Abstract
We present a novel method utilizing “saltless” pH gradient weak cation exchange-hydrophilic interaction liquid chromatography directly coupled to electron transfer dissociation (ETD) mass spectrometry for the automated on-line high throughput characterization of hypermodified combinatorial histone codes. This technique, performed on a low resolution mass spectrometer, displays an improvement over existing methods with an ∼100-fold reduction in sample requirements and analysis time. The scheme presented is capable of identifying all of the major combinatorial histone codes present in a sample in a 2-h analysis. The large N-terminal histone peptides are eluted by the pH and organic solvent weak cation exchange-hydrophilic interaction liquid chromatography gradient and directly introduced via nanoelectrospray ionization into a benchtop linear quadrupole ion trap mass spectrometer equipped with ETD. Each polypeptide is sequenced, and the modification sites are identified by ETD fragmentation. The isobaric trimethyl and acetyl modifications are resolved chromatographically and confidently distinguished by the synthesis of mass spectrometric and chromatographic information. We demonstrate the utility of the method by complete characterization of human histone H3.2 and histone H4 from butyrate-treated cells, but it is generally applicable to the analysis of highly modified peptides. We find this methodology very useful for chromatographic separation of isomeric species that cannot be separated well by any other chromatographic means, leading to less complicated tandem mass spectra. The improved separation and increased sensitivity generated novel information about much less abundant forms. In this method demonstration we report over 200 H3.2 forms and 70 H4 forms, including forms not yet detected in human cells, such as the remarkably highly modified histone H3.2 K4me3K9acK14acK18acK23acK27acK36me3. Such detail provided by our proteomics platform will be essential for determining how histone modifications occur and act in combination to propagate the histone code during transcriptional events and could greatly enable sequencing of the histone component of human epigenomes.
Eukaryotic nuclear DNA is nominally compacted into chromatin fibers by use of nucleosomes consisting of a 146-bp section of DNA wrapped around a core of histone proteins (1). Dynamic post-translational modifications (PTMs)1 of the histones, primarily in the accessible N-terminal region or histone “tail,” are an important but not fully understood component of dynamic gene regulation, epigenetic inheritance of cellular memory, genomic stability, and other nuclear mechanisms (2–7). An overwhelming number of studies point to the existence of a histone code of biological logic written on these proteins through these PTMs that are read by a diverse array of “effector” proteins leading to distinct biological events (3). Many single PTM sites on various histone proteins have been decidedly linked to specific physiological processes, such as histone H3 Lys-9 trimethylation (H3K9me3), which is associated with heterochromatin formation (one mode of gene silencing). Nevertheless what effect multiple modifications occurring in combination may have on modulating the histone code signal remains to be determined. Significant progress has been made toward understanding histone modifications using antibody-based histone modification detection methods and by bottom up mass spectrometry (4–6). However, these efforts are fundamentally incapable of maintaining the connectivity between sites of modification over long amino acid sequences and thus do not provide information on how these modifications occur and function in concert. There are, however, several lines of recent evidence that indicate the biological significance of the combinatorial aspects of the histone code (2, 7, 8), thus prompting research into the sequence analysis of long range histone PTM patterns.
The technologies capable of determining such long range patterns of PTMs, electron capture dissociation (ECD) (9) and electron transfer dissociation (ETD) (10) MS, are still relatively new. These have enabled top and middle down gas phase sequencing for combinatorial histone PTM analysis. For example, Kelleher and co-workers (11–14) have published several studies detailing the analysis of all core histones using ECD on a high resolution Fourier transform mass spectrometer. As histones H2B and H2A are modestly modified and histone H4 has limited complexity in comparison with histone H3, fairly thorough analysis of these proteins could be accomplished by a pure top down approach. However, the analysis of histone H3 has proven to be a significantly more difficult analytical problem and has only resulted in a limited survey by a sole top down approach (15). Generally ECD analysis of histones requires large amounts of fairly pure sample and potentially long instrument acquisition times (several minutes to hours) to produce a single useful ECD spectrum given the sample complexity. The sensitivity of bottom up analyses has revealed more diverse PTMs on H2A, H2B, and H4 than top down approaches have revealed (5, 16–19). ETD experiments have been shown previously to be compatible with on-line chromatography methods and have limits of detection and dynamic ranges similar to those of bottom up MS. Therefore, ETD analysis of histones would seem to be a better fit for top or middle down MS, and thus improved methods for top or middle down analysis of histones remain a priority. In support, ETD has been used recently by a few groups to sequence histone proteins and peptides (20, 21). However, as all of these on-line analyses have been performed using standard reverse phase (RP) HPLC, only limited analyses or analyses of the less complicated histones, H2A or H4, have been performed due in part to low chromatographic resolution.
The quality of any LC-MS analysis, as measured by dynamic range, sensitivity, and specificity, is highly dependent on the quality of the chromatography. This becomes critical in the case of modified histone peptides where the sample is a complex mixture of a wide concentration range of large peptides with identical amino acid sequences modified in slightly different ways, resulting in many isobaric structural isomers. Separation of these physically similar modified histone forms (especially the highly modified histone H3) by any method has proved difficult and non-routine (22). Chromatographic methods traditionally used in proteomics analyses (RP-HPLC) achieve only marginal separation of large histone modified peptides resulting in complicated middle down MS analyses of highly mixed precursor ion tandem mass spectra (several isomeric but uniquely modified species fragmented at once), highlighting the need for chromatographic resolution of histone forms prior to mass spectrometric interrogation (20, 23). Off-line chromatographic separations using weak cation exchange mechanisms have been demonstrated recently to do a reasonably good job of fractionating out the differently modified PTM forms (24–27). These methods, however, rely on non-volatile mobile phase additives that render them inadaptable to an on-line LC-MS method. As a consequence, each of the many resulting LC fractions from the up-front separation have to be further purified and separately analyzed by MS afterward. Although such methods served as excellent discovery platforms, this process is extremely time-consuming, leads to sample loss, and inherently reduces the chromatographic resolution prohibiting extensive studies of the relevance and dynamics of the modified forms discovered (24, 27).
Here we present the first on-line nanoflow weak cation exchange hydrophilic interaction liquid chromatography (WCX-HILIC) LC-MS/MS analysis method for the high throughput characterization of complex mixtures of hypermodified combinatorial histone codes. The chromatographic separation is performed on a WCX-HILIC PolyCAT A stationary phase (polyaspartic acid); however, our mechanism of elution is different from that reported previously (27). The ionic strength gradient (i.e. salt elution) used by off-line methods has been replaced with a pH gradient that protonates the stationary phase to remove the cation exchange interaction. This change in elution strategy leads to a similar chromatographic profile as an ionic strength gradient but renders the method “mass spectrometry-friendly” and results in dramatically improved analysis time, throughput, sample consumption, and dynamic range. Whereas previous methods required 50–100 h of manual MS data acquisition time and over 100 μg of sample to systematically characterize a single histone extract (20, 24, 27), the method presented here can achieve this with less than 1 μg of sample in as little as a couple hours with an overall improvement in data quality. Because of the improved chromatographic resolution and the inherent concentration of minor forms at the point of ionization of an on-line nanoflow LC-MS method, our dynamic range and limits of detection are significantly improved. Furthermore the selectivity of the chromatography means that isobaric modifications, most importantly trimethylation and acetylation, can be confidently distinguished and assigned by supplementing the ETD MS/MS with retention time data. Although this is the first work to distinguish between acetylation and trimethylation in such a manner, it should not be surprising that a modification that removes a positive charge can be resolved from a modification that permanently fixes a positive charge by cation exchange mechanisms. Thus, high resolution mass spectrometry as used previously (12–15, 20, 21, 25, 27) is not a strict requirement. We demonstrate using histones H3.2 and H4 from butyrate-treated HeLa cells (butyrate is a deacetylase inhibitor, and this results in a wide range and more complex mixture of potential forms ideal for methodological testing) that our method achieves a high quality comprehensive characterization of combinatorial histone codes using our nanoflow LC method in combination with ETD on a widely available ion trap instrument.
EXPERIMENTAL PROCEDURES
Sample Preparation
HeLa S3 cells were grown and harvested as described previously (15). In some instances, cells were treated with 10 mm sodium butyrate overnight to increase histone acetylation. After nuclei isolation, histones were acid-extracted according to standard protocols (28). Histones were then separated by RP-HPLC into the constituent family members (H2A, H2B, H3.1, H3.2, H3.3, H4, and H1) on a 4.6 × 250-mm C8 column (Grace Davidson, Deerfield, IL) using a System Gold (Beckman Coulter, Fullerton, CA) HPLC instrument to deliver a gradient at 0.8 ml/min from 30% B to 60% B in 100 min (A, 5% acetonitrile and 0.2% TFA; B, 95% acetonitrile and 0.18% TFA). Histone H3.2 was selected and diluted in 100 mm ammonium acetate (pH = 4) and digested with Glu-C protease (Roche Applied Science) at a protein:enzyme ratio of 10:1 for 5 h at room temperature after which the reaction was quenched by freezing at −80 °C. The resulting 1–50-amino acid peptide of the H3.2 histone protein was then further RP-HPLC-purified as described before (5) (1% B/min gradient, same solvents and solvent system as above except a 2.1 × 250-mm column and 0.2 ml/min flow rate was used). Histone H4 was enzymatically digested using Asp-N (Roche Applied Science) (5:1 ratio, 100 mm ammonium bicarbonate (pH = 8.0) for 6 h at 37 °C). The resulting 1–23-amino acid peptide of histone H4 was purified by solid phase extraction using a C18 stop and go extraction tip (29) by loading in 0.1% acetic acid and eluting in 50% MeCN in 0.1% acetic acid. Eluted protein was evaporated to near dryness and diluted into the HILIC A mobile phase before loading on capillary HILIC columns.
