Abstract
Although it is widely accepted that dynamic cross-talk between gut epithelia and microorganisms must occur to achieve gut homeostasis, the critical mechanisms by which gut–microbe interactions are regulated remain uncertain. In this issue of Genes & Development, Buchon and colleagues (pp. 2333–2344) revealed that the reaction of the gut to microorganisms is not restricted to activating immune systems, but extends to integrated responses essential for gut tissue homeostasis, including self-renewal and the differentiation of stem cells. Further investigation of the connection between immune response and stem cell regulation at the molecular level in the microbe-laden mucosal epithelia will accelerate our understanding of the regulatory mechanisms of gut homeostasis and of the pathogenesis of diseases such as chronic inflammatory diseases and colorectal cancers.
Keywords: Intestinal stem cell, proliferation, gut microbiota, JAK–STAT, JNK, pathogenic bacteria
The gut epithelia of all metazoan organisms harbor complex microbial communities (Hooper and Gordon 2001). Due to the high diversity of microorganisms, gut–microbe interactions are biologically complex and dynamic, and are dictated by the number and identity of the contacting microorganisms. As an adaptation to the ever-changing nature of gut environments, metazoan gut epithelia have evolved to possess subtle yet efficient means to achieve immune and cellular homeostasis by sensing beneficial and/or deleterious conditions imposed by microbial populations (Ha et al. 2005a,b, 2009a,b; Bischoff et al. 2006; Zaidman-Remy et al. 2006; Nehme et al. 2007; Ryu et al. 2008). Not surprisingly, many immune and cellular deregulatory diseases, including chronic inflammatory disease and cancers, are frequently found in the gut epithelia. However, the exact molecular mechanisms by which immune and cellular homeostasis occurs in microbe-laden gut epithelia have yet to be fully elucidated. Research in this direction has been hampered by the fact that gut–microbe interactions take place in live organisms and therefore can only be interpreted meaningfully in vivo. Recently, the Drosophila genetic model has been demonstrated to be an efficient tool for the analysis of the microbe-induced signaling pathways that link microorganisms and corresponding gut cell physiology (Ha et al. 2005a,b, 2009a,b; Bischoff et al. 2006; Zaidman-Remy et al. 2006; Nehme et al. 2007; Ryu et al. 2008).
Global analyses of the bacterial-modulated gut responses
Two recent large-scale studies provided a global view of the bacterial-modulated gut responses that occur during different gut–microbe interactions (Buchon et al. 2009a; Cronin et al. 2009). In a study conducted by Buchon et al. (2009a), global gut response was examined at the transcriptional level during infectious gut–microbe interactions. This type of gut–microbe interaction was achieved by oral ingestion of a high dose of Erwinia carotovora carotovora-15 (Ecc15), whose infection is usually nonlethal and noninvasive when hosts are equipped with adequate dual oxidase (DUOX)-dependent reactive oxygen species (ROS) immunity (Ha et al. 2005a). Therefore, this experimental setting revealed the integrated host response to nonlethal bacteria. Under these conditions, many host genes involved in stress response, cell growth, wound repair, and development were found to be induced. Alternatively, Cronin et al. (2009) conducted a genome-wide in vivo Drosophila RNAi screen to identify genes involved in host response during pathogenic gut–microbe interactions. Cronin et al. (2009) used the lethal pathogen Serratia marcescens Db11, which normally kills the host within a week (Nehme et al. 2007). It is well known that this bacterium passes through the gut and enters the body, thus inducing both gut immunity and systemic immunity, both of which are therefore believed to be required for maximum host resistance to this pathogen (Nehme et al. 2007). Using a gut-specific silencing technique, Cronin et al. (2009) demonstrated that multiple genes involved in proteolysis, transport, stress response, immune response, growth, wound repair, and cell death play crucial roles in the gut for better host resistance against the lethal S. marcescens infection. In sum, these two studies demonstrated that gut reaction to nonlethal or lethal bacteria is not restricted to immune activation, but extends to integrated responses involving diverse aspects of gut cell physiology.
