Abstract
Several well-characterized fungal proteins act as prions, proteins capable of multiple conformations, each with different activities, at least one of which is self-propagating. Through such self-propagating changes in function, yeast prions act as protein-based elements of phenotypic inheritance. We report a prion that makes cells resistant to the glucose-associated repression of alternative carbon sources, [GAR+] (for “resistant to glucose-associated repression,” with capital letters indicating dominance and brackets indicating its non-Mendelian character). [GAR+] appears spontaneously at a high rate and is transmissible by non-Mendelian, cytoplasmic inheritance. Several lines of evidence suggest that the prion state involves a complex between a small fraction of the cellular complement of Pma1, the major plasma membrane proton pump, and Std1, a much lower-abundance protein that participates in glucose signaling. The Pma1 proteins from closely related Saccharomyces species are also associated with the appearance of [GAR+]. This allowed us to confirm the relationship between Pma1, Std1, and [GAR+] by establishing that these proteins can create a transmission barrier for prion propagation and induction in Saccharomyces cerevisiae. The fact that yeast cells employ a prion-based mechanism for heritably switching between distinct carbon source utilization strategies, and employ the plasma membrane proton pump to do so, expands the biological framework in which self-propagating protein-based elements of inheritance operate.
Keywords: Prion, yeast, glucose repression, PMA1, STD1
The stable inheritance of biological information and phenotype across generations is a fundamental property of living systems. Prions, self-perpetuating and heritable protein conformations that cause multiple phenotypes, represent an unusual mechanism of information transfer that occurs via protein instead of nucleic acid (Wickner 1994). Prion proteins can assume at least two conformations and each conformation alters protein function, resulting in different phenotypes (Wickner et al. 2004; Shorter and Lindquist 2005). When in the self-templating or prion conformation, prion proteins acquire characteristics normally restricted to nucleic acids. The first prion protein identified, the mammalian protein PrP, can behave as a transmissible pathogen and causes a neurodegenerative disease in its prion form (PrPSc) (Prusiner 1998). Prion proteins in fungi, which are functionally unrelated to PrP and to each other, act as non-Mendelian elements of inheritance by switching to the self-perpetuating, cytoplasmically transmissible prion conformation (Wickner 1994).
Four prions have been extensively characterized in fungi: [PSI+], [URE3], [Het-s], and [RNQ+]. [PSI+] (Cox 1965) is the prion form of the translation termination factor Sup35, which causes nonsense suppression (Stansfield et al. 1995; Patino et al. 1996; Paushkin et al. 1996). [URE3] (Lacroute 1971) is an altered form (Wickner 1994) of the nitrogen catabolite repressor Ure2 (Courchesne and Magasanik 1988). [RNQ+] controls the ability of a cell to acquire other prions (Derkatch et al. 2000, 2001; Sondheimer and Lindquist 2000). [Het-s], found in Podospora anserina, causes heterokaryon incompatibility with certain mating partners (Rizet 1952; Coustou et al. 1997).
These four fungal prions, as well as several recently identified prions ([SWI+], [MCA], [OCT+], and [MOT+]) (Du et al. 2008; Alberti et al. 2009; Nemecek et al. 2009; Patel et al. 2009), share key genetic and physical characteristics despite their disparate functions (Chien et al. 2004; Shorter and Lindquist 2005). Their phenotypes appear spontaneously at higher frequencies than those caused by genetic mutations. They are dominant, show non-Mendelian segregation following meiosis, and are also transmissible by cytoduction (cytoplasmic transfer). Physically, they form a self-templating amyloid conformation in the [PRION+] state. Furthermore, their inheritance is linked to the activities of chaperones, proteins that mediate conformational changes in other proteins. Transient changes in chaperone levels, particularly Hsp104, are sufficient to eliminate the prions permanently from cells. This occurs because chaperones alter the prion conformations and transmission to daughter cells. Once the prion template is gone, cells are “cured” of the elements (Uptain and Lindquist 2002; Shorter and Lindquist 2005). Another unusual feature is that transient overexpression of the prion protein causes permanent inheritance of the prion phenotype. This is because the protein∷protein interactions involved in prion formation are more likely to occur at higher protein concentrations (Chernoff et al. 1993; Ter-Avanesyan et al. 1993; Wickner 1994; Serio et al. 2000; Sondheimer and Lindquist 2000; Derkatch et al. 2001; Uptain and Lindquist 2002; Shorter and Lindquist 2005). The yeast prions also share a distinctive feature with mammalian prions: a strong transmission barrier across species. Even subtle differences in amino acid sequence can reduce the ability of prion proteins from one species to convert the homolog from other species, even though the homologous protein is itself capable of forming a prion on its own (Aguzzi et al. 2007; Chen et al. 2007).
The precise nature of the mammalian prion template is not known, but all of the well-characterized fungal prions, as well as the newly discovered prions and prion domains (Du et al. 2008; Alberti et al. 2009; Nemecek et al. 2009; Patel et al. 2009), are self-templating amyloid amyloids. The simple and robust character of self-templating amyloids provides a compelling framework for protein-based inheritance (Glover et al. 1997; Shorter and Lindquist 2005). Indeed, in many cases the amyloid has been shown to be the sole determinant needed for prion formation: Recombinant amyloid fibers alone are sufficient to convert [prion−] cells to [PRION+] cells (Maddelein et al. 2002; King and Diaz-Avalos 2004; Tanaka et al. 2004; Brachmann et al. 2005; Patel and Liebman 2007; Alberti et al. 2009). Amyloid structure is therefore commonly held to be a critical feature of all naturally occurring systems for protein-based inheritance. Indeed, a recent genome-wide screen for new prion domains in yeast began by examining proteins likely to be amyloidogenic (Alberti et al. 2009).
Here, we took a different approach. We searched the literature for Saccharomyces. cerevisiae phenotypes with prion-like inheritance patterns. One was described many years ago in a screen for cells with an alteration in carbon source utilization (Ball et al. 1976). The basis of the screen was the extreme preference of S. cerevisiae for glucose as a carbon source. In glucose media, cells repress genes necessary to process other carbon sources such as glycerol (Santangelo 2006). Glucosamine, a nonmetabolizable glucose mimetic, induces a similar repression. Therefore, yeast cells cannot use glycerol as a carbon source if even small amounts of glucosamine are present (Hockney and Freeman 1980; Nevado and Heredia 1996). Some cells spontaneously acquire the ability to use glycerol in the presence of glucosamine, presumably due to defects in glucose repression. Some of these exhibit dominant, non-Mendelian inheritance (Ball et al. 1976). Further, the phenotype is neither carried by the mitochondrial genome nor by a plasmid (Kunz and Ball 1977). Employing a variety of methods, we show here that this factor, [GAR+] (for “resistant to glucose-associated repression,” with capital letters indicating dominance and brackets indicating its non-Mendelian character), exhibits all of the genetic characteristics of a yeast prion, and we use a broad range of biochemical and genetic methods to identify proteins that play a key role in [GAR+] inheritance.
