell proliferation and protect eye progenitor cells from apoptosis. Genetic epistasis experiments suggest that Hth and Tsh execute these functions with Yki, in part by up-regulating the microRNA bantam. A physical interaction between Hth and Yki can be detected in cell culture, and we show that Hth and Yki are bound to a DNA sequence ∼14 kb upstream of the bantam hairpin in eye imaginal disc cells, arguing that this regulation is direct. These data suggest that the Hippo pathway uses different DNA-binding transcription factors depending on the cellular context. In the eye disc, Hth and Tsh provide spatial information to this pathway, promoting cell proliferation and survival in the progenitor domain.
Keywords: Drosophila eye development, Hippo signaling pathway, Homothorax, Yorkie, apoptosis, cell proliferation
The coordination between cell proliferation and differentiation is critical for animal development and organogenesis. The development of the Drosophila compound eye is an excellent model system to address how these two processes are coordinated, in part because the transition from proliferation to differentiation can be visualized in a single eye imaginal disc as the morphogenetic furrow (MF) sweeps across the eye disc epithelium (Ready et al. 1976). Consequently, individual eye discs isolated from the third instar larval stage display the entire range of differentiation states, from uncommitted naïve progenitor cells in the anterior of the disc to fully committed photoreceptors in the posterior of the disc.
While much is known about how the network of retinal differentiation genes, together with secreted signals derived from the MF, choreograph the formation of the many cell types present in the differentiated eye (Desplan 1997; Treisman and Heberlein 1998; Silver and Rebay 2005; Morante et al. 2007), much less is known about the uncommitted progenitor cells prior to differentiation. These cells must accomplish at least two tasks. First, they must proliferate and survive in order to generate sufficient cells before the differentiation program begins. Second, they have to keep the differentiation program in check prior to the arrival of the MF. In the developing eye disc, retinal progenitor cells reside anterior to the MF, where the process of retinal differentiation begins. These naïve progenitor cells express two transcription factors—Homothorax (Hth), a TALE-homeodomain protein (Rieckhof et al. 1997), and Teashirt (Tsh), a zinc (Zn) finger transcription factor (Fasano et al. 1991)—both of which have important functions in other contexts during development. In the eye, hth is activated by Wingless (Wg), expressed from the periphery of eye field (Pichaud and Casares 2000), and is repressed by Decapentaplegic (Dpp), expressed from the MF (Bessa et al. 2002). In vivo, Hth and Tsh interact physically with each other and function together to repress the later-acting retinal differentiation genes, such as eyes absent (eya) and sine oculus (so), thus maintaining anterior eye disc cells in an undifferentiated state (Bessa et al. 2002). However, it has remained an open question whether Hth and Tsh also promote cell proliferation and survival of these progenitor cells and, if they do, what pathways and target genes these two transcription factors regulate to mediate these functions.
Recently, the Hippo tumor suppressor pathway has been shown to be an important cell proliferation control pathway in Drosophila and mammals (Dong et al. 2007; Pan 2007; Zhang et al. 2009). Activation of the Hippo kinase leads to the phosphorylation of a second kinase, Warts, which in turn phosphorylates and induces the cytoplasmic retention of Yorkie (Yki), a transcriptional coactivator that does not have its own DNA-binding domain (Huang et al. 2005; Dong et al. 2007; Oh and Irvine 2008; J Zhang et al. 2008). In its unphosphorylated state, Yki is nuclear and participates in the activation of growth- and survival-promoting target genes. One such Yki target gene is the microRNA (miRNA) bantam, which in turn represses the translation of the proapoptotic gene hid (Hipfner et al. 2002; Brennecke et al. 2003; Nolo et al. 2006; Thompson and Cohen 2006). In the wing disc, where it has been best studied, Yki regulates its target genes by binding to Scalloped (Sd), a TEAD/TEF family transcription factor (Goulev et al. 2008; Wu et al. 2008; L Zhang et al. 2008; Zhao et al. 2008). Although the Hippo signaling pathway controls growth in all known tissues, including the eye, Sd's expression pattern and requirement during development may be more limited (Campbell et al. 1992; Srivastava et al. 2004; L Zhang et al. 2008). For example, an enhancer trap into the sd locus, which reports sd's expression pattern, is not active in anterior eye disc cells (Campbell et al. 1992), and sd-null clones survive well in the eye imaginal disc but not in the wing pouch (L Zhang et al. 2008). These and other observations suggest that nuclear Yki may promote cell survival and proliferation in other tissues by interacting with transcription factors in addition to Sd.
Here we show that Hth and Tsh work together to promote cell proliferation and survival in the anterior eye disc. Genetic epistasis experiments suggest that Tsh and Hth work via the Hippo signaling pathway to execute these functions. Specifically, we provide evidence that bantam expression is up-regulated in anterior eye disc cells, and that this up-regulation is hth-dependent. Further, bantam and yki are both necessary for the proliferation-promoting functions of Hth and Tsh. Finally, we show that Hth and Yki are bound at the bantam locus in eye disc cells and that Yki and Hth can be coimmunoprecipitated when coexpressed. Together, these results provide strong evidence that Hth and Tsh, together with Yki, promote cell proliferation and survival of eye progenitor cells by directly up-regulating the bantam miRNA. Thus, the transcriptional regulation of hth provides spatial specificity to the Hippo pathway, ensuring that anterior eye disc cells, but not cells posterior to the MF, remain in a state of active proliferation.
Results
Hth and Tsh are required for cell survival and wild-type proliferation in the eye progenitor domain
The anterior progenitor domain of the eye imaginal disc expresses Hth and Tsh, with Tsh expression extending closer to the MF than Hth (Fig. 1A–E). As noted previously (Bessa et al. 2002), hthP2 mutant clones are rarely recovered anterior to the MF, but can be recovered posterior to the MF (Fig. 1F). In contrast, neutral control clones made in parallel are recovered throughout the eye disc (Fig. 1G). This indicates that the absence of hth results in poor survival of progenitor cells. The existence of hthP2 mutant clones posterior to the MF suggests that hthP2 mutant cells can divide and survive long enough to be “fixed” by the passage of the MF, after which hth is no longer required for survival. Loss-of-function tsh clones are also at a growth disadvantage in the progenitor domain, although in this case we had to use RNAi knockdown of tsh in a genetic background that was null for the highly related and functionally redundant gene tiptop (tio) to see an effect (Supplemental Fig. 1; Fasano et al. 1991; Laugier et al. 2005).
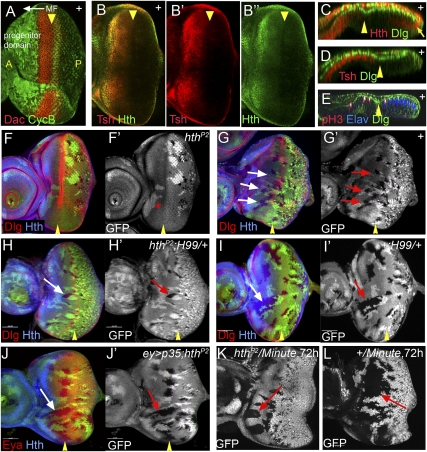
Figure 1.
hth is required for cell survival in the eye progenitor domain. (A) A third instar eye disc stained for CycB (green) and Dac (red, which is expressed anterior to the MF) to highlight the progenitor domain and the initiation of differentiation, respectively. The position and direction of the MF is indicated. (B) A third instar eye disc stained for Hth (green) and Tsh (red). (C–E) Confocal cross-section of third instar eye discs stained for Dlg (green, all panels), Hth (red, C), Tsh (red, D), pH3 (red, E), and Elav (blue, E). The position of MF is indicated (yellow arrowheads). Hth is present in the anterior of the main epithelium, throughout the peripodial epithelium, and in posterior margin cells (arrow). Tsh is expressed only in the main epithelium, and extends closer to the MF compared with Hth. Dlg is used here as a cell membrane marker. (F,G) Survival of hthP2 (F) or neutral clones (G) (marked by the absence of GFP) in eye discs stained for Hth (blue) and Dlg (red, which marks the MF, arrowheads). hthP2 clones are not recovered anterior to the MF, but twin spots (bright patches in F′,G′) are recovered. (H–J) A few hthP2 clones (marked by the absence of GFP, green/white) are recovered anterior to the MF in a H99/+ background (H) or when the apoptosis inhibitor p35 is expressed (J), but not as readily as neutral control clones (I). (K,L) Comparison of the growth of hthP2 Minute+ (K) and hth+ Minute+ (L) clones made in a Minute+/Minute− background.