Bacterial-modulated intestinal stem cell activation
One of the integrated responses is the repair of gut cell injury by intestinal stem cells (ISCs). ISCs have long been believed to be involved in the repair process of gut cell injury. Fly gut epithelia are similar to mammalian gut epithelia in that they possess an instinctive self-renewal program (Casali and Batlle 2009). In the adult midgut, the entire population of epithelial enterocytes (ECs) is replaced in about a week (Micchelli and Perrimon 2006). This cellular homeostatic event is ensured by the presence of ISCs residing underneath the ECs (Micchelli and Perrimon 2006; Ohlstein and Spradling 2006). The discovery of midgut ISCs and their proliferation and differentiation program have led to analyses of the essential signaling pathways that affect stem cell regulation. The cellular organization of midgut epithelia is extremely simple when compared with the mammalian counterpart (Micchelli and Perrimon 2006; Ohlstein and Spradling 2006). Absorptive ECs and secretory enteroendocrine (EE) cells are two major differentiated cell types derived from undifferentiated ISC daughter cells known as enteroblasts (EBs). ISCs are the only dividing cells, and their self-renewal is known to be maintained by Wnt signaling (Lin et al. 2008). ISCs give rise to two daughter cells that are initially identical, but one of them rapidly becomes the Delta-expressing ISC and the other becomes the EB with Notch signaling activation. Subsequently, depending on the intensity of the notch signaling, EBs can differentiate into either ECs or EE cells (Micchelli and Perrimon 2006; Ohlstein and Spradling 2007). Because the gut epithelial lining acts as the frontline barrier to the external environment, it is likely that the ISC division rate is controlled by sensing the surrounding signaling rather than by operation at a default rate. Indeed, experimental epithelial injury induced by feeding damaging agents or expressing apoptotic genes triggers faster ISC division (Amcheslavsky et al. 2009). Importantly, recent studies further demonstrate that more physiologically relevant signals such as those produced by different types of gut–microbe interactions are also involved in determination of ISC activity (Buchon et al. 2009a; Chatterjee and Ip 2009; Cronin et al. 2009; Jiang et al. 2009). These findings, together with the genome-wide information regarding diverse gut responses to microorganisms, have made it possible to dissect the regulatory signaling pathways of ISCs under ever-changing microbial environments.
JAK–STAT (Janus kinase–signal transducers and activators of transcription) pathway in bacterial-modulated intestinal stem cell activation
The work of Buchon et al. (2009b) in this issue of Genes & Development, together with the two aforementioned recent works (Cronin et al. 2009; Jiang et al. 2009), demonstrates that JAK and STAT signaling, previously known to be involved in diverse biological processes including tissue damage repair and regeneration (Agaisse and Perrimon 2004), are involved in bacterial-modulated stem cell regulation. Buchon et al. (2009a) showed previously that essential genes involved in the regulation of the JAK–STAT pathway, such as unpaired (Upd) gene products (cytokine-like ligands for JAK–STAT signaling activation), are induced in the gut following Ecc15 ingestion. Additionally, these gut–microbe interactions also resulted in increased ISC division (Buchon et al. 2009a; Chatterjee and Ip 2009). Given that the JAK–STAT pathway is involved in a variety of processes related to the physiological homeostasis of cells and that tissue damage associated with microbes can activate this signaling (Agaisse and Perrimon 2004), Buchon et al. (2009a) postulated that activated JAK–STAT signaling may induce replenishment of epithelial cells injured during gut–microbe interactions through ISC activation. Buchon et al. (2009b) and two other research groups (Cronin et al. 2009; Jiang et al. 2009) further linked the two distinct microbe-induced responses, Upd–JAK–STAT activation and stem cell activation, by showing that microbe-induced Upd and subsequent JAK–STAT pathway activation are essential for stem cell division, as evidenced by the presence of cells double-positive for phospho-histone H3 (marker for dividing cells) and escargot (marker for ISCs and EBs). Furthermore, JAK–STAT signaling is shown to be required for EC differentiation, possibly by inducing Delta and stimulating Notch signaling (Jiang et al. 2009). Importantly, depletion of Upd in ECs or of JAK–STAT signaling pathway potential in progenitor cells is sufficient to produce a high lethality during both nonlethal and lethal gut–microbe interactions (Buchon et al. 2009b; Jiang et al. 2009), suggesting that the JAK–STAT-mediated epithelial cell renewal program acts as an essential part of the host responses to microorganisms. Interestingly, ISC-specific overactivation of the JAK–STAT pathway also leads to host lethality in response to pathogenic S. marcescens infection (Cronin et al. 2009), suggesting that optimal JAK–STAT activation is required for host survival.