Results
[GAR+] shows non-Mendelian, infectious inheritance
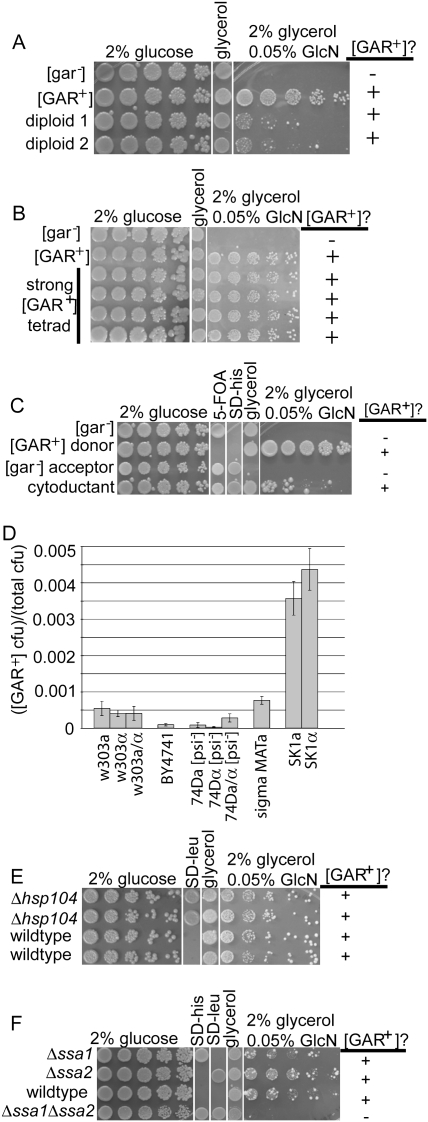
We obtained cells able to use glycerol as a carbon source despite the presence of glucosamine, as did Ball and colleagues (Ball et al. 1976; Kunz and Ball 1977), by selecting for cells that could grow in 2% glycerol in the presence of 0.05% glucosamine. Colonies appeared at a frequency of approximately five in 104 cells in the W303 genetic background, well above the predicted mutational frequency (Supplemental Fig. S1). Some recessive mutations allow growth on glycerol in the presence of glucosamine (see Supplemental Table S1; Ball et al. 1976), but the novel phenotypes described by Ball and colleagues (Ball et al. 1976; Kunz and Ball 1977) were dominant. Therefore, we first crossed our cells to wild-type cells. All diploids exhibited an unstable semidominant phenotype (Fig. 1A). Specifically, a mixed population was produced in which some diploids showed “strong” phenotypes (large colonies) and others showed “weak” phenotypes (small colonies) (Supplemental Fig. S2A). Cells with weak phenotypes invariably converted to strong over ∼25 generations (data not shown). Notably, both mammalian and fungal prions exhibit “strong” and “weak” strains (Aguzzi et al. 2007).
Figure 1.
[GAR+] shares the genetic characteristics of yeast prions. (A) Mating of [gar−] MATa to [GAR+] MATα in the W303 background. Resultant diploids show semidominant [GAR+] with a mixed population of large colonies (“strong”) and small colonies (“weak”). All spot tests shown are fivefold dilutions. Diploids are selected prior to plating to ensure that they are a pure population. (B) Tetrad spores from the “strong” [GAR+]. Diploids in A show non-Mendelian segregation of [GAR+]. (C). Cytoduction shows cytoplasmic inheritance of [GAR+]. The [GAR+] donor is 10B URA3+ his3− ρ+ kar1-1 and the acceptor is W303 ura3− HIS3+ ρ0 KAR1. The [GAR+] donor is therefore capable of growing on glycerol but the [gar−] acceptor is not; “mixed” cells were selected for growth on glycerol ([GAR+] cytoplasm) and SD-his 5-FOA ([gar−] nucleus and counterselection against the [GAR+] nucleus). (D). [GAR+] frequency in various laboratory strains. Data are shown as mean ± standard deviation (n = 6). (E). Tetrad spores from a [GAR+] diploid with the genotype hsp104∷LEU2/HSP104. Δhsp104 spores are still [GAR+]. (F). Tetrad spores from a [GAR+] diploid with the genotype ssa1∷HIS3/SSA1 ssa2∷LEU2/SSA2. Δssa1Δssa2 spores are no longer [GAR+].
In yeast, chromosomally inherited traits show 2:2 segregation following meiosis. Both strong and weak [GAR+] phenotypes, however, exhibited non-Mendelian 4:0 [GAR+]:[gar−] non-Mendelian segregation (Fig. 1B). That is, all meiotic progeny exhibited a capacity to grow on glucose in the presence of glucosamine. Spores produced from cells with weak phenotypes generally converted to strong phenotypes (Supplemental Fig. S2B, bottom). We named the responsible genetic element responsible for this trait [GAR+].
To determine whether [GAR+] is transmissible by cytoduction (that is, “infectious”), we used a mutant defective in nuclear fusion (kar1-1). During mating, kar1 cells fuse but nuclei do not (Conde and Fink 1976). Selecting for a particular nucleus and cytoplasm of interest after mating accomplishes cytoplasmic exchange without the transfer of nuclear material. We mated a [GAR+] strain carrying the nuclear markers URA3+ his3− and the cytoplasmic marker ρ+ to a kar1-1 [gar−] strain that was ura3− HIS3+ and ρ0. We then selected for cells containing the nucleus originally associated with [gar−] cells and the cytoplasm originally associated with [GAR+] cells. All 10 strains tested were [GAR+] (Fig. 1C). Thus, [GAR+] exhibits an “infectious,” nonnuclear pattern of inheritance.
[GAR+] appears at high frequency in a variety of genetic backgrounds
We next asked whether [GAR+] was an oddity of specific strains or could appear in diverse genotypes. Cells able to use glycerol in the presence of glucosamine appeared at a frequency of approximately nine in 105 cells in the BY background, approximately one in 104 cells in 74D, approximately five in 104 cells in W303, and approximately seven in 104 cells in Sigma. In the SK1 background, [GAR+] appeared at the astonishingly high rate of approximately one in 103 cells (Fig. 1D). In comparison, the frequency of heritable phenotypic change due to genetic mutation is generally approximately one in 106 haploid cells (Ohnishi et al. 2004).
We tested dozens of variants from each background for dominance. All exhibited the semidominant pattern observed in W303 (Fig. 1B; data not shown). [GAR+] cells of the 74D background did not sporulate, preventing us from testing segregation pattern. In W303 and W303/BY hybrids, [GAR+] only delayed sporulation (data not shown). In every tetrad tested from these backgrounds (>25 of each genotype), [GAR+] showed 4:0 [GAR+]:[gar−] segregation (Fig. 1B; data not shown). Together, these data establish that yeast strains of diverse genetic backgrounds commonly switch carbon utilization strategies in a heritable way by acquiring a non-Mendelian element of inheritance.
[GAR+] is curable by transient changes in chaperone protein levels
The inheritance of prions is based on self-perpetuating changes in protein conformations. In contrast to other non-Mendelian elements, a hallmark of prion phenotypes is the ability of transient changes in the expression of chaperones to cause a heritable loss of the phenotype. Other yeast prions, as well as 18 of 19 newly identified protein domains with prion-forming capability, require Hsp104 for propagation (Chernoff et al. 1995; Derkatch et al. 1997; Moriyama et al. 2000; Shorter and Lindquist 2004; Jones and Tuite 2005; Du et al. 2008; Alberti et al. 2009; Patel et al. 2009). To test the influence of Hsp104 on [GAR+], we crossed [GAR+] cells to cells carrying a knockout of hsp104 and sporulated them. Hsp104 was not required for [GAR+] inheritance: Δhsp104 segregants remained [GAR+] (Fig. 1E). [GAR+] was also not curable by growth on guanidinium hydrochloride, which inhibits Hsp104's ATPase activity (Ferreira et al. 2001; Jung and Masison 2001), nor by overexpression of HSP104 (data not shown).
