Abstract
Comparative proteomics is a powerful analytical method for learning about the responses of biological systems to changes in growth parameters. To make confident inferences about biological responses, proteomics approaches must incorporate appropriate statistical measures of quantitative data. In the present work we applied microarray-based normalization and statistical analysis (significance testing) methods to analyze quantitative proteomics data generated from the metabolic labeling of a marine bacterium (Sphingopyxis alaskensis). Quantitative data were generated for 1,172 proteins, representing 1,736 high confidence protein identifications (54% genome coverage). To test approaches for normalization, cells were grown at a single temperature, metabolically labeled with 14N or 15N, and combined in different ratios to give an artificially skewed data set. Inspection of ratio versus average (MA) plots determined that a fixed value median normalization was most suitable for the data. To determine an appropriate statistical method for assessing differential abundance, a -fold change approach, Student's t test, unmoderated t test, and empirical Bayes moderated t test were applied to proteomics data from cells grown at two temperatures. Inverse metabolic labeling was used with multiple technical and biological replicates, and proteomics was performed on cells that were combined based on equal optical density of cultures (providing skewed data) or on cell extracts that were combined to give equal amounts of protein (no skew). To account for arbitrarily complex experiment-specific parameters, a linear modeling approach was used to analyze the data using the limma package in R/Bioconductor. A high quality list of statistically significant differentially abundant proteins was obtained by using lowess normalization (after inspection of MA plots) and applying the empirical Bayes moderated t test. The approach also effectively controlled for the number of false discoveries and corrected for the multiple testing problem using the Storey-Tibshirani false discovery rate (Storey, J. D., and Tibshirani, R. (2003) Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445). The approach we have developed is generally applicable to quantitative proteomics analyses of diverse biological systems.
Quantitative proteomics experiments hold the promise of being able to interrogate entire proteomes and ultimately to identify those proteins that differ in their abundance between two or more experimental states. Metabolic labeling, such as 14N:15N labeling is a popular approach used to compare the ratios of observed peptides in two proteomes in a single LC-MS run. Despite the popularity of this approach, considerable issues remain with how experiments are designed, including the evaluation of the statistical significance of the proteomics data.
These issues include but are not limited to experimental design including the choice of biological and technical replicates (1, 2), sample pooling and preparation (2), peptide identification (3, 4) and quantitation (5), accounting for intersample variation via normalization within and between experiments (6–9), the “missing data problem” (1, 10–12), selecting a robust statistical analysis with often very few replicates, and the “multiple testing problem” (13). Many of these issues are not unique to proteomics, and the microarray literature has made a great number of advances in areas such as experimental design, normalization, statistical analysis, and multiple testing adjustments. In fact, there are many parallels between proteomics and transcriptomics for which robust methods already exist, the most pertinent of which is that two-color microarrays, which utilize red and green fluorescent labels, has strong similarities with 14N and 15N metabolic labeling of proteins. In contrast, there are also a number of issues that are unique to proteomics, such as peptide identification and quantitation and incomplete proteome coverage (the missing data problem) where each experimental run will inevitably differ in the peptides that are identified.
Although some of these issues have been discussed in the literature, a number of important issues have been neglected or not comprehensively addressed. Here we focus on four key areas in metabolically labeled quantitative proteomics: normalization, experimental design and linear models, empirical Bayes moderated t test, and adjustments for multiple testing.
The raw data obtained from proteomics experiments must be normalized to produce more accurate estimates of the underlying biological effects being measured. Normalization removes aberrant signals resulting from interexperimental variation possibly due to inevitable differences in sample processing and experimental runs that may be separated by days or weeks. In the field of microarray analysis, there have been considerable advances in methods to detect biases in experimental data and for removing such biases with a range of sophistication from simple scaling (14) to non-parametric quantile normalization (15). Despite these advances, normalization of two-dimensional PAGE-MS- (7, 10, 16–18) and LC-MS-based (6, 8, 19–21) proteomics data tends to be simple, global normalization where ratios are multiplied by a fixed constant to ensure that the medians or means are similar.
To address this, we inspected ratio versus average (MA)1 plots (14, 22) and identified a strong non-linear bias (i.e. a curved skew) that affected the 14N:15N ratios to a different but predictable extent dependent upon protein signal:noise ratio (S/N) values. We applied lowess normalization, a popular method from the -omics literature, that determines a factor by which to adjust the ratios using a sliding window across a range of S/N levels. In this way, a curved line is fitted through the data, thereby adjusting the 14N:15N ratios so that they are symmetrically distributed throughout the range of S/N intensities. Taking this approach, the size of the skew could be evaluated enabling a judgment to be made as to whether leaving it uncorrected would lead to a bias in the types of proteins that are identified as differentially abundant.
Following normalization, the data can be used for determining the magnitude of effect due to the treatment. In the current literature this has typically been achieved by applying a Student's t test to the ratios from each protein. A number of disadvantages, unique to proteomics experiments, arise as a result of using a Student's t test. First, many proteins have missing observations, thereby decreasing the number of degrees of freedom (df) that causes the t test to have low power. Subsequently fewer observations lead to difficulties in accurately determining the measurement error, making the denominator of the t test unreliable. Second, more complex experimental designs, such as using two different extraction buffers, cannot be appropriately represented by the t test. Third, an underlying assumption of the t test is that each observation is independent. However, this assumption is violated by having technical replicates and thus causes the t test to underestimate the true measurement variance and therefore overestimate the strength of the statistic.
We adopted, from microarray analysis, the widely used method of linear models to construct a statistical representation that mirrors the experimental design. Linear models are a mathematical framework that break down the observed expression ratios from one protein from each individual to an effect that is shared across samples due to each experimental parameter (e.g. growth temperature, extraction buffer, or biological replicate) and incorporate an error term unique to each individual. Linear models are simple to construct using design matrices that describe the treatment applied to each sample coupled with a robust mathematical framework to estimate the unknown effect sizes for each protein. It is important to understand that the linear model that represents a “simple” experiment consisting of a number of replicate measurements of the same treatment corresponds to an averaging of all observed ratios and is thus identical to the numerator from the Student's t test. However, as the experimental design becomes more complex, the linear model can accommodate these demands. The linear model can be used to estimate the biological effect of interest after accounting for the effects due to differing extraction buffers and importantly adjust for the non-independence between technical replicates. To demonstrate the use of a linear model in a typical experiment, the results from fitting an appropriately specified linear model are compared and contrasted to a Student's t test.
In addition to estimating the magnitude of the effect size using linear models, an empirical Bayes moderated t test (39) was used to perform statistical analysis. Instead of testing each protein in isolation from all others, the moderated t test “borrows strength” from all other proteins, improving the error estimates of each individual protein. The error estimate from each protein is replaced by a pooled estimate, i.e. one that is adjusted toward the population estimate. This shrinks or expands the error estimates for proteins with high or low variance, respectively. Although not immediately obvious, this prevents proteins that, by random chance, have tiny variance from becoming those proteins with the strongest statistical evidence for differential abundance. In addition, the moderated t test augments the degrees of freedom for each protein dependent on how variable the entire set of proteins is, thereby allowing for the statistical estimates of differential abundance for proteins with large numbers of missing observations.
Applied to proteomics, the multiple testing problem describes the repeated application of a statistical test to a set of protein (identification or abundance) measurements. This may result in 5% of all measurements having p values less than 0.05 due purely to chance, thereby producing many false positives (13). Our overall goal was to identify the largest number of differentially abundant proteins with robust statistical evidence, taking care to avoid an unacceptable level of false positives. To achieve this we applied the positive false discovery rate (FDR) procedure (23) by adopting an approach from the microarray field that is described by Käll et al. (4). In this approach, if a list of proteins is selected, all with an FDR <5%, then at most 5% of these proteins are false discoveries; the same is not true for a set of proteins that all have a p value <0.05. To compare with other methods for correcting for multiple testing, proteins were identified using either an FDR, Bonferroni correction (13), or no adjustment.
Commercially produced software packages for processing quantitative proteomics data tend to be high quality and relatively easily used (e.g. Refs. 9 and 24) but can be expensive and limited in their applicability (supplemental Table S1). Alternatively publicly available solutions incur no cost but tend to require the end user to assemble a large number of interacting components (e.g. a relational database and scripts for importing and manipulating the data) that are typically written in a number of programming languages (e.g. Ref. 25) (supplemental Table S1). To overcome these obstacles we developed a logical analytical pipeline by implementing a single, free, widely supported and used software environment (R). All steps of the analysis from data import to visualization and generation of results were integrated by extending the widely used limma library from the Bioconductor suite of packages.