The absence of hthP2 clones anterior to the MF is reminiscent of cell competition, where cells that have a growth disadvantage relative to their neighbors are eliminated (Simpson and Morata 1981; Morata and Martin 2007). At least one mechanism leading to the elimination of cells is apoptosis (de la Cova et al. 2004; Moreno and Basler 2004; Morata and Martin 2007). We carried out two experiments to test if hthP2 clones were eliminated by apoptosis in the anterior eye disc. When hthP2 clones were generated in a heterozygous Df(3L)H99/+ background, which removes one copy of the three proapoptotic genes hid, reaper, and grim, small hthP2 mutant clones were recovered anterior to the MF (Fig. 1H), although this rescue is not fully penetrant compared with neutral clones made side-by-side (Fig. 1I). Similar partial rescue was observed when hthP2 clones were generated in eye discs that express the baculovirus anti-apoptotic protein p35 (Fig. 1J). These results indicate that the poor survival of hthP2 clones in the anterior eye disc is, at least in part, because they are eliminated by apoptosis.
Another way to counteract the elimination of cells as a result of cell competition is to give them a growth advantage relative to their neighbors. In Drosophila, this can be accomplished by generating wild-type clones in a Minute (M)/+ background (Simpson and Morata 1981; Morata and Martin 2007). In the M(3)95A/+ background, hthP2 Minute+ clones were recovered anterior to the MF (Fig. 1K), demonstrating that hth is not essential for cells to survive and proliferate in the anterior eye disc. However, the size of these hthP2 Minute+ clones was dramatically smaller than the size of neutral (hth+ Minute+) clones generated in parallel in the same M/+ background (Fig. 1L). Thus, although hth is not absolutely required for progenitor eye disc cells to divide, their ability to proliferate is compromised in the absence of hth. Together with the partial rescue of hthP2 clones observed when apoptosis was blocked, these results suggest that hth carries out at least two functions in the eye progenitor domain: It increases cell survival by blocking apoptosis, and it promotes cell proliferation.
Coexpression of Hth and Tsh results in overgrowths
Previous work established that Hth works together with the Zn finger transcription factor Tsh to repress retinal determination genes in Drosophila (Bessa et al. 2002). Normally, hth is expressed in the anterior progenitor cells of the eye disc as well as in the peripodial cell layer (Fig. 1C,D). Although hth is repressed by Dpp derived from the MF, it is also expressed at the very posterior margin of the eye disc (Fig. 1B,C). Unlike hth, tsh is not expressed in the peripodial cell layer, nor is it expressed in posterior margin cells (Fig. 1B–D). In fact, tsh's restriction to the main epithelium of the eye disc helps to distinguish between these two tissue layers (Bessa and Casares 2005).
If both Tsh and Hth were required to promote proliferation in eye progenitor cells, we would expect that ectopic expression of Tsh would only be able to induce overgrowths in cells that already express hth. Consistent with this prediction, when Tsh was expressed ectopically in clones, posterior margin cells and peripodial cells could be induced to overgrow (Fig. 2A). In contrast, Tsh+ clones posterior to the MF in the main epithelium did not overgrow and instead differentiated into photoreceptor clusters with apparently normal morphology (Fig. 2A). Thus, there is a strong correlation between Tsh and Hth coexpression and their ability to induce overgrowths. Consistently, when both Hth and Tsh are coexpressed in clones (hereafter referred to as Hth + Tsh clones), they overgrow regardless of where they occur in the eye disc (Fig. 2F).
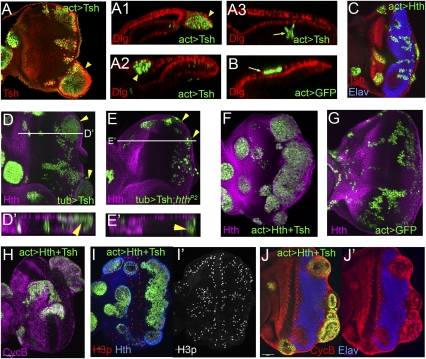
Figure 2.
Hth and Tsh function together to induce proliferation. All panels show eye discs with positively marked (GFP+) clones. (A,B) Eye discs containing clones that ectopically express Tsh (A) or GFP (B), stained for GFP (green, to mark the clones) and Dlg (red). Compared with neutral clones (B), Tsh+ clones result in overgrowths (arrowheads) when they arise in the posterior margin (A1) or in the peripodial epithelium (A2), but not when they arise in the main epithelium (A3). (C) Eye disc containing clones that ectopically express Hth (marked with GFP, green), stained for Tsh (red) and Elav (blue). These clones do not result in overgrowths. (D,E) MARCM clones that ectopically express Tsh (D) or that ectopically express Tsh and are also mutant for hth (E), stained for Hth (purple) and GFP (to mark the clones). Tsh+ clones result in overgrowths (arrowheads in D), but Tsh+; hthP2 clones do not (arrowheads in E). D′ and E′ show confocal cross-sections indicated by the lines in D and E. (F,G) Eye discs containing Hth + Tsh clones (F) or neutral clones (G) stained for Hth (purple) and GFP (to mark the clones). Hth + Tsh clones always overgrow compared with control clones made in parallel. (H–J) Eye discs containing Hth + Tsh clones up-regulate CycB (purple, H) and pH3 (red/ white in I,I′), and repress Elav (blue, J,J′).
As an additional experiment to test whether Hth and Tsh are both required to induce overgrowths, we used mosaic analysis with a repressible cell marker (MARCM) to generate hthP2 clones that simultaneously express Tsh. These Tsh+; hthP2 clones never overgrow, regardless of where they are located in the disc (Fig. 2E [cf. Tsh+; hth+ MARCM clones generated in parallel], D). These data strongly support the idea that Hth and Tsh must be coexpressed to induce proliferation.
We next examined the effect of Hth + Tsh expression on cell cycle and differentiation markers. The G2 cyclin Cyclin B (CycB) is normally expressed in proliferating anterior progenitor cells and in a row of cells posterior to the MF that corresponds to the second mitotic wave. In Hth + Tsh clones posterior to the MF, CycB expression is up-regulated (Fig. 2H). Similarly, staining for phosphorylated histone 3 (pH3), a marker for cells in mitosis, indicates that the cells in Hth + Tsh clones are actively dividing (Fig. 2I). Finally, we examined Elav, a marker for neural differentiation. In agreement with previous results showing that the retinal determination genes eya and so are repressed by Hth + Tsh (Bessa et al. 2002), Elav is repressed in Hth + Tsh-expressing clones (Fig. 2J). Together, these results indicate that when Hth and Tsh are coexpressed in the eye disc, they promote proliferation and block differentiation, mimicking the two main properties of anterior progenitor cells, which normally express these transcription factors.