We next tested the Hsp70 proteins Ssa1 and Ssa2 (Werner-Washburne et al. 1987), mutations in which affect the inheritance of other prions (Sweeny and Shorter 2008). These mutations are also a good measure of general chaperone sensitivity, as they induce production of most chaperone proteins (Oka et al. 1997). Strikingly, all Δssa1Δssa2 meiotic products lost the ability to grow on glycerol in the presence of glucosamine (Fig. 1F). Was this due to curing of the [GAR+] genetic element, or did the Δssa1Δssa2 mutations simply mask the phenotype? To test this, we restored SSA1 and SSA2 to the glucosamine-sensitive Δssa1Δssa2 progeny by mating them back to wild-type [gar−] cells (see Supplemental Fig. S3A for diagram of cross). Restoring Hsp70 function did not result in the reappearance of the [GAR+] phenotype (data not shown). However, when the cells were plated on medium with glucosamine, colonies able to grow on glycerol could be recovered at normal frequencies (Supplemental Fig. S3B). Thus, a transient change in chaperone proteins was sufficient to cure cells of [GAR+] and this curing was reversible, both hallmarks of prion biology (Wickner 1994). [GAR+] therefore exhibits all of the distinguishing genetic characteristics of yeast prions.
[GAR+] is regulated by the Rgt2/Snf3 glucose signaling pathway
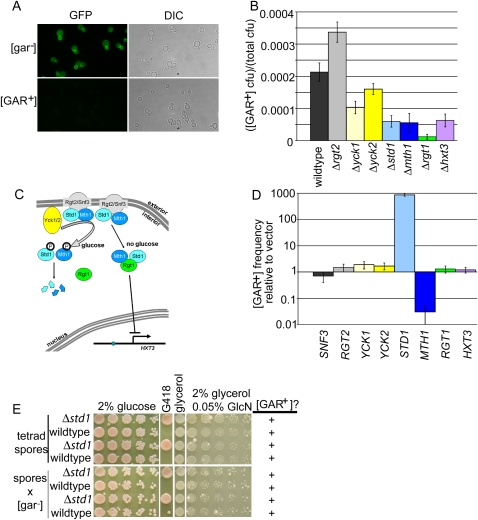
We performed gene expression profiling to identify transcriptional consequences of [GAR+]. In glucose-grown cultures tested just prior to the diauxic shift, only one gene showed a detectable difference between [gar−] cells and [GAR+] cells on our arrays, but that gene was very strongly affected. Hexose Transporter 3 (HXT3) was ∼36-fold down-regulated in [GAR+] cells compared with [gar−] cells (Supplemental Fig. S4). No other transcript exhibited more than a twofold change. We used an Hxt3-GFP fusion protein under the control of the endogenous HXT3 promoter to examine protein levels. Hxt3-GFP was easily visible at the plasma membrane in late log phase [gar−] cells, but extremely difficult to detect in [GAR+] cells (Fig. 2A). The loss of HXT3 expression (Δhxt3) alone did not allow cells to use glycerol in the presence of glucosamine (Fig. 2B), and thus does not explain the [GAR+] phenotype. However, it led us to hypothesize that the causal agent of [GAR+] is a regulator of HXT3 expression.
Figure 2.
The Snf3/Rgt2 glucose signaling pathway affects [GAR+]. (A) Hxt3-GFP signal in [gar−] and [GAR+] cells (S288c background) by fluorescence microscopy. (B) Frequency of [GAR+] in knockouts of members of the Snf3/Rgt2 glucose signaling pathway. Δsnf3 is completely resistant to glucosamine, and therefore [GAR+] frequency could not be measured. Furthermore, the frequency of spontaneous glucosamine-resistant colonies in the Δrgt1, Δstd1, and Δmths1 strains was close to the rate of genetic mutation, and therefore these colonies might not carry the actual [GAR+] element. Overall, this pathway is enriched for genes that alter [GAR+] frequency when knocked out relative to the library of nonessential genes (P = 8 × 10−6, Fisher's exact test). (C) The Snf3/Rgt2 glucose signaling pathway. (Adapted with permission from Moriya and Johnston 2004; ©2004 National Academy of Sciences, USA.) (D) Measurement of [GAR+] frequency following overexpression of Snf3/Rgt2 pathway members. Data are shown as mean ± standard deviation (n = 6). STD1 strongly induces conversion to [GAR+] and MTH1 blocks it. (E, top) Tetrad spores from a [GAR+] diploid with the genotype std1∷kanMX/STD1. (Bottom) Spores from top crossed to a [gar−] strain with a wild-type STD1 allele.
To define the protein(s) required for [GAR+] inheritance, we took advantage of two things. First, transient overexpression of each of the known prion proteins dramatically increases the frequency at which the corresponding prion appears (Uptain and Lindquist 2002). Second, the [GAR+] determinant exerts a strong effect on HXT3 expression, and HXT3 predominantly controlled by the Snf3/Rgt2 pathway (Kim et al. 2003; Santangelo 2006). When glucose is present, transmembrane glucose sensors Snf3 and Rgt2 transmit a signal to the Yck1 and Yck2 complex, which then phosphorylates Mth1 and Std1, marking them for degradation (Fig. 2C; Moriya and Johnston 2004). When glucose is not present, Mth1 and Std1 accumulate and interact with Rgt1. This complex then binds to the HXT3 promoter and represses transcription of HXT3 (Lakshmanan et al. 2003).
We tested each gene in the Snf3/Rgt2 regulatory pathway for induction of [GAR+] when overexpressed from a plasmid with a strong constitutive promoter, GPD (Fig. 2D). In every strain test, STD1 caused an extraordinary increase in the appearance of colonies able to grow on glycerol in the presence of glucosamine. In W303, for example, the increase was ∼900-fold over empty vector; more than one in 10 cells in these cultures converted to [GAR+]. This is at the high end of prion inductions obtained by analogous experiments with other proteins (Masison and Wickner 1995; Derkatch et al. 1996). While no other gene in this pathway induced [GAR+], overexpression of the STD1 paralog MTH1 blocked its appearance, further confirming the importance of members of this pathway in [GAR+] biology.
Transient STD1 overexpression induces [GAR+] but is not required for maintenance
Next, we asked if transient expression of STD1 was sufficient to create a heritable change in phenotype, a defining feature of prion biology. When ∼100 cells that had lost the overexpression plasmid were tested, all retained the [GAR+] phenotype (confirmed by marker loss) (data not shown). Thus, STD1 is not simply a dynamic regulator of glucose repression. Rather, its transient overexpression induces a new, heritable state of carbon utilization.
These data suggested that Std1 is the determinant of the [GAR+] prion, but further data indicated it could not be the sole determinant. First, most prion phenotypes mimic loss-of-function phenotypes of their prion determinants. However, Δstd1 strains derived from a [gar−] background were not able to grow on glycerol in the presence of glucosamine (Fig. 2B; data not shown). Furthermore, Δstd1 cells derived from a [GAR+] background were able to do so, indicating that they kept the prion (data not shown). Finally, such cells were able to pass the [GAR] element onto progeny in tester crosses for inheritance of the prion element (Fig. 2E). Therefore, [GAR+] maintenance does not require STD1. This makes [GAR+] highly unusual among yeast prions in that its transient inducing agent is not required for propagation.
We next examined all other members of the Rgt2/Snf3 pathway. None behaved as would be expected for the causal agent of [GAR+]. All knockouts were capable of propagating [GAR+] (Supplemental Fig. S5). Cells with rgt1 knockouts did not exhibit the prion phenotype, but they maintained it in a “cryptic” form. It reappeared when cells were crossed to [gar−] RGT1 cells. Therefore, RGT1 is required for the manifestation of the [GAR+] phenotype but is not necessary for its propagation.