Chromatography and Mass Spectrometry
A P2000 laser tip puller (Sutter Instruments, Novato, CA) was used to pull a 75-μm-inner diameter × 360-μm-outer diameter fused silica capillary to a tip. This tip was packed with 3-μm-diameter, 300-Å-pore size, PolyCAT A resin (PolyLC, Columbia, MD) to a length of ∼10 cm forming an integrated chromatography column and nanospray ionization emitter. Approximately 0.5–1.0 μg of histone was pressure bomb-loaded onto the capillary HILIC column at about 5 μl/min. Using an Agilent 1200 HPLC pump (Agilent Technologies, Santa Clara, CA) with a precolumn flow split giving ∼100–200 nl/min flow rate to the column, the histone modified forms were eluted from the column by a linear gradient (solvent A was 75% acetonitrile (Mallinckrodt-Baker, Inc., Phillipsburg, NJ), 20 mm propionic acid (Fluka puriss. pro analysis, ≥99.5% (gas chromatography); Sigma-Aldrich), adjusted to pH 6.0 using ammonium hydroxide (ACS reagent grade, Sigma-Aldrich), and solvent B was 25% acetonitrile adjusted to pH 2.5 with formic acid). Several other buffer systems for the B mobile phase were tried during method development as noted under “Results.” The column eluent was introduced into an LTQ-ETD ion trap mass spectrometer (Thermo Scientific, Waltham, MA) or an LTQ-Orbitrap XL (Thermo Scientific) (data in Fig. 1 only) by nanoelectrospray ionization. Every cycle a full mass spectrum was acquired from 300 to 2000 m/z followed by narrower mass range full mass spectrum to select a given charge state of the histone for data-dependent selection. Five to ten data-dependent tandem MS (MS2) ETD normal rate scans with three microscans each were acquired for each cycle based on the single charge state scan with a minimum signal threshold of 40,000, an automatic gain control target value of 3e4 for MS2, maximum analyte ion injection time of 100 ms, an isolation width of 1.5 m/z, a reagent ion injection automatic gain control value of 1e6, and a reaction time of 80 ms. This results in a cycle time of about 4–9 s for a top 10-based analysis. Tandem mass spectra obtained from all experiments were analyzed using in house-developed software (30) and manually validated. Briefly an in-house mixed integer linear optimization (MILP) computational framework is utilized for the identification and quantification of the modified histone forms using the LC-MS and ETD tandem mass spectrometry data. A first MILP model enumerates the entire set of post-translational modifications that are consistent with the experimentally observed precursor mass. Given this set of modified histone forms, an MILP superposition problem is then solved to determine the relative fractions of the modified forms that are present in the mixed ETD tandem mass spectrum. When viewed in the m/z versus retention time plane, the corresponding annotations reveal a complete separation of the modified forms with respect to the number and position of acetylation modifications. The algorithm utilizes this chromatographic information to resolve ambiguously assigned acetylation modifications in the predictions and to infer the modification states of partially assigned spectra.
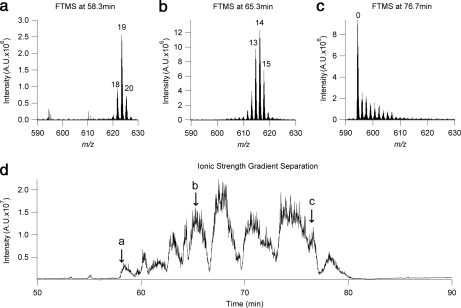
Fig. 1.
A complex mixture of histone H3.2-(1–50) modified forms derived from butyrate-treated HeLa cells separated by on-line WCX-HILIC chromatography using an ammonium acetate-based ionic strength gradient from A (75% ACN, 20 mm propionic acid adjusted to pH 6.0) to B (25% ACN, 500 mm ammonium acetate adjusted to pH 6.0 using ammonium hydroxide) at a rate of 1% B/min. a, b, and c, full MS at 58.3 min, 65.3 min, 76.7 min, respectively; d, total ion chromatogram of the separation. Labels a, b, and c indicate the retention time of the spectra in Fig. 1 a, b, and c, respectively. A.U., arbitrary units
RESULTS
Histone H3 Analysis
Our initial efforts to develop an effective on-line LC-MS method for the analysis of complex mixtures of modified histone forms involved an ionic strength gradient from the “A” mobile phase as described under “Experimental Procedures” above to a “B” mobile phase of 500 mm ammonium formate, 25% acetonitrile adjusted to pH 6.0 with ammonium hydroxide at a gradient velocity of 1% B/min. Butyrate-treated histone H3.2-(1–50) was analyzed. This approach succeeded in demonstrating the possibility of developing an on-line WCX-HILIC method for the analysis of histone modified forms and served as a basis for further development. As can be seen in Fig. 1, an effective separation was achieved, and many histone modified forms were resolved similarly to previous off-line HILIC-based methods. In general, the pattern of elution appears to be consistent with a separation primarily affected by acetylation state but is also influenced by other PTM differences between histone forms. In Fig. 1, a, b, and c, full mass spectra of the 19-, 14-, and 0-methyl equivalent forms (624.1, 616.3, and 594.5 m/z, respectively) are shown. Although this initial effort was a remarkable improvement in many ways by achieving on-line separation and analysis, several drawbacks of such a method quickly became apparent. Maintaining a stable and robust nanospray at such high salt concentrations was elusive, and thus the analysis generally required constant supervision and analyst intervention to achieve complete data sets. It also became apparent, not surprisingly, that such high salt concentrations were having dramatic ionization suppression effects and medium term deleterious effects on the mass spectrometer, necessitating frequent source cleaning.
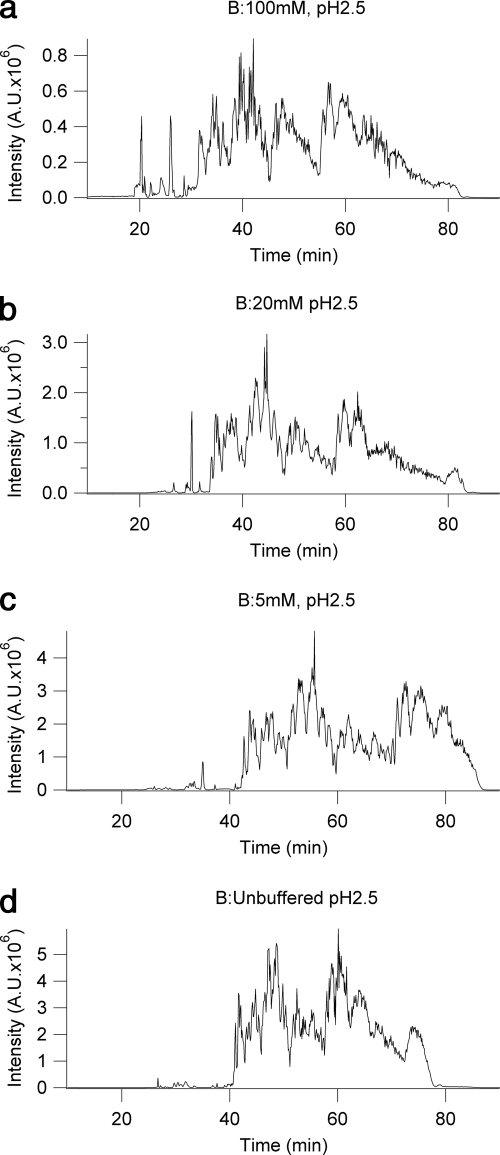
Because of the inherent drawbacks of the high salt concentrations necessary to an ionic strength gradient in conjunction with mass spectrometry, efforts to reduce the ammonium formate concentration were undertaken. Using otherwise identical conditions, several modified B mobile phases with pH adjusted to 2.5 with formic acid and ammonium formate concentrations of 100, 20, 5, and 0 mm were tested, and the results were compared to optimize the efficacy of the method. Butyrate-treated histone H3.2-(1–50) was used, and each of these analyses used the same quantity of the same sample to allow for valid comparison between analyses. By supplementing our ionic strength and organic solvent gradient with a pH gradient we found it possible to significantly reduce the salt concentration necessary to elute the modified histone forms. The experiments performed to optimize the B mobile phase shown in Fig. 2 resulted in the selection of the unbuffered pH gradient as seen in Fig. 2d. The unbuffered gradient appeared to have similar or slightly better resolution of analytes than the other experiments and exhibited the best ionization characteristics. The 100 mm combined ionic strength and pH gradient suffered significant ionization suppression effects. This effect was lessened on the 20 and the 5 mm gradients; these results point to a general trend toward better signal at lower ionic strength. Although the time over which the analytes elute is longer with greater added ionic strength, this likely results from effects on the effective gradient strength that are easily compensated for by reducing the gradient velocity.
Fig. 2.
A complex mixture of histone H3.2-(1–50) modified forms derived from butyrate-treated HeLa cells separated by on-line pH gradient WCX-HILIC chromatography using various concentrations of ammonium acetate in the B mobile phase: 100 mm ammonium formate (a), 20 mm ammonium formate (b), 5 mm ammonium formate (c), and unbuffered, ∼0.5% formic acid (d). In all cases A consisted of 75% ACN and 20 mm propionic acid adjusted to pH 6.0, and B contained 25% ACN and was adjusted to pH 2.5 using formic acid. A gradient of 1% B/min was used. A.U., arbitrary units.
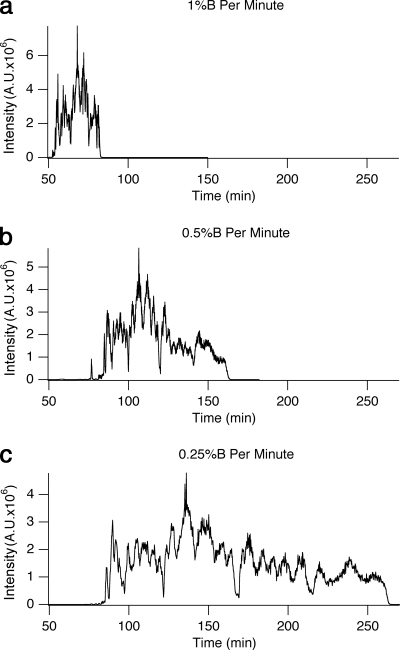
After deciding on an unbuffered formic acid pH gradient using the 0 mm ammonium formate, pH 2.5, 25% acetonitrile as the B mobile phase, we investigated the effects of gradient rate by analyzing H3.2-(1–50) at gradient velocities of 1, 0.5, and 0.25% B/min. The various gradient velocities tried resulted in nearly proportional scaling of elution profiles as shown in Fig. 3. Although the signal intensity is slightly reduced for any one spectrum when the gradient is elongated, significantly more spectra may be acquired and averaged. There was improved resolution of modified forms in the longer elution profiles. In our judgment, the trade-off between analysis time and data quality at different gradient velocities is fairly linear and should thus be adapted to each given purpose. Thus, the elution strategy and gradient presented in Fig. 3c seem to provide a near maximum chromatographic separation and ionization with the option of shortening the gradient for higher throughput as needed as in Fig. 3, a and b.
Fig. 3.
A complex mixture of histone H3.2-(1–50) modified forms derived from butyrate-treated HeLa cells separated by on-line pH gradient WCX-HILIC chromatography using various gradient velocities. In all cases A consisted of 75% ACN, 20 mm propionic acid adjusted to pH 6.0, and B was 25% ACN, ∼0.5% formic acid (pH 2.5). a, 1% B/min; b, 0.5% B/min; c, 0.25% B/min. A.U., arbitrary units.
