Abstract
Heparan sulfate proteoglycans are key regulators of complex molecular networks due to the interaction of their sugar chains with a large number of partner proteins, which in humans number more than 200 (Ori, A., Wilkinson, M. C., and Fernig, D. G. (2008) The heparanome and regulation of cell function: structures, functions and challenges. Front. Biosci. 13, 4309–4338). We developed a method to selectively label residues involved in heparin binding that matches the requirements for medium/high throughput applications called the “Protect and Label” strategy. This is based on the protection against chemical modification given by heparin/heparan sulfate to the residues located in the heparin-binding site. Thus, analysis of fibroblast growth factor-2 bound to heparin and incubated with N-hydroxysuccinimide acetate showed that lysines involved in the sugar binding are protected against chemical modification. Moreover following release from heparin, the protected lysine side chains may be specifically labeled with N-hydroxysuccinimide biotin. After protein digestion, the biotinylated peptides were readily isolated and identified by MALDI-Q-TOF mass spectrometry. The analysis of labeled peptides obtained from three well characterized heparin-binding proteins with very different heparin-binding sites, fibroblast growth factor-2, platelet factor-4, and pleiotrophin demonstrates the success of this new approach, which thus provides a rapid and reliable procedure to identify heparin-binding sites.
Heparan sulfate proteoglycans are a ubiquitous component of the extracellular space of complex organisms (1). They are characterized by their size and their plasticity that derives mainly from the complex and dynamically regulated structure of the glycosaminoglycan (GAG)1 moiety.
They participate in the structural organization of the extracellular space (2, 3) and play an active role in molecular networks driving complex biological phenomena such as development (4–6), inflammation and immune response (7, 8), and disease (9). Heparan sulfate proteoglycans exert their functions by interacting with a vast number of protein partners and so regulating their activity (8, 10).
In the last decade, a number of techniques have been used to investigate the interaction between proteins and GAGs (for reviews, see Refs. 11–15). However, despite the identification of more than 200 human heparin-interacting proteins (10, 16), our understanding of the structural features mediating the interaction remains quite poor (10, 13). Some models have been proposed, but they are based on the structural features of only restricted groups or families of heparin-binding proteins (HBPs) (11, 17, 18). Important limitations for modeling of heparin-binding sites (HBSs) are that many interactions are described solely at a qualitative level and that a three-dimensional structure of the sugar-protein complex is available for less than 10% of the annotated interactions (10). The available data derive mainly from x-ray crystallography (e.g. Refs. 19 and 20), NMR spectroscopy (e.g. Ref. 21), and quantitative biophysics with site-directed mutagenesis (e.g. Ref. 22) or HBP-derived synthetic peptides (e.g. Ref. 23). Although providing detailed structural and kinetic information about the interaction, all these methods are limited to the study of single protein-sugar interactions, and they cannot be translated into a high throughput format.
In 1989 Chang (24) used a lysine-reactive chromophoric reagent to investigate the HBS of antithrombin III (ATIII). ATIII was modified in the presence or absence of heparin, and the colored peptides generated by tryptic digestion were analyzed by RP-HPLC to obtain important insights into the heparin-ATIII interaction. N-Hydroxysuccinimide (NHS) esters have a strong and selective reactivity toward primary amines, in particular ε-amines on lysines, and their relative stability in neutral/weakly basic aqueous solution makes them compounds of choice for the investigation of HBSs, which are characterized by a high content of basic residues.
We developed a rapid and reliable method for the localization of HBSs that can represent an important complement to any structural investigation of heparin-protein interactions because of its low requirement in terms of sample quantity and handling time. The new method, called the “protect and label” strategy, is based on the protection against chemical modification given by heparin/HS to residues located in the HBSs. Thus, an acetyl NHS ester was used to protect residues exposed on the protein surface but not involved in heparin binding, and a second reagent, NHS-biotin, was used to selectively label residues in the HBS. The identification by mass spectrometry of the biotinylated peptides, derived from enzymatic digestion of the HBP, provides a fingerprint of sugar binding on the protein surface.
EXPERIMENTAL PROCEDURES
Heparin-binding Proteins
Recombinant fibroblast growth factor-2 (FGF-2) (Swiss-Prot accession number P09038) (residues 1–154) was expressed in C41 Escherichia coli cells using a pET-14b system (Novagen, Merck Chemical Ltd., Nottingham, UK) as described previously (25). Pleiotrophin (PTN), also known as heparin-binding growth-associated molecule or heparin affin regulatory peptide (Swiss-Prot accession number P21246) (residues 33–168), was produced as described previously (26). Platelet factor-4 (PF-4) (Swiss-Prot accession number P02776) (residues 32–101) was purchased from Athens Research and Technology Inc. (Athens, GA).
SDS-PAGE and Western Blot
Samples were separated on 15% (w/v) polyacrylamide-SDS gels. Gels were silver-stained according to Heukeshoven and Dernick (27). Samples separated by polyacrylamide-SDS gels were transferred to HybondTM nitrocellulose membrane (GE Healthcare) using a semidry system (Trans-Blot SD, Bio-Rad). Membranes were blocked with PBS-BSA (8.1 mm Na2HPO4, 1.2 mm KH2PO4, 150 mm NaCl, 2.7 mm KCl, pH 7.0 (PBS) supplemented with 5 mg/ml BSA (Sigma-Aldrich)) for 30 min at room temperature. The membranes were then incubated in PBST-BSA (PBS supplemented with 0.5% (v/v) Tween 20 (PBST) and 5 mg/ml BSA) with Strep-TactinTM-horseradish peroxidase (IBA GmbH, Göttingen, Germany) (1:7500) for 30 min at room temperature. After at least five washes of 5 min with PBST, the presence of horseradish peroxidase was revealed with the SuperSignal West Pico Chemiluminescent substrate (Pierce).
Peptide Map
Digested proteins were applied to an Agilent (Warrington, UK) Zorbax Eclipse Plus C18 RP-HPLC column (100 × 2.1 mm, 3.5 μm) equilibrated in 0.08% (v/v) TFA using a Beckman Coulter (High Wycombe, UK) System GoldTM HPLC system. Peptides were separated with a 95-min gradient of 0–64% (v/v) ACN in 0.08% (v/v) TFA. Elution was monitored at 214 nm.
Desalting
Proteins were desalted with a 5-ml HiTrapTM desalting column (GE Healthcare) using an ÄKTATM HPLC system (GE Healthcare). Proteins were isocratically eluted with the selected buffer at a flow rate of 0.5 ml/min. Elution was monitored at 214 and 280 nm.