To develop and test our methods, 14N and 15N metabolically labeled cultures of the marine bacterium Sphingopyxis alaskensis were grown at the same temperature and compared after being combined in different ratios. Comparisons of metabolically labeled cultures grown at two different temperatures were also performed. A total of 1,736 high confidence protein identifications representing 54% of the coding capacity of the genome were made, and 1,172 proteins with high quality data were quantified. Following the application of lowess normalization, linear models to each protein, an empirical Bayes moderated t test, and the Storey-Tibshirani positive FDR correction for multiple testing (23), we identified 217 differentially abundant proteins with an FDR <5%. The approach we have developed is applicable to quantitative proteomics in many biological systems.
EXPERIMENTAL PROCEDURES
Microbial Growth and Physiological Characterization
S. alaskensis was grown in artificial sea water (ASW) medium (26) at 10 and 30 °C with rotary shaking at 100 rpm as described previously (27). Colony-forming units were measured by the drop plate method on Väätänen nine-salt solution as described previously (26, 27). Protein yields were measured using a Bradford assay with BSA as a standard (28). Scanning electron microscopy was performed with cells grown at 10 or 30 °C to midlogarithmic phase. Cells were fixed with 2% (w/v) glutaraldehyde for 2 h and filtered through a 0.2-μm membrane filter. The filters were washed with 75, 50, and 25% ASW that contained 0.04% (w/v) MOPS for 3 min in each washing step. Cells were dehydrated in a graded series of 30, 50, 70, 80, 90, and 95% ethanol for 10 min in each step. The final dehydration step was performed three times with 100% ethanol for 10 min each. Samples were critical point-dried with carbon dioxide using a BAL-TEC CPD 030 critical point dryer and were sputter-coated with chromium to ∼0.5-nm thickness using a K575X Peltier cooled high resolution sputter coater (Emitech). Coated samples were viewed under vacuum at an accelerating voltage of 15 kV and magnifications from 2,500× to 35,000× on a Hitachi S-3400N automated vacuum pump scanning electron microscope. From the electron micrographs of each growth temperature, 100 cells were selected using a 36-point sampling grid used in unbiased stereology to visually compare and measure cell biovolume (V) using the equation V = π/4 × W2 × (L − W/3) where W is width and L is length of the cell (29).
Metabolic Labeling
Cells grown at 10 and 30 °C were metabolically labeled during growth in unlabeled (14NH4Cl) and labeled (99% enriched 15NH4Cl) media where all other sources of nitrogen had been eliminated. Cells grown in 15NH4Cl ASW were labeled to 99% 15N incorporation in 10 generations of growth. 14N and 15N cells were combined after cell harvest at midlogarithmic growth phase of OD 0.3 (λ = 433 nm) (27).
30 Versus 30 °C Experiment
To assess the ability of our methods to identify subtle changes in protein abundance, cells were grown at 30 °C with 14N or 15N, and cell pellets were combined at OD 0.3 (λ = 433 nm) in 0.8:1, 1:1, and 1.2:1 ratios in triplicate (total of nine experiments).
10 Versus 30 °C Experiment
To assess differential abundance due to temperature, cells were grown at 10 or 30 °C comprising six biological replicates and a total of 20 experiments. Four biological replicates (A–D) with two experimental replicates and two MS instrumental replicates per sample, representing 16 experiments of 14N:15N and inverse 15N:14N, 10 and 30 °C cell pellets, were combined 1:1 from cultures harvested at OD 0.3 (λ = 433 nm) (supplemental Fig. S1). An additional two biological replicates (E and F) with two MS instrumental replicates per sample, providing four experiments, were cultured as above; proteins were extracted separately from the 14N and 15N, 10 and 30 °C samples; and protein extracts were combined 1:1 (14N:15N and inverse 15N:14N, 10 and 30 °C) based on protein concentration (supplemental Fig. S2).
Protein Extraction and Gel-based Fractionation
To enhance proteome coverage, two different extraction buffers were used to extract proteins. Proteins from experiments A, B, E, and F were extracted in a 10 mm Tris-HCl, pH 8.0, 1 mm EDTA, and 1 mm PMSF buffer by sonication on ice as described previously (30). Proteins from experiments C and D were extracted in 8 m urea, 1 mm EDTA, and 1 mm PMSF buffer by sonication on ice. One-dimensional PAGE protein separation followed by nanocapillary LC-MS/MS was used to separate 1 mg of whole cell lysate by 12% SDS-PAGE as described previously (31, 32). Briefly the entire lane was excised into 22 slices and cut into 1-mm3 pieces, and proteins were reduced in 10 mm DTT at 37 °C for 1 h, alkylated with 25 mm iodoacetamide at 37 °C for 1 h in the dark, washed twice with deionized water and once with 10 mm NH4HCO3, dehydrated with ACN, and dried in vacuo. The gel pieces were rehydrated with 10 mm NH4HCO3 and 20 ng μl−1 trypsin, incubated at 4 °C for 1 h, and then digested overnight at 37 °C. Peptides were extracted with two changes of ACN and dried in vacuo.
LC-MS/MS
Digests were rehydrated in 0.1% formic acid and 0.05% heptafluorobutyric acid. The digested peptides from each fraction for the 30 versus 30 °C experiments were separated by on-line nano-LC using an Applied Biosciences microgradient system. Samples (2.5 μl) were concentrated and desalted on a micro-C18 precolumn with H2O:ACN (98:2, 0.1% (v/v) formic acid) at 20 μl min−1. After a 4-min wash, the column was switched in line with a fritless nanocolumn (75 μm × ∼10 cm) containing C18 medium (5-μm, 200-Å Magic, Michrom Bioresources, Inc., Auburn, CA) (33). Peptides were eluted using a linear gradient of 98:2 H2O:ACN (with 0.1% (v/v) formic acid) to 38:62 H2O:ACN (with 0.1% (v/v) formic acid) at ∼300 nl min−1 over 75 min and electrosprayed directly using high voltage (1.8 kV) into a three-dimensional ion trap (Thermo Electron, LCQ Deca XP+) mass spectrometer. A survey scan of 350–1800 (m/z) was collected followed by data-dependent acquisition of MS/MS spectra at 35% normalized collision energy of the most intense parent ion from the MS scan. Activation was set at q = 0.25 with an activation time of 30 ms, and a minimum of 5 × 106 counts for MS was required. Dynamic exclusion was enabled where after a maximum of three repeated MS/MS scans the parent ion was excluded for 1.5 min. Highly abundant singly charged ions of 391.25, 445.6, and 463.50 ± 1.5 m/z were excluded.
Peptides from cultures grown for the 10 versus 30 °C experiments were separated using nano-LC on an Ultimate 3000 HPLC and autosampler system (Dionex). Samples (2 μl) were concentrated and desalted onto a micro-C18 precolumn (500 μm × 2 mm; Michrom Bioresources, Inc.) with H2O:ACN (98:2, 0.05% (v/v) heptafluorobutyric acid) at 20 μl min−1. After a 4-min wash, the precolumn was switched (Valco 10-port valve, Dionex) in line with a fritless nanocolumn (75 μm × ∼10 cm) containing C18 medium (5-μm, 200-Å Magic, Michrom Bioresources, Inc.) (33). Peptides were eluted using a linear gradient of 98:2 H2O:ACN (with 0.1% (v/v) formic acid) to 55:45 H2O:ACN (with 0.1% (v/v) formic acid) at 250 nl min−1 over 75 min. High voltage (1.8 kV) was applied to a low volume tee (Upchurch Scientific, Oak Harbor, WA), and the column tip positioned ∼0.5 cm from the heated capillary (T = 200 °C) of an LTQ (Thermo Electron) mass spectrometer. Positive ions were generated by electrospray, and the LTQ was operated in data-dependent acquisition mode. A survey scan of 350–1750 (m/z) was collected followed by two MS/MS scans where the first and second most intense precursor ions from the MS trace were sequentially isolated and fragmented using CID at 35% normalized collision energy with an activation set at q = 0.25 and activation time of 30 ms with a minimum signal required at 2,000 counts. Dynamic exclusion was enabled where after a maximum of two repeated MS/MS scans the parent ion was excluded for 3 min.
