Abstract
Dosage compensation in Drosophila melanogaster males is achieved via targeting of male-specific lethal (MSL) complex to X-linked genes. This is proposed to involve sequence-specific recognition of the X at ∼150–300 chromatin entry sites, and subsequent spreading to active genes. Here we ask whether the spreading step requires transcription and is sequence-independent. We find that MSL complex binds, acetylates, and up-regulates autosomal genes inserted on X, but only if transcriptionally active. We conclude that a long-sought specific DNA sequence within X-linked genes is not obligatory for MSL binding. Instead, linkage and transcription play the pivotal roles in MSL targeting irrespective of gene origin and DNA sequence.
Keywords: Dosage compensation, transcription, Drosophila, spreading
X-chromosome dosage compensation is a model for investigating eukaryotic gene regulation in the context of chromatin organization. Dosage compensation serves to equalize expression of X-linked genes in most heterogametic species. In Drosophila melanogaster, it functions to increase transcription of genes on the single male X chromosome approximately twofold to equal the output of both female X chromosomes. Drosophila dosage compensation is mediated by a chromatin-modifying enzyme, the male-specifc lethal (MSL) complex, composed of at least five proteins (MSL1, MSL2, MSL3, MOF, and MLE) and two noncoding roX RNAs. This complex specifically recognizes the single male X chromosome to acetylate histone H4 at Lys 16 (H4K16ac) (Turner et al. 1992; Bone et al. 1994; Hilfiker et al. 1997; Akhtar and Becker 2000; Smith et al. 2000), thereby up-regulating transcription.
How is this transcriptional increase targeted exclusively to the X chromosome? The identification of two classes of elements, roX RNAs and chromatin entry sites (CES), has provided insight into how the MSL complex distinguishes the X from autosomes. roX1 and roX2 were discovered as genetically redundant RNA components of the MSL complex (Amrein and Axel 1997; Meller et al. 1997; Meller and Rattner 2002). In the absence of both roX RNAs, the MSL complex binds in a sparse and poorly defined pattern on the male polytene X chromosome and also displays ectopic localization to the chromocenter (Meller and Rattner 2002). The genes encoding roX RNAs are normally located on the X chromosome, but when transposed to autosomes they attract the MSL complex to their new location. Strikingly, MSL binding not only occurs at the cytological location of the transgene, but can spread long distances into flanking chromatin, up to a megabase in either direction (Kelley et al. 1999; Park and Kuroda 2001). Although roX transgenes can potently attract MSL binding to autosomes, a roX mutant X chromosome can still recruit the MSL complex within the same nucleus, indicating that there must be additional elements that signal X identity. Residual MSL-binding sites seen in msl3, mof, or mle mutants have been proposed to play this role as CES for the initial binding of the MSL complex (Kelley et al. 1999).
The CES were mapped recently by chromatin immunprecipitation (ChIP) coupled with microarray analysis (ChIP–chip) in msl3 mutants or under conditions that favored detection of only the highest MSL complex occupancy (Alekseyenko et al. 2008; Straub et al. 2008). A consensus sequence comprised of a GA-rich MRE (MSL recognition element) has been identified that, when tested at ectopic locations, is both necessary and sufficient to autonomously recruit MSL complex (Alekseyenko et al. 2008). Thus, CES appear to mediate the initial targeting of the MSL complex to the X chromosome, yet they are only approximately twofold enriched on X. Estimates suggest that there are 150–300 CES on X, so the majority of the ∼1000 genes bound by the MSL complex are not thought to contain an entry site. Instead, these genes may be recognized via spreading from nearby entry sites. Recent studies implicate the transcription-dependent trimethylated H3K36 (H3K36me3) chromatin mark as important for optimal levels of MSL binding in its complete pattern (Alekseyenko et al. 2006; Gilfillan et al. 2006; Larschan et al. 2007; Bell et al. 2008; Sural et al. 2008). However, hypothetical sequence determinants within each X-chromosomal gene have historically been thought to play an essential role. To date, the question of sequence specificity has been addressed mainly through gene-by-gene analysis of X segments inserted on autosomes, often yielding autonomous MSL binding without providing a clear candidate sequence for further study (Pirrotta et al. 1985; Scott and Lucchesi 1991; Baker et al. 1994; Qian and Pirrotta 1995; Roseman et al. 1995; Kind and Akhtar 2007). Therefore, it is not clear how many individual gene analyses would be required to derive additional X-specific motifs, if they exist. Also lacking is an understanding of whether genes transposed from the X may be influenced by MSL binding of their homologs on the endogenous X chromosome.