Protection and Labeling of HBS
AF-heparin beads (Tosoh Biosciences GmbH, Stuttgart, Germany; binding capacity of 4 mg of ATIII/ml of resin) were preferred to others based on polysaccharide matrix (e.g. Sepharose) to minimize potential unspecific interactions that could increase the noise level of the method. The loading capacity for FGF-2 was estimated at 15 mg/ml of resin. Furthermore the same resin has been successfully used in a cross-linking method (28).
Step 1: Binding
A 20-μl slurry of AF-heparin beads was packed in a minicolumn consisting of a P10 tip with a plastic air filter placed at its end to retain the resin. A 5-ml sterile syringe was used to pack the minicolumn and dispense buffers. The heparin column was equilibrated with 4 × 50 μl of PB 150 buffer (17.9 mm Na2HPO4, 2.1 mm NaH2PO4, 150 mm NaCl, pH 7.8) and loaded with either 1.2 nmol of FGF-2, 2.6 nmol of PF-4, or 1 nmol of PTN dissolved in 200 μl of PB 150 or buffer of compatible ionic strength. Loading was repeated three times to ensure complete binding. The minicolumn was then washed with 4 × 50 μl of PB 150.
Step 2: Acetylation of Exposed Lysines
The heparin column with bound protein was first equilibrated by a quick wash with 20 μl of PB 150 containing 50 mm sulfo-NHS-acetate (Pierce) and then incubated with 20 μl of PB 150 containing 50 mm sulfo-NHS-acetate for 5 min at room temperature to ensure complete blocking (acetylation) of exposed lysines. To stop the reaction, the column was extensively washed with 4 × 50 μl of PB 150. Acetylated protein was eluted from heparin with 40 μl of 2 m NaCl buffered with PB buffer (44.75 mm Na2HPO4, 5.25 mm NaH2PO4, pH 7.8).
Step 3: HBS Lysine Biotinylation
Eluted protein was diluted with 200 μl of PB buffer (to give a final NaCl concentration of 0.33 m) and concentrated with a 5-kDa-molecular mass cutoff centrifugal filter (Sartorius Ltd., Epsom, UK). After adjusting the volume to 37.2 μl with PB, biotinylation was performed by the addition of 2.8 μl of 145 mm NHS-biotin (Pierce) in DMSO (final concentration, 10 mm) and incubation for 30 min at room temperature. The reaction was quenched with 4 μl of 1 m Tris, pH 7.5. The excess reagent was removed by four cycles of sequential dilution with 200 μl of 10 times diluted PB (4.48 mm Na2HPO4, 0.52 mm NaH2PO4, pH 7.8) and concentration with a 5-kDa-molecular mass cutoff centrifugal filter. The sample was then dried by centrifugal evaporation.
Step 4: Protein Digestion
The dried sample was dissolved with 25 μl of 8 m urea, 400 mm NH4HCO3, pH 7.8 and 2.5 μl of 45 mm DTT and incubated for 15 min at 56 °C. Proteins were carbamidomethylated with 2.5 μl of freshly made 0.1 m iodoacetamide for 15 min at room temperature in the dark. Samples were diluted with 70 μl of HPLC grade water and digested overnight at 37 °C with 1.5 μg of chymotrypsin (Sigma-Aldrich).
Step 5: Biotinylated Peptide Purification
A 40-μl slurry of Strep-Tactin Sepharose beads (IBA GmbH) were packed in a minicolumn (see above), which was equilibrated with 4 × 50 μl of 500 mm urea, 25 mm NH4HCO3. Digested protein was diluted to a final volume of 400 μl with HPLC grade water and applied to the column. Loading was repeated three times to ensure complete binding. Unbound peptides were removed by 3 × 50-μl washes with 0.5 m urea, 25 mm NH4HCO3 followed by 2 × 50-μl washes with HPLC grade water. Biotinylated peptides were eluted with two lots of 20 μl of 80% (v/v) ACN, 20% (v/v) TFA, 5 mm biotin.
Steps 6 and 7: Identification of Labeled Peptides
The eluate was concentrated by rotary evaporation, desalted using a C18 ZipTipTM (Millipore Ltd., Watford, UK) according to manufacturer's instructions and analyzed by MALDI-Q-TOF mass spectrometry.
MALDI-Q-TOF Mass Spectrometry
Samples (1 μl) were spotted 1:1 (v/v) in 5 mg/ml α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich) in 50% (v/v) ACN, 50% (v/v) HPLC grade water. Data-dependent analysis was performed on the top 10 most intense peptides in the parent spectrum, the instrument switching to the next peptide for MS/MS after a minimum of 100 counts had been achieved. Collision gas was argon, laser energy was fixed at 250, the instrument was SYNAPTTM MALDI HDMS (Waters Corp., Manchester, UK) where CID can occur in either the trap or the transfer collision cell. A ramp profile in the trap collision cell of 34–44 V for m/z 700–750 up to 151–161 V for m/z 3350–3400 was used. Depending on the m/z of the peptide selected for MS/MS the corresponding energy ramp was applied. MS/MS spectra were processed using the MassLynx v4.0 software (Waters Corp.). Peak lists were generated using the MaxEnt3 algorithm applying a 5% base peak intensity cutoff. Data analysis was performed using the MS-tag tool of the Protein Prospector package v.5.2.2 using the following parameters: digest, chymotrypsin; maximum missed cleavages, 5; possible modifications, acetyl (Lys), biotin (Lys), carbamidomethyl (Cys), and carboxymethyl (Cys); parental ion tolerance, 100 ppm; fragment ion tolerance, 300 ppm; nonspecific cleavage, at one terminus. The UniProt accession number of the protein analyzed was used as a presearch parameter. All other settings were used at default values. In the case of modified parental ions (e.g. sodium adduct ions), the MS-product tool of the Protein Prospector package v.5.2.2 was used using default settings.
RESULTS
Method Development
First the kinetics of protein modification by NHS reagents were investigated. The signal/noise ratio of the protect and label procedure was found to depend on the level of modification (acetylation) of lysines exposed on the protein surface (Fig. 1, step 2). Despite the widespread use of NHS-derived reagents in chemistry and biochemistry, the kinetics of reaction with biomolecules in aqueous solution have been only partially elucidated (29–31). A series of experiments using two different model proteins were performed to establish the optimal NHS reagent concentration and incubation time that ensured a high level of lysine modification (supplemental Figs. S1 and S2).
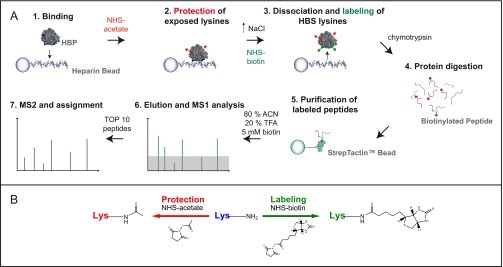
Fig. 1.