Protein Identification and Quantitation
MS/MS spectra were interrogated against the completed S. alaskensis genome database (containing 3,208 proteins) for protein identification using the SEQUEST search algorithm in the Bioworks BioBrowser (version 3.3) software package. The search parameters used were as follows: monoisotopic precursor and fragment mass type; fully enzymatic trypsin (KR) enzyme with allowance for one missed cleavage; and variable acrylamide, carbamidomethyl, and oxidation modifications. For data files from the LCQ Deca XP+ a 1.2-Da peptide tolerance and 0.6-Da fragment ion tolerance were used, whereas for LTQ data files a 0.8-Da peptide tolerance and 0.6-Da fragment ion tolerance were used. Identifications were filtered using DTASelect (34) based on the following parameters: ΔCN of at least 0.08 and a minimum XCorr of 2.1 for +1, 2.7 for +2, and 3.2 for +3 charged peptides. The MS/MS spectra were also interrogated against a decoy database (randomized S. alaskensis) with <1% false positive identification rate in DTASelect (35). Ambiguous peptides mapping to more than one protein were removed. To determine relative protein abundances, MS survey scans were analyzed for each experiment using RelEx software (version 0.92) (36). The quantitation parameters used were as follows: four scans before peak, four scans after peak, 0.15 threshold factor, apply Savitsky-Golay filter with seven points, apply S/N filter at 5, apply regression filter with 0.8 minimum correlation at 1 and 0.7 minimum correlation at 10, and 99% incorporation of 15N. In cases where there were multiple identifications of the same peptide the retention time (or scan number) of the highest scoring peptide identification (determined by DTASelect) was used by RelEx to determine the extracted ion chromatogram m/z and elution time of the precursor ions and also the extracted ion chromatogram of the labeled ions. The software then extracts ion intensities over a range of MS scans (100) so even if the identification occurred before or after the maximum the software would still likely integrate over the entire elution profile. RelEx chooses the peak closest to the time of the MS/MS spectrum (if more than one integration peak appears above the threshold) (34, 36). Only proteins with two or more unique peptides were considered for quantitation.
Data Processing
Raw data produced by RelEx were imported into R (version 2.5.1) (37), an open source statistical analysis program, using custom code. Because there was no R software for analyzing proteomics data, we developed a library of computer code that extends the limma (version 2.1) library (38) in R/Bioconductor (39) (see “Statistical Analysis of Differential Abundance”). This code is available from M. J. C. upon request. Peptide ratios were log2-transformed, S/N values were log10-transformed, and values were averaged to obtain the average ratio of 14N:15N and the average S/N for each protein. Because RelEx does not output absolute abundance estimates for each metabolically labeled sample, we used S/N as a proxy for the amount of abundance (22).
Intraexperimental Normalization
For the 30 versus 30 °C experiments that were combined in different ratios, the distribution of protein ratios was graphed for data obtained from each LC-MS run to determine whether protein ratios were symmetrically distributed about the expected ratio of either 1.0, 0.8, or 1.2 for the 1:1, 0.8:1, and 1.2:1 data sets, respectively. In addition, an MA plot (Ref. 14) was used to detect any non-linear skew in the ratios over a range of S/N values. For the 30 versus 30 °C experiments, median normalization was performed at the protein level according to Yang et al. (14) to ensure all medians were 0. For the 10 versus 30 °C experiments, lowess normalization (40) was performed. Normalization was performed at the protein level because if normalization was performed at the peptide level first followed by averaging then the non-linear skew in the 10 versus 30 °C data sets was not completely removed.
Interexperiment Normalization
Following intraexperiment normalization, protein ratios from all LC-MS runs from either the 30 versus 30 °C or 10 versus 30 °C experiments were visualized using box-and-whisker plots (box plots) and overlaid density plots to reveal the shapes and extents of the distributions.
Linear Modeling
To estimate the magnitude of the effect of changing temperature on protein abundance in the 10 versus 30 °C experiments, we adopted a linear modeling approach, which is widely used in DNA microarray experiments (39). The effect size due to changing temperature and extraction buffer was determined by fitting the following linear model to the abundance ratios for each protein (Equation 1),
where yi is the normalized 14N:15N log2 ratio for a protein from experiment i, 1 ≤ i < 20; βtemp is an unknown coefficient representing the average log2 -fold change (FC) between 10 versus 30 °C; Itemp is an indicator variable of +1 when the 10 °C sample was labeled with 14N and the 30 °C sample was labeled with 15N or −1 in the inverse labeled condition; βbuffer is an unknown coefficient estimating the average log2 FC due to different extraction buffers; Itemp is an indicator variable that is 0 when a Tris extraction buffer was used or 1 when a urea buffer was used; and εi is the residual error for experiment i. Thus the observed 14N:15N ratio (yi) of each protein is a combination of the 10 versus 30 °C effect (βtemp), the buffer effect (βbuffer), and a residual error (εi). In practice, the two indicator variables are represented as a design matrix (supplemental Table S2).
Prior to fitting the linear model (Equation 1), the magnitude of correlation between technical replicates from within the same biological replicate was determined according to Smyth (39). The values of the coefficients βtemp and βbuffer were estimated by least squares regression (39), accounting for the replicate correlation calculated above, in addition to determining the standard error of each protein and the residual df.
To demonstrate the advantages of using a fully specified linear model (Equation 1), we also fitted a linear model, which is identical to the Student's t test (Equation 2),
that only corrects for the 14N:15N label reversal and treats all observations as independent where any observed variation is due to the effect of changing temperature.
Statistical Analysis of Differential Abundance
Following the estimation of the average FC, standard error, and df of each protein by Equation 1 or 2, we calculated a one-sample, two-tailed t statistic for each protein from Equation 1 (hereafter the unmoderated t test) and from Equation 2 (hereafter the Student's t test). p values were calculated using standard lookup tables and the residual df obtained from the linear model fit.
Using the estimates obtained from Equation 1, we calculated an empirical Bayes moderated t statistic (hereafter the moderated t test) for each protein (39). p values were calculated using the moderated t statistic and df obtained following the empirical Bayes procedure. To demonstrate the benefits of the moderated t statistic, we compared the moderated t test to the unmoderated t test, both of which were calculated using estimates from Equation 1.
Multiple Testing Correction
p values from each of the three methods were adjusted for multiple testing by both the Bonferroni correction and the Storey-Tibshirani FDR (23). The posterior error probability of each individual protein (4) was not assessed. The assumptions underlying the FDR method (23) were tested by plotting histograms of unadjusted p values and by confirming that the proportion of truly null hypotheses (π0) was accurately estimated (see Figs. 1 and 3 from Ref. 23).
Fig. 1.
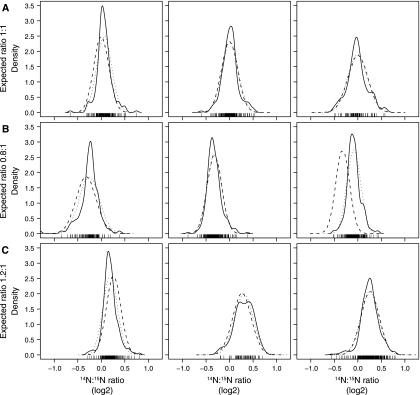
Unnormalized density distribution of the 30 versus 30 °C data set. Three 1:1 14N:15N experiments (A), three 0.8:1 experiments (B), and three 1.2:1 experiments (C) were plotted using 14N:15N ratio (x axis, log2 scale) and probability density (y axis). Experimentally observed distribution of the 14N:15N ratios, solid line; normal distribution with the mean and S.D. of the observed data, dotted line; normal distribution with the expected mean and observed S.D., dashed line.
Fig. 3.
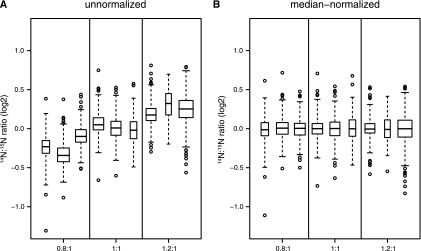
Box and whisker plots of un- and median-normalized data from the 30 versus 30 °C data set. The median ratio from each experiment is shown as a thick black line surrounded by a box representing the interquartile range, which contains the median ± 25% of the data with whiskers that extend at most two standard deviations from the median and outlying observations shown as open circles. The horizontal width of each box is proportional to the number of proteins in each experiment. The distribution of protein ratios in the 30 versus 30 °C data set before (A) and after (B) median normalization shows that the expected shift (0.8:1 and 1.2:1) can be successfully removed by median normalization.