For these reasons, we took a distinct approach to the question of specificity. Here, we take sequences that never normally attract the MSL complex, and ask whether they can acquire MSL binding simply by being transcriptionally active and physically linked to the X chromosome. We find that active autosomal genes consistently become targets of the MSL complex when inserted on X, even when introduced as part of a large, 65-kb autosomal segment into a region of X normally devoid of MSL binding. Taken together, our data argue against the idea that each X-chromosomal gene possesses special MSL recruiting signals. Instead, we propose that after initial attraction of the MSL complex by a CES or roX RNA gene, transcription of active genes serves as the main guiding feature for MSL complex binding.
Results and Discussion
Autosomal genes attract MSL proteins in the male X environment
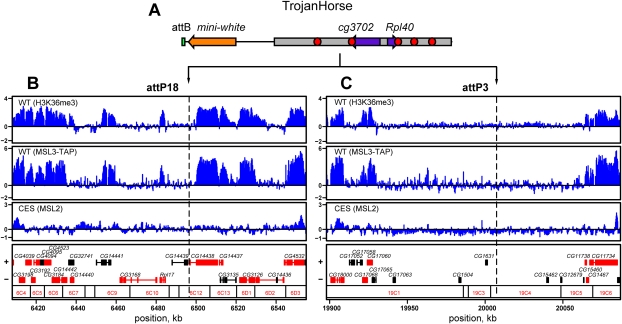
To ask whether autosomal genes can effectively recruit MSL complex, we created a construct that we named TrojanHorse, consisting of a 14-kb fragment from chromosome arm 2L encompassing two small genes, Rpl40 and cg3702 (Fig. 1A). These genes were chosen because they are expressed at all stages of development and are associated with H3K36me3, both hallmarks of typical MSL targets on the X. We altered the sequence of the 14-kb fragment by incorporating five 0.2-kb tags of non-Drosophila origin (Fig. 1A, red circles) into the 3′-untranslated regions (UTRs) of Rpl40 and cg3702, and the nontranscribed regions of TrojanHorse. This design allowed us to clearly discriminate between transcripts derived from the endogenous Rpl40 and cg3702 genes and their transposed copies on the X. Additionally, these tags enabled direct assessment of MSL protein and histone modification profiles across the TrojanHorse insertion on the X chromosome. We placed the TrojanHorse construct in two precise genomic positions (Fig. 1B,C) using the recently developed attB/attP phage phiC31 integration system (Groth et al. 2004; Bischof et al. 2007). One insertion site (attP18 at cytological position 6C12) was in a gene-rich region encompassing numerous MSL gene targets. In contrast, the second landing site (attP3 at 19C4) was intentionally chosen to be distant from MSL targets and active genes, with the nearest MSL target 60 kb proximal to the TrojanHorse insertion (Fig. 1C). Thus, in the second case, the two constitutively expressed autosomal genes were inserted on X within a large, MSL-depleted region.
Figure 1.
TrojanHorse strategy. (A–C) TrojanHorse transposon (A), containing two genes, cg3702 and Rpl40, on a 14-kb DNA segment from chromosome arm 2L (gray), was inserted into two genomic locations on chromosome X: gene-rich attP18 (B) and gene-poor attP3 (C). Shown are previously published ChIP–chip profiles for H3K36me3 generated from male S2 cells (WT, top), MSL3-TAP from embryos (WT, middle), or MSL2 from msl3 mutant embryos (CES, bottom) (Larschan et al. 2007; Alekseyenko et al. 2008). The locations of the attP18 and attP3 insertion sites are marked by dashed lines. The nearest flanking CES serve to delimit map borders. Log2 of immunoprecipitated (IP) to input (IN) signals ratio is shown on the Y-axis. Genomic coordinates (Release dm1), banding, and genetic maps are shown below. Genes are color-coded as follows: transcribed S2 cells are shown in red, and nontranscribed in S2 cells are shown in black (Alekseyenko et al. 2006).