Identification of genes that modify the frequency of [GAR+] appearance
We conducted genome-wide screens for affecters of [GAR+] induction. We screened the S. cerevisiae haploid deletion library (Giaever et al. 2002) for mutants that were incapable of inducing [GAR+] (Supplemental Table S2), caused a high frequency of appearance of [GAR+] (Supplemental Table S3), or that themselves exhibited an ability to grow on glycerol in the presence of glucosamine (Supplemental Table S1). Four of the eight members of the Snf3/Rgt2 pathway showed a phenotype in this screen (P = 8 × 10−6; Fisher's exact test). Δsnf3 grows on glycerol with glucosamine (Supplemental Table S1), and Δstd1, Δmth1, and Δrgt1 exhibited lower than normal [GAR+] induction (Fig. 2B; Supplemental Table S2). However, none of these genes were required for the maintenance of [GAR+] in strains already carrying the element (Supplemental Fig. S5).
Finally, we screened a library of ∼5000 ORFs (∼85% of yeast ORFs) on a galactose-inducible single-copy plasmid (Leonardo et al. 2002) to find genes that induce [GAR+] following overexpression. STD1 was the only clone that caused strong [GAR+] induction, ∼1000-fold when retested under the regulation of the GPD promoter. A second gene, DOG2, caused a 10-fold induction (Supplemental Fig. S6).
Pma1 associates with Std1 and is a component of [GAR+]
Since neither the deletion nor the overexpression screen identified a protein that by itself could embody the [GAR+] prion, we turned to biochemical methods. STD1 had been implicated in [GAR+] in three ways: (1) The highly specific down-regulation of HXT3 pointed to members of the Rgt2/Snf3 glucose signaling pathway, (2) transient STD1 overexpression caused huge increases in [GAR+] appearance, and (3) deletion of std1 reduced the spontaneous appearance of [GAR+] to the frequency of genetic mutations. We hypothesized, therefore, that Std1 might physically interact with an unknown propagating agent.
We sought proteins that interacted with Std1 by coimmunoprecipitation with an HA-tagged derivative. A high-molecular-weight (HMW) band was recovered from [GAR+] protein lysates but not from [gar−] lysates (Supplemental Fig. S7). Mass spectrometry analysis identified the protein as Pma1, a large, highly abundant P-type ATPase with 10 transmembrane domains that is the major controller of membrane potential and cytoplasmic pH (Morsomme et al. 2000). When the same assay was performed with isogenic Δstd1 cells, Pma1 was not detected. Notably, if Pma1 is indeed a constituent of the prion, we would not have identified it in our genetic screens. It is essential (Serrano et al. 1986), and therefore it is absent from the deletion library. Moreover, it is already the most abundant membrane protein in yeast and is notoriously difficult to overexpress (Eraso et al. 1987).
Transient overexpression of STD1 induced [GAR+] and transient overexpression of its paralog, MTH1, inhibited [GAR+] conversion. We therefore asked whether Pma1 exhibited heritable differences in association with Std1 and Mth1 in [gar−] and [GAR+] cells. As a multipass transmembrane protein, Pma1 is intractable to most methods of analyzing protein complexes, but it migrates as an oligomeric species when digitonin lysates are separated on Blue Native gels (Gaigg et al. 2005). Most Pma1 in [GAR+] and [gar−] cells migrated as heterogenous HMW complexes, but a smaller fraction migrated as two distinct complexes of (very roughly) 600 and 700 kDa (Fig. 3A, top). The lower bands (especially the 600-kDa species) were associated with Std1 in [GAR+] cells but with Mth1 in [gar−] cells (Fig. 3A, bottom). Std1 is much less abundant than Pma1. Consistent with the fact that only a small fraction of Pma1 is associated with Std1 in [GAR+] cells, Pma1 showed a minor but statistically significant change in protease sensitivity between [gar−] and [GAR+] cells (Supplemental Fig. S8).
Figure 3.
Pma1 is involved in [GAR+]. (A) Native gel of Pma1, Std1, and Mth1 in [gar−] and [GAR+]. Either Std1 (left) or Mth1 (right) was tagged with six tandem HA tags and samples were processed as described below from [gar−] and [GAR+] strains of each background. (Bottom right) Total, supernatant (sup.), digitonin soluble (det. sol.), and digitonin-insoluble (insol.) fractions were run on SDS gels and probed for Pma1 and Std1 or Mth1 as a fractionation control. No differences in Pma1, Std1, or Mth1 levels or localization were detected between [gar−] and [GAR+]. (Top right) Blots of the total fraction were stained with Ponceau Red to confirm equal amounts of starting material. (B) Measurement of [GAR+] frequency in knockout mutants of genes previously shown to affect (Δsur4, Δlst1) (Roberg et al. 1999; Eisenkolb et al. 2002) or not affect (Δlcb3, Δlcb4, Δdpl1, Δatg19) (Gaigg et al. 2005; Mazon et al. 2007) attributes of wild-type Pma1. Graph represents the mean ± standard deviation (n = 6). (C) Mutants in phosphorylation sites at the C terminus of Pma1 affect [GAR+] frequency. Starting strain is haploid, [gar−], genotype pma1∷kanMX with p316-PMA1. p314-PMA1 carrying wild-type PMA1 or mutants of interest were transformed into the starting strain and then p316-PMA1 plasmid selected against by growth on 5-FOA. Graph represents the mean ± standard deviation (n = 6). P-values are the binomial distribution of the mean. (D) Pma1 mutants that increase [GAR+] frequency show decreased levels of Hxt3-GFP. Graph represents the mean ± standard deviation (n ≥ 6) and P-values were determined using the χ2 test. Strain background is a hybrid of W303 and S288C.
Next, we asked whether mutations that affect Pma1 oligomerization and trafficking to the plasma membrane alter [GAR+] frequency. Mutants that affect phospholipid synthesis and protein trafficking but not Pma1 oligomerization—LCB3, LCB4, DPL1, and ATG19 (Lee et al. 2002; Mazon et al. 2007)—did not change the appearance of [GAR+] (Fig. 3B; Supplemental Fig. S9A). Mutants that do affect Pma1 oligomerization and trafficking— SUR4 and LST1 (Roberg et al. 1999; Lee et al. 2002)—decreased the appearance of [GAR+] (Fig. 3B; Supplemental Fig. S9A). These genes were not, however, required for [GAR+] maintenance (Supplemental Fig. S9B).
We explored the relationship between Pma1, [GAR+], and the Rgt2/Snf3 glucose signaling pathway. Carbon sources regulate Pma1's phosphorylation state (Lecchi et al. 2005), its ATPase activity (Serrano 1983), and its conformation (Miranda et al. 2002) through residues S899, S911, and T912 in the C-terminal tail, which faces the cytosol (Eraso et al. 2006; Lecchi et al. 2007). We mutated S899, S911, and T912 to alanine, which cannot be phosphorylated, or to aspartic acid, which mimics constitutive phosphorylation. (Phosphorylated S911 and T912 are commonly observed in glucose media and the nonphosphorylated forms when cells are starved of glucose [Lecchi et al. 2007].) S899 mutations and S911D and/or T912D mutations had no effect on [GAR+] frequency. However, S911A and S911A/T912A increased the frequency of [GAR+] appearance by several-fold (Fig. 3C). Notably, these same mutants also reduced levels of an Hxt3-GFP reporter, both a readout for the Rgt2/Snf3 pathway and the only change in gene expression detected in [GAR+] cells (Fig. 3D). These results indicate that Pma1 affects glucose signaling to regulate HXT3. In any case, the fact that such subtle mutations in the Pma1 protein affect [GAR+] induction confirms that Pma1 plays a key role in [GAR+] biology.