How does the host mount an adequate rate of epithelial cell renewal in the face of constantly variable local microbial environments? A dose-dependent assay with pathogenic Pseudomonas entomophila showed that the extent of epithelial renewal is correlated up to a certain threshold with the microbial density. However, epithelial renewal is abolished by a high lethal dose of P. entomophila, which is reminiscent of the response to excess doses of tissue-damaging agents such as paraquat or sodium dodecyl sulfate (Buchon et al. 2009b). Furthermore, gut–microbe interactions using avirulent or attenuated P. entomophila mutant strains revealed that the epithelial renewal rate is also proportional to the degree of microbial virulence and gut pathology. Interestingly, by comparing germ-free and conventional animals, Buchon et al. (2009b) elegantly showed that the routine microbial contact with commensal microbiota can also activate the basal level of Upd expression and subsequent epithelial cell turnover through the JAK–STAT pathway at a reduced rate. This result gives us an important clue to understanding the beneficial roles of symbiotic microbiota on gut physiology because gut–microbe interactions routinely take place in all metazoans even in the absence of infection. Flies lacking the ability to control normal gut microbiota due to the absence of microbe-controlling innate immunity (such as immune deficiency [IMD] pathway mutant flies lacking antimicrobial peptide [AMP] gene expression) or aging harbor an abnormal commensal community structure in terms of density and possibly also identity of gut microbes. Strikingly, these flies show a hyperproliferative state of ISCs and abnormal gut morphology, which could be abolished if put under germ-free conditions (Buchon et al. 2009b). This observation suggests that the lack of microbe-controlling capacity is not directly involved in loss of gut homeostasis, but, rather, that abnormal number and/or structure of the commensal community resulting from loss of microbe-controlling capacity is the direct cause of loss of gut homeostasis. These results also support the idea that indigenous gut microbiota influence host physiology such as gut cell homeostasis. Taken together, it is likely that epithelial cell renewal rate is highly dynamic and is adjusted in response to the state of local microbial burdens.
Notch and JNK (c-Jun NH2 terminal kinase) pathway in bacterial-modulated intestinal stem cell activation
Interestingly, in addition to the JAK–STAT pathway, Notch signaling and JNK signaling are also involved in the microbe-induced epithelial renewal program (Jiang et al. 2009). When Notch is depleted in the progenitor cells, the host fails to survive during sublethal pathogenic gut–microbe interactions. As Notch signaling is required for ISC differentiation but not ISC mitosis (Jiang et al. 2009), differentiation of the ISCs, not just mitosis, also appears to be an essential event for host resistance against pathogens. JNK signaling was shown recently to be required for gut regeneration by promoting ISC cytoprotection and proliferation in old flies (Biteau et al. 2008). Consistent with these findings, reduced JNK activity in ISCs leads to depletion of ISCs resulting in host death during gut–Ecc15 interactions (Buchon et al. 2009b), demonstrating the essential role of the JNK pathway in bacterial-induced epithelium renewal. However, JNK pathway activity is dispensable for ISC mitoses during interactions with sublethal doses of P. entomophila (Buchon et al. 2009b; Jiang et al. 2009), indicating that the JNK activation in ISCs can be co-opted depending on the nature of contacting microorganisms. Taken together, these recent advances provide a novel conceptual framework wherein gut–microbe interactions can act as a physiological signal to activate various signaling pathways—including Upd–JAK–STAT, JNK, and Notch signaling—for the control of stem cell activity and host physiology.