RESULTS
A Technical Replicate Data Set
To assess whether microarray-based analysis methods would be suitable for the analysis of LC-MS data and whether our system was sensitive enough to detect subtle differences in protein abundance, S. alaskensis cultures were grown at 30 °C, labeled with either 14N or 15N, and then combined in known ratios of 0.8:1, 1:1, and 1.2:1, in triplicate. Following averaging of the peptide ratios from each LC-MS run, we observed the distributions of protein ratios (Fig. 1, solid lines) and found that the non-log-transformed medians were 0.88 ± 0.1, 1.04 ± 0.13, and 1.21 ± 0.15 for experiments mixed 0.8:1, 1:1, and 1.2:1, respectively (Table I). This demonstrates the ability of our experimental methods to detect relatively subtle differences in protein abundance.
Table I. Protein quantitation for artificially biased data.
The mean 14N:15N ratios were within 0.02 of the median in all cases (data not shown). Bold values are averages.
| 14N:15N ratio | No. of proteins ≥ 1 peptidea | No. of proteins ≥ 2 peptidesb | Medianc | Mind | Maxe | S.D.f |
|---|---|---|---|---|---|---|
| 0.8:1 | 147 | 106 | 0.86 | 0.63 | 1.31 | 0.11 |
| 399 | 225 | 0.94 | 0.70 | 1.36 | 0.10 | |
| 469 | 264 | 0.79 | 0.55 | 1.30 | 0.09 | |
| 0.86 | 0.63 | 1.33 | 0.10 | |||
| 1:1 | 133 | 84 | 1.01 | 0.72 | 1.49 | 0.15 |
| 333 | 192 | 1.02 | 0.68 | 1.46 | 0.13 | |
| 368 | 212 | 1.04 | 0.67 | 1.68 | 0.12 | |
| 1.02 | 0.69 | 1.54 | 0.13 | |||
| 1.2:1 | 122 | 62 | 1.28 | 0.92 | 1.63 | 0.16 |
| 363 | 189 | 1.14 | 0.83 | 1.76 | 0.13 | |
| 585 | 379 | 1.20 | 0.68 | 1.74 | 0.16 | |
| 1.21 | 0.81 | 1.71 | 0.15 |
a The number of proteins identified in each experiment with at least one peptide.
b The number of proteins identified in each experiment with at least two peptides.
c The median of 14N:15N ratios.
d The minimum of 14N:15N ratios.
e The maximum of 14N:15N ratios.
f The S.D. of 14N:15N ratios.
We compared the observed distributions of protein ratios (Fig. 1, solid lines) with normal distributions with either the observed mean and S.D. (Fig. 1, dotted lines) or the observed S.D. and the expected mean (Fig. 1, dashed lines). In all but one case, the observed protein ratios overlap the normal distributions centered upon the expected protein ratio where one of the 0.8:1 samples was centered closer to 1:1. Furthermore we compared the distribution of observed ratios against expected ratios under a normal distribution using quantile-quantile plots (supplemental Fig. S3). These revealed that the majority of proteins are consistent with a normal distribution; however, they are heavy tailed. These heavy tails most likely reflect proteins that are naturally variable in their abundance. This pattern of protein abundance is similar to two-color microarray analysis of RNA abundance, suggesting that microarray normalization approaches are likely to be applicable to quantitative proteomics data.
Inspecting MA plots prior to normalization helps to determine whether a relationship or trend exists between the protein ratio and abundance levels. We did not detect such a trend in any of the 30 versus 30 °C data sets (Fig. 2A and supplemental Fig. S4) because the ratios were symmetrically distributed throughout the range of S/N. Box plots of unnormalized protein ratios are presented (Fig. 3A), confirming the observations from Fig. 1 that the median of each distribution is close to the expected ratios. We performed median normalization to remove the bias that we had introduced, thereby forcing the medians of each distribution of protein ratios to a log2 FC = 0, corresponding to a ratio of 1:1 (Figs. 2A and 3B). Because the distributions of median-normalized protein ratios had similar shapes (Fig. 1) and similar extents (Fig. 3B), any additional interexperiment normalization was not performed.
Fig. 2.
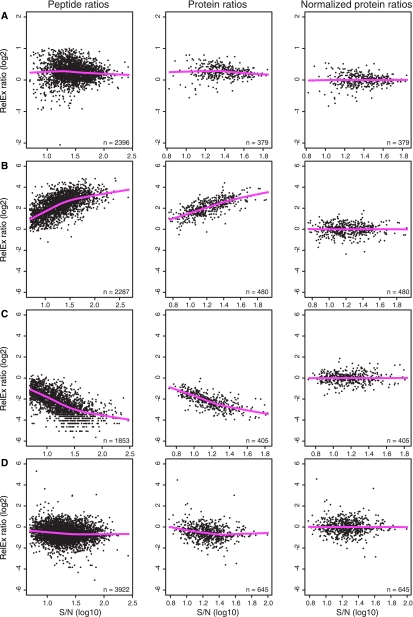
MA plots of peptides and proteins pre- and postnormalization. Unnormalized peptides (column 1) and proteins (column 2) and normalized proteins (column 3) were plotted in MA plots using the log2 14N:15N ratio of abundance (y axis), log10 S/N (x axis), and a non-linear, locally weighted regression (lowess) line (pink line). A, artificially biased 1.2:1 14N:15N experiment where the absence of a systematic trend in ratio versus S/N validated a median normalization of proteins across the experiment. B, a representative skewed data set created by combining 1:1 based on OD 10 °C 14N and 30 °C 15N. C, as for B but inversely labeled (10 °C 15N and 30 °C 14N). D, a representative non-skewed data set generated by combining 1:1 based on protein concentration 10 °C 14N and 30 °C 15N. Normalized protein ratios from B–D were derived from using lowess normalization.
A Biological Data Set
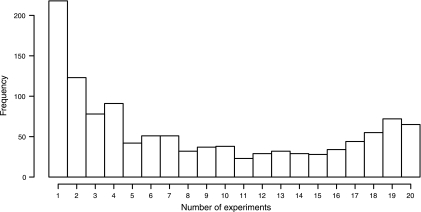
Over 400,000 tandem mass spectra were generated, resulting in 1,736 unique and high confidence protein identifications (p < 0.01, S/N > 5) with at least two peptides, giving a 54% genome coverage of S. alaskensis. An additional 399 proteins were detected by a single peptide (data not presented). From the list of 1,736 protein identifications, 1,172 proteins with ≥2 peptides were quantified (37% genome coverage) (supplemental Tables S3 and S4). Approximately 230 proteins were identified in only one experiment, and 65 of the most abundant proteins were detected in all 20 experiments (Fig. 4).
Fig. 4.
Protein detection frequency. The frequency of occurrence of 1,172 proteins obtained from six biological replicates and a total of 20 MS runs from the 10 versus 30 °C experiments is shown.
In four biological replicates (A–D) of 10 versus 30 °C cultures (representing 16 MS runs), the 14N- and 15N-labeled cells were combined based on equal OD, which unexpectedly produced a 2–16-fold non-linear skew. At low S/N values, the skew was less pronounced, becoming more extreme at higher S/N, always in favor of the 30 °C sample, both in the 14N:15N and 15N:14N inverse labeling conditions (Fig. 2, B and C, and supplemental Fig. S5). Venable et al. (22) reported a similar non-linear bias that was partly attributed to the signal intensity of some peptides that fell below the limit of quantitation. In contrast, we used a S/N > 5 cutoff to avoid measurements below the limit of quantitation. To determine whether the skew was due to combining samples based on OD, we performed two additional biological replicates (E and F) in duplicate that were combined 1:1 by protein concentration, and we observed no non-linear skew nor a deviation from the expected ratio of 1:1 (Fig. 2D and supplemental Fig. S6).
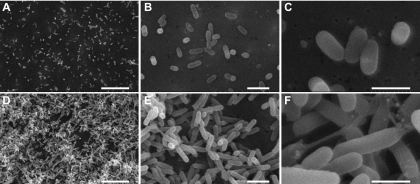
To examine the biological reason for the observed skew in the samples combined 1:1 based on OD, morphological examinations were performed on S. alaskensis cells grown at 10 and 30 °C. Scanning electron micrographs show that at 10 °C cells appear primarily as individuals, whereas at 30 °C cells tend to be clumped together connected by an extracellular matrix (Fig. 5). The biovolume of cells grown at 10 °C was 0.13 ± 0.03 μm3, whereas the cells were ∼1.4-fold larger (0.18 ± 0.03 μm3) at 30 °C. Spectrophotometric measurement of cells (OD) is based on the light-absorbing quality of the cells in solution and can be affected by cell size, the properties of the plasma membrane, the internal structure of the cell, and the presence of materials that absorb light (41). Cell clumping may cause inconsistent OD measurements and may result in OD not providing a true reflection of culture turbidity (42).