The unstructured N terminus of Pma1 is involved in [GAR+] propagation
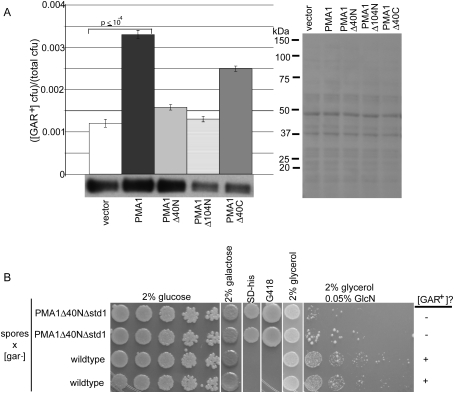
A characteristic of prions is that transient overexpression is sufficient for induction. However, Pma1 is the most abundant plasma membrane protein in yeast (Morsomme et al. 2000), and overexpression is not well tolerated (Eraso et al. 1987). We found that we could obtain a threefold increase in Pma1 protein levels with a CEN plasmid and a GPD promoter. This caused a corresponding increase in [GAR+] frequency (Fig. 4A). Introducing stop codons at amino acid positions 23 or 59 eliminated this effect (Supplemental Fig. S10). Thus, it is not the nucleic acid sequence but the Pma1 protein that contributes to [GAR+] induction. Finally, when the inducing GPD PMA1 plasmid was lost, the cells remained [GAR+]. Thus, a transient increase in PMA1 was sufficient to induce a heritable change in phenotype.
Figure 4.
Alterations to Pma1 affect [GAR+]. (A) [GAR+] induction by transient overexpression of PMA1 in a wild-type background. Data are shown as the mean of [GAR+] frequency ± standard deviation (n = 6). Western is total protein probed with αPma1 antibody and quantified using Scion Image. (Right) The blot was stained with Ponceau Red to confirm equal loading. (B) Propagation of [GAR+] is impaired in PMA1Δ40N Δstd1 double mutants. Tetrad spores from a [GAR+] diploid with the genotype GAL-PMA1Δ40N/PMA1 std1∷kanMX/STD1 were crossed to a [gar−] strain with wild-type PMA1 and STD1 alleles. PMA1Δ40N Δstd1 spores cannot propagate [GAR+] to wild-type [gar−] yeast. The few glucosamine-resistant colonies found in the PMA1Δ40N Δstd1 background exhibit standard, Mendelian inheritance of the glucosamine resistance phenotype and thus do not carry the [GAR+] element.
Pma1's N and C termini face the cytosol. The C terminus is predicted to be α-helical and the N terminus unstructured (Morsomme et al. 2000), the latter a characteristic of prions. An N-terminally truncated (Δ40) mutant of PMA1 did not increase [GAR+] appearance. although the protein was expressed at wild-type levels (Fig. 4A). A C-terminally truncated PMA1 did increase [GAR+] induction, even though its levels were reduced.
[GAR+] could be propagated through cells whose only source of Pma1 was a GAL1-regulated N-terminal deletion, PMA1Δ40N (Supplemental Fig. S11). Strikingly, however, it did not propagate through a double mutant of PMA1Δ40N and Δstd1, and it did not reappear when wild-type PMA1 and STD1 function were restored with crosses (Fig. 4B). (The few glucosamine-resistant colonies that remained were not [GAR+] but contained conventional recessive; data not shown.) Thus, once [GAR+] has been established, it is maintained in the absence of either Std1 or the N terminus of Pma1, but not in the absence of both.
[GAR+] is sensitive to a Pma1-dependent ‘species barrier’
Previously described yeast prion proteins exhibit changes in localization and solubility in the prion state (Uptain and Lindquist 2002) and affect the induction of other prions by cross-templating (Derkatch et al. 2000, 2001). There was no difference in localization of Pma1 or Std1 between [gar−] and [GAR+] (Supplemental Fig. S12). Neither formed a detectable SDS-resistant species in [GAR+] (Supplemental Fig. S13). Furthermore, the frequency of [GAR+] appearance did not change in backgrounds carrying [PSI+], [RNQ+], or [URE3], prions that broadly affect the appearance of amyloid-based prions (Supplemental Fig. S14). Analysis of protein extracts by two-dimensional (2D) gel electrophoresis did not reveal any proteins that changed solubility between [gar−] and [GAR+] (Supplemental Fig. S15). [GAR+] was not affected by Hsp104 expression (Fig. 1E). Whatever the manner by which Pma1 and Std1 contribute to the prion state, it is not likely by forming amyloid.
The extremely stable nature of amyloids allows them to be confirmed as prion determinants by “protein only” transformation (Maddelein et al. 2002; Tanaka et al. 2004). The lack of an identifiable amyloid in [GAR+] cells precluded the use of this procedure for [GAR+]. Instead, to rigorously test the relation between Pma1, Std1, and [GAR+], we performed a classic “transmission barrier” experiment. Small differences in amino acid sequence cause prions that originate in one species to fail in transmission to another (Santoso et al. 2000; Bagriantsev and Liebman 2004; Chen et al. 2007). If Pma1 and Std1 contribute to a transmission barrier for [GAR+], it would establish that they are integral to the propagating element.
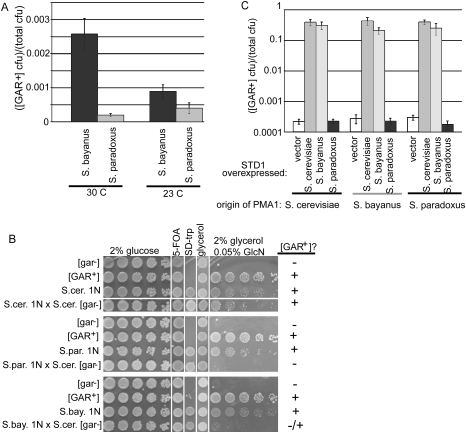
We chose to study a possible [GAR+] transmission barrier using Saccharomyces bayanus and Saccharomyces paradoxus, two closely related sensu stricto species that also exhibit glucose-mediated repression of the utilization of other carbon sources. First, we asked whether diploids of these species could also acquire the ability to use glycerol in the presence of glucosamine (Fig. 5A). They could, and they did so at a higher frequency than expected for mutation. Indeed, [GAR+] appeared in S. bayanus at an astonishingly high rate (greater than one in 1000 cells). Moreover, the [GAR+] phenotype was very stable in these cells. Thus, the ability to heritably switch carbon utilization strategies through this prion is broadly used.
Figure 5.
[GAR+] exhibits a Pma1-dependent species barrier. (A) [GAR+] frequency of S. bayanus and S. paradoxus cells grown at 30°C (left), the optimal growth temperature of S. paradoxus, or 23°C (right), the optimal growth temperature of S. bayanus. Data are shown as the mean of [GAR+] frequency ± standard deviation (n = 6). (B) Substitution of PMA1 from S. cerevisiae with PMA1 from S. bayanus or S. paradoxus prevents [GAR+] propagation. Starting strain is haploid, [GAR+], genotype pma1∷kanMX with p316-PMA1 S. cerevisiae as a covering plasmid. p314-PMA1 carrying PMA1 from S. cerevisiae (S.c., top), S. paradoxus (S.par., middle), or S. bayanus (S.bay., bottom) was transformed into the starting strain and p316-PMA1 S.c. selected against by replica plating to 5-FOA (S.c. 1N, S.p. 1N, or S.b. 1N). These haploids were mated to a wild-type S. cerevisiae [gar−] background, restreaked twice, and tested for [GAR+]. Representative data from three independent experiments are shown.