ROS in bacterial-modulated intestinal stem cell activation
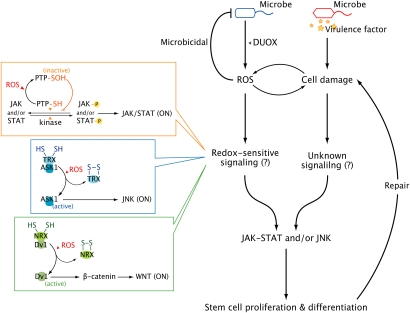
Several important questions remain to be clarified. One of the most important questions is how gut–microbe interactions act as physiological signals leading to ISC proliferation and differentiation. It is likely that stem cell activity is directly proportional to microbial burden because routine gut–microbe interactions produce basal Upd–JAK–STAT activity, whereas infectious and pathogenic interactions induce enhanced Upd–JAK–STAT activity (Buchon et al. 2009b; Jiang et al. 2009). In other words, variations in the degree of tissue damage that occur due to differences in density and/or virulence of contacting microbes may determine the strength of signal activation. Indeed, tissue-damaging chemical agents such as sodium dodecyl sulfate or dextran sodium sulphate can provoke Upd–JAK–STAT pathway activation and subsequent stem cell division in a dose-dependent manner (Buchon et al. 2009b). It is currently unknown how microbe-induced (or chemical-induced) tissue damage can be transformed into a physiological signal for stem cell activation. Much more work is necessary to determine how this occurs. Because high microbial burdens can induce both gut immune signaling and gut cell apoptosis, it is conceivable that immune signaling pathways such as IMD–AMP and DUOX–ROS signaling and/or the apoptosis signaling pathway play an instructive role in the activation of stem cell signaling. In this regard, it is important to note that neither Upd–JAK–STAT signaling nor ISC division requires the IMD pathway to respond to gut–microbe interactions (Jiang et al. 2009). The more likely scenario thus involves recently identified signaling pathways leading to microbe-induced ROS generation (such as Gαq-phospholipase Cβ-Ca2+ pathway and p38 mitogen-activated protein kinase pathway) (Ha et al. 2009a,b) or microbe-induced ROS themselves. Notably, flies lacking the ability to produce microbe-induced ROS generation (Gαq mutant flies or DUOX knockdown flies) or flies exhibiting enhanced antioxidant potential (either by treatment with antioxidants or by overexpression of ROS-removing enzymes to deplete microbe-induced ROS) showed a reduced level of microbe-induced ISC proliferation (Buchon et al. 2009b). It is not yet clear if ROS act as direct inducers of stem cell signaling or simply cause tissue damage that would signal to stem cells via other means (Fig. 1). Although ROS can lead to cellular apoptosis and tissue damage, it is well-established that physical and chemical tissue damage can also provoke ROS generation (Fig. 1). This reciprocal relationship between ROS and tissue damage makes it difficult to clarify the exact mechanism by which ROS induce stem cell activation. Nevertheless, these observations raise an intriguing possibility that the signaling pathways leading to ROS generation and/or ROS themselves are somehow involved in stem cell regulation. That ROS are rapidly produced in response to different external stimuli including microbial contact and mechanical injury suits the proposed role in the activation of stem cell signaling (Fig. 1).
Figure 1.
Hypothetical scheme for bacterial-modulated stem cell signaling. It is well-established that gut–microbe interactions induce different amounts of DUOX-dependent ROS production proportional to the microbial load and/or virulence (e.g., low chronic ROS production in the case of beneficial/routine interactions and high acute ROS production in the case of infectious/pathogenic interactions). Additionally, physical and chemical tissue damage may also directly induce ROS in a DUOX-dependent (as shown in zebrafish model) (Niethammer et al. 2009) and/or DUOX-independent manner. It remains to be elucidated whether microbe-induced ROS act as direct inducers of stem cell signaling or simply cause tissue damage that in turn signals to stem cells via other means. Given that tissue damage can also provoke ROS generation and that ROS can act both as cytotoxic molecules and signaling second messengers, one can speculate that both mechanisms are involved in stem cell signaling but are in operation in a context-dependent manner depending on spatial and temporal ROS concentration. See the text for more details about the putative redox-sensitive signaling modulation.
It had long been believed that ROS are an inevitable toxic waste produced by aerobic organisms that accelerate tissue injury when the amounts exceed certain thresholds. However, it is now generally accepted that various cells also produce appropriate levels of ROS intentionally in response to external stimuli through members of the NOX/DUOX family of NADPH oxidases to control biological processes (Rhee 2006). The key aspect is that the effects of ROS depend on their local concentration. ROS at high levels are involved in senescence and apoptosis, whereas ROS at low levels function as signaling molecules to mediate essential biological processes including cell proliferation, migration, differentiation, and gene expression. Recently, NOX activity has been found to be dynamically modulated during embryonic stem cell proliferation and differentiation, suggesting that ROS plays a role in stem cell regulation (Haneline 2008). Exogenous application of low concentrations of H2O2 ranging from 1 to 10 nM can lead to the differentiation of embryonic stem cells to contracting cardiomyocytes (Sauer and Wartenberg 2005). Furthermore, application of 100 nM H2O2 has been shown to promote the proliferation of stem cell-derived cardiomyocytes (Buggisch et al. 2007). Although it may be premature to draw a direct parallel between mammalian embryonic stem cells and Drosophila ISCs, these studies strongly suggest in principle that NOX/DUOX-dependent ROS can directly intervene in stem cell signaling pathways.