Hth + Tsh function with the Hippo pathway
In order to identify which pathways Hth and Tsh function with to promote proliferation, we carried out several genetic tests using mutations that either activate or inactivate pathways previously implicated in growth control in the eye. We tested the Wg, Notch, and Jak–Stat signaling pathways, all implicated in tissue growth regulation in Drosophila (Lee and Treisman 2001; Kenyon et al. 2003; Dominguez et al. 2004; Firth and Baker 2005; Ekas et al. 2006; Tsai et al. 2007). With the exception of Wg, which is required for hth expression in the progenitor domain (Pichaud and Casares 2000), manipulation of these pathways had no effect on hth or tsh expression (data not shown). Moreover, none of these pathways were required for ectopic Hth + Tsh-induced overgrowths (Supplemental Fig. 2). Based on these data, these three pathways are unlikely to mediate the proliferation and survival functions executed by Hth and Tsh in the anterior eye disc.
In contrast to these results, we found that Hth and Tsh require components of the Hippo pathway to carry out their proliferation-inducing functions. First, although wtsP2 clones proliferate well throughout the eye disc and lead to modest overgrowths, wtsP2 hthP2 double-mutant clones behave like hthP2 clones: They fail to survive in the anterior of the eye disc (Fig. 3A,B). Similarly, although ectopic expression of Yki results in overgrowths throughout the eye disc, Yki+, hthP2 clones do not survive anterior to the MF (Fig. 3C,D). These results argue that the inability of hth mutant clones to survive anterior to the MF cannot be rescued by activating the Hippo pathway. Conversely, they show that even when the Hippo pathway is in its growth-promoting state (Yki+ or wts−), it cannot induce proliferation in the eye progenitor domain in the absence of hth.
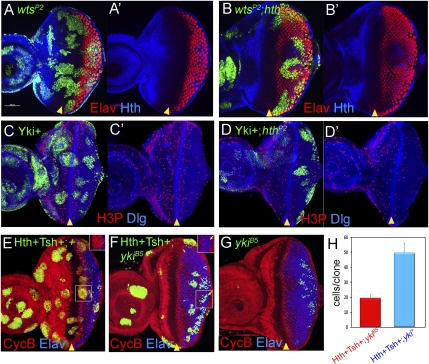
Figure 3.
Codependency of the Hippo pathway and Hth + Tsh to induce proliferation in the eye progenitor domain. A–G all show eye discs with positively marked (GFP+) MARCM clones. (A,B) Eye discs with wtsP2 (A) and wtsP2 hthP2 (B) clones, stained for GFP (green, to mark the clones), Hth (blue), and Elav (red). wtsP2 clones grow well throughout the eye disc, whereas wtsP2 hthP2 clones are not recovered in the progenitor domain. (C,D) Eye discs with Yki+ (C) or Yki+; hthP2 (D) clones stained for GFP (green, to mark the clones), pH3 (red), or Dlg (blue). Yki+ clones grow well throughout the eye disc, whereas Yki+; hthP2 clones are not recovered in the progenitor domain. (E–G) Eye discs with Hth + Tsh (E), Hth + Tsh; ykiB5 (F), or ykiB5 (G) clones stained for GFP (green, to mark the clones), CycB (red), and Elav (blue). In E and F, the insets show only the Elav and CycB stains of the boxed regions. Note that Hth + Tsh clones, but not Hth + Tsh; ykiB5 clones, repress the differentiation marker Elav and up-regulate CycB. (H) The number of cells present in Hth + Tsh; ykiB5 clones (red bar; n = 55) and Hth + Tsh clones (blue bar; n = 58).
To provide further genetic support for these conclusions, we tested if the overgrowths produced by Hth + Tsh require yki. As described above, Hth + Tsh clones overgrow no matter where they are produced in the eye disc (Fig. 3E,H). In contrast, Hth + Tsh; ykiB5 clones generated in parallel grow much smaller and are rarely recovered anterior to the MF (Fig. 3F,H). Unlike Hth + Tsh clones, Hth + Tsh; ykiB5 clones do not repress Elav, suggesting that they are unable to block differentiation (Fig. 3F). Hth + Tsh; ykiB5 clones do, however, grow better than ykiB5 clones (Fig. 3G), suggesting that not all of the growth-promoting functions of Hth + Tsh may require yki. These conclusions are consistent with another manipulation of the Hippo pathway that, like removing yki, causes cells to proliferate poorly. Clones that overexpress the Hpo kinase grow poorly, especially in the anterior of the eye disc (Supplemental Fig. 3A). Overexpressing Hpo+ can suppress most, but not all, of the growth-promoting effects of ectopic Hth + Tsh expression (Supplemental Fig. 3B,C).
Thus, in the eye progenitor domain, the growth-promoting effects observed when the Hippo pathway is compromised require hth. One scenario that could explain these observations is if hth or tsh were transcriptional targets of the Hippo pathway. This is ruled out, however, because manipulating the activity of the Hippo pathway does not affect the patterns of Hth and Tsh expression in the eye disc (Supplemental Fig. 4A; data not shown). We also tested if Sd, the only previously described transcription factor in the Hippo pathway, was required for proliferation the anterior eye disc. In contrast to hthP2 clones, sd-null clones were recovered in the anterior eye disc, arguing that Sd is not required for eye progenitor cell proliferation or survival (Supplemental Fig. 5A,B). Moreover, we found that Hth + Tsh can induce overproliferation in the eye disc in the absence of sd (Supplemental Fig. 5C,D). Together, these data suggest a model in which, like Sd and Yki in the wing pouch, Hth + Tsh and Yki directly regulate Hippo pathway targets in the anterior eye disc. Below, we present biochemical and genetic data that further support this hypothesis.
Hth and Tsh regulate bantam in eye progenitor cells
Because the overgrowth-inducing property of Hth + Tsh depends on yki and the ability of Yki+ clones to grow in the eye progenitor domain depends on hth, we considered the possibility that they work together to regulate common targets in the anterior eye disc. Furthermore, because hthP2 clones can be partially rescued by blocking apoptosis, we focused on the known Yki target bantam, which encodes a miRNA that blocks the translation of the proapoptotic gene hid (Brennecke et al. 2003). Based on these findings, we tested whether Hth + Tsh activate bantam in the anterior progenitor domain of the eye disc.
To monitor bantam, we used a bantam sensor in which a green fluorescent protein (GFP) transgene contains two perfect copies of a sequence that is complementary to the bantam miRNA in its 3′ untranslated region (UTR) (Brennecke et al. 2003). As this transgene is transcribed ubiquitously via a tubulin promoter, GFP levels are inversely proportional to the levels of the bantam miRNA. In early third instar eye discs, the bantam sensor is expressed at lower levels in the anterior regions compared with the posterior regions (Fig. 4A,B). Double staining with Hth and Tsh demonstrates that where Tsh and Hth levels are high, bantam sensor levels are low (indicating high bantam levels). At a later stage of eye disc development (mid–late third instar), these relationships remain the same, but now the sensor is expressed at especially high levels in differentiating photoreceptors (Fig. 4C,D). This high level of sensor expression is similar to that observed in ban-null clones (data not shown), suggesting that it represents the complete absence of bantam. The inverse correlation between Hth + Tsh and bantam sensor levels suggests that bantam is up-regulated by these factors in the anterior of the eye disc.
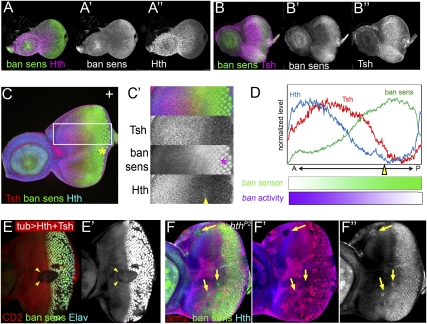
Figure 4.