The protect and label strategy. A, the protect and label strategy consists of three main steps. The first is the modification/blocking of exposed lysines (steps 1 and 2). HBP is bound to heparin. The protein is then modified by incubation with sulfo-NHS-acetate. This results in the acetylation of primary amines exposed on the protein surface, the N terminus and ε-amines of lysines. The second is the labeling of lysines protected by heparin/HS (step 3). The protein is then dissociated from the sugar by increasing the ionic strength of the solution. The following incubation with NHS-biotin leads to selective labeling of lysines involved in the interaction with heparin that have not been blocked in the previous step. The third is the identification of labeled peptides (steps 4–7). The modified protein is enzymatically digested. Biotinylated peptides are purified by affinity chromatography using a Strep-Tactin minicolumn. Following elution captured peptides are identified by MALDI-Q-TOF mass spectrometry. B, the reactions of protection (acetylation) and labeling (biotinylation) mediated by NHS esters are schematized.
To demonstrate the ability of heparin to selectively protect lysines present in the HBS, a series of pilot experiments were performed using FGF-2 as a model protein. FGF-2 was acetylated according to the conditions chosen in the absence or in the presence of heparin beads (under “Experimental Procedures” see “Protection and Labeling of HBS” steps 1 and 2). After buffer exchange, heparin binding activity was assessed by application of the modified proteins to a heparin minicolumn. The same amount of native FGF-2 was applied to the minicolumn as a control. A further sample, obtained by exposing FGF-2 acetylated in the presence of heparin beads to NHS-biotin (under “Experimental Procedures” see “Protection and Labeling of HBS” step 3), was also applied to the heparin minicolumn. This last sample was included because it represented the final product of the protect and label strategy. Lysine acetylation clearly impaired the binding of FGF-2 to heparin (Fig. 2A, sample 2). However, when the acetylation was performed in the presence of heparin beads, the protein retained almost completely its binding activity (Fig. 2A, sample 3). This suggests a selective protection given by the sugar chain to lysines involved in heparin binding. Furthermore the retained binding activity of sample 3 (acetylated in the presence of heparin beads) could be abolished by further exposure to NHS-biotin (Fig. 2A, sample 4).
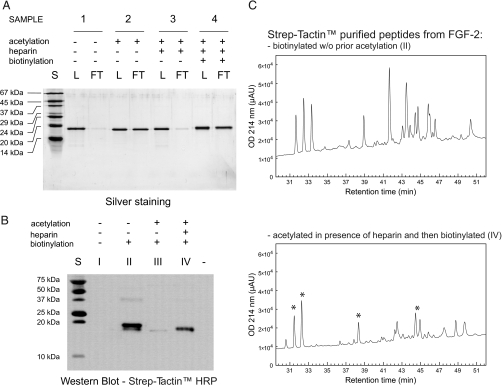
Fig. 2.
Heparin/HS protect residues involved in their binding against chemical modification. A, silver stain. S, standard; L, of the protein loaded onto the microcolumn; FT, of the unbound fraction. 1, FGF-2; 2, acetylated FGF-2 (protein was incubated with 50 mm sulfo-NHS-acetate for 5 min at room temperature); 3, FGF-2 acetylated in the presence of heparin beads (under “Experimental Procedures” see “Protection and Labeling of HBS” steps 1 and 2); 4, FGF-2 acetylated in the presence of heparin beads (same as 3) and then biotinylated after dissociation from the sugar with NHS-biotin at a final concentration of 10 mm for 30 min at room temperature. All samples were buffer-exchanged to the same buffer (PB 150) by gel filtration chromatography before application to the heparin minicolumn. The heparin minicolumn was packed using 20 μl of AF-heparin beads and equilibrated in PB 150. B, Western blot. I, FGF-2; II, FGF-2; III, acetylated FGF-2 (protein was incubated with 50 mm sulfo-NHS-acetate for 5 min at room temperature); IV, FGF-2 acetylated in the presence of heparin beads (under “Experimental Procedures” see “Protection and Labeling of HBS” steps 1 and 2). Samples were buffer-exchanged to PB 150 as in (A). Samples II–IV were then biotinylated with NHS-biotin at a final concentration of 10 mm for 30 min at room temperature. Protein concentrations were measured after the desalting step, and the same amount of protein was subjected to biotinylation (samples II–IV) and then SDS-PAGE. C, biotinylated peptides from enzymatic digestion of samples II and IV were purified with a Strep-Tactin minicolumn using 80% (v/v) ACN, 20% (v/v) TFA as elution buffer and analyzed by RP-HPLC. Asterisks indicate peaks deriving from peptides selectively protected from acetylation by heparin and, therefore, subsequently biotinylated. HRP, horseradish peroxidase; μAU, microabsorbance units; w/o, without.
After demonstrating that heparin can effectively prevent the modification of residues crucial for its binding, the next step was to show that those residues can be efficiently labeled (biotinylated) after dissociation of the protein from the sugar (Fig. 1, step 3). FGF-2 was acetylated in the absence or presence of heparin beads and buffer-exchanged. Both the samples were then biotinylated as in step 3 of the protect and label strategy (see “Experimental Procedures”). Native and fully biotinylated FGF-2 (same conditions as above but without prior acetylation) were used as a negative and positive control, respectively. Biotinylated proteins were identified by Western blot. The acetylation condition chosen guaranteed a high level of protection against further biotinylation (Fig. 2B, sample III). However, the presence of heparin during the acetylation step partially prevented the modification of residues that can, therefore, be biotinylated after dissociation from the sugar (Fig. 2B, sample IV). To establish whether the reduced protection was a generalized phenomenon involving all lysines exposed on the protein surface, biotinylated peptides derived from sample II (fully biotinylated FGF-2) and sample IV (protected and labeled FGF-2) were compared by RP-HPLC (Fig. 2C). As expected, a generally reduced recovery of biotinylated peptides was observed in sample IV. Importantly the difference between the elution profiles indicates that the presence of heparin is selectively affecting the protection of a restricted number of residues (Fig. 2C).