Fig. 4 shows the analysis of histone H3.2 with several masses and times selected and the PTM combinations determined. Fig. 4a is the base peak chromatogram of the analysis showing the regions separated by degree of acetylation. Fig. 4, b–d, shows the MS1 from which data-dependent MS2 scans are selected. Fig. 4, e–g, shows the annotated ETD MS2 scans selected from Fig. 4, b–d. Of particular interest and indicative of the utility of the method is the analysis of the minor 21-methyl equivalent component selected from Fig. 4b and analyzed in Fig. 4e that can unambiguously be characterized, using both ETD MS2 and retention time, as histone H3 K4me3K9acK14acK18acK23acK27acK36me3, which is modified to full occupancy at each lysine except K37, which has never been reported modified in any other work to date (5). In this case the distinction between trimethylations and acetylations is trivial as five acetyls are required chromatographically, and there are only five known sites of acetylation possible. This assignment is made even more apparent by the close elution of K4me2- and K36me2-containing homologous forms and the absence of K9me2 or K27me2 forms (or even me1 or unmodified forms) in the chromatographic region.
Fig. 4.
An analysis of the combinatorial histone codes of histone H3.2 from butyrate-treated HeLa cells. a, a base peak chromatogram of the WCX-HILIC separation. b, c, and d, full MS at 86, 161, and 197 min, respectively, showing relatively few co-eluting forms. The peaks labeled with the number of methyl equivalences necessary to account for the mass shift were selected for tandem mass spectrometry using electron transfer dissociation shown in e–g. e, ETD spectrum of the 627.3 m/z or 21-methyl equivalent precursor ion at 86 min. f, ETD spectrum of the 608.6 m/z precursor ion at 162 min. g, ETD spectrum of the 605.5 m/z precursor ion at 196 min. A.U., arbitrary units.
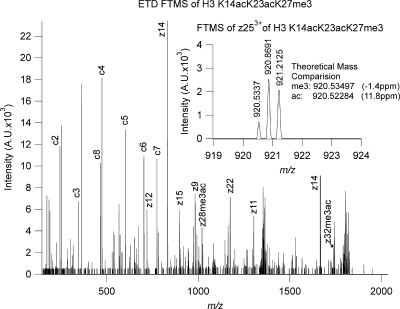
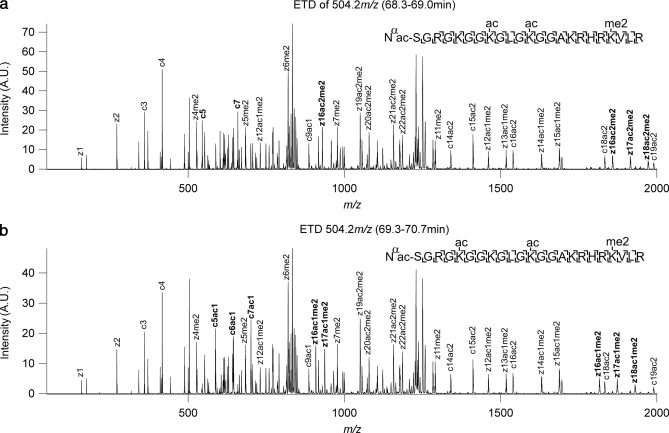
The evidence for chromatographic resolution of trimethyl from acetyl species is strong based on the physical separation mechanisms (cation exchange of differentially charged species; trimethyl is a fixed charge, and acetyl is mostly uncharged) and the empirical observation of trends (consistent apparent degree of acetylation within chromatographic regions; apparent acetylated species eluting far from analogous me2, me1, and me0 forms; apparent trimethyl species eluting near and in a consistent order with respect to analogous me2, me1, and me0 forms; and even consistent regions of mass-retention time space that distinguish MS2 unambiguously assigned acetyl positional isomers). However, to further validate our use of chromatographic information in conjunction with ETD MS2 to distinguish trimethylations from acetylations we performed a parallel experiment on an LTQ-Orbitrap equipped with ETD. Shown in Fig. 5 is the ETD MS2 data analyzed in the high mass accuracy Orbitrap FTMS analyzer of the species analyzed in Fig. 4f (K14acK23acK27me3). The mass error analysis (−1.4 ppm for me3 compared with 11.8 ppm for ac) is based on external calibration and only improves when internal calibration based on the known unmodified fragment ions is used. Although acquisition of such Orbitrap-ETD data on the time scale necessary for the method is not routine or particularly sensitive, the accurate mass data further validate our results, and many of the lysine residue assignments (the only residue both trimethylated and acetylated) have been validated by such means.
Fig. 5.
The ETD MS2 spectrum of the K14acK23acK27me3 species from Fig. 4f analyzed in a parallel experiment on an LTQ-Orbitrap equipped with ETD fragmentation. The inset accurate mass error analysis of the z25 ion (−1.4 ppm for me3 versus 11.8 ppm for ac) indicates that the K27me3 is properly assigned based on chromatographic information in combination with low resolution mass spectral information. A.U., arbitrary units.
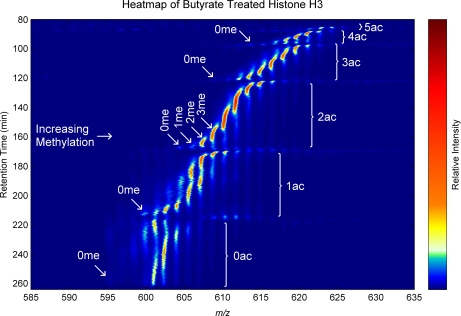
Presented in Fig. 6 is a liquid chromatography-mass spectrometry heat map of the MS1 data from the same analysis as Fig. 4. As mentioned above the histone codes separate primarily by degree of acetylation, secondarily by the specific acetylation isomeric state or theme, and then by methylation state and location. At any given nominal mass or methyl equivalent, several peaks are resolved and can be queried free of what otherwise would be isobaric interferences (see Fig. 7). However, on even more detailed analysis, several partially resolved peaks frequently make up each apparent peak elution at a given mass, often stepping through variations of methyl placements on a given acetylation theme (see Fig. 8). Using this approach, we comprehensively identified and relatively quantified over 200 modified histone H3.2 forms (see Table I) in a single LC-MS/MS analysis.
Fig. 6.
A mass spectral heat map of the on-line pH gradient WCX-HILIC LC-MS separation of histone H3.2-(1–50) modified forms derived from butyrate-treated HeLa cells shown in Fig. 4 as a chromatogram. There are six major groups of ions that represent different degrees of acetylation. At the top right of the figure in a narrow band around 85 min and 618–627 m/z is the five-acetyl group. At the lower left in a broad band from ∼215 to 260 min and 594 to 610 m/z is the unacetylated group. The degree of acetylation decrements with increasing retention time. Within each of these are several different isobaric methyl equivalent groups. Each methyl equivalent group often contains multiple structural isomers that are revealed in the tandem MS and usually have distinct retention times, yet there is a complex mixture of isomers that elute closely enough to appear approximately as a continuous signal in the first dimension MS.
Fig. 7.
A complex mixture of histone H3.2-(1–50) modified forms derived from butyrate-treated HeLa cells separated by on-line WCX-HILIC with an A mobile phase consisting of 75% ACN, 20 mm propionic acid adjusted to pH 6.0 and B consisting of 25% ACN, ∼0.5% formic acid (pH 2.5) using a 0.25% B/min gradient. a, a SIC of the ninth charge state of the 10-methyl equivalent state at ∼610 m/z. b, an ETD tandem MS spectrum of the major 10-methyl equivalent form eluting at 147 min showing it to be K9me2K14acK23acK27me2. c, an ETD tandem MS spectrum of the second most abundant 10-methyl equivalent form eluting at 170 min showing it to be K9me3K14acK27me2K36me2. A.U., arbitrary units.
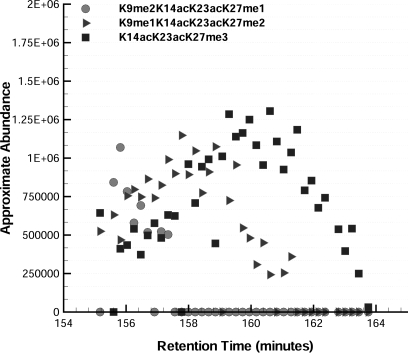
Fig. 8.
Three isomeric H3.2 histone codes eluting in close proximity but with distinct retention times: K9me2K14acK23acK27me1 at ∼156 min, K9me1K14acK23acK27me2 at ∼158 min, and K14acK23acK27me2 at ∼160 min. The plot is derived from the consensus of multiple ions as to the ratio of the ETD spectrum that each form represents multiplied by the precursor ion intensity. This demonstrates partial chromatographic resolution of very closely related isomers that shows that these forms are distinct species and improves confidence of assignment through retention time correlation of structural changes.
Table I. A listing of all of the combinatorial histone codes found and further validated in the butyrate-treated histone H3.2 analysis shown in Figs. 4–7.
The annotated form is shown in column 1. An approximate percent abundance (Approx. percent abund.) is shown in column 2. This value is based on the sum of all precursor ion intensities that exhibit each histone code multiplied by an approximation of the fraction of the resulting ETD spectrum that the histone code represents based on the ratios of fragment ions. This information is presented for qualitative purposes only; however, we expect to be able to use such an approach in semiquantitative analyses with further validation. “BQL” (below quantitation limit) indicates that the histone code was detected, but information is insufficient for approximation of abundance. The m/z of the precursor ion is given in column 3, and in column 4 the number of “methyl equivalents” with which the m/z value correlates is given where an acetylation counts as three methyl equivalents. In column 5 the retention time of the histone code is given.