ROS in the modulation of signaling pathways
At present, it is unclear how ROS modulate stem cell regulation, but the mechanism by which ROS participate in signaling for cell proliferation and differentiation via JAK–STAT and JNK pathways is relatively well-established (Fig. 1). Specifically, because ROS are highly reactive and diffusible molecules that can act as intracellular messengers, they are able to transiently modulate a variety of signaling pathways through oxidative inactivation of critical redox-sensitive signaling proteins harboring low-pKa cysteine residues (Rhee et al. 2000; Rhee 2006). It is interesting to note that the oxidation of essential cysteine controls the “on–off” state of protein activity through a reversible reaction that is reminiscent of protein phosphorylation used for the activity modulation of signaling molecules. One of the best-known redox control mechanisms is the ROS-mediated inhibition of protein tyrosine phosphatases (PTPs) that contain a redox-sensitive low-pKa cysteine residue in the active site (Kwon et al. 2004). Indeed, numerous important signaling molecules are phosphorylated by tyrosine kinases and dephosphorylated by PTPs, and oxidative inactivation of PTPs leads to a shift in equilibrium toward increased tyrosine phosphorylation and subsequent signaling activation. The JAK–STAT pathway can also be controlled in such a way (Fig. 1). In mammalian epithelial cells, it has been shown that virus-induced ROS activate STAT3 through inducing an imbalance in favor of cellular tyrosine kinases over PTPs, which results in increased net phosphorylation and activation of signaling (Liu et al. 2004). In Drosophila, PTP 61F has been identified as a regulator of dephosphrylation of JAK and/or STAT (Baeg et al. 2005). Loss of PTP 61F results in an increase in JAK–STAT signaling via enhancement of the phosphorylation of JAK and STAT. Therefore, it will be interesting to investigate whether PTP 61F is under redox control during various gut–microbe interactions. Other important signaling pathways involved in stem cell regulation, such as the JNK and Wnt pathways, can also be under redox control through redox-sensing thioredoxin (TRX) family protein members (Fig. 1). The TRX protein family is a key component of redox regulation conserved throughout prokaryotes and eukaryotes, attesting to its importance as signaling regulators (Eklund et al. 1991). In the case of redox-modulated JNK signaling, the reduced form of TRX associates with apoptosis signal-regulating kinase 1 (ASK1, an upstream mitogen-activated protein kinase kinase kinase for JNK) and keeps it inactive, which means that JNK signaling is inactive in the absence of ROS (Junn et al. 2000). Upon signal-induced ROS production, TRX is oxidized and changes its conformation, which leads to the dissociation of ASK1 from TRX, resulting in ASK1 self-activation and subsequent JNK activation (Fig. 1; Junn et al. 2000). A similar mechanism is used for the redox-based control of Wnt signaling. In this case, nucleoredoxin (NRX), a member of the TRX family, acts as a redox-sensing protein. In the normal state, reduced NRX binds to Dishevelled (Dvl) and blocks Wnt/β-catenin signaling (Funato et al. 2006). When signal-induced ROS are produced, ROS oxidize NRX, which in turn dissociates from Dvl. Freed Dvl leads to the activation of Wnt/β-catenin signaling (Fig. 1; Funato et al. 2006). Taken together, it can be stated with reasonable certainty that signaling cascades involved in stem cell regulation—such as JAK–STAT, JNK, and Wnt signaling pathways—are redox-controlled, at least in some cellular context.