The last step of the implementation of the method was the optimization of a high efficiency protocol for the purification and identification of labeled peptides derived from the enzymatic digestion of the product of the protect and label strategy (Fig. 1, steps 4–7). The high affinity of the avidin-biotin interactions allows the use of stringent conditions for the purification of biotinylated biomolecules; however, it limits the yield of recovery that is achievable. An array of capture supports and rationally designed elution buffers were compared in a model experimental setting to maximize the efficiency of recovery of biotinylated peptide and ensure compatibility with the subsequent mass spectrometric analysis (supplemental Fig. S3). Strep-Tactin Sepharose was identified as the best affinity capture support (data not shown). Regarding the composition of the elution buffer, the peptide recovery was enhanced by increasing the concentration of both acid (TFA) and solvent (ACN) up to the extreme condition where water was excluded from the buffer composition (80% ACN, 20% TFA) (supplemental Fig. S3B). Further improvement was achieved by the addition of a large excess of competitor (5 mm biotin) to the elution buffer (supplemental Fig. S3B).
Identification of HBSs by the Protect and Label Strategy
The protect and label strategy was then evaluated for its ability to correctly identify HBSs in well characterized heparin-binding proteins. Labeled peptides were identified by MALDI-Q-TOF mass spectrometry. The 10 most intense ions for each sample were automatically selected in the quadrupole, and MS/MS data were acquired to assign chemical modifications introduced during the protect and label procedure (Fig. 1, steps 6 and 7, and supplemental spectra). Furthermore fragmentation spectra allowed a further control on the affinity purification of labeled peptides. A modification-specific marker ion at m/z 310.16, assigned as Lys(biotin)-NH3, was in fact observed in all the fragmentation spectra analyzed. This is consistent with the ion at m/z 126.1 previously described as a marker for lysine acetylation (biotin − acetyl = 184) (32) and with the presence in most of the spectra of the ions m/z 227.08 and 84.08 corresponding to biotin and Lys-NH3, respectively.
The test proteins were chosen to represent the variety of the heparin interactors as highlighted by the heparin interactome characterized so far (10). Two growth factors were selected, FGF-2 and PTN. The first is a prototypical member of the FGF family of growth factors and one of the most studied HBPs, whereas the second is characterized by the presence of a thrombospondin type-1 repeat (TSR) motif responsible for the interaction with the sugar (33). PF-4 was added as an example of a chemokine that interacts with heparin as an oligomer and that possesses a bipartite HBS (18).
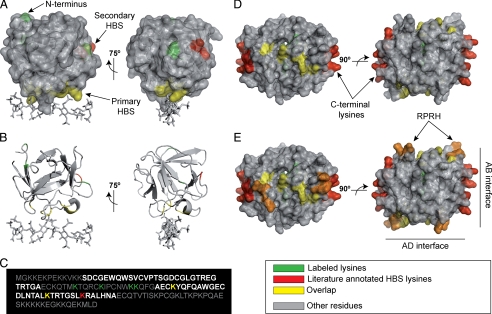
The structure of FGF-2 is described as β-trefoil, comprising a 3-fold repeat of a four-stranded antiparallel β-sheet (34). The main HBS of FGF-2 has been extensively characterized by different approaches including site-directed mutagenesis, synthetic peptides, x-ray crystallography, and NMR spectroscopy (19, 22, 34–37). It consists of residues Lys35, Asn36, and the cluster of basic amino acids located in the region 128–144 (22) (Table I). These residues form an extremely basic surface including strand I/strand II loop (Lys35 and Asn36), strand X/strand XI loop, strand XI, and strand XI/XII loop (Fig. 3B). The biotinylated peptides identified by the protect and label strategy mapped to three areas on the protein surface (Table II) (Fig. 3A). These include the previously described HBS (Table II, peptides 1, 2, 6, 7, 9, and 10), a region located in strand VI/strand VII loop (Table II, peptide 5), and the beginning of strand I (Table II, peptides 3, 4, and 8). The loop identified by peptide 5 is located on the same surface of the sequence 115YRSRKYSSWY124 described by Baird et al. (36) and the sequence 117SRKYTSWYVA126 described by Kinsella et al. (23) as a possible secondary HBS. No evidence has been published to date regarding the involvement of strand I in heparin binding. MS/MS spectra allowed the assignment of modified residues and the identification of peptides deriving from unspecific cleavage (Table II, peptide 4) or other chemical modifications (e.g. deamination (Table II, peptide 1) and sodium adduct (Table II, peptide 9)). Only in the case of the ion at m/z 1371.71 (Table II, peptide 5) was a double assignment necessary. Fragment ions deriving from both the isobaric peptides 83LAMK(biotin)EDGRLL92 (MH+ monoisotopic mass, 1371.7123) and 27K(biotin)DPK(biotin)RLY33 (MH+ monoisotopic mass, 1371.6912) were in fact assigned in the MS/MS spectrum (supplemental Spectrum S5).
Table I. HBS of protect and label strategy test proteins.
The HBSs and the methods used to identify these are given. SDM, site-directed mutagenesis; x-ray, x-ray crystallography; SP, synthetic peptide; MM, molecular modeling.
| Name | Swiss-Prot accession no. | Molecular mass | pI | HBS | Method | Ref. |
|---|---|---|---|---|---|---|
| Da | ||||||
| FGF-2 | P09038 | 17,253.8 | 9.58 | HBS-1: 35KN36, Arg90, 128KRTGQYKLGSKTGPGQK144 | SDM, x-ray | 19, 22 |
| HBS-2: 102FFFERLESNNYNTYRSRKYSSWYVALKR129 | SP | 36 | ||||
| PF-4 | P02776 | 10,844.9 | 8.93 | HBS-1: 77KNGR80, 92KKIIKK97 | NMR, SDM, MM | 18, 21 |
| HBS-2: 51RPRH54, Lys62, Thr56 | NMR, SDM | 21 | ||||
| PTN | P21246 | 18,942 | 9.66 | HBS-1: 97AECKYQFQAWGECDLNTALKTRTGSLKRALHNA129 | SP | 38 |
Fig. 3.
Labeled peptides and heparin-binding site: test proteins. A, FGF-2 (residues 25–153) is shown using overlaid schematic and surface representations (Protein Data Bank code 1FQ9). Heparin-derived octasaccharide is shown as sticks. B, FGF-2 (residues 25–153) is shown using schematic representation only (Protein Data Bank code 1FQ9). Heparin-derived octasaccharide is shown as sticks. C, primary structure of PTN (residues 1–167). D, tetrameric PF-4 (residues 38–101) is shown using overlaid schematic and surface representations (Protein Data Bank code 1F9Q). Previously described HBS lysines are shown in red (see Table I). Lysines labeled by the protect and label strategy are shown in green (see Table II). Yellow indicates an overlap between labeled amino acids and previously described HBS residues. Other residues are shown in gray. E, tetrameric PF-4 is shown as in C but in addition the HBS sequence 51RPRH54 of each monomer is highlighted in orange. Structures are represented using PyMol.
Table II. Protect and label strategy labeled peptides.