If specific DNA sequences at each gene are required to complete the MSL pattern on X, then TrojanHorse genes, originating from an autosome, should not acquire MSL binding. Alternatively, if linkage to the X chromosome and the active state are sufficient, TrojanHorse genes should be bound and acquire H4K16ac. Therefore, we performed ChIP assays to determine the binding profiles for two MSL subunits, MSL2 and TAP-tagged MSL3, within TrojanHorse placed at either attP18 or attP3 (Fig. 2B; Supplemental Fig. 1A). Independent of the integration site, we observed pronounced binding of MSL proteins to the tags located at the 3′ ends of both cg3702 and Rpl40 of TrojanHorse in mixed sex embryos (Fig. 2B, red and blue bars; Supplemental Fig. 1A) and male larvae (Supplemental Fig. 2A; data not shown). In contrast, the untranscribed regions of TrojanHorse did not attract the MSL complex. Furthermore, MSL complex binding led to H4K16 acetylation of TrojanHorse chromatin. In agreement with the binding profiles for MSL2 and MSL3-TAP, H4K16ac was found to peak at the 3′-UTRs of cg3702 and Rpl40 in both embryos (Fig. 2C, red bars; Supplemental Fig. 1B, red and blue bars) and male larvae (Supplemental Fig. 2B). Notably, the histone H4 acetylation profile was broader than that of the MSL complex, consistent with recent studies (Gelbart et al. 2009; Schwaiger et al. 2009). Finally, we confirmed that the two genes within TrojanHorse were indeed transcribed in larvae and male and female embryos by mapping RNA polymerase II and histone methylation marks expected for active genes: dimethylated H3K4 (H3K4me2) at the 5′ ends, and H3K36me3 biased toward the 3′ ends (Fig. 2D, red bars; Supplemental Figs. 1C,D, 2C–E). Taken together, our results demonstrate that transcribed and H3K36me3-marked autosomal genes can become typical MSL targets when placed on the male X, favoring a sequence-independent model for spreading. Our data also indicate that targeting can occur in the apparent absence of a nearby CES, as the closest identified entry sites are located ∼54 kb and 80 kb proximal to attP18 and attP3, respectively. Furthermore, attP3 is 60 kb away from the nearest active gene cluster or MSL-bound region, indicating that targeting can occur over long distances in cis.
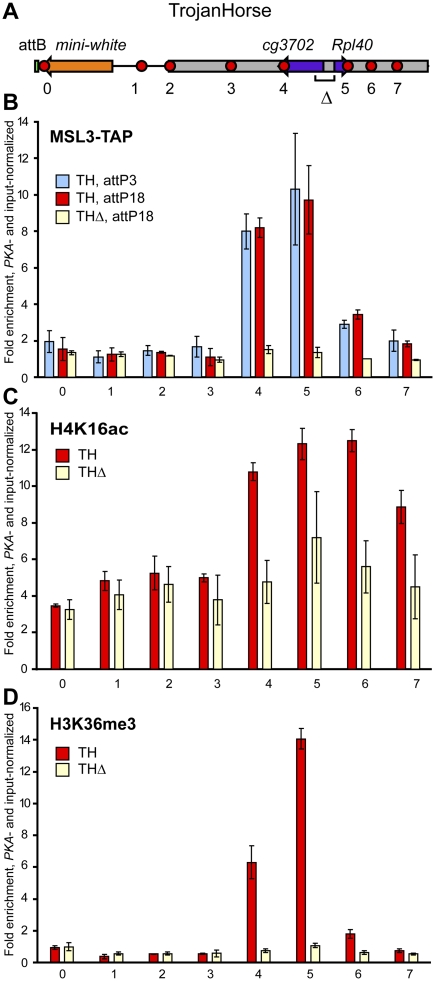
Figure 2.
Autosomal genes attract the MSL complex in a transcription-dependent manner on the male X. (A) Structure of the TrojanHorse transposon integrated on the X chromosome. Sequences from chromosome 2L are shown in gray. Positions of qPCR probes are depicted as red circles. Δ denotes sequences deleted in the “promoterless” version of the TrojanHorse, TrojanHorseΔ. Fold enrichment of MSL3-TAP (B), H4K16ac (C), and H3K36me3 (D) across TrojanHorse (red bars) and TrojanHorseΔ (yellow bars), inserted at attP18. MSL3-TAP fold enrichment at TrojanHorse inserted at attP3 is shown as blue bars. Enrichment values are PKA-normalized and input-normalized. The 3′ ends of genes from 2L contained within TrojanHorse, cg3702, and Rpl40 (probes 4 and 5), are bound and acetylated by the MSL complex only when transcribed.