Fig. 5.
Scanning electron microscope images of S. alaskensis. Cells grown at 10 °C (A–C) and 30 °C (D–F) are shown. Scale bars in A and D, 10 μm; in B and E, 2 μm; and in C and F, 1 μm.
Differences in cell clumping and light absorption do not explain why there was a curved skew and not a simple shift in the distribution such as when 14N and 15N 30 °C samples were combined at 0.8:1 or 1.2:1. The skew may be caused by the inherent sampling bias in LC-MS where higher abundance peptides are preferentially selected. As a result, for the high abundance peptides, the probability of detecting a peptide from a 30 °C sample is higher than a peptide from 10 °C, thereby exaggerating the skew at higher abundance. At lower S/N, the lower abundance proteins from either the 10 or 30 °C samples have a similar chance of being sampled, thus minimizing the extent of the skew. It is in these situations that we must rely on normalization to attempt to correct the skew and recover the true difference in protein abundance due to temperature.
Normalization
In two-color microarrays, the different spectral properties of red and green cyanine dyes causes non-linear skews, which are usually removed using lowess normalization (14). The main assumption is that the majority of genes/proteins are not differentially expressed and should be centered about the 1:1 ratio throughout the range of abundance. Applying lowess normalization to 10 versus 30 °C data sets removed the non-linear trend and produced protein ratios with a median of 0 (Fig. 2, B and C, and supplemental Figs. S5 and S6). The distributions of lowess-normalized protein ratios were very similar across experiments; thus we did not perform any additional interexperiment normalization.
Although the lowess normalization appeared to effectively remove the systematic bias in protein ratios, we further evaluated whether the large skew compromised the ability to score relative protein abundance. Four samples that had been combined based on equal OD (A1_1, A2_1, B1_1, and B2_1) were compared with the samples that had been combined based on equal protein concentration (E_1, E_2, F_1, and F_2). The 607 proteins that were shared between these eight data sets were sorted into five groups by examining their average FC values (Table II). The majority (groups 1–3 representing 67.5%) of normalized protein abundances were positively correlated (i.e. equivalent trends with growth temperature in both data sets). An additional 23.9% (group 4) had opposing trends but with small FCs (<1.5) and could represent proteins with “normal” levels of variation. Only 8.6% (group 5) were judged poorly correlated as their normalized quantities were opposing and above a 1.5-FC. The overall agreement between the two data sets is a strong indication that the normalization effectively removed skew in the data while preserving the biological effect due to temperature.
Table II. Comparing postnormalization protein quantities for experiments that that were combined by optical density (A and B) or protein concentration (E and F).
| Category | Condition | No. of proteins | Correlation |
|---|---|---|---|
| 1 | Both proteins FCa > 1.5 in the same directionb | 37 | Positive |
| 2 | One protein FC > 1.5 and FC < 1.5 in the other protein, both in the same direction | 101 | Positive |
| 3 | Both proteins FC < 1.5 in the same direction | 272 | Positive |
| 4 | Both proteins FC < 1.5 in different directions | 145 | Uncertain |
| 5 | Both proteins FC > 1.5 in different directions | 52 | Negative |
a FC is expressed as a 14N:15N value.
b Direction refers to the relative increased abundance of proteins toward either the 14N or 15N label.
Identifying Differentially Abundant Proteins
Initial proteomics studies focused on identifying those proteins with the largest -fold change, citing cutoffs of 1.5- or 2-fold changes as being significant. More recently, studies have used simple statistical tests such as the Student's t test to identify proteins with p < 0.05. We compared these two methods with a more robust framework for statistical analysis, the linear model. In addition, we demonstrate that the empirical Bayes moderated t test, a more powerful test for differential abundance, can be successfully interfaced with the linear model.
-Fold Change Approach
Using an FC approach, 278 and 84 proteins with FC > 1.5 and 2, respectively, were differentially abundant (Table III).
Table III. -Fold change approach outcomes.
a The number of proteins with 14N:15N ratios greater than the FC threshold.
b The number of rejected proteins below the FC threshold.
c The expected number of false positives (FP), which by using an FC threshold approach cannot be determined (—).
Linear Models as a Framework for Statistical Analysis
All 10 versus 30 °C experiments were each subjected to different combinations of experimental parameters, including whether the 30 °C sample was labeled with 14N or 15N, the extraction buffer that was used, and from which biological replicate the sample originated (A–F). To accurately represent this experimental design, a linear model that precisely described each sample was constructed (Equation 1). To account for the correlation due to technical replicates from the same biological replicate, the correlation between technical replicates was estimated to be 0.672 on average (see “Experimental Procedures”). We note that such a large amount of correlation will undoubtedly cause the Student's t test to overestimate the statistical significance of many proteins, and thus we expect to obtain smaller but more accurate t statistics using Equation 1. Using this value and the experiment label (A–F) as a blocking variable (supplemental Table S2), the ratios from each protein were fitted to the linear model (Equation 1), and the coefficients for the effect of temperature (βtemp) and the extraction buffer (βbuffer) on protein abundance were estimated by least squares regression (see “Experimental Procedures”). The coefficient estimates can be directly interpreted as the average 14N:15N FC (log2) due to their respective experimental parameter; thus, proteins with |βtemp| > 0.585 (log2) or >1.0 (log2) represent proteins with a 1.5- or 2-FC due to growth temperature, respectively. Finally because this model has two parameters, only proteins with n ≥ 2 from the same extraction buffer can be fitted.
Evaluating the Utility of Linear Modeling Using an Unmoderated Student's t Test
From the 1,172 quantified proteins, 954 were detected at least twice and could be analyzed by the Student's t test with the data fitted to the simple linear model (Equation 2) (Table III). This approach tested the null hypothesis that the average abundance ratio due to the effect of temperature is 0 with (n − 1) df where n is the number of observations for each protein. Using p < 0.05, 325 proteins had significant changes in abundance due to temperature (Table III). Similarly 830 proteins detected at least three times or only two times from the same extraction buffer (i.e. no estimate for βbuffer but a valid estimate for βtemp) could be fitted to the full linear model (Equation 1) (Table IV). Using p < 0.05, the Student's t test estimated 144 significantly changed proteins.
Table IV. Comparing methods of significance testing with the 10 versus 30 °C data set.
Three methods for estimating differential protein abundance were compared.
| Student's t testa |
Unmoderated t testb |
Moderated t testc |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| pd | Bonfe | FDRf | p | Bonf | FDR | p | Bonf | FDR | |
| Ng | 954 | 954 | 954 | 830 | 830 | 830 | 1,172 | 1,172 | 1,172 |
| <0.05h | 325 | 56 | 272 | 144 | 14 | 58 | 214 | 11 | 45 |
| E(FP)i | 48 | 0.05 | 14 | 42 | 0.05 | 3 | 59 | 0.05 | 2 |
| FDR (%)j | 15 | 0.1 | 5 | 29 | 0.4 | 5 | 28 | 0.5 | 5 |
a Student's t test where each protein abundance measurement was treated as independent using a simple linear model accounting for 14N and 15N label reversal only.
b Unmoderated t test with the full linear model.
c Empirical Bayes moderated t test with the full linear model.
d p, the number of differentially abundant proteins with statistic <0.05 for unadjusted p values.
e Bonf, the number of differentially abundant proteins with statistic <0.05 for Bonferroni method-corrected p values.
f FDR, the number of differentially abundant proteins with statistic <0.05 for FDR-corrected q values.
g The number of proteins analyzed from the 1,172 quantified proteins; proteins analyzed by the Student's t test had at least two observations, the unmoderated t test had at least three observations or two observations from the same extraction buffer, and the moderated t test had at least one observation.
h Number of differentially abundant proteins with statistic <0.05.
i Expected false positives.
j FDR was calculated by dividing E(FP) by the number of differentially abundant proteins with statistic <0.05.
The Empirical Bayes Moderated t Test
We calculated a moderated t statistic for each protein using the same initial estimates of FC, residual error, and df parameters from fitting the linear model (Equation 1) that the unmoderated t test used. Because the moderated t test provides improved error estimates and an increase in the number of residual df, it can be applied to proteins with even one observation (39, 43). From the 1,172 quantified proteins, we identified 214 proteins with q < 0.2 (Table IV and supplemental Table S4).