| Histone code | Approx. percent abund. | m/z | Methyl eq | Retention time |
|---|---|---|---|---|
| min | ||||
| K9me2K14acK23acK27me3 | 9.0 | 611.6 | 11 | 148 |
| K9me2K14acK23acK27me2 | 6.4 | 610.1 | 10 | 149 |
| K9me1K14acK23acK27me3 | 4.6 | 610.1 | 10 | 150 |
| K9me3K27me2 | 4.4 | 602.3 | 5 | 239 |
| K14acK23acK27me3 | 3.2 | 608.5 | 9 | 161 |
| K9me3K14acK23acK27me2 | 3.1 | 611.6 | 11 | 143 |
| K9me2K27me3 | 3.1 | 602.3 | 5 | 240 |
| K9me3K27me1 | 3.1 | 600.7 | 4 | 250 |
| K9me2K23acK27me3 | 3.0 | 607.0 | 8 | 186 |
| K9me2K27me2 | 2.8 | 600.7 | 4 | 250 |
| K9me2K23acK27me2 | 2.7 | 605.4 | 7 | 196 |
| K9me2K14acK23acK27me2K36me2 | 2.4 | 613.2 | 12 | 128 |
| K9me3K14acK27me2 | 2.2 | 607.0 | 8 | 179 |
| K9me2K14acK18acK23acK27me3 | 2.2 | 616.3 | 14 | 112 |
| K9me3K23acK27me1 | 1.9 | 605.4 | 7 | 195 |
| K9me2K14acK18acK23acK27me2K36me2 | 1.9 | 617.9 | 15 | 105 |
| K9me1K14acK23acK27me2 | 1.7 | 608.5 | 9 | 159 |
| K9me3K14acK23acK27me1 | 1.7 | 610.1 | 10 | 147 |
| K23acK27me2 | 1.6 | 602.3 | 5 | 210 |
| K9me2K14acK27me3 | 1.6 | 607.0 | 8 | 179 |
| K4me1K9acK14acK18acK23acK27me2K36me2 | 1.5 | 621.0 | 17 | 90 |
| K14acK23acK27me2 | 1.4 | 607.0 | 8 | 166 |
| K9me1K14acK18acK23acK27me3 | 1.3 | 614.7 | 13 | 116 |
| K9me2K14acK18acK23acK27me2 | 1.2 | 614.7 | 13 | 116 |
| K14acK18acK23acK27me3 | 1.2 | 613.2 | 12 | 124 |
| K9me1K23acK27me3 | 1.2 | 605.4 | 7 | 198 |
| K9me3K23acK27me2 | 1.2 | 607.0 | 8 | 189 |
| K9me2K27me2K36me1 | 1.1 | 602.3 | 5 | 236 |
| K9me2K14acK18acK23acK27me1K36me2 | 1.1 | 616.3 | 14 | 113 |
| K4me1K9me2K14acK18acK23acK27me2K36me2 | 1.0 | 619.4 | 16 | 98 |
| K9me3K14acK18acK23acK27me2K36me2 | 0.98 | 619.4 | 16 | 102 |
| K9me3K14acK23acK36me2 | 0.85 | 611.6 | 11 | 160 |
| K9me3K14acK23acK27me3 | 0.75 | 613.2 | 12 | 131 |
| K9me2K14acK23acK27me1 | 0.71 | 608.5 | 9 | 157 |
| K9me1K14acK18acK23acK27me2K36me2 | 0.70 | 616.3 | 14 | 110 |
| K9me1K23acK27me2 | 0.68 | 603.8 | 6 | 206 |
| K9me1K14acK23acK27me3K36me1 | 0.67 | 611.6 | 11 | 152 |
| K9me3K14acK23acK27me2K36me2 | 0.65 | 614.7 | 13 | 123 |
| K4me1K9acK14acK18acK23acK27me1K36me2 | 0.59 | 619.4 | 16 | 92 |
| K9me2K23acK27me1 | 0.58 | 603.8 | 6 | 205 |
| K9me3K27me2K36me1 | 0.58 | 603.8 | 6 | 225 |
| K9me3K14acK23acK27me2K36me1 | 0.55 | 613.2 | 12 | 127 |
| K9me3K14acK18acK23acK27me2K36me2 | 0.53 | 619.4 | 16 | 98 |
| K9me3K27me1K36me1 | 0.52 | 602.3 | 5 | 234 |
| K9me2K27me2K36me2 | 0.51 | 603.8 | 6 | 225 |
| K9acK14acK18acK23acK27me2K36me2 | 0.51 | 619.4 | 16 | 93 |
| K9me2K14acK27me2K36me2 | 0.50 | 608.5 | 9 | 173 |
| K9me3K14acK27me2K36me2 | 0.49 | 610.1 | 10 | 171 |
| K9me1K14acK18acK23acK27me3K36me1 | 0.48 | 616.3 | 14 | 109 |
| K9me3K14acK18acK23acK27me1K36me2 | 0.44 | 617.9 | 15 | 104 |
| K4me1K9acK14acK18acK23acK27me3 | 0.39 | 619.4 | 16 | 93 |
| min | ||||
| K9me3K14acK27me2K36me1 | 0.39 | 608.5 | 9 | 172 |
| K9me3K14acK36me2 | 0.38 | 607.0 | 8 | 181 |
| K9me2K27me3K36me1 | 0.37 | 603.8 | 6 | 227 |
| K9me1K14acK18acK23acK27me3K36me2 | 0.35 | 617.9 | 15 | 104 |
| K4me1K9me3K14acK18acK23acK27me1K36me2 | 0.32 | 619.4 | 16 | 98 |
| K4me1K9me1K14acK23acK27me2K36me3 | 0.31 | 614.7 | 13 | 122 |
| K9me2K14acK27me2 | 0.31 | 605.4 | 7 | 185 |
| K9me2K14acK18acK23acK27me2K36me3 | 0.30 | 619.4 | 16 | 99 |
| K9acK14acK18acK23acK27me2K36me3 | 0.30 | 621.0 | 17 | 90 |
| K9me3K14acK23acK27me1K36me2 | 0.28 | 613.2 | 12 | 128 |
| K9me3K14acK23acK27me1K36me1 | 0.27 | 611.6 | 11 | 133 |
| K14acK23acK27me1 | 0.27 | 605.4 | 7 | 197 |
| K9acK14acK18acK23acK27me3K36me1 | 0.26 | 619.4 | 16 | 92 |
| K9me1K14acK18acK23acK36me2 | 0.24 | 613.2 | 12 | 119 |
| K4me1K9acK14acK18acK23acK27acK36me2 | 0.24 | 622.5 | 18 | 86 |
| K4me1K9me3K14acK18acK23acK27me2K36me2 | 0.24 | 621.0 | 17 | 92 |
| K9me3K14acK18acK23acK27me2K36me1 | 0.23 | 617.9 | 15 | 105 |
| K9me3K27me3 | 0.22 | 603.8 | 6 | 227 |
| K9me2K14acK23acK27me3K36me1 | 0.21 | 613.2 | 12 | 126 |
| K9me2K14acK18acK23acK27acK36me2 | 0.20 | 619.4 | 16 | 95 |
| K9me2K14acK27me2K36me1 | 0.20 | 607.0 | 8 | 183 |
| K9me3K27me1K36me2 | 0.18 | 603.8 | 6 | 226 |
| K9me2K14acK23acK36me1 | 0.17 | 608.5 | 9 | 164 |
| K4me1K9me2K14acK18acK23acK27me2K36me1 | 0.16 | 617.9 | 15 | 105 |
| K9me3K14acK27me3 | 0.16 | 608.5 | 9 | 175 |
| K14acK18acK23acK27me2 | 0.16 | 611.6 | 11 | 121 |
| K4me1K9me2K14acK18acK23acK36me3 | 0.16 | 617.9 | 15 | 104 |
| K9me2K14acK23acK27me1K36me2 | 0.16 | 611.6 | 11 | 133 |
| K4me1K9me2K14acK18acK23acK27me3 | 0.15 | 617.9 | 15 | 105 |
| K9me3K14acK27me1 | 0.15 | 605.4 | 7 | 185 |
| K9me3K23acK36me2 | 0.14 | 607.0 | 8 | 182 |
| K4me2K9me2K14acK18acK23acK27me1K36me3 | 0.14 | 621.0 | 17 | 94 |
| K4me1K9me2K14acK18acK23acK27me3K36me2 | 0.14 | 621.0 | 17 | 94 |
| K9me2K14acK18acK23acK27me1 | 0.14 | 613.2 | 12 | 124 |
| K9me1K14acK18acK23acK27acK36me3 | 0.13 | 619.4 | 16 | 96 |
| K9me2K14acK18acK23acK27me3K36me2 | 0.13 | 619.4 | 16 | 98 |
| K9me2K14acK23acK27me3K36me2 | 0.13 | 614.7 | 13 | 123 |
| K9me3K27me2K36me2 | 0.13 | 605.4 | 7 | 221 |
| K9me1K14acK18acK23acK27me1K36me1 | 0.12 | 613.2 | 12 | 125 |
| K4me1K9acK14acK18acK23acK27me3K36me2 | 0.12 | 622.5 | 18 | 88 |
| K9me3K14acK18acK23acK27me3 | 0.11 | 617.9 | 15 | 107 |
| K9acK14acK18acK23acK27me1K36me3 | 0.11 | 619.4 | 16 | 92 |
| K23acK27me3 | 0.11 | 603.8 | 6 | 207 |
| K4me1K14acK18acK23acK27acK36me3 | 0.10 | 619.4 | 16 | 95 |
| K9me1K14acK18acK23acK27me1K36me2 | 0.10 | 614.7 | 13 | 122 |
| K14acK23acK36me2 | 0.099 | 607.0 | 8 | 178 |
| K4me1K14acK18acK23acK27me3K36me1 | 0.099 | 616.3 | 14 | 107 |
| K9me3K14acK23acK27me3K36me2 | 0.097 | 616.3 | 14 | 123 |
| K9me3K14acK23acK36me3 | 0.096 | 613.2 | 12 | 126 |
| K9me2K14acK27me3K36me1 | 0.093 | 608.5 | 9 | 173 |
| K4me1K9me2K14acK18acK23acK27me2K36me3 | 0.091 | 621.0 | 17 | 95 |
| K9me3K14acK27me1K36me1 | 0.089 | 607.0 | 8 | 184 |
| K4me1K9me1K14acK18acK23acK27me2K36me3 | 0.089 | 619.4 | 16 | 99 |
| K4me1K9me1K14acK18acK23acK27me3 | 0.085 | 616.3 | 14 | 111 |
| K14acK23acK27me3K36me1 | 0.082 | 610.1 | 10 | 155 |
| K9me3K14acK27me1K36me2 | 0.081 | 608.5 | 9 | 173 |
| K9me1K14acK18acK23acK27me2K36me3 | 0.079 | 617.9 | 15 | 105 |
| K14acK23acK27me1K36me1 | 0.078 | 607.0 | 8 | 165 |
| K4me1K9acK14acK18acK23acK27acK36me3 | 0.076 | 624.1 | 19 | 85 |
| min | ||||
| K4me1K9me1K14acK18acK23acK27me3K36me2 | 0.074 | 619.4 | 16 | 100 |
| K9me1K14acK18acK23acK27me2 | 0.069 | 613.2 | 12 | 118 |
| K9me3K14acK27me3K36me2 | 0.067 | 611.6 | 11 | 170 |
| K9me1K14acK18acK23acK27acK36me2 | 0.065 | 617.9 | 15 | 97 |
| K9me2K14acK27me3K36me2 | 0.064 | 610.1 | 10 | 171 |
| K4me1K9me2K14acK18acK23acK27me1K36me2 | 0.062 | 617.9 | 15 | 103 |
| K9me2K14acK18acK23acK36me1 | 0.061 | 613.2 | 12 | 119 |
| K4me2K9me3K14acK18acK23acK27me1K36me3 | 0.061 | 622.5 | 18 | 88 |
| K4me1K9me3K14acK23acK27me1K36me3 | 0.059 | 616.3 | 14 | 123 |
| K9me2K14acK18acK23acK27me3K36me1 | 0.057 | 617.9 | 15 | 107 |