Control of bantam expression by Hth + Tsh. (A,B) Early to mid-third instar eye discs stained for the bantam sensor (green) and Hth (purple, A) or Tsh (purple, B). (C) Late third instar eye disc stained for Tsh (red), ban sensor (green), and Hth (blue). C′ shows a blow-up and individual channels for the region boxed in C. (D) Quantification of the Hth, Tsh, and ban sens stains in the regions boxed in C. bantam activity (purple gradient) is inferred to be the inverse for what is observed for the ban sensor (green gradient). (E) Eye disc containing clones expressing Hth + Tsh (absence of red, CD2) stained for the ban sensor (green/white in C,C′) and Elav (blue in C). Hth + Tsh clones repress both the very high levels of the ban sensor in the Elav+ cells and the intermediate ban sensor levels that are present close to the MF (arrowheads). (F) Eye disc containing hthP2 clones made in a H99/+ background stained for β-gal (red, absence of arm-lacZ expression marks the clones), Hth (blue), and the ban sensor (green). An increase in ban sensor levels compared with surrounding cells can be seen in three of three hthP2 clones that survived in the progenitor domain (arrows).
Both loss-of-function and gain-of-function experiments support the idea that bantam is regulated by Hth + Tsh in eye progenitor cells. In clones that coexpress Hth + Tsh, the levels of the bantam sensor are reduced, suggesting that bantam is up-regulated (Fig. 4E). Conversely, hthP2 clones anterior to the MF [made in a Df(3L)H99/+ background to allow their survival] show increased levels of the bantam sensor (Fig. 4F). The levels observed in hthP2 clones are not as high as those in differentiated photoreceptors, suggesting that some bantam expression remains in the absence of hth. Thus, hth boosts the levels of bantam above a hth-independent baseline. In contrast to these effects on bantam, Hth + Tsh clones in eye discs had only a weak effect or no effect on other previously described Hippo pathway targets, cycE, diap1, and expanded (Supplemental Fig. 6). Taken together, these results suggest Hth + Tsh are required for high bantam levels that are normally present in the eye progenitor domain.
bantam is required for ectopic Hth + Tsh-induced overgrowths
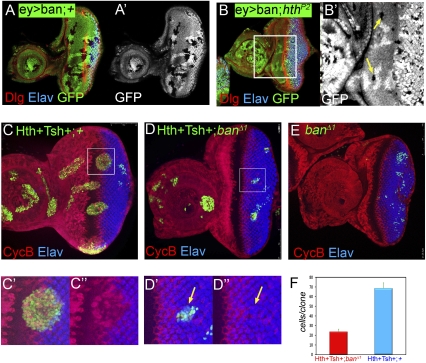
Based on the observations that Hth + Tsh regulate bantam, we carried out two additional genetic tests to determine if this regulation was relevant to eye progenitor cells. First, we reasoned that because bantam carries out its functions in part by repressing the translation of the proapoptotic gene hid, and thereby protects cells from apoptosis, hthP2 clones might survive in the anterior eye disc if bantam expression was provided independently of hth. Consistent with this idea, hthP2 clones could be recovered in the progenitor domain when bantam was expressed throughout the eye disc (Fig. 5A,B). This rescue was incomplete, similar to that observed when p35 was used to rescue hthP2 clones.
Figure 5.
Mutual dependence of bantam and Hth + Tsh for cell proliferation and survival. (A,B) Eye discs expressing bantam via eyeless-Gal4 (ey > ban) containing neutral clones (A) or hthP2 clones (B) stained for GFP (a clone marker), Elav (blue), and Dlg (red). A few hthP2 clones surviving in the progenitor domain are observed in B (arrows). (C–E) Eye discs containing Hth + Tsh clones (C), Hth + Tsh; banΔ1 clones (D), or banΔ1 clones (E) stained for CycB (red) and Elav (blue). C′, C″, D′, and D″ show enlargements of the boxed regions in C and D, respectively. Hth + Tsh; banΔ1 clones (D) grow more poorly compared with Hth + Tsh clones (C) made in parallel. Note also that Hth + Tsh; banΔ1 clones fail to repress Elav and do not activate CycB. (F) The number of cells present in Hth + Tsh; banΔ1 clones (red bar, n = 44) and Hth + Tsh clones (blue bar, n = 69).
Second, we tested whether bantam was necessary for Hth + Tsh-induced overgrowths. As shown above, Hth + Tsh clones overgrow, up-regulate CycB, and repress Elav, regardless of where they originate in the eye disc. In contrast, Hth + Tsh; banΔ1 clones generated in parallel are much smaller, fail to survive in the anterior eye disc, and do not alter Elav or CycB expression levels (Fig. 5C-F). banΔ1 clones survive even more poorly than Hth + Tsh; banΔ1 clones (Fig. 5E). From these data, we conclude that, like yki, bantam is required for Hth + Tsh-induced overgrowths. In addition, these data suggest that Hth + Tsh have functions in addition to up-regulating bantam.
Hth and Yki can be copurified from S2 cells
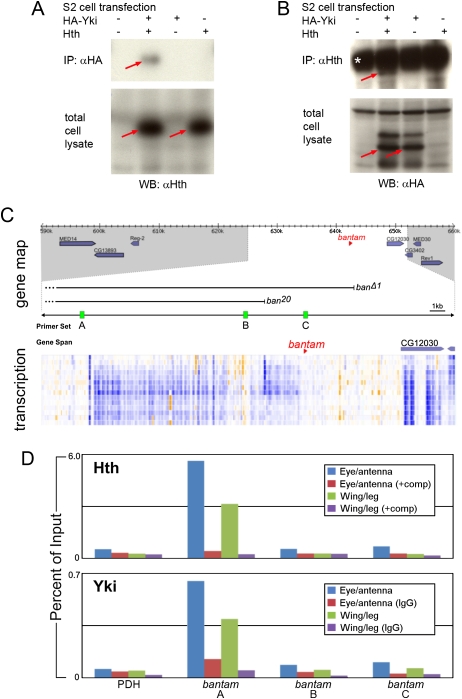
Previous results demonstrated that Hth and Tsh can directly interact with each other in eye discs (Bessa et al. 2002). Here, we tested the idea that Hth and Yki may interact physically with each other to regulate target genes. We tested if Hth and Yki can bind to each other by transfecting S2 cells with HA-tagged Yki and Hth. In cotransfected cells, Hth was coimmunoprecipitated with HA-Yki (Fig. 6A,B). These results suggest that Hth and Yki can interact physically with each other when coexpressed, consistent with the idea that they function together in vivo.
Figure 6.