Comparing the Moderated t Test with an Unmoderated t Test
Given that the moderated t test was capable of analyzing 342 (41%) more proteins than the unmoderated t test (Table IV) and that there were an additional 70 (49%) differentially abundant proteins in the moderated t test relative to the unmoderated t test, the observed increase in the number of differentially abundant proteins may be expected by chance. To perform an unbiased comparison between the unmoderated and the moderated t tests, the moderated t test was restricted to the same 830 proteins tested in the unmoderated t test. Accordingly 168 proteins were determined to be significantly differentially abundant, 24 more than the unmoderated t test (data not shown).
Comparing Multiple Hypothesis Testing Approaches
To correct for multiple hypothesis testing, the popular yet conservative method of Bonferroni correction was compared with the FDR proposed by Storey and Tibshirani (23) using data from all three t tests (Table IV). The model-based assumptions of the Storey-Tibshirani FDR procedure (23) are that the p values are derived from two distributions: those from the null hypothesis, which are uniformly distributed, and those from the alternative hypothesis that have p values closer to 0. Accordingly histograms of unadjusted p values (supplemental Fig. S7) were plotted, and as expected, an accumulation of small p values with a uniform distribution of p values from p > 0.5 was observed. The shapes of the distributions were compatible with the assumptions required for using the Storey-Tibshirani FDR (23). Thus for each protein, a q value was calculated (supplemental Table S4). The resulting list of proteins with q < 0.05 have at most 5% false positives within that list, whereas the list of proteins with p < 0.05 have a false positive rate of 5% from within the entire set of proteins (23). The q value is in fact a measure of what many think the p value represents.
From the moderated t test, 214, 11, and 45 proteins were identified with unadjusted p < 0.05, Bonferroni p value correction, or FDR q < 0.2, respectively (Table II). The expected number of false positive proteins is 59, 0, and 2, respectively (Table III). Given the distribution of p values (supplemental Fig. S7) we found that the FDR gives the most realistic impression of the true number of differentially expressed proteins.
DISCUSSION
The objective of this work was to examine approaches for normalization and select an appropriate significance test that maximized the final list of protein candidates from quantitative proteomics analyses balanced with acceptable and interpretable levels of false discoveries. Ultimately this will enable effective biological interpretation of the data. We tested our methods in two experimental systems with either very little biological variation (30 versus 30 °C) or large amounts of variation (10 versus 30 °C) and described a strategy for determining an appropriate set of normalizations, statistical analysis, and multiple testing adjustments. We concluded that linear modeling of the data coupled with an empirical Bayes, moderated t test, and FDR correction for multiple testing was a beneficial strategy for this experiment. We expect this approach will be of use for a wide range of metabolically labeled and indeed other types of quantitative proteomics experiments.
Normalization
Normalization of quantitative proteomics data or indeed of high throughput biological data in general greatly assists in reducing differences between data sets caused by experimental artifacts, revealing the true underlying biological differences. Experimental artifacts that may contribute to differences in 14N:15N ratios include pipetting errors at various stages of sample processing, sample quality, and various unpredictable or potentially uncontrollable factors. For microarrays, normalization is typically performed to control for intraexperiment variation and subsequently for interexperiment variation (14). Determining which normalization procedures to use requires careful assessment of the data that are generated.
We showed that density plots and box plots were useful for determining whether data are centered about the expected 1:1 ratio and that MA plots were useful for identifying non-linear relationships that then warrant removal by normalization. Fixed value median normalization was suitable for the 30 versus 30 °C data sets because of the absence of systematic non-linear trends (Table I and Fig. 3). Lowess normalization was useful for the OD-based 10 versus 30 °C proteomics data sets because there was a strong systematic non-linear trend associated with the 2–16-fold skew (Fig. 2). Additional interexperimental normalization was not required. However, we caution the need to inspect box plots or overlaid density plots to ensure that the ratio distributions do indeed have a similar shape and extent. As illustrated by the use of these two specific approaches, normalization needs to be considered on a case-by-case basis. The point should also be made that to avoid overmanipulation of data normalization should be kept to a minimum and only used when there is good evidence that it is required (e.g. from MA plots). Furthermore determining the reason for any skewing of data can lead to additional insight into cell biology.
Linear Models as a Framework for Statistical Analysis
As the future of proteomics is likely to involve increasingly sophisticated experimental designs that incorporate multiple experimental parameters and combinations of biological and technical replicates, we anticipate that a more flexible framework for data analysis, such as linear models, will become of increasing importance and value. We stress that even simple experimental designs can be effectively represented by a linear model; indeed our implementation of the Student's t test used a simple linear model that considered all samples as independent observations. Although we were not interested in the experimental parameter of extraction buffer per se, including it in the linear model allowed for additional variance to be assigned to this parameter, thereby reducing the residual variance and increasing our power to detect the effect that was truly of interest, temperature.
Comparing the results from the linear model (Equation 1) using the unmoderated t statistic with the Student's t statistic (Equation 2) revealed a striking difference in the number of differentially expressed proteins (Table IV). Despite just a 15% increase in the number of proteins able to be analyzed by the Student's t test, there were an additional 185 (129%) differentially abundant proteins from the Student's t test. This marked enrichment exists over a number of different p value thresholds and is due to ignoring the correlation between technical replicates; i.e. values obtained from technical replicates will have lower variance than those from independent samples. This artificially decreases the overall measurement variance and causes overestimation of statistical significance in the Student's t test. Furthermore the increased number of apparently “significant” proteins is likely to adversely affect biological conclusions drawn from the data. Accounting for the correlation between arbitrary experimental effects is therefore critical.
Significance Testing
Significance testing was absent in the early development and publication of quantitative proteomics studies. Its purpose is to create a list of confident protein abundances to enable robust biological interpretations based on the observed changes between test samples. An appropriate test should maximize the number of proteins with significant changes in abundance while balancing the number of false discoveries and false negatives. The four significance testing approaches (FC, Student's t test, unmoderated t test, and moderated t test) combined with three multiple testing correction approaches (none, Bonferroni, and Storey-Tibshirani FDR (23)) provided markedly different outcomes, highlighting (as for normalization) the need to adopt a considered approach to data treatment.
-Fold Change
A commonly used method for identifying differentially abundant proteins is FC. By this approach, proteins with an FC larger than a defined cutoff (e.g. 1.5- or 2-fold) are classified as differentially abundant. Although this is an intuitively simple approach, there are a number of limitations that have been widely discussed for microarray work (44) that are relevant to quantitative proteomics. The FC approach assumes that all proteins have the same variance (or standard error of measurement). However, for many reasons this may not be the case. Proteins with low abundance that are close to the detection limit of the mass spectrometer tend to have higher variability than those with higher cellular abundances. In addition, if a highly abundant protein (e.g. 10,000 copies per cell) such as a ribosomal or cell structure protein increases 1.4-fold, this represents a large increase in the balance of protein synthesis and has implications for nutrient and energy utilization. Whereas an equivalent 1.4-fold increase for a protein (e.g. gene regulatory protein) with 10 copies per cell may have negligible impact on energy balance. It is clear that the biological roles of individual proteins must be considered when judging protein abundance differences (see “Statistical Versus Biological Significance” below). Another factor that is not effectively dealt with by only considering FC is the number of observations. Clearly a single measurement is not as reliable as an FC for a protein that is quantified in 20 of 20 experiments.
One of the most important limitations of basing assessments on FC is the lack of statistical confidence defining the probability of differential abundance. Taking this approach, the risk of making false biological conclusions is therefore high.
Comparing the Empirical Bayes Moderated t Test with an Unmoderated t Test
The empirical Bayes moderated t test is superior to the Student's t test for high throughput studies where typically there are thousands of measurements made with only a few replicates (the “large p, small n” paradigm) that is compounded by the inherent missing data problem in proteomics. The empirical Bayes moderated t statistic estimates prior parameters from the observed protein ratios and consequently improves the error estimate for each individual protein by borrowing information from the other proteins (39, 43). The benefit of this approach is largest for proteins with few observations and even allows the estimation of p values for proteins with a single measurement (39, 43), although users may still wish to filter out proteins with few observations because the numerator of the t statistic may be poorly estimated.