For each sample, peptides are sorted in a descending order according to the intensity of their parental ion. Labeled peptides were identified using the tools MS-tag and MS-product of the package Protein Prospector v5.2.2. The “Matched HBS” column reports an overlap between the identified peptide and a previously characterized HBS as defined in Table I. Assigned MS/MS spectra are available as supplemental data. na, not assigned.
| Peptide | m/z observed | m/z theoretical | Error | Sequence | Residues | Matched HBS | Supplemental spectrum |
|---|---|---|---|---|---|---|---|
| ppm | |||||||
| FGF-2 | |||||||
| 1 | 1981.89 | 1981.92 | −15.1 | KDPK(acetyl)RLYC(carboxymethyl)K(biotin)NGGFF − NH3 | 27–40 | HBS-1 | S1 |
| 2 | 1261.65 | 1261.67 | −15.9 | VALK(biotin)RTGQY | 125–133 | HBS-1 | S2 |
| 3 | 1187.61 | 1187.62 | −8.4 | K(acetyl)DPK(biotin)RLY | 27–33 | S3 | |
| 4 | 902.48 | 902.49 | −11.1 | PK(biotin)RLY | 29–33 | S4 | |
| 5 | 1371.7 | 1371.71/1371.69 | 6.4/7.3 | LAMK(biotin)EDGRLL/K(biotin)DPK(biotin)RLY | 83–92/27–33 | S5 | |
| 6 | 2075.95 | 2076.08 | −62.6 | K(biotin)LGSK(biotin)TGPGQK(biotin)AIL | 134–147 | HBS-1 | S6 |
| 7 | 1998.92 | 1998.95 | −15.0 | KDPK(acetyl)RLYC(carboxymethyl)K(biotin)NGGFF | 27–40 | HBS-1 | S7 |
| 8 | 2166.9 | 2167.03 | −60.0 | K(acetyl)DPK(biotin)RLYCK(biotin)NGGFF | 27–40 | HBS-1 | S8 |
| 9 | 2097.94 | 2098.06 | −57.2 | K(biotin)LGSK(biotin)TGPGQK(biotin)AIL + Na+ | 134–147 | HBS-1 | S9 |
| 10 | 1891.95 | 1892.01 | −31.7 | K(biotin)LGSK(acetyl)TGPGQK(biotin)AIL | 134–147 | HBS-1 | S10 |
| PF-4 | |||||||
| 11 | 1441.71 | 1441.71 | −0.9 | K(biotin)NGRK(biotin)IC(carboxymethyl)L | 77–84 | HBS-1 | S11 |
| 12 | 1423.7 | 1423.71 | −7.9 | K(biotin)NGRK(biotin)IC(carboxymethyl)L − H20 | 77–84 | HBS-1 | S12 |
| 13 | 2078.94 | 2079.05 | −52.9 | K(biotin)NGRK(biotin)IC(carboxymethyl)LDLQAPL | 77–90 | HBS-1 | S13 |
| 14 | 3019.96 | na | S14 | ||||
| 15 | 2060.94 | 2061.04 | −48.5 | K(biotin)NGRK(biotin)IC(carboxymethyl)LDLQAPL − H20 | 77–90 | HBS-1 | S15 |
| 16 | 2241.9 | 2242.12 | −97.3 | K(biotin)NGRK(biotin)IC(carbamidomethyl)LDLQAPLY | 77–91 | HBS-1 | S16 |
| 17 | 1852.94 | 1852.97 | −16.2 | K(biotin)NGRKIC(carboxymethyl)LDLQAPL | 77–90 | HBS-1 | S17 |
| 18 | 1894.94 | 1894.98 | −21.1 | K(acetyl)NGRK(biotin)IC(carboxymethyl)LDLQAPL | 77–90 | HBS-1 | S18 |
| 19 | 1746.85 | 1746.86 | −5.7 | EVIK(biotin)AGPHC(carbamidomethyl)PTAQL | 59–72 | HBS-2 | S19 |
| 20 | 2015.95 | 2016.04 | −44.6 | K(biotin)NGRKIC(carboxymethyl)LDLQAPLY | 77–91 | HBS-1 | S20 |
| PTN | |||||||
| 21 | 1942.83 | 1942.89 | −30.9 | K(biotin)TQRC(carbamidomethyl)K(biotin)IPC(carbamidomethyl)NW | 81–91 | S21 | |
| 22 | 1359.61 | 1359.62 | −7.4 | RC(carbamidomethyl)K(biotin)IPC(carbamidomethyl)NW | 84–91 | S22 | |
| 23 | 1758.81 | 1758.82 | −5.7 | K(biotin)TQRC(carbamidomethyl)K(acetyl)IPC(carbamidomethyl)NW | 81–91 | S23 | |
| 24 | 1752.78 | 1752.79 | −5.7 | K(acetyl)K(biotin)QFGAEC(carbamidomethyl)K(biotin)Y | 92–101 | HBS-1 | S24 |
| 25 | 1936.81 | 1936.85 | −20.7 | K(biotin)K(biotin)QFGAEC(carbamidomethyl)K(biotin)Y | 92–101 | HBS-1 | S25 |
| 26 | 1958.81 | 1958.85 | −20.4 | K(biotin)K(biotin)QFGAEC(carbamidomethyl)K(biotin)Y + Na+ | 92–101 | HBS-1 | S26 |
| 27 | 1774.77 | 1774.79 | −11.3 | K(acetyl)K(biotin)QFGAEC(carbamidomethyl)K(biotin)Y + Na+ | 92–101 | HBS-1 | S27 |
| 28 | 1342.63 | 1342.58 | 37.2 | RC(carbamidomethyl)K(biotin)IPC(carbamidomethyl)NW − NH3 | 84–91 | S28 | |
| 29 | 1980.8 | na | S29 | ||||
| 30 | 1498.81 | 1498.85 | −26.7 | K(biotin)TRTGSLK(acetyl)RAL | 116–126 | HBS-1 | S30 |
PF-4 belongs to the CXC family of chemokines. The monomeric structure consists of a triple stranded antiparallel β-sheet overlaid by a C-terminal α-helix, whereas the N-terminal region is essentially disordered (18, 21). PF-4 exists as a tetramer in solution. Dimerization occurs through the antiparallel association of the first strands to form a larger β-sheet (18). The tetramer results from the further assembly of two dimers mediated by contacts between N-terminal regions (18). The HBS has been characterized by site-directed mutagenesis coupled to NMR spectroscopy (21) and by molecular modeling (18). The HBS consists of a ring of positively charged residues around the surface of the tetramer formed by two pairs of lysines located within the C-terminal α-helix (92KKIIKK97), two arginines from the N terminus/strand I loop (51RPRH54), Lys77 and Arg80 from the strand II/strand III loop (77KNGR80), and the residues Lys62 and Thr56 (18, 21) (Table I) (Fig. 3D). Peptides including biotinylated Lys77 (Table II, peptides 11–13, 15–17, and 20) and Lys62 (Table II, peptide 19) were identified in agreement with published data. Lys81 was found biotinylated in five cases in association with labeled Lys77 (Table II, peptides 11–13, 15, and 16) and in one case as a single biotinylated peptide (Table II, peptide 18).