Transcription is essential for MSL targeting
Despite the clear association of transposed autosomal genes with MSL protein in the male X environment, it remained to be determined whether their transcription or some other features were required for the observed complex binding. In order to address this question, we created a “promoterless” version of TrojanHorse where the 1.2-kb region, including the promoters and 5′ ends of both cg3702 and Rpl40, was deleted (Figs. 1A, 2A). This TrojanHorseΔ, otherwise identical to TrojanHorse, was placed in the same attP18 site by attP/attB recombination. We confirmed by quantitative PCR (qPCR) analysis that transcription of both genes in TrojanHorseΔ was dramatically reduced (at least 25-fold), approaching the detection limit (data not shown). In addition, the genes exhibited background levels of H3K36me3 in TrojanHorseΔ (Fig. 2D). We then asked whether transcriptionally inactive genes showed any changes in binding patterns of MSL and other proteins. Indeed, MSL3-TAP no longer bound nontranscribed cg3702 and Rpl40 (Fig. 2B), resulting in decreased H4K16ac levels throughout TrojanHorseΔ (Fig. 2C). This direct comparison clearly supports the importance of transcription for MSL recognition of its targets.
Newly targeted genes are dosage-compensated
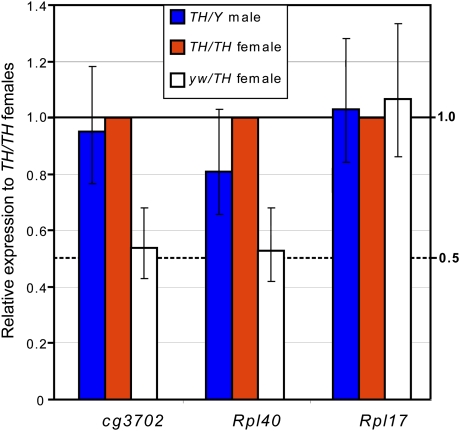
To test whether the genes within the intact TrojanHorse were not only associated with MSL complex and marked with H4K16ac but also were subject to dosage compensation, we directly measured the degree of up-regulation of cg3702 and Rpl40 in the TrojanHorse context. As shown in Figure 3, these genes are transcribed and dosage-compensated in male larvae so that expression of one copy in males is approximately equal to two copies in females, and about twofold higher than in heterozygous females. Likewise, up-regulation is observed in male embryos (Supplemental Fig. 3). However, in this case the dosage compensation appears less complete, perhaps due to maternal deposition of the transcripts leading to an underestimate of the male/female zygotic transcription ratio. Taken together, these data indicate that the MSL complex bound to TrojanHorse retains all its functions: It acetylates the underlying chromatin and increases transcriptional output, resulting in dosage compensation of both Rpl40 and cg3702.
Figure 3.
TrojanHorse genes are dosage-compensated. Shown are relative transcript levels observed for hemizygous TrojanHorse/Y male larvae (blue bars) and hemizygous +/TrojanHorse female larvae (white bars), both normalized to values obtained for homozygous TrojanHorse/TrojanHorse female larvae. Error bars denote 95% confidence intervals. Importantly, the relative expression of cg3702 and Rpl40 in males does not differ significantly from that observed in homozygous females, and is twice as high as in hemizygous females, suggesting a near complete dosage compensation at cg3702 and Rpl40 and linear dependence of expression to gene dosage, confirming the sensitivity of our assay. Under such normalization, relative enrichment values for the endogenous X-linked Rpl17 gene are close to unity, as is predicted for a fully compensated flanking gene whose dosage remains the same in TrojanHorse/+ and TrojanHorse/TrojanHorse females.