In the unbiased comparison of the moderated versus the unmoderated t statistic where the protein ratios were both fitted to the same 830 proteins using the same fully specified linear model the moderated t test generated 24 extra significant proteins. This suggests that the moderated t test has more power to detect differential protein abundance than the unmoderated t test. The moderated t statistic is, in many cases, more conservative than the unmoderated t statistic: for example, the protein Sala_1422 had a small FC of 1.38, only two observations, and an unmoderated t statistic of 39.3 (p = 0.016) due to a small standard error estimate, whereas the moderation of standard errors resulted in a far more conservative moderated t statistic of 1.057 (p = 0.327) (supplemental Table S4). Taken together, these data demonstrate that compared with an unmoderated t test the moderated t test has more power to detect significant changes in protein abundance while being more conservative in estimating significance.
False Discovery Rates and Correcting for Multiple Testing
A goal was to identify the largest number of differentially expressed proteins in a statistically rigorous fashion; thus we adopted the q value of Storey and Tibshirani (23). Using a q value provides a more direct way of interpreting significance than a p value. In the context of quantitative proteomics, p values control the rate at which proteins with no change in abundance are deemed significant, whereas q values control the rate of significantly changed proteins being false. For example, if the FDR threshold has been set at 5% (q value of 0.05), then from a list of 100 proteins with significant differential abundance, there will be a tolerated error of five false positive proteins. The same cannot be said for a set of 100 significant proteins that have a maximum p value of 0.05; this is because the number of false positives is calculated from the entire data set of proteins tested. However, choosing a list of proteins with p < 0.05 without correcting for multiple testing may produce a large number of false positives (13, 23, 45). The importance of FDRs for protein identification (3, 4) and quantitative proteomics analyses (5, 10, 17, 19) has recently been addressed.
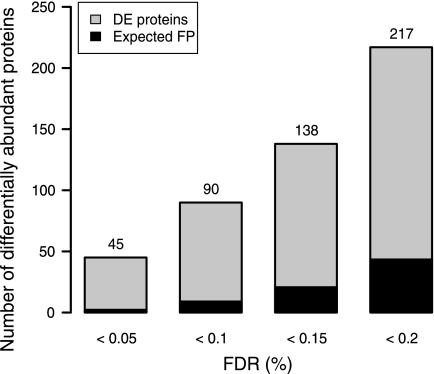
Determining a suitable statistical threshold is a trade-off between the false positive and false negative rates. Using the moderated t test, we compared the use of no correction to the Bonferroni and FDR methods. The Bonferroni method clearly demonstrated a strong control over the false positive rate at the expense of identifying very few differentially abundant proteins (Table IV), and as has been found previously (13), we found the Bonferroni method to be far too conservative for proteomic applications. We note that instead of the FDR of each individual protein that we adopted a local FDR can also be used (4, 46). The FDR method (q < 0.05) identified 4 times as many differentially abundant proteins as the Bonferroni method with just two false positives expected from a list of 45 proteins. Using FDR thresholds that are frequently used in microarray studies, 90, 138, and 217 differentially abundant proteins were identified with q < 0.1, 0.15, or 0.2, respectively (Fig. 6). Lastly it is important to note that the proteins with p < 0.05 had an FDR of 28%. This serves to highlight that using a significance threshold of q < 0.2 (20% FDR), which may seem to be a large error value, provides a more conservative and better informed outcome than an uncorrected p < 0.05.
Fig. 6.
Number of differentially abundant proteins passing q value thresholds. FDR q value thresholds of <0.05, 0.1, 0.15, and 0.2 were applied to the 10 versus 30 °C data set after significance testing using a moderated t test. There was a linear increase in the number of significantly changed proteins as q value increased and, as a result, a linear increase of the number of expected false positives (FP). DE, differentially expressed.
Statistical Versus Biological Significance
As discussed above (under “-Fold Change”) there are good reasons to distinguish statistical significance from biological significance, the most important reason being that without having confidence in the proteomics outcomes (statistical significance) it is not possible to draw confident inferences about the biology. From the 278 proteins with FC > 1.5, approximately half (n = 135) had a q value >0.2. This illustrates the potential difficulties that would be created for interpreting the biology if half of the proteins are in fact not reliably associated with the test conditions being examined (in this case, temperature).
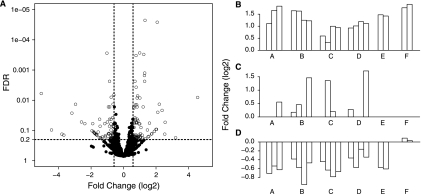
One method for exploring this difference is via a volcano plot where the relationship between FC and statistical significance (q value) can be examined (Fig. 7A). Proteins that have a q value <0.2 and an FC > 1.5 are differentially abundant by both the statistical and FC approaches. Proteins that satisfy both statistical and FC criteria (Fig. 7B), the FC criteria only (Fig. 7C), or the statistical criteria only (Fig. 7D) have been highlighted. The majority of proteins with large FCs but insignificant changes (Fig. 7C) arise from proteins with fewer than five observations. Furthermore in these cases the large FC may be associated with high variance and in cases that involved few measurements are less likely to be indicative of a consistent and important biological change. Importantly the statistical approach is capable of identifying proteins that have small but consistent changes in abundance (Fig. 7D) that would have been overlooked using an FC thresholding approach.
Fig. 7.
Assessing statistical versus biological relevance. A, volcano plot of all 1,172 quantified proteins from the 10 versus 30 °C data set displaying the relationship between statistical significance and FC of each protein. The log2 FC (x axis) was plotted against the −log10 q value (y axis). A q value threshold of 0.2 (dashed horizontal line) and >1.5-FC (vertical dashed lines) are shown where open circles are differentially abundant proteins with small q values and large FCs. B–D, normalized FC values of three representative proteins were plotted across all 10 versus 30 °C experiments (A–F) to illustrate experimental variance. B, a representative protein with statistically significant differential abundance (q < 0.2) and a large FC value (i.e. the protein satisfies both statistical and FC criteria). C, a representative protein with a large FC that is not significantly differentially abundant (q > 0.2). D, a representative protein with statistically significant differential abundance (q < 0.2) and small FC.
Conclusion
Complex quantitative proteomics experiments represent an analytical challenge for computing the probability of differential protein abundance while correctly accounting for an experimental design that can include label swapping, different extraction buffers, and biological and technical replicates. The data processing and analysis work flow combining MA plotting, linear data models, lowess normalization, and use of an empirical Bayes moderated t test in a single analysis environment (R) is novel in its application in quantitative proteomics. We found that the optimum normalization approach is dependent upon the individual experiment and must be assessed on a case-by-case basis by inspection of MA plots. Fitting the data to a properly specified linear model accounting for correlation due to technical replication using an empirical Bayes moderated t test with a Storey-Tibshirani FDR (23) was a successful approach for maximizing quantitative proteomics data while controlling false discoveries and correcting for multiple testing. The normalization and statistical testing approach provided rigorous data processing and evaluation of differential abundance in a high throughput quantitative proteomics data set and is generally applicable to global quantitative proteomics analyses.
Supplementary Material
Acknowledgments
We thank Peter Little and Rohan Williams for helpful discussions on the application of microarray statistics to proteomics, Matt DeMaere and David Wilkins for assistance with data processing, and John Yates III for DTASelect and RelEx. We also acknowledge the useful comments that we received during the review process. Mass spectrometric analysis for the work was performed at the Bioanalytical Mass Spectrometry Facility, University of New South Wales.
Footnotes
* This work was supported in part by the Australian Research Council by grants from the Australian Government Systemic Infrastructure Initiative and Major National Research Facilities Program (University of New South Wales (UNSW) node of the Australian Proteome Analysis Facility) and by the UNSW Capital Grants Scheme.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- MA
- ratio versus average
- ASW
- artificial sea water
- FC
- -fold change
- FDR
- false discovery rate
- S/N
- signal:noise ratio
- LTQ
- linear quadrupole ion trap.