PTN is a heparin-dependent growth factor characterized by the presence of two TSR-I motifs. It consists of two disordered basic clusters at its N and C termini and two β-sheets containing the TSR-I motifs (N-TSR, residues 45–71; C-TSR, residues 97–129) (33, 38). The two TSR motifs have been proposed as possible HBSs (33); however, Hamma-Kourbali et al. (38) recently demonstrated the involvement of only the C-TSR in heparin binding (Table I). Labeled peptides mapped lysines located at the beginning (Table II, peptides 24–28) and in the center (Table II, peptide 30) of the C-TSR (Fig. 3C). Biotinylated lysines deriving from the region between the two TSR domains (Table II, peptides 21–23 and 28) (Fig. 3C) were also identified.
DISCUSSION
One of the main limitations on the study of heparin/HS-protein interactions is the lack of rapid and reliable methods for the identification of HBSs. A new strategy for the identification of HBSs was developed. Reaction conditions able to ensure a high level of protein modification by NHS reagents were established. A high efficiency protocol for the recovery of labeled (biotinylated) peptides was optimized. Automated tandem mass spectrometry and partially manually curated data interpretation allowed the assignment of labeled residues in 28 of the 30 peptides selected for the three samples analyzed. Data obtained were used to evaluate the ability of the new method to correctly predict HBSs in well characterized heparin-binding proteins.
The results show that (i) heparin can effectively prevent the modification of residues in a protein that are crucial for its binding to the polysaccharide (Fig. 2A, sample 3), (ii) further exposure to NHS-biotin after dissociation of the protein from the sugar can abolish the retained heparin binding activity (Fig. 2A, sample 4), (iii) the residues that remain unmodified during acetylation in the presence of heparin can be effectively labeled (biotinylated) (Fig. 2B, sample IV), and (iv) the labeling is not a generalized phenomenon but is restricted to a specific set of lysines (Fig. 2C). Thus, the application of the protect and label strategy (steps 1–3) allows the specific labeling of lysines crucial for heparin binding that, by definition, will represent the HBS.
The protect and label strategy allowed the correct localization of the main HBS of all the test proteins, providing a strong rationale for the validity of the method. Some important points highlight the advantages and limitations of the new method. In the case of FGF-2, the canonical HBS was successfully mapped by the biotinylated peptides, including the loop between strand I and strand II (34CKNGGFF40) even though the contribution of the lysine to the binding is relatively minor (22). The second region identified, strand VI/strand VII loop (83LAMKEDGRLL92), is adjacent to a proposed secondary heparin-binding site (23, 36, 39) of approximately millimolar affinity. The region including residues Lys119, Tyr120, and Trp123 has been proposed also as a secondary receptor-binding site (40). Thus the FGF-2 mutants K119E, Y120A, and W123A displayed decreased mitogenic activity on Swiss 3T3 fibroblasts when compared with wild type FGF-2 (40). These residues were shown to be responsible for a secondary binding event between FGF-2 and its receptor that is dependent on the presence of heparin (40). Residue Lys86 and residues Arg118 and Lys119 are located in two adjacent loops (distance of Lys86 Cα to Lys119 Cα, 9.3 Å) (Fig. 3, A and B), forming a basic patch on the protein surface that could better describe the secondary binding site proposed with the use of synthetic peptides. More difficult is the interpretation of the signal deriving from the N terminus of the protein. The N terminus of FGF-2 is a highly disordered and mobile region (34, 41). Its involvement in heparin binding has been excluded by deletion mutagenesis (39) because mutants lacking 23 (N14 FGF-2) and 49 (N41 FGF-2) N-terminal amino acids displayed unaltered affinity for heparin compared with a 146-amino acid form (FGF-2-(11–157)) as judged by heparin affinity chromatography (39). However, this analysis is qualitative, and both the mutants were less efficient in stimulating DNA synthesis in BALB/c3T3 cells when compared with FGF-2-(11–157) (39), supporting a functional role for these residues. Alternatively, labeling of the N terminus may arise due to protection deriving from a conformational change induced upon binding to heparin.
All the lysines that are affected by major chemical shift changes upon binding of heparin to a tetrameric form of PF-4 (21) were successfully identified by the protect and label strategy with the exception of the two pairs of lysine located in the C-terminal helix. The sequence 92KKIIKK97 was first proposed as the center of the HBS of PF-4 and also used for the development of a heparin-binding consensus sequence by Cardin and Weintraub (17). However, investigation by NMR spectroscopy and site-directed mutagenesis revealed a minor contribution of this region to sugar binding (21). Side chain resonances of residues Arg53, His54, Thr56, Lys62, and Arg80 were more perturbed upon heparin binding than those of C-terminal lysines (21). Furthermore substitution of all four lysines to alanines reduced the affinity of the mutant PF-4 for heparin only by a factor 4 as assessed by a fluorescence binding assay (21). In agreement with these data, the protect and label strategy supports the AD interface of the PF-4 tetramer (Fig. 3E) as the core of the heparin-binding region with C-terminal lysines involved in secondary and more transitory contacts, which are unable to ensure protection of these residues against chemical modification.
Finally the selective labeling confirmed recent data (38) indicating the C-TSR domain as the main HBS of PTN (Fig. 3C). However, the biotinylation of other residues including Lys81, Lys86, Lys92, and Lys93, located at the boundaries between N-TSR and C-TSR, suggest a possible involvement of this area, which has not been investigated previously, in heparin binding.
In summary, the examples cited above indicate that the protect and label strategy can represent a useful tool for the localization of unknown HBSs in heparin-binding proteins. The method displayed a great sensitivity for the detection of heparin-binding surfaces together with a good level of specificity despite the high lysine content of the protein tested (14 for FGF-2, eight for PF-4, and 28 for PTN) with the possible exception of the highly flexible N-terminal region of FGF-2. The main limitation of the technique remains the identification of regions involved in the sugar binding that do not contain lysines (e.g. the 51RPRH54 region of PF-4; Fig. 3E). The protect and label strategy can be considered as an in vitro docking that, because of its low requirements in terms of material (nanomolar range) and sample handling time (2 bench days), can find applications as a hypothesis generator for further mutagenesis studies or as a complementary technique for structural studies, e.g. in silico docking.