Larger autosomal segments also recruit MSLs within the male X-chromosomal context
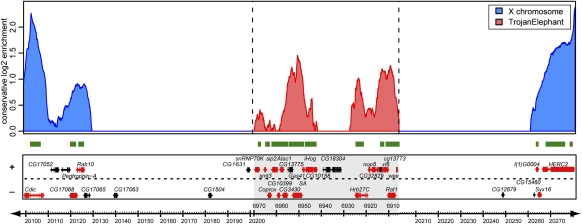
To exclude the possibility that the two genes in the TrojanHorse construct were somehow exceptional in their ability to attract MSL complex, we tested a more extensive set of active autosomal genes by engineering a larger A-to-X transposition. We created TrojanElephant by inserting a 65-kb region from chromosome 2L, containing 20 genes from cg13773 to snRNP70K into the attB-P[acman] vector via recombineering (Venken et al. 2006). None of the TrojanHorse or TrojanElephant genes attracted MSL complex when located in their endogenous positions on autosomes (Supplemental Fig. 4). Therefore, we inserted the TrojanElephant construct at the previously characterized attP3 site on the X, which is a gene-poor region. Several possible results were hypothesized: (1) Within the 65-kb segment, the MSL complex might target all H3K36me3-positive genes. (2) MSL binding might be detected over the active genes closest to the ends of the 65-kb TrojanElephant, with a gradual decrease in binding closer to the center. (3) The MSL complex might skip genes in TrojanElephant altogether.
To determine which of these scenarios occurs in vivo, we analyzed MSL binding across TrojanElephant in third-instar larvae by ChIP followed by qPCR (Supplemental Fig. 5) or ChIP and sequencing (ChIP-seq) (Fig. 4). The data obtained from both experiments clearly demonstrated the ability of the MSL complex to faithfully recognize the transcribed and H3K36me3-marked genes within the transposed material, with no evidence for skipping any of the active genes. Therefore, if the MSL complex spreads linearly from the closest identified flanking CES, it can travel at least 83 kb. The fact that all active autosome-derived genes were bound by MSL complex argues strongly against the idea that each X-chromosomal gene possesses special MSL recruiting signals. Instead, we propose that after the initial attraction of the MSL complex by a CES or roX RNA gene, transcription of active genes serves as the main guiding feature for MSL complex binding.
Figure 4.
Active genes within the large TrojanElephant transposon recruit MSL complex. The ChIP-seq tag profile (Solexa) was obtained from male TrojanElephant; MSL3-TAP; msl31 larvae. MSL binding shows a broad distribution along wild-type MSL targets on the X (blue), and pronounced MSL binding within active but not inactive TrojanElephant sequences transposed from 2L (red). The gene map is shown below, and genes are color-coded as in Figure 1. Green bars represent H3K36me3-positive regions measured in S2 cells (Larschan et al. 2007).
Our results are consistent with early transgenic studies, indicating that single genes could become dosage-compensated when inserted on X (Scholnick et al. 1983; Spradling and Rubin 1983; Sass and Meselson 1991). However, our work still needs to be reconciled with results from established transposition stocks, in which much larger inserts of autosomal material onto X (∼1.5 Mb and ∼5.8 Mb), failed to acquire dosage compensation (Roehrdanz et al. 1977) or cytologically visible MSL binding (Fagegaltier and Baker 2004; Oh et al. 2004), or showed binding only a few kilobases into the insertion by ChIP analysis (Sun and Birchler 2009). One possibility is that stable stocks carrying large spontaneous or X-ray-induced rearrangements may display exceptional behavior, as they have been preselected for viability and thus possibly for the maintenance of chromosome of origin regulation. Alternatively, these results may indicate that distance to the nearest functional CES is critical for MSL targeting, but that we did not reach this critical distance in our current experiments. It may be that once a certain size is attained, any DNA insertion will tend to associate with its chromosome of origin rather than be generally localized within the X-chromosome three-dimensional territory, thus excluding spreading. Testing these possibilities may be within reach in the near future, as improvements in transgenic technology allow larger insertions at predefined breakpoints to be obtained.
Our data allow further refinement of the two-step model for MSL complex recruitment to the male X chromosome. In the first step, MSLs are thought to bind 150–300 CES containing MRE motifs on X, and ignore autosomes. In the second step, MSLs recognize the active genes on the X irrespectively of their sequence and origin through a transcription-dependent mechanism. Trimethylation of H3K36 is partially responsible for this sequence-independent step. Here we speculate on the coexistence of two modes of MSL spreading on the X: (1) long-range spreading from the roX genes, and (2) local distribution of the MSL complex from the CES scattered throughout the X (with a median distance of ∼100 kb). It is only in the context of the X chromosome that both roX genes and CES are found in cis to each other, making possible both correct chromosome identification and efficient spreading. The enigmatic nature of roX spreading remains to be understood, including whether features of active genes might be recognized directly by roX RNAs.