Molecular interactions between Hth, Yki, and the bantam locus. (A,B) Western blots of extracts from S2 cells that were transfected with the indicated expression plasmids. Blots were probed with anti-Hth (A) or anti-HA (B). The top panels show blots of immunoprecipitations with anti-HA (A) or anti-Hth (B), and the bottom panels show blots of total cell lysates. In A, Hth is coimmunoprecipitated with HA-Yki. In B, HA-Yki is coimmunoprecipitated with Hth. In A, the red arrows point to Hth detected in total cell lysates and anti-HA immunoprecipitate. In B, the red arrows point to HA-Yki detected in total cell lysates and anti-Hth immunoprecipitate. The asterisk in B indicates cross-reaction from the IgG heavy chain in the anti-Hth antiserum. (C) The top section shows a gene map in the vicinity of bantam; the regions deleted in two ban deletion mutants (banΔ1 and ban20) are indicated (Hipfner et al. 2002; Yang et al. 2009). The middle section reproduces data from the modENCODE project measuring the amount of transcription around the bantam locus; each row represents a different developmental time point (from embryogenesis to second instar) and provides a measure of amount of mRNA encoded by that genomic position; blue indicates high expression, and orange indicates low expression (see http://www.modencode.org for details). Regions A, B, and C indicate the positions of three of 11 primers used for ChIP experiments; based on the modENCODE transcription map, region A is close to an apparent start of bantam transcription and was the only amplicon that gave a positive ChIP signal. (D) The results of ChIP experiments using anti-Hth (top) and anti-Yki (bottom) antibodies to precipitate chromatin prepared from the indicated tissues. In the anti-Hth experiment, “comp” refers to the addition of a competitor peptide that specifically blocks binding to Hth (see the Materials and Methods for details). In the anti-Yki experiments, the control was unprogrammed IgG. Immunoprecipitated chromatin was assayed for the presence of regions A, B, or C or for a negative control locus, PDH. Data are presented as a percentage of the signal obtained relative to input chromatin. For both anti-Hth and anti-Yki, a significant signal was obtained only for region A.
Hth and Yki bind to the bantam locus
We next used chromatin immunoprecipitation (ChIP) to ask if Hth and Yki bind to the bantam locus in eye–antenna imaginal discs. Using this assay, we identified a region ∼14 kb upstream of the bantam hairpin that was immunoprecipitated by both anti-Hth and anti-Yki (Fig. 6D). Based on modENCODE data (http://modencode.oicr.on.ca/cgi-bin/gbrowse/fly), this site appears to be close to a start site for a primary bantam transcript (Fig. 6C). Although binding was stronger in the eye–antenna disc, the same region was immunoprecipitated from chromatin isolated from wing and leg imaginal discs (Fig. 6D). Two other regions at the bantam locus failed to be immunoprecipitated from either imaginal disc tissue, nor did a negative control region from pyruvate dehydrogenase (PDH). These data argue that Hth and Yki regulation of bantam is direct. Although the available antibodies against Tsh failed to work in ChIP experiments (data not shown), the observation that Hth and Tsh bind to each other in vivo (Bessa et al. 2002) suggests that Tsh is also a direct regulator of bantam.
Discussion
Coordination of cell proliferation, survival, and differentiation in eye development
We provide evidence that Hth and Tsh promote cell proliferation and survival in the undifferentiated progenitor cells of the eye imaginal disc. Further, our experiments suggest that these transcription factors carry out these functions with the coactivator Yki, a downstream component of the Hippo signaling pathway. Sd, which acts with Yki to mediate the regulation of Hippo targets elsewhere in the fly, does not seem to be required in this context. Finally, our data suggest that at least one direct Hth–Yki target in the eye disc is the miRNA bantam, which has been shown previously to both promote proliferation and prevent apoptosis in other tissues (Brennecke et al. 2003; Nolo et al. 2006; Thompson and Cohen 2006). Together, these data suggest a model in which a Hth–Tsh–Yki complex functions in eye progenitor cells to promote proliferation and block apoptosis, at least in part by up-regulating the levels of the miRNA bantam (Fig. 7). Below we discuss the roles of both Hth + Tsh and the Hippo pathway in tissue growth control.
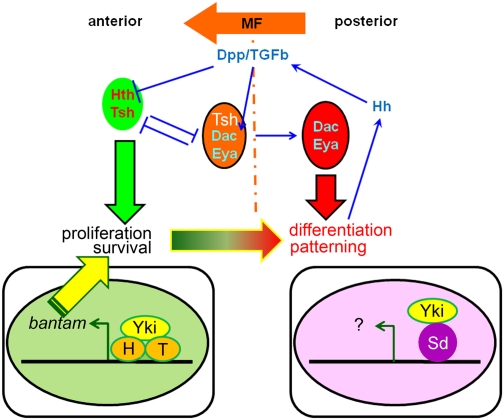
Figure 7.
Model for the switch from proliferation to differentiation in the eye disc. Hth and Tsh are expressed in the progenitor domain where they repress retinal differentiation genes such as dac and eya and also up-regulate bantam expression in conjunction with Yki. Signals such as Dpp from the MF repress hth and tsh, allowing dac and eya to execute the differentiation program. In this region of the eye disc, Yki may work with other transcription factors such as Sd to regulate a different subset of Hippo pathway target genes.
Role of Hth and Tsh in eye progenitor cells
Our evidence, together with previous findings (Bessa et al. 2002; Bessa and Casares 2005), suggests that Hth and Tsh function as partners to carry out two main functions in anterior eye disc cells: They repress the expression of the later-acting retinal determination factors (Bessa et al. 2002), and they promote cell proliferation (this study). That these functions require hth is supported by both loss-of-function studies as well as gain-of-function studies. For example, hthP2 clones fail to survive anterior to the MF, and Tsh's ability to induce overgrowths when ectopically expressed is abolished in the absence of hth. The involvement of Tsh is supported by gain-of-function experiments and our previous finding that Hth and Tsh directly interact with each other in vivo (Bessa et al. 2002). Carrying out loss-of-function genetics for tsh is difficult because this gene is located proximal to the standard Flp recombination targets (FRTs) used to generate mitotic recombination. In addition, the highly related gene tio, which is closely linked to tsh, functions redundantly with tsh in several instances, including some aspects of eye development (Fasano et al. 1991; Laugier et al. 2005). Nevertheless, we found that knocking down tsh using RNAi in a tio-null background results in poor survival in the progenitor domain. Taken together, these data provide a compelling argument for Hth + Tsh functioning together to promote cell survival in the anterior eye disc.
A functional relationship between Hth and Tsh also exists in other tissues in Drosophila, most notably in both wing and leg imaginal discs, where they are coexpressed in cells that will give rise to the proximal domains of these appendages. In both wings and legs, Tsh has the capacity to regulate hth when expressed in clones, and both tsh and hth have the ability to suppress distal appendage development when misexpressed (Rieckhof et al. 1997; Azpiazu and Morata 2000; Casares and Mann 2000; Wu and Cohen 2000, 2002; Zirin and Mann 2004). However, in these tissues, and unlike the eye disc, Hth + Tsh expression is not correlated with proliferation, which occurs uniformly throughout these discs. Consistently, the expression pattern exhibited by the bantam sensor does not correlate with Hth or Tsh in the leg or wing (Brennecke et al. 2003; Nolo et al. 2006; Thompson and Cohen 2006). The special relationship between proliferation and Hth + Tsh in the eye may be due in part to the Drosophila Pax6 homolog Eyeless (Ey), which is critical for eye identity (Gehring 1996). Moreover, Ey is found in a complex with Hth in vivo and participates with Hth and Tsh in the repression of retinal determination genes (Bessa et al. 2002). Thus, it may also be the case that Ey directly participates in the regulation of bantam together with Hth and Yki.
hth promotes, but is not essential for, cell proliferation and survival
Although hthP2 clones fail to survive in the eye progenitor domain, our data demonstrate that hth is not absolutely required for cells in this domain to proliferate. The effects we observed on the bantam sensor are consistent with the idea that hth promotes, but is not essential for, cells to proliferate in the eye progenitor domain. In hthP2 clones, bantam sensor levels increased above those normally observed in progenitor cells, but not as high as the levels observed in differentiated photoreceptors. Thus, if the level in photoreceptors represents the complete absence of bantam, these data imply that hth only up-regulates bantam over a basal hth-independent level. Moreover, the levels of the bantam sensor in other tissues, such as the wing disc, rarely approach those observed in photoreceptors, suggesting that most cells have some bantam expression, and that bantam regulators, such as hth, only serve to modulate bantam levels.