In 2004 Vivès et al. (28) reported a new method for defining amino acid residues involved in heparin binding based on cross-linking between HBP and heparin beads and N-terminal sequencing of the captured peptides. This method, recently updated to an “in-solution” version (42), shares advantages with the protect and label strategy, such as the low requirement of material and its rapidity, as well as drawbacks, such as being based on NHS chemistry. However, the cross-linking reaction occurs upon binding of the protein to a preactivated and hence modified sugar, whereas the protect and label strategy introduces chemical modification on protein-heparin complexes assembled in pseudophysiological conditions. Furthermore the combination of the selective labeling of peptides with MS/MS sequencing techniques allows the translation of the protect and label strategy into a medium/high throughput format where a semipurified mixture of HBPs are analyzed at the same time, and their HBSs are mapped due to the unique mass signatures deriving from the labeled peptides. The application of the method could possibly be expanded to other electrostatic interactions (protein-GAG, protein-protein, and protein-nucleic acid) provided there is the involvement of lysines in at least one of the binding partners. Finally the development of new protein modification reagents able to selectively target different amino acids, e.g. guanido-specific compounds (43), could further expand the applicability of protect and label strategies.
Supplementary Material
Acknowledgments
We thank Dr. Liam Brady (Waters Corp.), Dr. D. A. Ward and M. C. Prescott (Functional Proteomics facility, University of Liverpool) for MS data acquisition and Professor J. E. Turnbull for reviewing the manuscript.
Footnotes
* This work was supported by the European Union Marie Curie Early Stage Training Programme (MolFun) (to A. O.), the Cancer and Polio Research Fund and the North West Cancer Research Fund (to A. O., M. C. W., and D. G. F.), and “Agence National de la Recherche” Grant P002754 and “Institut National du Cancer” Grant PL06-093 (to J. C.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- GAG
- glycosaminoglycan
- HBS
- heparin-binding site
- HBP
- heparin-binding protein
- HS
- heparan sulfate
- NHS
- N-hydroxysuccinimide
- TSR
- thrombospondin type-1 repeat
- ATIII
- antithrombin III
- RP
- reverse phase
- FGF
- fibroblast growth factor
- PTN
- pleiotrophin
- PF-4
- platelet factor-4.
REFERENCES
- 1.Medeiros G. F., Mendes A., Castro R. A., Baú E. C., Nader H. B., Dietrich C. P. (2000) Distribution of sulfated glycosaminoglycans in the animal kingdom: widespread occurrence of heparin-like compounds in invertebrates. Biochim. Biophys. Acta 1475, 287–294 [DOI] [PubMed] [Google Scholar]
- 2.Bishop J. R., Schuksz M., Esko J. D. (2007) Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 3.Rodgers K. D., San Antonio J. D., Jacenko O. (2008) Heparan sulfate proteoglycans: a GAGgle of skeletal-hematopoietic regulators. Dev. Dyn 237, 2622–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin X. (2004) Functions of heparan sulfate proteoglycans in cell signaling during development. Development 131, 6009–6021 [DOI] [PubMed] [Google Scholar]
- 5.Häcker U., Nybakken K., Perrimon N. (2005) Heparan sulphate proteoglycans: the sweet side of development. Nat. Rev. Mol. Cell Biol 6, 530–541 [DOI] [PubMed] [Google Scholar]
- 6.Bülow H. E., Hobert O. (2006) The molecular diversity of glycosaminoglycans shapes animal development. Annu. Rev. Cell Dev. Biol 22, 375–407 [DOI] [PubMed] [Google Scholar]
- 7.Parish C. R. (2006) The role of heparan sulphate in inflammation. Nat. Rev. Immunol 6, 633–643 [DOI] [PubMed] [Google Scholar]
- 8.Handel T. M., Johnson Z., Crown S. E., Lau E. K., Proudfoot A. E. (2005) Regulation of protein function by glycosaminoglycans—as exemplified by chemokines. Annu. Rev. Biochem 74, 385–410 [DOI] [PubMed] [Google Scholar]
- 9.Fuster M. M., Esko J. D. (2005) The sweet and sour of cancer: glycans as novel therapeutic targets. Nat. Rev. Cancer 5, 526–542 [DOI] [PubMed] [Google Scholar]
- 10.Ori A., Wilkinson M. C., Fernig D. G. (2008) The heparanome and regulation of cell function: structures, functions and challenges. Front. Biosci 13, 4309–4338 [DOI] [PubMed] [Google Scholar]
- 11.Hileman R. E., Fromm J. R., Weiler J. M., Linhardt R. J. (1998) Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. BioEssays 20, 156–167 [DOI] [PubMed] [Google Scholar]
- 12.Powell A. K., Yates E. A., Fernig D. G., Turnbull J. E. (2004) Interactions of heparin/heparan sulfate with proteins: appraisal of structural factors and experimental approaches. Glycobiology 14, 17R–30R [DOI] [PubMed] [Google Scholar]
- 13.Forster M., Mulloy B. (2006) Computational approaches to the identification of heparin-binding sites on the surfaces of proteins. Biochem. Soc. Trans 34, 431–434 [DOI] [PubMed] [Google Scholar]
- 14.Yu W., Hill J. S. (2006) Mapping the heparin-binding domain of human hepatic lipase. Biochem. Biophys. Res. Commun 343, 659–665 [DOI] [PubMed] [Google Scholar]
- 15.Imberty A., Lortat-Jacob H., Pérez S. (2007) Structural view of glycosaminoglycan-protein interactions. Carbohydr. Res 342, 430–439 [DOI] [PubMed] [Google Scholar]
- 16.Fang J., Dong Y., Salamat-Miller N., Middaugh C. R. (2008) DB-PABP: a database of polyanion-binding proteins. Nucleic Acids Res 36, D303–D306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardin A. D., Weintraub H. J. (1989) Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis 9, 21–32 [DOI] [PubMed] [Google Scholar]
- 18.Lortat-Jacob H., Grosdidier A., Imberty A. (2002) Structural diversity of heparan sulfate binding domains in chemokines. Proc. Natl. Acad. Sci. U.S.A 99, 1229–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faham S., Hileman R. E., Fromm J. R., Linhardt R. J., Rees D. C. (1996) Heparin structure and interactions with basic fibroblast growth factor. Science 271, 1116–1120 [DOI] [PubMed] [Google Scholar]