Materials and methods
Plasmids and constructs
Complete maps and details of cloning are available on request. Briefly, to generate the TrojanHorse construct, a genomic region of 2L chromosome from 4,218,616 to 4,232,796 base pairs (bp) was PCR-amplified from BACR11D14. Five tags ∼200 bp long encompassing various fragments of EGFP, firefly luciferase, and Renilla luciferase were incorporated within TrojanHorse via standard PCR mutagenesis and cloning procedures. TrojanHorseΔ was obtained from TrojanHorse by PCR mutagenesis and deleting the sequences from 4,226,410 to 4,227,694 bp. Both TrojanHorse and TrojanHorseΔ harbor a single attB site and a mini-white genetic transformation marker. To construct TrojanElephant, a fragment of 2L spanning 6,971,319–6,906,389 bp was used and placed into the attB-P[acman] vector via recombineering from BACR20B21 (R5.18 coordinates are used throughout). Details of fly genetics and transgenesis can be found in the Supplemental Material.
Chromatin preparation for ChIP and ChIP-seq
Preparation of chromatin for ChIP from mixed sex embryos (ChIP) or sexed larvae (ChIP and ChIP-seq) was performed as described previously (Larschan et al. 2007; Alekseyenko et al. 2008). Commercial antibodies used for ChIP were from Abcam (H3K36me3, ab9050; H3K4me2, ab7766; H3, ab1791; RNA polymerase II (4H8), ab5408) and Upstate Biotechnologies (H4K16ac, 07-329). Quantitation of ChIP DNA (three biological replicates) by qPCR (in duplicates) was done as described previously (Larschan et al. 2007). Error bars correspond to standard deviations among three independent experiments. Sequences of real-time PCR primers used can be found in Supplemental Table 1. To prepare large batches of chromatin from TrojanElephant; MSL3-TAP; msl31 mixed sex larvae (second and third instars), the animals were separated from food and pupae by washing in 20% sucrose. Up to 20 g of larvae were used per chromatin prep. For ChIP-seq, ∼10,000 TrojanElephant/Y; MSL3-TAP; msl31 male larvae were collected. Details of ChIP-seq analysis algorithms and data accessibility can be found in the Supplemental Material.
RNA extraction and quantification
Total RNA was isolated from single larvae (six independent preps per genotype), pools of 30–200 sexed embryos (at least three replicates), or 10 age-matched sexed adults (three replicates per genotype) by a standard Trizol extraction, followed by RNA purification and removal of genomic DNA with RNeasy Plus kit (Qiagen). Vilo cDNA synthesis kit (Invitrogen) was used to reverse-transcribe RNA using random primers. To quantify transcript levels, real-time PCR was performed on an ABI 7000 machine. qPCR primer sequences are provided in Supplemental Table 1.
Acknowledgments
We are grateful to M. Gelbart, E. Larschan, and J. Luo for critical comments on the manuscript. Many thanks to S. Peng and M. Tolstorukov for excellent help and advice on statistics and bioinformatics, and J. Bateman, T. Wu, J. Bischof, F. Karch, K. Basler, M. Markstein, P. Graham, and P. Schedl for key fly stocks. This work was supported by the National Institutes of Health GM45744 to M.I.K, and GM67825 to P.J.P.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1840409.
Supplemental material is available at http://www.genesdev.org.