If eye progenitor cells have the capacity to proliferate in the absence of hth, how important is the proliferation-promoting function of hth? Although normal eyes can develop in animals in which hthP2 clones are generated, this is likely due to the ability of neighboring wild-type cells to compensate in this mosaic situation. In contrast, when wild-type and heterozygous cells are killed (using the EGUF method [ey-Gal4/UAS-flp/GMF-hid]) (Stowers and Schwarz 1999), we find that the remaining hthP2 tissue is unable to produce normal-sized eyes (Supplemental Fig. 7). This experiment suggests that the proliferation-promoting functions of hth in the eye progenitor domain are critical for normal eye development, likely by providing a sufficient pool of progenitor cells prior to differentiation.
The role of Yki partners in Hippo pathway specificity
The Hippo pathway has emerged recently as an important regulator of cell proliferation and survival in both vertebrates and invertebrates (Pan 2007; Zhang et al. 2009). In Drosophila, this pathway appears to regulate proliferation in nearly all tissues. For example, wts− clones or Yki+ clones have the capacity to induce overgrowths throughout the body (Harvey et al. 2003; Udan et al. 2003; Wu et al. 2003; Huang et al. 2005). As Yki and its mammalian ortholog Yap are transcriptional coactivators that do not have their own DNA-binding domain, they are thought to partner with DNA-binding transcription factors to regulate gene expression. Prior to this work, the only transcription factor proposed to work directly with Yki was Sd, a member of the TEAD/TEF family of DNA-binding proteins (Goulev et al. 2008; Wu et al. 2008; L Zhang et al. 2008; Zhao et al. 2008). However, unlike other components of the Hippo pathway, the available data suggest that Sd plays a more limited role in cell proliferation and survival in Drosophila (Campbell et al. 1992; Goulev et al. 2008; Wu et al. 2008; L Zhang et al. 2008; Zhao et al. 2008). In contrast to its essential role in the wing pouch, sd− clones survive well in other tissues, including the region of the wing disc that will give rise to the notum of the fly (Supplemental Fig. 5; Wu et al. 2008; L Zhang et al. 2008). Similarly, sd-null clones grow well in the eye progenitor domain. Thus, unlike in the wing pouch, sd is not required for cell survival and proliferation in the eye progenitor domain.
In contrast to the survival of sd clones in this domain, hthP2 clones fail to survive in the eye progenitor domain. Thus, analogous to sd in the wing pouch, hth is required for cells to survive and proliferate in the anterior eye imaginal disc. This observation suggests that hth could play an analogous role to sd in this progenitor domain, a view that is supported by our results. This evidence includes (1) Hth can interact with Yki when coexpressed in S2 cells, (2) Hth + Tsh regulate the Yki target bantam, and (3) Hth and Yki are both bound to the same region of the bantam locus in eye discs. Genetically, we show that the Hippo pathway is unable to induce overgrowths in the eye progenitor domain without hth, and that Hth + Tsh cannot induce overgrowths in the absence of Yki. These results suggest that Hth + Tsh comprise the DNA-binding transcription factors that function with Yki to regulate proliferation and survival genes, such as bantam. Thus, analogous to Sd in the wing pouch, Hth + Tsh are transcription factors used by the Hippo signaling pathway in eye progenitor cells.
The finding that Hth + Tsh play an analogous role in the eye progenitor domain as Sd does in the wing pouch has several implications for how the Hippo pathway is regulated in vivo. For one, the use of different DNA-binding transcription factors to regulate Hippo target genes suggests a previously unknown degree of specificity available to this pathway. Hth, a TALE family homeodomain protein, and Tsh, a Zn finger protein, are likely to bind very different target DNA sequences than Sd, a TEAD/TEF domain DNA-binding factor. Accordingly, we find that ectopic Hth + Tsh clones in the eye disc do not consistently up-regulate diap1 or expanded, known Sd–Yki targets in the wing disc (Supplemental Fig. 6; Harvey et al. 2003; Jia et al. 2003; Udan et al. 2003; Wu et al. 2003).
These results also imply that the transcriptional regulation of hth, tsh, and sd has the potential to change the output of the Hippo pathway. Because hth and tsh are transcriptionally repressed by signals coming from the MF, these factors are not available to work with the Hippo pathway posterior to the MF. However, loss of Hippo kinase activity can lead to proliferation of differentiated cells posterior to the MF. In these cells, sd is expressed (Campbell et al. 1992), suggesting that Yki may use this transcription factor in this context. Analogously, loss of Hippo kinase activity can cause overgrowths in the notum as well as in the wing pouch. As sd− clones grow well in the notum, but not in the wing pouch, these data suggest that the notum overgrowths may be mediated by a transcription factor other than Sd. hth clones also survive well in the notum, implying that yet another transcription factor or factors may work with Yki in this tissue. In sum, we suggest that Yki, and thus the Hippo pathway, may be able to work with multiple transcription factors to regulate target genes. In principle, the use of several transcription factors that are themselves developmentally regulated allows the Hippo pathway to be interpreted in different ways in different contexts.
Although our data suggest that the Hippo pathway uses Hth + Tsh to up-regulate bantam, they also suggest that both Hth + Tsh and Yki have additional, independent targets. For example, the loss of Hippo kinase activity leads to the up-regulation of diap1 throughout the eye disc (Wu et al. 2003). Because diap1 is not affected when Hth + Tsh are coexpressed, the Hippo pathway has the capacity to regulate some genes independently of Hth + Tsh, even in the eye progenitor domain. Moreover, at least when Yki is ectopically expressed, sd appears to be required in all regions of the eye disc for diap1 activation (Wu et al. 2008). Thus, although it has not been shown that sd is required for endogenous diap1 expression in this tissue, these data, together with those presented here, suggest that Yki may use both Sd and Hth + Tsh to regulate gene expression in the eye disc. In fact, L Zhang et al. (2008) suggest that sd is also a modifier of bantam expression in the eye disc and that sd is required for normal-sized eyes. However, these clones, which used RNAi to knock down Sd, grew well in the eye progenitor domain. In addition, the smaller eyes observed by L Zhang et al. (2008) when sd was knocked down could be due to the earlier embryonic expression of the Gal4 driver used in these experiments. In contrast, when generated during larval stages, hth− clones, but not sd− clones, fail to survive in the eye progenitor domain, arguing that, at least post-embryonically, gene regulation by Hth + Tsh, not Sd, is critical for cell survival in this tissue. This conclusion is also supported by our finding that Hth + Tsh can induce proliferation in the absence of sd (Supplemental Fig. 5).
As shown previously (Bessa et al. 2002), Hth + Tsh play a key role in blocking eye differentiation by repressing the retinal determination genes eya and so. The available data do not yet resolve whether this repression works independently of the Hippo pathway. On the one hand, the loss of Hippo kinase activity leads to overgrowths without blocking differentiation, arguing that nuclear Yki promotes proliferation without changing cell fate (Harvey et al. 2003; Udan et al. 2003; Wu et al. 2003). Consistently, we find that wts− or Yki+ clones do not alter Elav expression in differentiated photoreceptors. Curiously, however, ectopic expression of Hth + Tsh did not block differentiation in the absence of Yki. Although these data could be interpreted to suggest that Yki is directly required for repressing differentiation, they could alternatively suggest that repression requires cell proliferation. Consistently, Hth + Tsh were also unable to block differentiation in the absence of bantam. These observations raise the possibility that the absence of bantam or yki indirectly inhibits Hth + Tsh's ability to repress differentiation by compromising the proliferation of these cells, although other indirect affects are also possible.