| K9acK14acK18acK23acK27acK36me3 | 0.057 | 622.5 | 18 | 86 |
| K4me1K9acK14acK18acK23acK36me3 | 0.050 | 619.4 | 16 | 93 |
| K14acK23acK27me2K36me1 | 0.049 | 608.5 | 9 | 167 |
| K9me3K14acK18acK23acK27me2 | 0.045 | 616.3 | 14 | 110 |
| K4me1K9me3K14acK18acK23acK27me1K36me3 | 0.043 | 621.0 | 17 | 98 |
| K9me3K14acK18acK23acK27me1K36me3 | 0.042 | 619.4 | 16 | 95 |
| K4me1K9acK14acK18acK23acK27me2K36me3 | 0.040 | 622.5 | 18 | 88 |
| K4me2K9me1K14acK18acK23acK27me3K36me2 | 0.039 | 621.0 | 17 | 94 |
| K14acK23ac | 0.038 | 603.8 | 6 | 216 |
| K4me2K9acK14acK18acK23acK27me2K36me2 | 0.038 | 622.5 | 18 | 87 |
| K4me1K9me1K14acK27me3K36me2 | 0.036 | 610.1 | 10 | 171 |
| K9me1K14acK18acK23acK27me3K36me3 | 0.035 | 619.4 | 16 | 95 |
| K4me2K9acK14acK18acK23acK36me3 | 0.034 | 621.0 | 17 | 89 |
| K9me2K14acK27me2K36me3 | 0.033 | 610.1 | 10 | 171 |
| K4me1K9me1K14acK18acK23acK27me1K36me3 | 0.032 | 617.9 | 15 | 102 |
| K9me3K23acK27me1K36me1 | 0.030 | 607.0 | 8 | 179 |
| K4me1K9me3K14acK18acK23acK36me3 | 0.029 | 619.4 | 16 | 100 |
| K4me1K9me1K14acK18acK23acK27me3K36me1 | 0.029 | 617.9 | 15 | 106 |
| K4me3K9me3K14acK18acK23acK36me3 | 0.028 | 622.5 | 18 | 86 |
| K4me1K9me3K14acK18acK23acK27me2K36me1 | 0.025 | 619.4 | 16 | 100 |
| K9me3K14acK23acK27me2K36me3 | 0.025 | 616.3 | 14 | 122 |
| K4me1K9me2K14acK18acK27me3K36me3 | 0.025 | 617.9 | 15 | 168 |
| K4me3K9me2K14acK18acK23acK27me1K36me3 | 0.024 | 622.5 | 18 | 88 |
| K4me1K9acK14acK18acK23acK27me3K36me1 | 0.024 | 621.0 | 17 | 90 |
| K4me1K9me3K14acK18acK23acK27me2K36me3 | 0.022 | 622.5 | 18 | 98 |
| K4me1K9me1K14acK27me2K36me3 | 0.022 | 610.1 | 10 | 170 |
| K9me3K23acK27me1K36me3 | 0.022 | 610.1 | 10 | 177 |
| K4me3K9me2K14acK18acK23acK27me2K36me1 | 0.022 | 621.0 | 17 | 89 |
| K4me2K9acK14acK18acK23acK27acK36me2 | 0.021 | 624.1 | 19 | 86 |
| K4me1K9me2K14acK23acK27me2K36me2 | 0.021 | 614.7 | 13 | 122 |
| K4me1K9me2K14acK27me2K36me2 | 0.021 | 610.1 | 10 | 172 |
| K4me1K9me2K14acK27me2K36me3 | 0.021 | 611.6 | 11 | 167 |
| K9me3K23acK27me2K36me2 | 0.021 | 610.1 | 10 | 177 |
| K4me2K9me2K14acK18acK23acK27me2K36me2 | 0.021 | 621.0 | 17 | 97 |
| K9me3K14acK18acK23acK36me3 | 0.021 | 617.9 | 15 | 102 |
| K4me1K14acK18acK23acK27me2K36me3 | 0.020 | 617.9 | 15 | 103 |
| K23ac | 0.020 | 599.2 | 3 | 213 |
| K4me2K9acK14acK18acK23acK27me2K36me3 | 0.020 | 624.1 | 19 | 85 |
| K4me1K9me2K14acK18acK23acK27me3K36me3 | 0.018 | 622.5 | 18 | 87 |
| K9me3K27me3K36me1 | 0.017 | 605.4 | 7 | 227 |
| K4me2K9me2K14acK18acK23acK27me2K36me3 | 0.016 | 622.5 | 18 | 88 |
| K9me3K27me3K36me2 | 0.015 | 607.0 | 8 | 220 |
| K4me1K9acK14acK18acK23acK27me2K36me1 | 0.015 | 619.4 | 16 | 93 |
| K4me3K9acK14acK18acK23acK27me1K36me3 | 0.015 | 624.1 | 19 | 85 |
| K4me1K9me1K14acK23acK27me3K36me2 | 0.014 | 614.7 | 13 | 119 |
| K4me1K9me2K14acK23acK27me3K36me1 | 0.013 | 614.7 | 13 | 116 |
| K4me2K9me1K14acK18acK23acK27me2K36me3 | 0.012 | 621.0 | 17 | 91 |
| K9me3K14acK18acK23acK27acK36me1 | 0.012 | 619.4 | 16 | 95 |
| K4me1K9me2K14acK23acK27me3K36me2 | 0.011 | 616.3 | 14 | 120 |
| min | ||||
| K4me1K9me2K14acK27me3K36me2 | 0.011 | 611.6 | 11 | 170 |
| K4me3K9me2K14acK18acK23acK27me2K36me2 | 0.010 | 622.5 | 18 | 86 |
| K4me3K9me2K14acK18acK23acK27acK36me2 | 0.010 | 624.1 | 19 | 85 |
| K9me1K14acK23acK27me1K36me1 | 0.010 | 608.5 | 9 | 166 |
| K9me2K27me3K36me2 | 0.009 | 605.4 | 7 | 221 |
| K4me1K9me3K14acK18acK23acK27ac | 0.009 | 619.4 | 16 | 95 |
| K4me3K9me1K14acK18acK23acK27me2K36me2 | 0.007 | 621.0 | 17 | 88 |
| K4me1K9me2K27me3K36me1 | 0.007 | 605.4 | 7 | 221 |
| K9me1K14acK18acK23acK27me1K36me3 | 0.006 | 616.3 | 14 | 107 |
| K9me3K14acK18acK23acK27me2K36me3 | 0.005 | 621.0 | 17 | 91 |
| K4me1K9me1K27me2K36me3 | 0.004 | 605.4 | 7 | 221 |
| K4me2K9me2K14acK18acK23acK27me3K36me2 | 0.004 | 622.5 | 18 | 93 |
| K4me1K9me1K27me3K36me2 | 0.004 | 605.4 | 7 | 220 |
| K4me2K9me3K14acK18acK23acK27me2K36me2 | 0.003 | 622.5 | 18 | 92 |
| K9me2K14acK18acK23acK27me2K36me1 | 0.002 | 616.3 | 14 | 113 |
| K4me3K9me2K14acK18acK23acK27me2K36me3 | 0.002 | 624.1 | 19 | 86 |
| K4me1K9me2K27me2K36me3 | 0.002 | 607.0 | 8 | 220 |
| K4me1K9me2K27me2K36me2 | 0.002 | 605.4 | 7 | 220 |
| K4me2K9me2K14acK18acK23acK27me3K36me1 | 0.002 | 621.0 | 17 | 90 |
| K4me1K9me2K14acK18acK23acK27me3K36me1 | 0.002 | 619.4 | 16 | 103 |
| K23acK27me2K36me1 | BQL | 603.8 | 6 | 210 |
| K27me1 | BQL | 596.0 | 1 | 219 |
| K4me1K9me2K14acK23acK27me1K36me2 | BQL | 613.2 | 12 | 123 |
| K4me1K9me3K14acK23acK27me2K36me2 | BQL | 616.3 | 14 | 122 |
| K4me1K9me3K14acK23acK27me2K36me3 | BQL | 617.9 | 15 | 122 |
| K4me2K9me3K14acK18acK23acK27me1 | BQL | 617.9 | 15 | 87 |
| K4me2K9me3K14acK18acK23acK27me3K36me3 | BQL | 625.7 | 20 | 85 |
| K4me3K9me1K14acK18acK23acK27me3K36me2 | BQL | 622.5 | 18 | 88 |
| K4me3K9me2K14acK18acK23acK27me3K36me3 | BQL | 625.7 | 20 | 85 |
| K4me3K9me3K14acK18acK23acK27me3K36me3 | BQL | 627.2 | 21 | 84 |
| K9me2 | BQL | 597.6 | 2 | 256 |
| K9me2K14acK23acK27me2K36me1 | BQL | 611.6 | 11 | 144 |
| K9me2K27me1 | BQL | 599.2 | 3 | 219 |
| K9me2K36me1 | BQL | 599.2 | 3 | 230 |
In general, multiple species were resolved chromatographically at each mass. In the single ion chromatogram (SIC) of the ninth charge state of the 10-methyl equivalent in Fig. 7a, at least five chromatographically distinct peaks are observed. The major 10-methyl form at 147 min is K9me2K14acK23acK27me2 as determined from the ETD spectrum in Fig. 7b. The second most abundant 10-methyl form at 170 min is K9me3K14acK27me2K36me2 as shown in Fig. 7c. The peak at 120 min is clearly distinguished from the four other peaks by being unequivocally unmodified at both K9 and at K36 (data not shown). It can be concluded that the earlier peak at 89 min is K9me2 and K14ac and unmodified at K36; however, further assignment is not possible. Although the signal is relatively low, the ETD evidence for the last peak at 214 min indicates that it has the same nominal mass and indistinguishable ETD spectra on a unit resolution instrument as the major peak at 147 min. Based on the retention time shift to later in the gradient where only K23 monoacetylated species are otherwise observed, the peak at 214 min is likely a minor modified form where an acetylation on K14 has been replaced with a trimethylation. Such apparent pairs of trimethylated and acetylated forms appear throughout the data but most frequently on K9 for which acetylation has been reported previously (31). Further validation of these novel results on a high resolution mass spectrometer is clearly needed; however, the retention time alone seems to be capable of distinguishing between acetylation and trimethylation, and such observations would likely be difficult to impossible to validate even on a high resolution mass spectrometer without using an on-line separation as presented here.