References
- Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP–chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes & Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrein H, Axel R. Genes expressed in neurons of adult male Drosophila. Cell. 1997;88:459–469. doi: 10.1016/s0092-8674(00)81886-3. [DOI] [PubMed] [Google Scholar]
- Baker BS, Gorman M, Marin I. Dosage compensation in Drosophila. Annu Rev Genet. 1994;28:491–521. doi: 10.1146/annurev.ge.28.120194.002423. [DOI] [PubMed] [Google Scholar]
- Bell O, Conrad T, Kind J, Wirbelauer C, Akhtar A, Schubeler D. Transcription-coupled methylation of histone H3 at lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila melanogaster. Mol Cell Biol. 2008;28:3401–3409. doi: 10.1128/MCB.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone JR, Lavender J, Richman R, Palmer MJ, Turner BM, Kuroda MI. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes & Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- Fagegaltier D, Baker BS. X chromosome sites autonomously recruit the dosage compensation complex in Drosophila males. PLoS Biol. 2004;2:e341. doi: 10.1371/journal. pbio.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Larschan E, Peng S, Park PJ, Kuroda MI. Drosophila MSL complex globally acetylates H4 Lys16 on the male X chromosome for dosage compensation. Nat Struct Mol Biol. 2009;16:801–803. doi: 10.1038/nsmb.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan GD, Straub T, de Wit E, Greil F, Lamm R, van Steensel B, Becker PB. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes & Dev. 2006;20:858–870. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Kind J, Akhtar A. Cotranscriptional recruitment of the dosage compensation complex to X-linked target genes. Genes & Dev. 2007;21:2030–2040. doi: 10.1101/gad.430807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Meller VH, Rattner B. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 2002;21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell. 1997;88:445–457. doi: 10.1016/s0092-8674(00)81885-1. [DOI] [PubMed] [Google Scholar]
- Oh H, Bone JR, Kuroda MI. Multiple classes of MSL binding sites target dosage compensation to the X chromosome of Drosophila. Curr Biol. 2004;14:481–487. doi: 10.1016/j.cub.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Park Y, Kuroda MI. Epigenetic aspects of X-chromosome dosage compensation. Science. 2001;293:1083–1085. doi: 10.1126/science.1063073. [DOI] [PubMed] [Google Scholar]
- Pirrotta V, Steller H, Bozzetti MP. Multiple upstream regulatory elements control the expression of the Drosophila white gene. EMBO J. 1985;4:3501–3508. doi: 10.1002/j.1460-2075.1985.tb04109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S, Pirrotta V. Dosage compensation of the Drosophila white gene requires both the X chromosome environment and multiple intragenic elements. Genetics. 1995;139:733–744. doi: 10.1093/genetics/139.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehrdanz RL, Kitchens JM, Lucchesi JC. Lack of dosage compensation for an autosomal gene relocated to the X chromosome in Drosophila melanogaster. Genetics. 1977;85:489–496. doi: 10.1093/genetics/85.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman RR, Swan JM, Geyer PK. A Drosophila insulator protein facilitates dosage compensation of the X chromosome mini-white gene located at autosomal insertion sites. Development. 1995;121:3573–3582. doi: 10.1242/dev.121.11.3573. [DOI] [PubMed] [Google Scholar]
- Sass H, Meselson M. Dosage compensation of the Drosophila pseudoobscura Hsp82 gene and the Drosophila melanogaster Adh gene at ectopic sites in D. melanogaster. Proc Natl Acad Sci. 1991;88:6795–6799. doi: 10.1073/pnas.88.15.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholnick SB, Morgan BA, Hirsh J. The cloned dopa decarboxylase gene is developmentally regulated when reintegrated into the Drosophila genome. Cell. 1983;34:37–45. doi: 10.1016/0092-8674(83)90134-4. [DOI] [PubMed] [Google Scholar]
- Schwaiger M, Stadler MB, Bell O, Kohler H, Oakeley EJ, Schubeler D. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes & Dev. 2009;23:589–601. doi: 10.1101/gad.511809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MJ, Lucchesi JC. Structure and expression of the Drosophila melanogaster gene encoding 6-phosphogluconate dehydrogenase. Gene. 1991;109:177–183. doi: 10.1016/0378-1119(91)90607-d. [DOI] [PubMed] [Google Scholar]
- Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol Cell Biol. 2000;20:312–318. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. The effect of chromosomal position on the expression of the Drosophila xanthine dehydrogenase gene. Cell. 1983;34:47–57. doi: 10.1016/0092-8674(83)90135-6. [DOI] [PubMed] [Google Scholar]
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Birchler JA. Studies on the short range spreading of the male specific lethal (MSL) complex on the X chromosome in Drosophila. Cytogenet Genome Res. 2009;124:158–169. doi: 10.1159/000207524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sural TH, Peng S, Li B, Workman JL, Park PJ, Kuroda MI. The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat Struct Mol Biol. 2008;15:1318–1325. doi: 10.1038/nsmb.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM, Birley AJ, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]