- 20.Jin L., Abrahams J. P., Skinner R., Petitou M., Pike R. N., Carrell R. W. (1997) The anticoagulant activation of antithrombin by heparin. Proc. Natl. Acad. Sci. U.S.A 94, 14683–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayo K. H., Ilyina E., Roongta V., Dundas M., Joseph J., Lai C. K., Maione T., Daly T. J. (1995) Heparin binding to platelet factor-4. An NMR and site-directed mutagenesis study: arginine residues are crucial for binding. Biochem. J 312, 357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson L. D., Pantoliano M. W., Springer B. A. (1994) Energetic characterization of the basic fibroblast growth factor-heparin interaction: identification of the heparin binding domain. Biochemistry 33, 3831–3840 [DOI] [PubMed] [Google Scholar]
- 23.Kinsella L., Chen H. L., Smith J. A., Rudland P. S., Fernig D. G. (1998) Interactions of putative heparin-binding domains of basic fibroblast growth factor and its receptor, FGFR-1, with heparin using synthetic peptides. Glycoconj. J 15, 419–422 [DOI] [PubMed] [Google Scholar]
- 24.Chang J. Y. (1989) Binding of heparin to human antithrombin III activates selective chemical modification at lysine 236. Lys-107, Lys-125, and Lys-136 are situated within the heparin-binding site of antithrombin III. J. Biol. Chem 264, 3111–3115 [PubMed] [Google Scholar]
- 25.Duchesne L., Gentili D., Comes-Franchini M., Fernig D. G. (2008) Robust ligand shells for biological applications of gold nanoparticles. Langmuir 24, 13572–13580 [DOI] [PubMed] [Google Scholar]
- 26.Delbé J., Vacherot F., Laaroubi K., Barritault D., Courty J. (1995) Effect of heparin on bovine epithelial lens cell-proliferation induced by heparin affin regulatory peptide. J. Cell. Physiol 164, 47–54 [DOI] [PubMed] [Google Scholar]
- 27.Heukeshoven J., Dernick R. (1985) Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis 6, 103–112 [Google Scholar]
- 28.Vivès R. R., Crublet E., Andrieu J. P., Gagnon J., Rousselle P., Lortat-Jacob H. (2004) A novel strategy for defining critical amino acid residues involved in protein/glycosaminoglycan interactions. J. Biol. Chem 279, 54327–54333 [DOI] [PubMed] [Google Scholar]
- 29.Smith J. S., Miller B. T., Knock S. L., Kurosky A. (1991) Biotinylated peptides/proteins. I. Determination of stoichiometry of derivatization. Anal. Biochem 197, 247–253 [DOI] [PubMed] [Google Scholar]
- 30.Smith G. P. (2006) Kinetics of amine modification of proteins. Bioconjug. Chem 17, 501–506 [DOI] [PubMed] [Google Scholar]
- 31.Abello N., Kerstjens H. A., Postma D. S., Bischoff R. (2007) Selective acylation of primary amines in peptides and proteins. J. Proteome Res 6, 4770–4776 [DOI] [PubMed] [Google Scholar]
- 32.Kim J. Y., Kim K. W., Kwon H. J., Lee D. W., Yoo J. S. (2002) Probing lysine acetylation with a modification-specific marker ion using high-performance liquid chromatography/electrospray-mass spectrometry with collision-induced dissociation. Anal. Chem 74, 5443–5449 [DOI] [PubMed] [Google Scholar]
- 33.Kilpelainen I., Kaksonen M., Kinnunen T., Avikainen H., Fath M., Linhardt R. J., Raulo E., Rauvala H. (2000) Heparin-binding growth-associated molecule contains two heparin-binding beta-sheet domains that are homologous to the thrombospondin type I repeat. J. Biol. Chem 275, 13564–13570 [DOI] [PubMed] [Google Scholar]
- 34.Zhang J. D., Cousens L. S., Barr P. J., Sprang S. R. (1991) Three-dimensional structure of human basic fibroblast growth factor, a structural homolog of interleukin 1 beta. Proc. Natl. Acad. Sci. U.S.A 88, 3446–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L. Y., Safran M., Aviezer D., Böhlen P., Seddon A. P., Yayon A. (1994) Diminished heparin binding of a basic fibroblast growth factor mutant is associated with reduced receptor binding, mitogenesis, plasminogen activator induction, and in vitro angiogenesis. Biochemistry 33, 10999–11007 [DOI] [PubMed] [Google Scholar]
- 36.Baird A., Schubert D., Ling N., Guillemin R. (1988) Receptor- and heparin-binding domains of basic fibroblast growth factor. Proc. Natl. Acad. Sci. U.S.A 85, 2324–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moy F. J., Safran M., Seddon A. P., Kitchen D., Böhlen P., Aviezer D., Yayon A., Powers R. (1997) Properly oriented heparin-decasaccharide-induced dimers are the biologically active form of basic fibroblast growth factor. Biochemistry 36, 4782–4791 [DOI] [PubMed] [Google Scholar]
- 38.Hamma-Kourbali Y., Bernard-Pierrot I., Heroult M., Dalle S., Caruelle D., Milhiet P. E., Fernig D. G., Delbé J., Courty J. (2008) Inhibition of the mitogenic, angiogenic and tumorigenic activities of pleiotrophin by a synthetic peptide corresponding to its C-thrombospondin repeat-I domain. J. Cell. Physiol 214, 250–259 [DOI] [PubMed] [Google Scholar]
- 39.Seno M., Sasada R., Kurokawa T., Igarashi K. (1990) Carboxyl-terminal structure of basic fibroblast growth factor significantly contributes to its affinity for heparin. Eur. J. Biochem 188, 239–245 [DOI] [PubMed] [Google Scholar]
- 40.Springer B. A., Pantoliano M. W., Barbera F. A., Gunyuzlu P. L., Thompson L. D., Herblin W. F., Rosenfeld S. A., Book G. W. (1994) Identification and concerted function of two receptor binding surfaces on basic fibroblast growth factor required for mitogenesis. J. Biol. Chem 269, 26879–26884 [PubMed] [Google Scholar]
- 41.Moy F. J., Seddon A. P., Campbell E. B., Böhlen P., Powers R. (1995) 1H, 15N, 13C and 13CO assignments and secondary structure determination of basic fibroblast growth factor using 3D heteronuclear NMR spectroscopy. J. Biomol. NMR 6, 245–254 [DOI] [PubMed] [Google Scholar]
- 42.Crublet E., Andrieu J. P., Vivès R. R., Lortat-Jacob H. (2008) The HIV-1 envelope glycoprotein gp120 features four heparan sulfate binding domains, including the co-receptor binding site. J. Biol. Chem 283, 15193–15200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q., Crosland E., Fabris D. (2008) Nested Arg-specific bifunctional crosslinkers for MS-based structural analysis of proteins and protein assemblies. Anal. Chim. Acta 627, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.