Hth + Tsh are also likely to regulate genes in addition to bantam to promote proliferation and survival in the eye progenitor domain. This is most strongly supported by our observation that ectopic expression of bantam only partially rescues the survival of hthP2 clones. In addition, we found that the overgrowths generated by ectopic expression of Hth + Tsh are only partially suppressed by the coexpression of Hpo, whose overexpression removes Yki from the nucleus. These data suggest that some of the Hth + Tsh targets that mediate growth and survival in the eye progenitor domain are regulated independently of Yki.
hth and tsh as focal points for the switch from proliferation to differentiation
In summary, these results suggest that the transcriptional regulation of hth and tsh along the anterior–posterior axis of the eye disc changes the output of the Hippo pathway. In the eye progenitor domain, where Hth and Tsh are both present, the pathway uses these transcription factors to promote proliferation and cell survival, at least in part by up-regulating bantam. Once hth and tsh are repressed by signals coming from the MF, the Hippo pathway may use other transcription factors, such as Sd, to regulate a different set of target genes. Thus, together with other functions carried out by these transcription factors, their regulation across the anterior–posterior axis coordinates the complex switch from proliferation to differentiation during eye development.
Materials and methods
Drosophila strains and genetics
Wild-type flies were Oregon R or yw. Standard methods were used to recombine and balance chromosomes bearing mutations, markers, or transgenes. The mutations and transgenes used were hthP2 (Rieckhof et al. 1997); tsh8, tsh1,LacZ, UAS-hth∷GFP, UAS-hth∷3′UTR (Noro et al. 2006); and UAS-tsh (Zirin and Mann 2004). For gain-of-function clones, act > y+ > Gal4 or tub > CD2,y+ > Gal4 flip-out cassettes were used (Struhl and Basler 1993). Heat shocks were given 48 h ± 12 h after egg laying (AEL). For mutant clones, standard FRTs (Xu and Rubin 1993) with dominant visible markers (ubi > GFP or arm>LacZ) were used. Heat shocks were given 60 h ± 12 h AEL. For MARCM clones (Lee and Luo 1999), unless otherwise indicated, tub > Gal4 was used to express UAS-GFP and other UAS transgenes. The stocks generally contain yw, hsFLP, tub > Gal4. Heat shocks were given 36 h ± 12 h AEL to allow for Gal80 to fade. FRT arms containing tub > Gal80 are from Bloomington Stock Center or from G. Struhl and M. Zecca. UAS-p35, UAS-DIAP1, and Df(3L)H99 are from L. Johnston. The Minute mutation was M(3)95A. For the sd experiments, the null allele sdΔB was used; for the sdΔB; Hth + Tsh MARCM experiment, act-Gal4 was used instead of tub-Gal4.
wtsP2 and diap1lacZ were obtained from L. Johnston. UAS-Yki and ykiB5 were from D.J. Pan (Huang et al. 2005). UAS-bantam and banΔ1 were from S.M. Cohen (Brennecke et al. 2003). bantamEP3622 was from Bloomington Stock Center and gave the same results as UAS-bantam. ey > Gal4 and GMR > Gal4 drivers were from Bloomington Stock Center. UAS-Hpo/dMST1 was from J. Jiang (Jia et al. 2003). tub > EGFP∷2xanti-bantam (bantam sensor) lines were constructed by Brennecke et al. (2003) and were obtained from L. Johnston.
Antibodies
Guinea pig anti-Hth (GP52), rabbit anti-Hth, guinea pig anti-Dll, and guinea pig anti-Tsh were described previously (Casares and Mann 2000; Zirin and Mann 2004; Estella and Mann 2008). Rabbit anti-CycB and rabbit anti-CycE were gifts from H. Richardson (Richardson et al. 1995). Rabbit anti-Histone H3 subunit, Ser-10 phosphorylated, anti-Caspase-3, and anti-GFP were from Upstate Biotechnologies. Monoclonal antibody against Dlg, Dacapo, Wg, Elav, Eya, Dac, and CycE were from the Developmental Studies Hybridoma Bank. Phalloidin-AlexaFluor555 conjugate was from Molecular Probes and was used at the recommended concentration with secondary antibodies. Rabbit anti-β-Gal was from Cappel. Mouse anti-rat CD2 was from Invitrogen. Guinea pig anti-Stat92E was from S. Hou (Hou et al. 1996). Anti-Hth (dG-20) polyclonal antibody and Hth competitor peptide (dG-20P) used for ChIP analysis were purchased from Santa Cruz Biotechnologies. Anti-HA (3F10) monoclonal antibody was purchased from Roche.
Immunohistochemistry
Imaginal discs were dissected and immunostained using standard procedures. Secondary antibodies used were AlexaFluor488, AlexaFluor555, and AlexaFluo647 conjugates from Molecular Probes and were used at 1:1000. After staining, discs were washed five times in PBST, 15 min each at room temperature, and were dissected onto glass slides in VectaShield.
Optical section single images or z-series were collected on either Zeiss AxioScope/ApoTome, Bio-Rad MRC1024 confocal microscope, or Leica SP5 LSM confocal system. Z-series were analyzed by ImageJ (http://rsbweb.nih.gov/ij). Other image analysis was done with Photoshop CS3.
S2 cell transfection, immunoprecipitation, and Western blot
S2 cells were from N. Senoo-Matsuda and maintained at room temperature in Drosophila Schneider medium with glutamate supplemented with 10% fetal bovine serum, 5mg/mL penicillin–streptomycin, and 2.5 mg/mL Bacto Peptone. pAc-HA-Yki plasmid was from D.J. Pan (Huang et al. 2005). pAc-Hth and pAc-GFP were made by J. Culi (Culi and Mann 2003). For each construct, 15 μg of plasmid DNA were transfected into S2 cells by Effectene (Qiagen). Cells were lysed in Noro's RIPA buffer (20 mM Tris at pH 7.5, 0.5% NP-40, 150 mM NaCl, 10 mM MgCl2). Lysates were passed through a 25-gauge needle five times and cleared by centrifugation. Immunoprecipitation in RIPA buffer was with either 1 μg/mL anti-HA (3F10) or 2 μL/mL GP52 antiserum. Protein A/G-agarose beads captured the immunoprecipitates and were washed five times with RIPA buffer. Immnoprecipitates were denatured in 1× SDS sample buffer.
Immunoprecipitates and control lysates were separated by 10% SDS-PAGE and blotted to PVDF membrane (Millipore). The blot was blocked in TBST (10 mM Tris at pH 7.6, 150 mM NaCl, 0.1% Tween-20) with 5% skim milk for 30 min at room temperature and was incubated with primary antibody (1:2000 for GP52 anti-Hth, 1:400 for anti-HA, 3F10) in TBST + 5% milk at 4°C overnight. After five 15-min washes at room temperature in TBST, blots were incubated for 2 h at room temperature with HRP-conjugated secondary antibody. The blot was washed five times for 15 min at room temperature in TBST, and was detected by ECL Plus (GE Healthcare).
ChIP
Mid-third instar larvae (96 h AEL) were dissected and imaginal discs were collected in PBS on ice. Dissection time was minimized to <1 h to process 80–120 animals. Discs were fixed with 1.8% formaldehyde. Chromatin preparation and immunoprecipitation were performed as described (McKay et al. 2009). Anti-Hth (dG-20) (1.5 μg) was used for each immunoprecipitation. Specificity for the anti-Hth antibody was confirmed by comparing the signal obtained from parallel immunoprecipitations carried out in the presence of the dG-20P competitor peptide, which specifically blocks antigen recognition (Santa Cruz Biotechnologies). Rabbit anti-Yki (D. Pan) was used at a final dilution of 1:300, and specificity was tested by parallel immunoprecipitations carried out with normal rabbit IgG (Santa Cruz Biotechnologies). Eleven real-time PCR amplicons surrounding the bantam hairpin (from ∼14 kb 5′ to ∼3 kb 3′) were used to quantify immunoprecipitated chromatin. Of these, only the primer set labeled A in Figure 6 produced a positive signal. Sequences of the primers are available on request.