The complexity of histone H3 means that in some cases very closely related histone codes may not fully resolve chromatographically even with our very effective separation method. Fig. 8 plots the elution of three isomeric histone codes eluting in close proximity but with distinct retention times: K9me2K14acK23acK27me1 at ∼156 min, K9me1K14acK23acK27me2 at ∼158 min, and K14acK23acK27me3 at ∼160 min. The plot in Fig. 8 is derived from the consensus of multiple ions as to the ratio of the ETD spectrum that each form represents multiplied by the precursor ion intensity. On a spectrum by spectrum basis, as the initially dominant K9me2-indicative ETD signal decreases relative to the rising K9me1 signal there is a correlated shift from K27me1- to K27me2-indicative ETD signal. This shift then continues with the K9me1 signal shifting to unmethylated K9 signal as the K27me2 signal is overtaken by K27me3-indicative signal. We can observe retention time differences between such species because our method is on line and has sufficient time resolution to observe shifts over time in mixed ETD spectra. The unique characteristic retention times make it unambiguous that there are multiple distinct chemical species. Without this information it is much more likely to misinterpret mixed spectra because of the non-orthogonal set of possible histone codes that can be used to explain a single mixed spectrum. Thus, we can more accurately decouple mixed spectra by use of chromatographically correlated changes and confidently assign the component histone codes. A complete list of the combinatorial histone codes identified and relatively quantitated (30) using the fragment ion relative ratios and precursor ion relative ratios approach (32) and then further validated from the analysis shown in Figs. 4–7 is given in Table I.
Histone H4 Analysis
The histone H4-(1–23) peptide is a significantly less complex sample to analyze because of fewer overall combinations of modifications and better ionization and fragmentation characteristics compared with the histone H3.2-(1–50) peptide. Our method is capable of separating positional isomers, greatly enhancing the capacity to detect and distinguish minor histone modified forms. As seen in Fig. 9a, the H4-(1–23) peptide modified forms separate primarily by degree of acetylation. The major peaks are all K20me2-containing forms, which have varying degrees of acetylation in different positions. Histone H4 has four sites of variable acetylation, and thus there are a limited number of modified forms for each degree of acetylation in a 1:4:6:4:1 (0ac:1ac:2ac:3ac:4ac) distribution. The K20me2 forms dominate, although other methylation states are observed. The four acetylated isomers of the monoacetylated (and dimethylated) form are easily distinguished near 80 min and have been assigned as marked. The most complex diacetyl (triacetyl including the Nα-ac) forms are shown in the SIC in Fig. 9b. These forms appear to group together chromatographically based on the most C-terminally acetylated residue; the most dominant peptide form is Nα-acK12acK16acK20me2. All six of these forms are well resolved. The ETD spectra for the Nα-acK8acK12acK20me2 form (labeled 8ac,12ac in Fig. 9b) and Nα-acK5acK12acK20me2 form (labeled 5ac,12ac in Fig. 9b) are shown in Fig. 10. These very closely related isomers are more than sufficiently resolved to yield spectra that exhibit one dominant form. Although the ETD spectra of the minor K16ac-containing forms shown in Fig. 10b (labeled 8ac,16ac and 5ac,16ac) exhibit some contribution from the earlier eluting Nα-acK12acK16acK20me2 major form they still dominate their respective ETD spectra (data not shown). The four possible triacetylated forms (for K20me2) are also resolved and give relatively pure ETD spectra with the Nα-acK8acK12acK16acK20me2 and Nα-acK5acK12acK16acK20me2 dominant in an ∼4:3 proportion (data not shown).
Fig. 9.
A WCX-HILIC separation of histone H4-(1–23) from HeLa cells treated with butyrate using a 0. 5% B/min gradient. a, the base peak chromatogram from 470 to 530 m/z representing the fifth charge state of histone H4-(1–23). The modified forms separate primarily by the number of acetylations as indicated but also by the number of methylations on K20 and the location of the acetylations. b, a SIC of 612 m/z correlating to the dimethylated and diacetylated forms of histone H4. The dimethylation is on lysine 20, and the two acetylations can be on any of four lysines (K5, K8, K12, and K16) leading to six different positional isomers. These positional isomers are chromatographically separated by the method. The sites of acetylation are labeled on the figure as determined by ETD mass spectrometry. A.U., arbitrary units.
Fig. 10.
ETD tandem mass spectra of two chromatographically resolved diacetylated-dimethylated structural isomers of histone H4 that differ by a shift of an acetyl group by three amino acids. a, the Nα-acK8acK12acK20me2 modified form of histone H4 showing full sequence coverage. The ions labeled in bold text differ from the spectrum below it. b, the Nα-acK5acK12acK20me2 modified form of histone H4 fully sequenced by ETD. The change from K8 acetylation to K5 acetylation is indicated by the bold ions. A.U., arbitrary units.
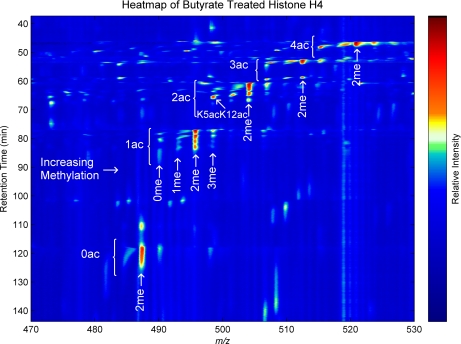
Fig. 11 is an MS heat map overview of the MS1 of the histone H4 separation. Note that certain combinatorial histone codes are clearly more dominant. The most intense peaks at each acetylation group are the K20me2 forms, and within these the acetylations and combinations of acetylations toward the C terminus are generally the most abundant. Interestingly there are strong deviations from these trends such as the relative strength of the unmethylated K5acK12ac form in the lower left of the diacetyl region. Because of the decreased complexity of H4 relative to H3, the heat map is less occupied, and isomers are more clearly resolved.
Fig. 11.
A mass spectral heat map of the on-line pH gradient WCX-HILIC LC-MS separation of histone H4-(1–23) modified forms derived from butyrate-treated HeLa cells. Similar to Fig. 6, the most acetylated (in this case 4ac) forms elute first (38–45 min), and the number of acetylations decrements with retention time.
An analysis of these data, as described above for histone H3, yields 70 H4 combinatorial histone codes (see Table II). Note the predominance of relatively few forms with the top four forms making up more than 50% of the detected histone codes.
Table II. A listing of all of the combinatorial histone codes found and further validated in the butyrate-treated histone H4 analysis shown in Figs. 8–10.
The annotated form is shown in column 1. An approximate percent abundance (Approx. percent abund.) is shown in column 2. This value is based on the sum of all precursor ion intensities that exhibit each histone code multiplied by an approximation of the fraction of the resulting ETD spectrum that the histone code represents based on the ratios of fragment ions. This information is presented for qualitative purposes only; however, we expect to be able to use such an approach in semiquantitative analyses with further validation. The m/z of the precursor ion is given in column 3, and in column 4 the number of methyl equivalents with which the m/z value correlates is shown where an acetylation counts as three methyl equivalents. In column 5 the retention time of the histone code is given.
| Histone code | Approx. percent abund. | m/z | Methyl eq | Retention time |
|---|---|---|---|---|
| min | ||||
| N-acK20me2 | 17.5 | 487.4 | 5 | 102.1 |
| N-acK12acK16acK20me2 | 15.9 | 504.2 | 11 | 62.7 |
| N-acK16acK20me2 | 13.1 | 495.7 | 8 | 78.0 |
| N-acK5acK8acK12acK16acK20me2 | 8.7 | 521.0 | 17 | 42.6 |
| N-acK12acK20me2 | 6.3 | 495.7 | 8 | 80.8 |
| N-acK5acK12acK16acK20me2 | 4.8 | 512.5 | 14 | 52.0 |
| N-acK8acK16acK20me2 | 3.1 | 504.1 | 11 | 64.6 |
| N-acK5acK8acK12acK16ac | 2.8 | 515.4 | 15 | 44.8 |
| N-acK5acK8acK16acK20me2 | 2.3 | 512.5 | 14 | 54.1 |
| N-acK5acK16acK20me2 | 2.3 | 504.2 | 11 | 66.4 |
| N-acK5acK12ac | 2.3 | 498.5 | 9 | 71.4 |
| K5acK8acK12acK16acK20me2 | 2.1 | 512.5 | 14 | 52.1 |
| K5acK8acK16acK20me2 | 1.8 | 504.2 | 11 | 64.1 |
| N-acK8acK20me2 | 1.7 | 495.7 | 8 | 83.2 |
| N-acK8acK12acK16acK20me2 | 1.6 | 512.5 | 14 | 50.3 |
| N-acK5acK20me2 | 1.5 | 495.9 | 8 | 85.2 |
| K8acK12acK16acK20me2 | 1.5 | 504.2 | 11 | 64.6 |
| K5acK16acK20me2 | 1.2 | 495.8 | 8 | 77.9 |
| N-acK8acK12acK20me2 | 1.1 | 504.2 | 11 | 68.9 |
| N-acK5acK12acK20me2 | 1.1 | 504.1 | 11 | 69.9 |
| K16acK20me2 | 1.0 | 487.3 | 5 | 94.4 |
| N-acK5acK8acK12acK16acK20me3 | 0.76 | 524.1 | 18 | 40.7 |
| N-acK5acK8acK12acK20me2 | 0.71 | 512.5 | 14 | 57.8 |
| N-acK5acK12acK16ac | 0.57 | 507.0 | 12 | 53.6 |
| N-acK5acK12acK20me1 | 0.56 | 501.3 | 10 | 70.2 |
| N-acK5acK8acK20me2 | 0.40 | 504.1 | 11 | 72.6 |
| N-acK12acK16acK20me3 | 0.38 | 507.0 | 12 | 62.3 |
| N-acK5acK8acK12ac | 0.37 | 506.9 | 12 | 59.1 |
| K12acK20me2 | 0.26 | 487.4 | 5 | 97.0 |
| N-acK16acK20me3 | 0.23 | 498.7 | 9 | 77.1 |
| N-acK8acK12acK16acK20me3 | 0.21 | 515.7 | 15 | 50.6 |
| K12acK16acK20me2 | 0.20 | 495.8 | 8 | 76.6 |
| N-acK5acK8acK12acK16acK20me1 | 0.19 | 518.4 | 16 | 43.9 |
| N-acK5acK8acK16ac | 0.16 | 506.9 | 12 | 55.7 |
| N-acK5acK12acK16acK20me3 | 0.15 | 515.5 | 15 | 51.0 |
| N-acR3me1K5acK8acK12acK16acK20me2 | 0.12 | 523.8 | 18 | 38.9 |
| K20me2 | 0.11 | 479.1 | 2 | 148.3 |
| N-acR3me2K5acK8acK12acK16acK20me2 | 0.073 | 526.7 | 19 | 39.3 |
| K8acK20me2 | 0.068 | 487.4 | 5 | 97.4 |
| N-acK5acK8acK12acK20me1 | 0.064 | 509.9 | 13 | 58.6 |
| N-acK12acK20me3 | 0.064 | 498.7 | 9 | 80.2 |
| N-acK12ac | 0.058 | 490.2 | 6 | 82.2 |
| N-acK20me3 | 0.053 | 490.3 | 6 | 98.7 |
| K5acK12ac | 0.049 | 490.3 | 6 | 82.5 |
| N-acR3me1K16acK20me2 | 0.046 | 498.7 | 9 | 76.2 |
| K5acK12acK16acK20me2 | 0.039 | 504.2 | 11 | 65.5 |
| N-acR3me1K8acK12acK16acK20me2 | 0.038 | 515.5 | 15 | 49.8 |
| N-acK5acK16ac | 0.036 | 498.6 | 9 | 69.0 |
| N-acK16ac | 0.029 | 490.2 | 6 | 79.3 |
| N-acK5acK12acK16acK20me1 | 0.027 | 510.0 | 13 | 52.7 |
| N-acK5acK8acK16acK20me3 | 0.020 | 515.4 | 15 | 53.5 |
| N-acK5acK8acK16acK20me1 | 0.016 | 509.9 | 13 | 54.8 |
| min | ||||
| N-acR3me1K12acK16acK20me2 | 0.013 | 507.2 | 12 | 60.5 |
| N-acK8acK12ac | 0.011 | 499.0 | 9 | 71.3 |
| K5acK8acK12acK16ac | 0.0097 | 507.4 | 12 | 53.3 |
| N-acK8acK12acK20me1 | 0.0086 | 501.7 | 10 | 70.0 |
| K5acK20me2 | 0.0075 | 487.4 | 5 | 97.8 |
| K5acK12acK16acK20me3 | 0.0054 | 507.1 | 12 | 64.2 |
| N-acR3me1K20me2 | 0.0053 | 490.3 | 6 | 97.8 |
| N-acK5acK16acK20me1 | 0.0047 | 501.6 | 10 | 67.4 |
| N-acR3me1K5acK8acK16acK20me2 | 0.0046 | 515.5 | 15 | 49.8 |
| N-acK12acK20me1 | 0.0043 | 493.2 | 7 | 81.9 |
| N-acK5acK20me1 | 0.0040 | 493.1 | 7 | 85.7 |