We asked whether the Pma1 proteins from S. bayanus and S. paradoxus can propagate [GAR+] in S. cerevisiae. Sequence differences between the species are slight (Supplemental Fig. S16): S. bayanus Pma1 and S. paradoxus Pma1 are 96% and 99% identical to S. cerevisiae Pma1, respectively. Most of these changes are in the N-terminal region, which is required for prion induction
First, we transformed S. bayanus or S. paradoxus PMA1 plasmids into an S. cerevisiae strain in which a deletion of the essential PMA1 gene was covered by a plasmid encoding S. cerevisiae Pma1. The S. cerevisiae PMA1 plasmid was then selected against. All cells grew at the same rate on glucose, indicating that the Pma1 protein from these species was fully functional in S. cerevisiae. However, when [GAR+] cells were selected by plating these cells to glycerol–glucosamine medium, the resultant phenotypes were weak, unstable, and appeared at a low frequency. When putative [GAR+] cells were passaged on nonselective medium and then plated back onto glucosamine-containing medium, many fewer cells with S. bayanus or S. paradoxus PMA1 maintained the resistant phenotype than cells with S. cerevisiae PMA1 (data not shown). Thus, in a background where the entire genome otherwise remains the same, changing the species of origin for Pma1 had a critical effect on [GAR+] induction and propagation.
Next, we asked whether the S. bayanus or S. paradoxus Pma1 proteins could propagate a [GAR+] state received from the S. cerevisiae protein. We performed another plasmid shuffle, this time starting with cells already carrying a strong S. cerevisiae [GAR+] element. We selected against the plasmid carrying the S. cerevisiae PMA1 after ∼25 generations. After another 25 generations, cells were tested for the ability to grow on glycerol in the presence of glucosamine. Most retained a strong [GAR+] phenotype. Thus, strains with S. bayanus and S. paradoxus PMA1 were capable of accepting and propagating [GAR+] from strains with S. cerevisiae PMA1 (Fig. 5B), at least after coexpression of both proteins for 25 generations.
Finally, we tested how efficiently [GAR+] elements from cells expressing S. bayanus or S. paradoxus PMA1 could be transmitted back to cells expressing only S. cerevisiae PMA1. Multiple [GAR+] strains carrying the three PMA1 genes were mated to wild-type [gar−] cells. Cells expressing PMA1 from S. paradoxus could not transmit [GAR+] at all, and cells expressing PMA1 from S. bayanus transmitted it very inefficiently. Controls expressing S. cerevisiae PMA1 transmitted [GAR+] efficiently (Fig. 5B). Thus, the PMA1 species of origin creates a strong transmission barrier for [GAR+] propagation.
Might Std1, the [GAR+] induction factor that is complexed with Pma1 in [GAR+] cells, create an induction barrier? Std1 is 81% identical between S. cerevisiae and S. bayanus but much more divergent in S. paradoxus (Supplementaql Fig. S17). We transiently overexpressed STD1 from each organism in [gar−] S. cerevisiae cells carrying each of the three Pma1 genes. STD1 alleles of S. cerevisiae and S. bayanus acted as general inducers. They increased the appearance of [GAR+] ∼1000-fold in strains producing the Pma1 protein of any of the three species (Fig. 5C). In contrast, S. paradoxus STD1 did not induce [GAR+] in any. Presumably, some other factor contributes to [GAR+] induction in S. paradoxus. Most importantly, however, this experiment demonstrates that Std1 creates a strong species barrier for [GAR+] induction, confirming its intimate involvement in the prion.
Discussion
The ability of cells to sense and adapt to nutrients is crucial to survival in highly competitive and rapidly fluctuating environments. Here, we describe a cytoplasmically inherited element, [GAR+], that is involved in the fundamental processes of glucose sensing and signaling and carbon source utilization. [GAR+] arises spontaneously in every S. cerevisiae strain tested, as well as sibling species separated by ∼5 million years of evolution (Kellis et al. 2004)—S. paradoxus and S. bayanus—at frequencies much higher than genetic mutations.
[GAR+] fulfills all of the genetic criteria established for prions: It is dominant (or at least semidominant). It exhibits non-Mendelian inheritance. It can be transferred via cytoplasmic exchange. Transient changes in the levels of chaperone proteins are sufficient to heritably cure cells of the [GAR+] state. Transient changes in the expression of proteinaceous determinants heritably induce [GAR+]. The non-Mendelian mechanism of inheritance that best describes [GAR+] is that of a prion.
In other ways, however, [GAR+] seems very different from previously described yeast prions. It has at least two components: the plasma membrane proton pump Pma1, and the glucose signaling factor Std1. Transient overexpression of either PMA1 or STD1 is sufficient to establish a heritable conversion to [GAR+], yet once [GAR+] is established, either is sufficient for propagation. Cells lacking std1 and also carrying a small deletion in the N terminus of Pma1 cannot propagate [GAR+] at all. Pma1 and Std1 associate in an oligomeric complex in [GAR+] cells, but this complex is barely detectable in [gar−] cells. The integral relationship between these proteins and the [GAR+] state was tested and confirmed by transmission barrier experiments. Substituting the PMA1 gene from S. bayanus or S. paradoxus for that of S. cerevisiae blocked propagation of [GAR+] to S. cerevisiae Pma1. Substituting Std1 from S. paradoxus eliminated its potency in [GAR+] induction.
What, then, is the nature of [GAR+]? It does not involve a detectable amyloid form, at least of the Pma1 or Std1 proteins. It also is not sensitive to overexpression or deletion of the general amyloid remodeling protein Hsp104. Hsp104 severs amyloid filaments to ensure orderly inheritance of prion templates to daughter cells. It is required for the propagation of all known prions as well as for 18 of 19 recently discovered prion candidates (Chernoff et al. 1995; Patino et al. 1996; Derkatch et al. 1997; Ness et al. 2002; Cox et al. 2003; Kryndushkin et al. 2003; Shorter and Lindquist 2004, 2006; Jones and Tuite 2005; Tipton et al. 2008; Alberti et al. 2009). Thus, the absence of dependence on Hsp104 makes it rather unlikely that [GAR+] involves any amyloid-based element.
One possibility is that [GAR+] inheritance and propagation result from heritable alterations in Rgt2/Snf3 signaling involving a self-sustaining feedback loop. Indeed, Std1 and its paralog, Mth1, are subject to many feedback mechanisms involving their own transcription and degradation (Lakshmanan et al. 2003; Moriya and Johnston 2004; Polish et al. 2005; Kim et al. 2006), and Std1 is found both in the nucleus and on the plasma membrane (Schmidt et al. 1999). Furthermore, Pma1 is very abundant and Std1 is extremely scarce (Morsomme et al. 2000). Our data suggest that only a small fraction of Pma1 contributes to [GAR+] and that Std1 is the limiting factor. This would be consistent with altered signaling, as only small amounts of the Std1 protein would be necessary to shift the activity of a fraction of Pma1. However, if [GAR+] is simply due to altered signaling, the mechanism that maintains it must be remarkably robust, as it has been maintained in a highly stable state in some of our strains for 6 years now, with repeated dilutions into log phase, storage in the freezer and refrigeration, transitions back to room temperature, and growth in liquid and on plates, in a wide variety of different media, through repeated rounds of growth into stationary phase (wherein most aspects of carbon metabolism undergo profound changes), and through starvation-induced meiosis.
Another possibility is that [GAR+] starts with a change in the association of Std1 and Pma1 that induces a conformational change in oligomeric species of each. These can then be maintained in the absence of either Std1 or the Pma1 N terminus, but not in the absence of both (Fig. 6). We do not exclude the possibility that another protein contributes to the [GAR+] state. Indeed, our observations that S. paradoxus acquires [GAR+] at a high frequency, but that the Pma1 and Std1 proteins of S. paradoxus do not reconstitute [GAR+] in S. cerevisiae, suggesting the involvement of another protein. (This protein might even form amyloid, but if so it does not require Hsp104 and has escaped detection in our genetic screens.)
Figure 6.