Acknowledgments
We thank S. Cohen, L. Fasano, G. Halder, J. Jiang, L. Johnston, R. da Silva, G. Struhl, D.J. Pan, H. Richardson, and M. Zecca for reagents and fly stocks, and L. Johnston and A. Tomlinson for comments on the manuscript. This work was supported by NIH grant GM 058575 awarded to R.S.M.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1820009.
Supplemental material is available at http://www.genesdev.org.
References
- Azpiazu N, Morata G. Function and regulation of homothorax in the wing imaginal disc of Drosophila. Development. 2000;127:2685–2693. doi: 10.1242/dev.127.12.2685. [DOI] [PubMed] [Google Scholar]
- Bessa J, Casares F. Restricted teashirt expression confers eye-specific responsiveness to Dpp and Wg signals during eye specification in Drosophila. Development. 2005;132:5011–5020. doi: 10.1242/dev.02082. [DOI] [PubMed] [Google Scholar]
- Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes & Dev. 2002;16:2415–2427. doi: 10.1101/gad.1009002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Campbell S, Inamdar M, Rodrigues V, Raghavan V, Palazzolo M, Chovnick A. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes & Dev. 1992;6:367–379. doi: 10.1101/gad.6.3.367. [DOI] [PubMed] [Google Scholar]
- Casares F, Mann RS. A dual role for homothorax in inhibiting wing blade development and specifying proximal wing identities in Drosophila. Development. 2000;127:1499–1508. doi: 10.1242/dev.127.7.1499. [DOI] [PubMed] [Google Scholar]
- Culi J, Mann RS. Boca, an endoplasmic reticulum protein required for wingless signaling and trafficking of LDL receptor family members in Drosophila. Cell. 2003;112:343–354. doi: 10.1016/s0092-8674(02)01279-5. [DOI] [PubMed] [Google Scholar]
- de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- Desplan C. Eye development: Governed by a dictator or a junta? Cell. 1997;91:861–864. doi: 10.1016/s0092-8674(00)80475-4. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Ferres-Marco D, Gutierrez-Avino FJ, Speicher SA, Beneyto M. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat Genet. 2004;36:31–39. doi: 10.1038/ng1281. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekas LA, Baeg GH, Flaherty MS, Ayala-Camargo A, Bach EA. JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development. 2006;133:4721–4729. doi: 10.1242/dev.02675. [DOI] [PubMed] [Google Scholar]
- Estella C, Mann RS. Logic of Wg and Dpp induction of distal and medial fates in the Drosophila leg. Development. 2008;135:627–636. doi: 10.1242/dev.014670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano L, Roder L, Core N, Alexandre E, Vola C, Jacq B, Kerridge S. The gene teashirt is required for the development of Drosophila embryonic trunk segments and encodes a protein with widely spaced zinc finger motifs. Cell. 1991;64:63–79. doi: 10.1016/0092-8674(91)90209-h. [DOI] [PubMed] [Google Scholar]
- Firth LC, Baker NE. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell. 2005;8:541–551. doi: 10.1016/j.devcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. The master control gene for morphogenesis and evolution of the eye. Genes Cells. 1996;1:11–15. doi: 10.1046/j.1365-2443.1996.11011.x. [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Hipfner DR, Weigmann K, Cohen SM. The bantam gene regulates Drosophila growth. Genetics. 2002;161:1527–1537. doi: 10.1093/genetics/161.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes & Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon KL, Ranade SS, Curtiss J, Mlodzik M, Pignoni F. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev Cell. 2003;5:403–414. doi: 10.1016/s1534-5807(03)00243-0. [DOI] [PubMed] [Google Scholar]
- Laugier E, Yang Z, Fasano L, Kerridge S, Vola C. A critical role of teashirt for patterning the ventral epidermis is masked by ectopic expression of tiptop, a paralog of teashirt in Drosophila. Dev Biol. 2005;283:446–458. doi: 10.1016/j.ydbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee JD, Treisman JE. The role of Wingless signaling in establishing the anteroposterior and dorsoventral axes of the eye disc. Development. 2001;128:1519–1529. doi: 10.1242/dev.128.9.1519. [DOI] [PubMed] [Google Scholar]
- McKay DJ, Estella C, Mann RS. The origins of the Drosophila leg revealed by the cis-regulatory architecture of the Distalless gene. Development. 2009;136:61–71. doi: 10.1242/dev.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J, Desplan C, Celik A. Generating patterned arrays of photoreceptors. Curr Opin Genet Dev. 2007;17:314–319. doi: 10.1016/j.gde.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morata G, Martin FA. Cell competition: The embrace of death. Dev Cell. 2007;13:1–2. doi: 10.1016/j.devcel.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117:117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Noro B, Culi J, McKay DJ, Zhang W, Mann RS. Distinct functions of homeodomain-containing and homeodomain-less isoforms encoded by homothorax. Genes & Dev. 2006;20:1636–1650. doi: 10.1101/gad.1412606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes & Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pichaud F, Casares F. homothorax and iroquois-C genes are required for the establishment of territories within the developing eye disc. Mech Dev. 2000;96:15–25. doi: 10.1016/s0925-4773(00)00372-5. [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Richardson H, O'Keefe LV, Marty T, Saint R. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development. 1995;121:3371–3379. doi: 10.1242/dev.121.10.3371. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Rebay I. Signaling circuitries in development: Insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- Simpson P, Morata G. Differential mitotic rates and patterns of growth in compartments in the Drosophila wing. Dev Biol. 1981;85:299–308. doi: 10.1016/0012-1606(81)90261-x. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Simmonds AJ, Garg A, Fossheim L, Campbell SD, Bell JB. Molecular and functional analysis of scalloped recessive lethal alleles in Drosophila melanogaster. Genetics. 2004;166:1833–1843. doi: 10.1534/genetics.166.4.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Treisman JE, Heberlein U. Eye development in Drosophila: Formation of the eye field and control of differentiation. Curr Top Dev Biol. 1998;39:119–158. doi: 10.1016/s0070-2153(08)60454-8. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Yao JG, Chen PH, Posakony JW, Barolo S, Kim J, Sun YH. Upd/Jak/STAT signaling represses wg transcription to allow initiation of morphogenetic furrow in Drosophila eye development. Dev Biol. 2007;306:760–771. doi: 10.1016/j.ydbio.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Wu J, Cohen SM. Proximal distal axis formation in the Drosophila leg: Distinct functions of teashirt and homothorax in the proximal leg. Mech Dev. 2000;94:47–56. doi: 10.1016/s0925-4773(00)00311-7. [DOI] [PubMed] [Google Scholar]
- Wu J, Cohen SM. Repression of Teashirt marks the initiation of wing development. Development. 2002;129:2411–2418. doi: 10.1242/dev.129.10.2411. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xu S, Xia L, Wang J, Wen S, Jin P, Chen D. The bantam microRNA is associated with drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet. 2009;5:e1000444. doi: 10.1371/journal.pgen.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Smolen GA, Haber DA. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 2008;68:2789–2794. doi: 10.1158/0008-5472.CAN-07-6205. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yue T, Jiang J. Hippo signaling pathway and organ size control. Fly (Austin) 2009;3:68–73. doi: 10.4161/fly.3.1.7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes & Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin JD, Mann RS. Differing strategies for the establishment and maintenance of teashirt and homothorax repression in the Drosophila wing. Development. 2004;131:5683–5693. doi: 10.1242/dev.01450. [DOI] [PubMed] [Google Scholar]