| K8acK16acK20me3 | 0.0038 | 498.7 | 9 | 76.4 |
| N-acR3me1K5acK12acK16acK20me2 | 0.0021 | 515.5 | 15 | 49.7 |
| K5acK8acK12acK16acK20me3 | 0.0020 | 515.5 | 15 | 51.7 |
| N-acK8acK16acK20me3 | 0.0017 | 507.1 | 12 | 61.2 |
| K5acK8acK12acK16acK20me1 | 0.0012 | 510.0 | 13 | 52.7 |
| K8acK16acK20me2 | 0.0006 | 496.0 | 8 | 76.2 |
| K5acK16acK20me3 | 0.0003 | 498.1 | 9 | 76.4 |
DISCUSSION
The method presented here provides a means to probe the biological meaning of the combinatorial histone code in a manner not previously accessible on a reasonable time scale. The application of this method directly to an array of biological states from cancer cells to stem cells to different tissue types and organisms will provide massive amounts of important new information about the combinatorial histone code in normal functioning biology and disease. Furthermore the application of this method in combination with molecular biology techniques, which would generally not be possible with less sensitive and lower throughput techniques, will provide a means of probing a very large space of important questions in biology. Just as genomics technologies have accelerated our understanding of biology and human disease, the information provided by the emerging epigenetics technologies should yield similar impact. The complexity of epigenetics makes understanding the mechanisms thereof significantly more difficult (33). Not only are the combinatorial histone codes of a single histone astoundingly complex, there are four histones with multiple variants (34–36) and two copies of each histone family member in each nucleosome, these are spaced across the entire genome, and the specific genomic context of the histone codes are important (37, 38). Additionally there are nucleotide-based epigenetic mechanisms such as DNA methylation (39). The payoff of solving the epigenetics conundrum, however, will be great given that epigenetics is essential to the biology of stem cells (40) and how cells differentiate and fail to maintain the required differentiated states as in cancer (41). Whereas genomics tells us what genes are available for expression and generally their propensity for expression, epigenetics directs the parts in a concerted manner to produce a viable differentiated cell. In fact epigenetics is the mechanism of multicellularity (37). How the histone code functions in concert is largely unknown; however, what is known indicates that how combinations combine, both on a single histone and between different histones within the same nucleosome, is important (8, 40, 42–48). This work is among the first works capable of accessing such information and the first to do so at a scale and throughput that allows for extensive probing of this information. We have high expectations, given the sensitivity and throughput of the method, for combining this method with approaches that probe complementary epigenetic information, such as genomic location, but result in significant fractionation and thus smaller total amounts of combinatorial histone codes to be elucidated.
This work represents a dramatic improvement in most important analytical metrics relative to previous efforts in the middle down analysis of hypermodified peptides in general and histones in particular. The selectivity of the chromatographic separation enables relatively pure tandem MS spectra of structural isomers compared with previous methods. Although mixed spectra are observed in the more complicated cases (i.e. H3), these are easier to deconvolve, and in the simpler cases (i.e. H4) only a few mixed spectra are observed. For comparison, the recent work investigating the combinatorial forms of histone H4 from human embryonic stem cells found that the H4 forms are only partially resolved requiring significant computation to identify forms, and some, particularly the diacetyl forms, were not sufficiently distinguishable to allow quantification (20). As shown in Fig. 9 these same forms are largely resolved by our method, and quantitative information can essentially be read off of the chromatogram.
The capacity of the method to chromatographically resolve trimethylations from acetylations greatly improves the quality of data achieved and confidence in assignments. Spectral complexity is reduced by the resolution of these isobaric species, and the confidence of PTM assignments is improved by the predictable relationship between modification state and relative retention time. The ability of the chromatography to resolve these modifications arises from the differential charge between the acetyl, which is primarily uncharged, and trimethyl, which is a fixed positive charge. This capacity to resolve these modifications has been validated by multiple lines of evidence, including high resolution mass spectrometry; however, the most useful indicator of this distinction is that analogous me0, me1, me2, and me3 species elute closely in numerical or reverse numerical order as seen in Fig. 8, and the acetyl analogs elute much earlier. The ability of this approach to distinguish such isobaric species may be widely applicable to other analyses using low resolution mass spectrometry given enough knowledge of the system to predict retention times or simply by the relative retention times of multiple peaks.
By adapting our method to the capillary nanoflow scale and using on-line ETD, we have dropped the sample requirements from >100 to <1 μg. With the improved chromatographic resolution and the inherent concentration of minor forms at the point of ionization of an on-line nanoflow LC-MS method, our dynamic range is significantly improved. This improvement in dynamic range and limit of detection has resulted in the identification of minor forms not previously reported in human cells (27). In cases where minor forms are not fully separated from more dominant forms our confidence in distinguishing secondary forms from artifacts is enabled by the retention time information provided by on-line LC-MS. Most importantly, improved data quality is achieved while dramatically improving the throughput and sensitivity relative to off-line methods. Throughput is further enhanced by being amenable to automation for replicate and multiple sample analyses, a practice generally necessary for biologically and statistically meaningful results.
Although this work is of a particular focus, the general chromatographic approach may prove useful for similar biological analytes. Recently several examples of other types of apparent combinatorial biological codes have emerged. For example RNA polymerase II has been shown to have a unique pattern of phosphorylation rather than a particular degree of phosphorylation to switch between active and inactive states (49). Interestingly many proteins that are phosphorylated are frequently found to be hyperphosphorylated, allowing for information encoded in combinatorial patterns. Yet these patterns of phosphorylation are not well studied. The high mobility group proteins exhibit modifications similar to histones and are found in association with histones in chromatin (50). Other examples include tubulin (51) and p53 (52). The rate of discovery of such combinatorial codes would seem to indicate that there are significantly more such examples yet to be discovered, and the importance of resolving and analyzing such largely isomeric biological codes will become greater (53).
Very little is known about the variation of the histone code and which modifications occur in combination across different biological states, cell and tissue types, and organisms. This is largely because of the limited throughput and sensitivity of previous approaches. A simple global (whole chromatin) analysis of such samples will provide a wealth of novel biological information and provide a basic set of combinatorial histone codes for further studies. When combined with techniques that themselves probe related biological properties the level of specificity and potential to gain insights into this important aspect of biology are even greater. Techniques such as chromatin immunoprecipitation (ChIP) (54) that probe for specific histone modifications can enrich for subsets of the histone code to be further probed for combinations of PTMs via this method while also providing the potential for genome localization by sequencing the associated DNA (as in ChIP-seq) (55). Such combinations of techniques with middle down MS methods require very good sensitivity because it is an even further subset of the chromatin being analyzed. Previous off-line approaches required large amounts of histones in the starting material to the point that even global analyses were near the limits of practical scalability and thus near impossible for minor fractions thereof. There are also many sample types that are of great interest and do not scale well, making this the first practical method for such analyses. These include cells of limited availability, such as oocytes, and more importantly clinical samples, such as tumors. Of even greater challenge would be the application of selective fractionation, ChIP, in combination with the described method on such inherently limited samples. Such analyses, however, have great scientific potential. For example, knowing the combinatorial histone code of many types of cancer tumors with information about their placement in the genome simultaneously would be truly ground-breaking information that could elucidate key epigenetic aspects of cancer biology (56, 57).
The characterization of histone modified forms is critical to furthering our understanding of fundamental aspects of biology. There is a consensus in the scientific community that there is a need for a deeper understanding of the molecular basis for and cataloguing of epigenetic control of gene expression. This has resulted in the call for a human epigenome project on the scale of the human genome project (41, 58, 59). The histone code, which this method reads the most challenging aspect thereof, is recognized as an important aspect of epigenetics and dynamic gene regulation. With the data quality and throughput achieved with this method, it represents one of the current best platforms for a significant aspect of such an undertaking particularly when combined with the aforementioned ChIP protocols. This method should greatly enable sequencing histone PTM patterns in a potential large scale epigenome project, the outcome of which will have a massive impact on our understanding of chromatin biology, particularly regarding cellular differentiation, stem cell biology, cancer biology, dynamic gene regulation, and epigenetic inheritance of phenotype.
Supplementary Material
Acknowledgments
We thank Andrew Alpert of PolyLC Inc. for technical advice.