Pma1 and the Rgt2/Snf3 glucose signaling pathway We propose that Pma1 acts as a part of the Rgt2/Snf3 signaling pathway. (A) In [gar−] glucose-grown cells, Pma1 associates with Mth1. The glucose signal is propagated through Snf3 and Rgt2 to Yck1 and Yck2, which phosphorylate Mth1 and Std1. This phosphorylation marks Mth1 and Std1 for degredation, leaving their interacting partner, Rgt1, free in the cytosol, where it does not repress transcription at the HXT3 locus. (B) Under [GAR+] conditions, HXT3 transcription is repressed, which resembles that of cells grown in a carbon source other than glucose. Pma1 associates with Std1, which somehow facilitates the repression of HXT3, possibly by altering the affinity of Std1 for Rgt1. Association with Std1 has been shown previously to facilitate the binding of Rgt1 to DNA (Lakshmanan et al. 2003). The association between Pma1 can either be transient or stable, but either way it aids in the establishment of an altered signaling pathway. This altered pathway is then maintained either by the contained association between Std1 and Pma1 or by a feedback loop within the signaling cascade itself.
Of course, models involving self-perpetuating signaling loops and conformational changes are not mutually exclusive. Associations between Pma1 and Std1 might result in a conformational change that alters signaling and sets up a robust feedback loop that helps maintain the association, either between those same molecules of Pma1 and Std1 or between other molecules and these proteins (Fig. 6). It will be of great interest to determine what might render such states stable enough to be so robustly heritable.
Another remaining question is the precise reason why cells carrying [GAR+] are able to grow on glycerol in the presence of glucosamine. We hypothesize that the [GAR+] phenotype involves altered signaling through a glucose-sensing pathway, likely through Std1's previously reported ability to interact with the DNA-binding protein Rgt1 (Fig. 6; Lakshmanan et al. 2003). Experiments investigating gene expression patterns over a much broader range of carbon sources and time points than examined here, as well as chromatin immunoprecipitation experiments with Std1 and Rgt1, may prove illuminating.
Whatever the mechanism may prove to be, Pma1, the major plasma membrane ATPase, and Std1, a much rarer and poorly understood signaling protein, contribute to a prion-like phenotypic state that heritably alters fundamental decisions about carbon source utilization. This heritable element, [GAR+], has all of the definitive characteristics of a prion. It has been stated that prion-mediated epigenetic states are simply diseases of yeast (Nakayashiki et al. 2005). Our findings that such an element controls something as fundamental to yeast biology as glucose repression, and that this element spontaneously arises at high frequency in diverse strains and sibling species, suggests that such epigenetic switches are actually integral to yeast biology. Clearly, self-propagating protein-based elements (prions) that can stably perpetuate biological states across generations operate over a much broader mechanistic landscape than supposed previously.
Materials and methods
Yeast strains and genetic manipulations
Strain construction and manipulation followed standard yeast techniques. A list of strains and plasmids used in this study is available in Supplemental Tables S1 and S2. Unless otherwise stated, data shown are from genetic background W303. Fivefold dilutions were used for all spotting assays. Media used were yeast peptone-based medium containing the designated carbon source (YPD, YPglycerol, and YPgalactose), synthetic medium lacking a particular amino acid (SD), or glycerol glucosamine medium (GGM; 1% yeast extract, 2% peptone, 2% glycerol, 0.05% D-[+]-glucosamine [Sigma G4875]).
[GAR+] frequency assays and isolation of [GAR+]
Cultures for [GAR+] frequency assays were grown overnight in 2% glucose, either YPD or SD, subcultured in the same, then grown to early exponential phase (OD600 = 0.2–0.4). Cultures were plated straight to GGM and diluted 10−4 for plating to YPD. To isolate [GAR+] for further study, colonies from GGM were restreaked once to GGM then used in downstream applications. Unless otherwise stated, error bars in [GAR+] frequency assays represent the standard deviation and P-values are the binomial distribution of the mean. In all assays for [GAR+] propagation, cells were passaged for >100 generations before testing for growth on glycerol in the presence of glucosamine. Sporulation was performed by growing to diauxic shift in YPD or SD, plating to sporulation plates (1% potassium acetate, 0.05% dextrose, 0.1% yeast extract, 0.01% complete amino acid mix [Bio101]), and incubating at 23°C until sporulated.
Genetic, biochemical, and cell biological analysis
Gene expression profiling, Western blotting, immunoprecipitation, fluorescent microscopy, Blue Native gel analysis, protease sensitivity analysis, and genetic screens were all performed using standard procedures. Detailed descriptions are available in the Supplemental Material.
Acknowledgments
We thank and acknowledge Neal Sondheimer, whose initial investigation first suggested that [GAR+] might be a prion-based phenotype (Sondheimer 2000). We thank Amy Chang for gifts of an antibody and strains, and general advice on working with Pma1; Tom Rapaport for a Sec61 antibody; and Eric Spooner for mass spectrometry. Whitehead Institute Center for Microarray Technology performed cDNA synthesis, labeling, and hybridization reactions for the microarrays. We thank members of the Lindquist laboratory for providing comments and advice. This work was supported by the Mathers Foundation (to S.L.), Du-Pont MIT Alliance (to S.L.) and NIH grant GM25874. J.C.S.B is supported by an NSF Graduate Research Fellowship and the Arthur Siegel Fellowship from Whitehead Institute for Biomedical Research.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1839109.
Supplemental material is available at http://www.genesdev.org.