REFERENCES
- 1.Chich J. F., David O., Villers F., Schaeffer B., Lutomski D., Huet S. (2007) Statistics for proteomics: experimental design and 2-DE differential analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 849, 261–272 [DOI] [PubMed] [Google Scholar]
- 2.Karp N. A., Spencer M., Lindsay H., O'Dell K., Lilley K. S. (2005) Impact of replicate types on proteomic expression analysis. J. Proteome Res 4, 1867–1871 [DOI] [PubMed] [Google Scholar]
- 3.Choi H., Nesvizhskii A. I. (2008) False discovery rates and related statistical concepts in mass spectrometry-based proteomics. J. Proteome Res 7, 47–50 [DOI] [PubMed] [Google Scholar]
- 4.Käll L., Storey J. D., MacCoss M. J., Noble W. S. (2008) Posterior error probabilities and false discovery rates: two sides of the same coin. J. Proteome Res 7, 40–44 [DOI] [PubMed] [Google Scholar]
- 5.Karp N. A., McCormick P. S., Russell M. R., Lilley K. S. (2007) Experimental and statistical considerations to avoid false conclusions in proteomic studies using differential in-gel electrophoresis. Mol. Cell. Proteomics 6, 1354–1364 [DOI] [PubMed] [Google Scholar]
- 6.Callister S. J., Barry R. C., Adkins J. N., Johnson E. T., Qian W. J., Webb-Robertson B. J., Smith R. D., Lipton M. S. (2006) Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J. Proteome Res 5, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreil D. P., Karp N. A., Lilley K. S. (2004) DNA microarray normalization methods can remove bias from differential protein expression analysis of 2D difference gel electrophoresis results. Bioinformatics 20, 2026–2034 [DOI] [PubMed] [Google Scholar]
- 8.Paoletti A. C., Parmely T. J., Tomomori-Sato C., Sato S., Zhu D., Conaway R. C., Conaway J. W., Florens L., Washburn M. P. (2006) Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl. Acad. Sci. U.S.A 103, 18928–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X. J., Yi E. C., Kemp C. J., Zhang H., Aebersold R. (2005) A software suite for the generation and comparison of peptide arrays from sets of data collected by liquid chromatography-mass spectrometry. Mol. Cell. Proteomics 4, 1328–1340 [DOI] [PubMed] [Google Scholar]
- 10.Chang J., Van Remmen H., Ward W. F., Regnier F. E., Richardson A., Cornell J. (2004) Processing of data generated by 2-dimensional gel electrophoresis for statistical analysis: missing data, normalization, and statistics. J. Proteome Res 3, 1210–1218 [DOI] [PubMed] [Google Scholar]
- 11.Pedreschi R., Hertog M. L., Carpentier S. C., Lammertyn J., Robben J., Noben J. P., Panis B., Swennen R., Nicolaï B. M. (2008) Treatment of missing values for multivariate statistical analysis of gel-based proteomics data. Proteomics 8, 1371–1383 [DOI] [PubMed] [Google Scholar]
- 12.Jung K., Gannoun A., Sitek B., Meyer H. E., Stuhler K., Urfer W. (2005) Analysis of dynamic protein expression data. REVSTAT Stat. J 3, 99–111 [Google Scholar]
- 13.Manly K. F., Nettleton D., Hwang J. T. (2004) Genomics, prior probability, and statistical tests of multiple hypotheses. Genome Res 14, 997–1001 [DOI] [PubMed] [Google Scholar]
- 14.Yang Y. H., Dudoit S., Luu P., Lin D. M., Peng V., Ngai J., Speed T. P. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 16.Corzett T. H., Fodor I. K., Choi M. W., Walsworth V. L., Chromy B. A., Turteltaub K. W., McCutchen-Maloney S. L. (2006) Statistical analysis of the experimental variation in the proteomic characterization of human plasma by two-dimensional difference gel electrophoresis. J. Proteome Res 5, 2611–2619 [DOI] [PubMed] [Google Scholar]
- 17.Meunier B., Bouley J., Piec I., Bernard C., Picard B., Hocquette J. F. (2005) Data analysis methods for detection of differential protein expression in two-dimensional gel electrophoresis. Anal. Biochem 340, 226–230 [DOI] [PubMed] [Google Scholar]
- 18.Karp N. A., Griffin J. L., Lilley K. S. (2005) Application of partial least squares discriminant analysis to two-dimensional difference gel studies in expression proteomics. Proteomics 5, 81–90 [DOI] [PubMed] [Google Scholar]
- 19.Oberg A. L., Mahoney D. W., Eckel-Passow J. E., Malone C. J., Wolfinger R. D., Hill E. G., Cooper L. T., Onuma O. K., Spiro C., Therneau T. M., Bergen H. R., 3rd (2008) Statistical analysis of relative labeled mass spectrometry data from complex samples using ANOVA. J. Proteome Res 7, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Listgarten J., Emili A. (2005) Statistical and computational methods for comparative proteomic profiling using liquid chromatography-tandem mass spectrometry. Mol. Cell. Proteomics 4, 419–434 [DOI] [PubMed] [Google Scholar]
- 21.Cairns D. A., Thompson D., Perkins D. N., Stanley A. J., Selby P. J., Banks R. E. (2008) Proteomic profiling using mass spectrometry—does normalising by total ion current potentially mask some biological differences? Proteomics 8, 21–27 [DOI] [PubMed] [Google Scholar]
- 22.Venable J. D., Dong M. Q., Wohlschlegel J., Dillin A., Yates J. R. (2004) Automated approach for quantitative analysis of complex mixtures from tandem mass spectra. Nat. Methods 1, 39–45 [DOI] [PubMed] [Google Scholar]
- 23.Storey J. D., Tibshirani R. (2003) Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 25.Xia Q., Wang T., Park Y., Lamont R. J., Hackett M. (2007) Differential quantitative proteomics of Porphyromonas gingivalis by linear ion trap mass spectrometry: non-label methods comparison, q-values and LOWESS curve fitting. Int. J. Mass Spectrom 259, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eguchi M., Nishikawa T., Macdonald K., Cavicchioli R., Gottschal J. C., Kjelleberg S. (1996) Responses to stress and nutrient availability by the marine ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol 62, 1287–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fegatella F., Lim J., Kjelleberg S., Cavicchioli R. (1998) Implication of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol 64, 4433–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 29.Krambeck C., Krambeck H. J., Overbeck J. (1981) Microcomputer-assisted biomass determination of plankton bacteria on scanning electron micrographs. Appl. Environ. Microbiol 42, 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fegatella F., Ostrowski M., Cavicchioli R. (1999) An assessment of protein profiles from the marine oligotrophic ultramicrobacterium, Sphingomonas sp. strain RB2256. Electrophoresis 20, 2094–2098 [DOI] [PubMed] [Google Scholar]
- 31.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 32.Lasonder E., Ishihama Y., Andersen J. S., Vermunt A. M., Pain A., Sauerwein R. W., Eling W. M., Hall N., Waters A. P., Stunnenberg H. G., Mann M. (2002) Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature 419, 537–542 [DOI] [PubMed] [Google Scholar]
- 33.Gatlin C. L., Kleemann G. R., Hays L. G., Link A. J., Yates J. R., 3rd (1998) Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem 263, 93–101 [DOI] [PubMed] [Google Scholar]
- 34.Tabb D. L., McDonald W. H., Yates J. R., 3rd (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P. (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 36.MacCoss M. J., Wu C. C., Liu H., Sadygov R., Yates J. R., 3rd (2003) A correlation algorithm for the automated quantitative analysis of shotgun proteomics data. Anal. Chem 75, 6912–6921 [DOI] [PubMed] [Google Scholar]
- 37.Ihaka R., Gentleman R. (1996) R: a language for data analysis and graphics. J. Comput. Graph. Stat 5, 299–314 [Google Scholar]
- 38.Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth G. K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol 3, Article3. [DOI] [PubMed] [Google Scholar]
- 40.Dudoit S., Yang Y. H., Callow M. J., Speed T. P. (2002) Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat. Sin 12, 111–139 [Google Scholar]
- 41.McGann L. E., Walterson M. L., Hogg L. M. (1988) Light scattering and cell volumes in osmotically stressed and frozen-thawed cells. Cytometry 9, 33–38 [DOI] [PubMed] [Google Scholar]
- 42.Meyers P. R., Bourn W. R., Steyn L. M., van Helden P. D., Beyers A. D., Brown G. D. (1998) Novel method for rapid measurement of growth of Mycobacteria in detergent-free media. J. Clin. Microbiol 36, 2752–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lonnstedt I., Speed T. P. (2002) Replicated microarray data. Stat. Sin 12, 31–46 [Google Scholar]
- 44.Gusnanto A., Calza S., Pawitan Y. (2007) Identification of differentially expressed genes and false discovery rate in microarray studies. Curr. Opin. Lipidol 18, 187–193 [DOI] [PubMed] [Google Scholar]
- 45.Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. A 57, 289–300 [Google Scholar]
- 46.Efron B., Tibshirani R., Storey J., Tusher V. (2001) Empirical Bayes analysis of a microarray experiment. J. Am. Stat. Assoc 96, 1151–1160 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.