DUOX-dependent ROS in gut homeostasis
In mouse gut epithelia, bacterial contact is believed to promote NOX/DUOX-dependent ROS generation to maintain cellular homeostasis. Specifically, interactions between gut epithelia and commensal microbes provoke ROS generation within 30 min, which mediates the oxidative inactivation of the E3-SCFβ-TrCP-dependent protein degradation machinery leading to IκB and β-catenin stabilization (Kumar et al. 2007; Lee 2008). This mechanism enables microbe-induced ROS to induce reduced NF-κB signaling and enhanced β-catenin signaling to achieve anti-inflammatory effects and proliferative stimulation in the host cells, respectively. In addition to bacteria-derived signals, tissue damage can also promote ROS production at injury sites in the epithelia of humans, zebrafish, and plants. Recently, the formation of an H2O2 gradient was observed around the wounded epithelial barrier of zebrafish, which acts as an essential beneficial signal by recruiting leukocytes to the site of the injury (Niethammer et al. 2009). In this system, DUOX was shown to be responsible for wound-induced ROS generation (Niethammer et al. 2009). In Drosophila, DUOX is known to be responsible for microbe-induced generation of ROS during routine and infectious gut–microbe interactions, and DUOX-generated ROS play critical roles for host defense (Ha et al. 2005a, 2009a). In humans, DUOX2 is known to be highly expressed in the gastrointestinal tract (Geiszt et al. 2003; Lambeth 2004; El Hassani et al. 2005). Recently, microbe-induced ROS were found to be generated in human intestinal Caco2 cells through DUOX2 activity (Ha et al. 2009b). Thus, NOX/DUOX-dependent ROS generation during tissue damage or microbial contact appears to be an evolutionarily conserved phenomenon. Interestingly, Buchon et al. (2009b) found that flies with reduced ROS levels as a result of the ingestion of antioxidant such as N-acetyl cysteine or glutathione exhibit reduced potential for Upd–JAK–STAT signaling during infectious gut–microbe interactions. Furthermore, flies with reduced ROS potential due to the lack of DUOX expression—as in the case of DUOX knockdown flies—suffer severe pathological conditions during routine and infectious gut–microbe interactions, including gut apoptosis and ultimately host death (Ha et al. 2005a, 2009a,b). Despite the severe tissue damage, DUOX knockdown flies showed reduced and/or delayed Upd–JAK–STAT signaling in response to Ecc15 (Buchon et al. 2009b), which suggests that ROS (at the right time and dose) play a role in the activation of stem cell signaling via this pathway. In this context, it is tempting to speculate that in addition to their direct microbicidal roles, microbe-induced DUOX activation and subsequent generation of ROS constitute the key mechanism that leads to amplification of stem cell signaling. These intriguing and exciting issues could be addressed in a live gut–microbe interaction model of Drosophila by taking advantage of the powerful genetic tools and newly available ROS detection techniques. Ultimate identification of an intracellular messenger that links immune response to stem cell regulation in the ever-changing microbe-laden mucosal epithelia will greatly accelerate our understanding of regulatory mechanisms involved in gut homeostasis and thereby deregulatory diseases such as chronic inflammatory diseases and colorectal cancers.
Acknowledgments
This work was supported by National Creative Research Initiative Program and in part by WCU program (R31-2008-000-10010-0) from the Korea Ministry of Education, Science and Technology.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1858709.
References
- Agaisse H, Perrimon N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol Rev. 2004;198:72–82. doi: 10.1111/j.0105-2896.2004.0133.x. [DOI] [PubMed] [Google Scholar]
- Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell. 2009;4:49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes & Dev. 2005;19:1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: Activation of host defense and stem cell proliferation. Cell Host Microbe. 2009a;5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes & Dev. 2009b doi: 10.1101/gad.1827009. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggisch M, Ateghang B, Ruhe C, Strobel C, Lange S, Wartenberg M, Sauer H. Stimulation of ES-cell-derived cardiomyogenesis and neonatal cardiac cell proliferation by reactive oxygen species and NADPH oxidase. J Cell Sci. 2007;120:885–894. doi: 10.1242/jcs.03386. [DOI] [PubMed] [Google Scholar]