References
- Aguzzi A, Heikenwalder M, Polymenidou M. Insights into prion strains and neurotoxicity. Nat Rev Mol Cell Biol. 2007;8:552–561. doi: 10.1038/nrm2204. [DOI] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo: [PSI+] AND [PIN+] form separate structures in yeast. J Biol Chem. 2004;279:51042–51048. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- Ball AJS, Wong DK, Elliott JJ. Glucosamine resistance in yeast. I. A preliminary genetic analysis. Genetics. 1976;84:311–317. doi: 10.1093/genetics/84.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Newnam GP, Chernoff YO. Prion species barrier between the closely related yeast proteins is detected despite coaggregation. Proc Natl Acad Sci. 2007;104:2791–2796. doi: 10.1073/pnas.0611158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- Conde J, Fink GR. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne WE, Magasanik B. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: Roles of the URE2 and GLN3 genes. J Bacteriol. 1988;170:708–713. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BS. PSI, a cytoplasmic suppressor of the super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- Cox B, Ness F, Tuite MF. Analysis of the generation and segregation of propagons: Entities that propagate the [PSI+] prion in yeast. Genetics. 2003;165:23–33. doi: 10.1093/genetics/165.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PS1] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI+] and [PIN+]: A two-prion system in yeast? EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: The story of [PIN+] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Du Z, Park K-W, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenkolb M, Zenzmaier C, Leitner E, Schneiter R. A specific structural requirement for ergosterol in long-chain fatty acid synthesis mutants important for maintaining raft domains in yeast. Mol Biol Cell. 2002;13:4414–4428. doi: 10.1091/mbc.E02-02-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eraso P, Cid A, Serrano R. Tight control of the amount of yeast plasma membrane ATPase during changes in growth conditions and gene dosage. FEBS Lett. 1987;224:193–197. doi: 10.1016/0014-5793(87)80446-5. [DOI] [PubMed] [Google Scholar]
- Eraso P, Mazón MJ, Portillo F. Yeast protein kinase Ptk2 localizes at the plasma membrane and phosphorylates in vitro the C-terminal peptide of the H+-ATPase. Biochim Biophys Acta. 2006;1758:164–170. doi: 10.1016/j.bbamem.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol. 2001;40:1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- Gaigg B, Timischl B, Corbino L, Schneiter R. Synthesis of sphingolipids with very long chain fatty acids but not ergosterol is required for routing of newly synthesized plasma membrane ATPase to the cell surface of yeast. J Biol Chem. 2005;280:22515–22522. doi: 10.1074/jbc.M413472200. [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu J-J, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- Hockney RC, Freeman RF. Gratuitous catabolite repression by glucosamine of maltose utilization in Saccharomyces cerevisiae. J Gen Microbiol. 1980;121:469–474. [Google Scholar]
- Jones GW, Tuite MF. Chaperoning prions: The cellular machinery for propagating an infectious protein? Bioessays. 2005;27:823–832. doi: 10.1002/bies.20267. [DOI] [PubMed] [Google Scholar]
- Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: A possible explanation for its effect in curing yeast prions. Curr Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kim J-H, Polish J, Johnston M. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol Cell Biol. 2003;23:5208–5216. doi: 10.1128/MCB.23.15.5208-5216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Brachet V, Moriya H, Johnston M. Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:167–173. doi: 10.1128/EC.5.1.167-173.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C-Y, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Kunz BA, Ball AJ. Glucosamine resistance in yeast II: Cytoplasmic determinants conferring resistance. Mol Gen Genet. 1977;153:169–177. doi: 10.1007/BF00264732. [DOI] [PubMed] [Google Scholar]
- Lacroute F. Non-mendelian mutation allowing ureidosuccinic acid uptake in yeast. J Bacteriol. 1971;106:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan J, Mosley A, Özcan S. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr Genet. 2003;44:19–25. doi: 10.1007/s00294-003-0423-2. [DOI] [PubMed] [Google Scholar]
- Lecchi S, Allen KE, Pardo JP, Mason AB, Slayman CW. Conformational changes of yeast plasma membrane H(+)-ATPase during activation by glucose: Role of threonine-912 in the carboxy-terminal tail. Biochem. 2005;44:16624–16632. doi: 10.1021/bi051555f. [DOI] [PubMed] [Google Scholar]
- Lecchi S, Nelson CJ, Allen KE, Swaney DL, Thompson KL, Coon JJ, Sussman MR, Slayman CW. Tandem phosphorylation of Ser-911 and Thr-912 at the C terminus of yeast plasma membrane H+-ATPase leads to glucose-dependent activation. J Biol Chem. 2007;282:35471–35481. doi: 10.1074/jbc.M706094200. [DOI] [PubMed] [Google Scholar]
- Lee MC, Hamamoto S, Schekman R. Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J Biol Chem. 2002;277:22395–22401. doi: 10.1074/jbc.M200450200. [DOI] [PubMed] [Google Scholar]
- Leonardo B, Aaron R, Gerald M, Joshua L. The FLEXGene repository: Exploiting the fruits of the genome projects by creating a needed resource to face the challenges of the post-genomic era. Arch Med Res. 2002;33:318–324. doi: 10.1016/s0188-4409(02)00372-7. [DOI] [PubMed] [Google Scholar]
- Maddelein M-L, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci. 2002;99:7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- Mazon MJ, Eraso P, Portillo F. Efficient degradation of misfolded mutant Pma1 by endoplasmic reticulum-associated degradation requires Atg19 and the Cvt/autophagy pathway. Mol Microbiol. 2007;63:1069–1077. doi: 10.1111/j.1365-2958.2006.05580.x. [DOI] [PubMed] [Google Scholar]
- Miranda M, Pardo JP, Allen KE, Slayman CW. Stalk segment 5 of the yeast plasma membrane H+-ATPase. Labeling with a fluorescent malemide reveals a conformational change during glucose activation. J Biol Chem. 2002;277:40981–40988. doi: 10.1074/jbc.M206793200. [DOI] [PubMed] [Google Scholar]
- Moriya H, Johnston M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci. 2004;101:1572–1577. doi: 10.1073/pnas.0305901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: Requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsomme P, Slayman CW, Goffeau A. Mutagenic study of the structure, function and biogenesis of the yeast plasma membrane H+-ATPase. Biochim Biophys Acta. 2000;1469:133–157. doi: 10.1016/s0304-4157(00)00015-0. [DOI] [PubMed] [Google Scholar]
- Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek J, Nakayashiki T, Wickner RB. A prion of the yeast netacaspase homolog (Mca1p) detected by a genetic screen. Proc Natl Acad Sci. 2009;106:1892–1896. doi: 10.1073/pnas.0812470106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ness F, Ferreira P, Cox BS, Tuite MF. Guanidine hydrochloride inhibits the generation of prion ‘seeds’ but not prion protein aggregation in yeast. Mol Cell Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevado J, Heredia CF. Galactose induces in Saccharomyces cerevisiae sensitivity of the utilization of hexose to inhibition by D-glucosamine. Can J Microbiol. 1996;42:6–11. doi: 10.1139/m96-002. [DOI] [PubMed] [Google Scholar]
- Ohnishi G, Endo K, Doi A, Fujita A, Daigaku Y, Nunoshiba T, Yamamoto K. Spontaneous mutagenesis in haploid and diploid Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2004;325:928–933. doi: 10.1016/j.bbrc.2004.10.120. [DOI] [PubMed] [Google Scholar]
- Oka M, Kimata Y, Mori K, Kohno K. Saccharomyces cerevisiae KAR2 (BiP) gene expression is induced by loss of cytosolic HSP70/Ssalp through a heat shock element-mediated pathway. J Biochem. 1997;121:578–584. doi: 10.1093/oxfordjournals.jbchem.a021624. [DOI] [PubMed] [Google Scholar]
- Patel BK, Liebman SW. “Prion-proof” for [PIN+]: Infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN+] J Mol Biol. 2007;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11:344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino MM, Liu J-J, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Polish JA, Kim J-H, Johnston M. How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics. 2005;169:583–594. doi: 10.1534/genetics.104.034512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizet G. Les phenomenes de barrage chez Podospora anserina: Analyse genetique des barrages entre les souches s et S. Rev Cytol Biol Veg. 1952;13:51–92. [Google Scholar]
- Roberg KJ, Crotwell M, Espenshade P, Gimeno R, Kaiser CA. LST1 is a SEC24 homologue used for selective export of the plasma membrane ATPase from the endoplasmic reticulum. J Cell Biol. 1999;145:659–672. doi: 10.1083/jcb.145.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso A, Chien P, Osherovich LZ, Weissman JS. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- Schmidt MC, McCartney RR, Zhang X, Tillman TS, Solimeo H, Wolfl S, Almonte C, Watkins SC. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4561–4571. doi: 10.1128/mcb.19.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- Serrano R. In vivo glucose activation of the yeast plasma membrane ATPase. FEBS Lett. 1983;156:11–14. doi: 10.1016/0014-5793(83)80237-3. [DOI] [PubMed] [Google Scholar]
- Serrano R, Kielland-Brandt MC, Fink GR. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature. 1986;319:689–693. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformer. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N. University of Chicago; Chicago: 2000. “The identification of novel prion elements in Saccharomyces cerevisiae.”. PhD thesis, [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1: An epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeny EA, Shorter J. Prion proteostasis: Hsp104 meets its supporting cast. Prion. 2008;2:135–140. doi: 10.4161/pri.2.4.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions of the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu Rev Microbiol. 2002;56:703–741. doi: 10.1146/annurev.micro.56.013002.100603. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M, Stone DE, Craig EA. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Ross ED, Pierce MM, Baxa U, Brachmann A, Shewmaker F. Prion genetics: New rules for a new kind of gene. Annu Rev Genet. 2004;38:681–707. doi: 10.1146/annurev.genet.38.072902.092200. [DOI] [PubMed] [Google Scholar]