- Casali A, Batlle E. Intestinal stem cells in mammals and Drosophila. Cell Stem Cell. 2009;4:124–127. doi: 10.1016/j.stem.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Ip YT. Pathogenic stimulation of intestinal stem cell response in Drosophila. J Cell Physiol. 2009;220:664–671. doi: 10.1002/jcp.21808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin SJ, Nehme NT, Limmer S, Liegeois S, Pospisilik JA, Schramek D, Leibbrandt A, Simoes Rde M, Gruber S, Puc U, et al. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science. 2009;325:340–343. doi: 10.1126/science.1173164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund H, Gleason FK, Holmgren A. Structural and functional relations among thioredoxins of different species. Proteins. 1991;11:13–28. doi: 10.1002/prot.340110103. [DOI] [PubMed] [Google Scholar]
- El Hassani RA, Benfares N, Caillou B, Talbot M, Sabourin JC, Belotte V, Morand S, Gnidehou S, Agnandji D, Ohayon R, et al. Dual oxidase2 is expressed all along the digestive tract. Am J Physiol Gastrointest Liver Physiol. 2005;288:G933–G942. doi: 10.1152/ajpgi.00198.2004. [DOI] [PubMed] [Google Scholar]
- Funato Y, Michiue T, Asashima M, Miki H. The thioredoxin-related redox-regulating protein nucleoredoxin inhibits Wnt–β-catenin signalling through dishevelled. Nat Cell Biol. 2006;8:501–508. doi: 10.1038/ncb1405. [DOI] [PubMed] [Google Scholar]
- Geiszt M, Witta J, Baffi J, Lekstrom K, Leto TL. Dual oxidases represent novel hydrogen peroxide sources supporting mucosal surface host defense. FASEB J. 2003;17:1502–1504. doi: 10.1096/fj.02-1104fje. [DOI] [PubMed] [Google Scholar]
- Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005a;310:847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- Ha EM, Oh CT, Ryu JH, Bae YS, Kang SW, Jang IH, Brey PT, Lee WJ. An antioxidant system required for host protection against gut infection in Drosophila. Dev Cell. 2005b;8:125–132. doi: 10.1016/j.devcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Ha EM, Lee KA, Park SH, Kim SH, Nam HJ, Lee HY, Kang D, Lee WJ. Regulation of DUOX by the Gαq-phospholipase Cβ-Ca2+ pathway in Drosophila gut immunity. Dev Cell. 2009a;16:386–397. doi: 10.1016/j.devcel.2008.12.015. [DOI] [PubMed] [Google Scholar]
- Ha EM, Lee KA, Seo YY, Kim SH, Lim JH, Oh BH, Kim J, Lee WJ. Coordination of multiple dual oxidase-regulatory pathways in responses to commensal and infectious microbes in Drosophila gut. Nat Immunol. 2009b;10:949–957. doi: 10.1038/ni.1765. [DOI] [PubMed] [Google Scholar]
- Haneline LS. Redox regulation of stem and progenitor cells. Antioxid Redox Signal. 2008;10:1849–1852. doi: 10.1089/ars.2008.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Gordon JI. Commensal host–bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junn E, Han SH, Im JY, Yang Y, Cho EW, Um HD, Kim DK, Lee KW, Han PL, Rhee SG, et al. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J Immunol. 2000;164:6287–6295. doi: 10.4049/jimmunol.164.12.6287. [DOI] [PubMed] [Google Scholar]
- Kumar A, Wu H, Collier-Hyams LS, Hansen JM, Li T, Yamoah K, Pan ZQ, Jones DP, Neish AS. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J. 2007;26:4457–4466. doi: 10.1038/sj.emboj.7601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J, Lee SR, Yang KS, Ahn Y, Kim YJ, Stadtman ER, Rhee SG. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc Natl Acad Sci. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- Lee WJ. Bacterial-modulated signaling pathways in gut homeostasis. Sci Signal. 2008;1:pe24. doi: 10.1126/stke.121pe24. [DOI] [PubMed] [Google Scholar]
- Lin G, Xu N, Xi R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455:1119–1123. doi: 10.1038/nature07329. [DOI] [PubMed] [Google Scholar]
- Liu T, Castro S, Brasier AR, Jamaluddin M, Garofalo RP, Casola A. Reactive oxygen species mediate virus-induced STAT activation: Role of tyrosine phosphatases. J Biol Chem. 2004;279:2461–2469. doi: 10.1074/jbc.M307251200. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Nehme NT, Liegeois S, Kele B, Giammarinaro P, Pradel E, Hoffmann JA, Ewbank JJ, Ferrandon D. A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 2007;3:e173. doi: 10.1371/journal.ppat.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Bae YS, Lee SR, Kwon J. Hydrogen peroxide: A key messenger that modulates protein phosphorylation through cysteine oxidation. Sci STKE. 2000;2000:PE1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- Sauer H, Wartenberg M. Reactive oxygen species as signaling molecules in cardiovascular differentiation of embryonic stem cells and tumor-induced angiogenesis. Antioxid Redox Signal. 2005;7:1423–1434. doi: 10.1089/ars.2005.7.1423. [DOI] [PubMed] [Google Scholar]
- Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, Oh BH, Ueda R, Mengin-Lecreulx D, Lemaitre B. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]