Footnotes
* This work was supported by an American Society for Mass Spectrometry research award and by Princeton University. This work was also supported by the National Science Foundation (CBET-0941143).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- PTM
- post-translational modification
- ETD
- electron transfer dissociation
- WCX
- weak cation exchange
- HILIC
- hydrophilic interaction liquid chromatography
- ECD
- electron capture dissociation
- RP
- reverse phase
- LTQ
- linear trap quadrupole
- MILP
- mixed integer linear optimization
- SIC
- single ion chromatogram
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2.Barlési F., Giaccone G., Gallegos-Ruiz M. I., Loundou A., Span S. W., Lefesvre P., Kruyt F. A., Rodriguez J. A. (2007) Global histone modifications predict prognosis of resected non small-cell lung cancer. J. Clin. Oncol 25, 4358–4364 [DOI] [PubMed] [Google Scholar]
- 3.Bönisch C., Nieratschker S. M., Orfanos N. K., Hake S. B. (2008) Chromatin proteomics and epigenetic regulatory circuits. Expert Rev. Proteomics 5, 105–119 [DOI] [PubMed] [Google Scholar]
- 4.Burlingame A. L., Zhang X., Chalkley R. J. (2005) Mass spectrometric analysis of histone posttranslational modifications. Methods 36, 383–394 [DOI] [PubMed] [Google Scholar]
- 5.Garcia B. A., Shabanowitz J., Hunt D. F. (2007) Characterization of histones and their post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol 11, 66–73 [DOI] [PubMed] [Google Scholar]
- 6.Su X., Ren C., Freitas M. A. (2007) Mass spectrometry-based strategies for characterization of histones and their post-translational modifications. Expert Rev. Proteomics 4, 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taverna S. D., Ilin S., Rogers R. S., Tanny J. C., Lavender H., Li H., Baker L., Boyle J., Blair L. P., Chait B. T., Patel D. J., Aitchison J. D., Tackett A. J., Allis C. D. (2006) Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sims J. K., Houston S. I., Magazinnik T., Rice J. C. (2006) A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J. Biol. Chem 281, 12760–12766 [DOI] [PubMed] [Google Scholar]
- 9.Zubarev R. A., Kelleher N. L., McLafferty F. W. (1998) Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc 120, 3265–3266 [Google Scholar]
- 10.Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang L., Smith J. N., Anderson S. L., Ma P., Mizzen C. A., Kelleher N. L. (2007) Global assessment of combinatorial post-translational modification of core histones in yeast using contemporary mass spectrometry. Lys4 trimethylation correlates with degree of acetylation on the same H3 tail. J. Biol. Chem 282, 27923–27934 [DOI] [PubMed] [Google Scholar]
- 12.Boyne M. T., 2nd, Pesavento J. J., Mizzen C. A., Kelleher N. L. (2006) Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J. Proteome Res 5, 248–253 [DOI] [PubMed] [Google Scholar]
- 13.Siuti N., Roth M. J., Mizzen C. A., Kelleher N. L., Pesavento J. J. (2006) Gene-specific characterization of human histone H2B by electron capture dissociation. J. Proteome Res 5, 233–239 [DOI] [PubMed] [Google Scholar]
- 14.Pesavento J. J., Kim Y. B., Taylor G. K., Kelleher N. L. (2004) Shotgun annotation of histone modifications: a new approach for streamlined characterization of proteins by top down mass spectrometry. J. Am. Chem. Soc 126, 3386–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas C. E., Kelleher N. L., Mizzen C. A. (2006) Mass spectrometric characterization of human histone H3: a bird's eye view. J. Proteome Res 5, 240–247 [DOI] [PubMed] [Google Scholar]
- 16.Tsubota T., Berndsen C. E., Erkmann J. A., Smith C. L., Yang L., Freitas M. A., Denu J. M., Kaufman P. D. (2007) Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell 25, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonenfant D., Coulot M., Towbin H., Schindler P., van Oostrum J. (2006) Characterization of histone H2A and H2B variants and their post-translational modifications by mass spectrometry. Mol. Cell. Proteomics 5, 541–552 [DOI] [PubMed] [Google Scholar]
- 18.Garcia B. A., Hake S. B., Diaz R. L., Kauer M., Morris S. A., Recht J., Shabanowitz J., Mishra N., Strahl B. D., Allis C. D., Hunt D. F. (2007) Organismal differences in post-translational modifications in histones H3 and H4. J. Biol. Chem 282, 7641–7655 [DOI] [PubMed] [Google Scholar]
- 19.Zhang K., Tang H., Huang L., Blankenship J. W., Jones P. R., Xiang F., Yau P. M., Burlingame A. L. (2002) Identification of acetylation and methylation sites of histone H3 from chicken erythrocytes by high-accuracy matrix-assisted laser desorption ionization-time-of-flight, matrix-assisted laser desorption ionization-postsource decay, and nanoelectrospray ionization tandem mass spectrometry. Anal. Biochem 306, 259–269 [DOI] [PubMed] [Google Scholar]
- 20.Phanstiel D., Brumbaugh J., Berggren W. T., Conard K., Feng X., Levenstein M. E., McAlister G. C., Thomson J. A., Coon J. J. (2008) Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A 105, 4093–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coon J. J., Ueberheide B., Syka J. E., Dryhurst D. D., Ausio J., Shabanowitz J., Hunt D. F. (2005) Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A 102, 9463–9468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindner H. H. (2008) Analysis of histones, histone variants, and their post-translationally modified forms. Electrophoresis 29, 2516–2532 [DOI] [PubMed] [Google Scholar]
- 23.Bonenfant D., Towbin H., Coulot M., Schindler P., Mueller D. R., van Oostrum J. (2007) Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol. Cell. Proteomics 6, 1917–1932 [DOI] [PubMed] [Google Scholar]
- 24.Taverna S. D., Ueberheide B. M., Liu Y., Tackett A. J., Diaz R. L., Shabanowitz J., Chait B. T., Hunt D. F., Allis C. D. (2007) Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc. Natl. Acad. Sci. U.S.A 104, 2086–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pesavento J. J., Bullock C. R., LeDuc R. D., Mizzen C. A., Kelleher N. L. (2008) Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J. Biol. Chem 283, 14927–14937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarg B., Helliger W., Talasz H., Förg B., Lindner H. H. (2006) Histone H1 phosphorylation occurs site-specifically during interphase and mitosis: identification of a novel phosphorylation site on histone H1. J. Biol. Chem 281, 6573–6580 [DOI] [PubMed] [Google Scholar]
- 27.Garcia B. A., Pesavento J. J., Mizzen C. A., Kelleher N. L. (2007) Pervasive combinatorial modification of histone H3 in human cells. Nat. Methods 4, 487–489 [DOI] [PubMed] [Google Scholar]
- 28.Shechter D., Dormann H. L., Allis C. D., Hake S. B. (2007) Extraction, purification and analysis of histones. Nat. Protoc 2, 1445–1457 [DOI] [PubMed] [Google Scholar]
- 29.Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 30.DiMaggio P. A., Jr., Young N. L., Baliban R. C., Garcia B. A., Floudas C. A. (2009) A mixed-integer linear optimization framework for the identification and quantification of targeted post-translational modifications of highly modified proteins using multiplexed ETD tandem mass spectrometry. Mol. Cell. Proteomics 10.1074/mcp.M900144-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thatcher K. N., LaSalle J. M. (2006) Dynamic changes in Histone H3 lysine 9 acetylation localization patterns during neuronal maturation require MeCP2. Epigenetics 1, 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pesavento J. J., Mizzen C. A., Kelleher N. L. (2006) Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal. Chem 78, 4271–4280 [DOI] [PubMed] [Google Scholar]
- 33.Strahl B. D., Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 34.Loyola A., Bonaldi T., Roche D., Imhof A., Almouzni G. (2006) PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 24, 309–316 [DOI] [PubMed] [Google Scholar]
- 35.Sarma K., Reinberg D. (2005) Histone variants meet their match. Nat. Rev. Mol. Cell Biol 6, 139–149 [DOI] [PubMed] [Google Scholar]
- 36.Allis C. D., Glover C. V., Bowen J. K., Gorovsky M. A. (1980) Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell 20, 609–617 [DOI] [PubMed] [Google Scholar]
- 37.Margueron R., Trojer P., Reinberg D. (2005) The key to development: interpreting the histone code? Curr. Opin. Genet. Dev 15, 163–176 [DOI] [PubMed] [Google Scholar]
- 38.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 39.Fuks F. (2005) DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev 15, 490–495 [DOI] [PubMed] [Google Scholar]
- 40.Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 41.Esteller M. (2006) The necessity of a human epigenome project. Carcinogenesis 27, 1121–1125 [DOI] [PubMed] [Google Scholar]
- 42.Ohm J. E., McGarvey K. M., Yu X., Cheng L., Schuebel K. E., Cope L., Mohammad H. P., Chen W., Daniel V. C., Yu W., Berman D. M., Jenuwein T., Pruitt K., Sharkis S. J., Watkins D. N., Herman J. G., Baylin S. B. (2007) A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet 39, 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraga M. F., Ballestar E., Villar-Garea A., Boix-Chornet M., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K., Iyer N. G., Pérez-Rosado A., Calvo E., Lopez J. A., Cano A., Calasanz M. J., Colomer D., Piris M. A., Ahn N., Imhof A., Caldas C., Jenuwein T., Esteller M. (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet 37, 391–400 [DOI] [PubMed] [Google Scholar]
- 44.Hirota T., Lipp J. J., Toh B. H., Peters J. M. (2005) Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438, 1176–1180 [DOI] [PubMed] [Google Scholar]
- 45.Fischle W., Tseng B. S., Dormann H. L., Ueberheide B. M., Garcia B. A., Shabanowitz J., Hunt D. F., Funabiki H., Allis C. D. (2005) Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 [DOI] [PubMed] [Google Scholar]
- 46.Briggs S. D., Xiao T., Sun Z. W., Caldwell J. A., Shabanowitz J., Hunt D. F., Allis C. D., Strahl B. D. (2002) Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418, 498. [DOI] [PubMed] [Google Scholar]
- 47.Fingerman I. M., Du H. N., Briggs S. D. (2008) Controlling histone methylation via trans-histone pathways. Epigenetics 3, 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trojer P., Li G., Sims R. J., 3rd, Vaquero A., Kalakonda N., Boccuni P., Lee D., Erdjument-Bromage H., Tempst P., Nimer S. D., Wang Y. H., Reinberg D. (2007) L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129, 915–928 [DOI] [PubMed] [Google Scholar]
- 49.Egloff S., Murphy S. (2008) Cracking the RNA polymerase II CTD code. Trends Genet 24, 280–288 [DOI] [PubMed] [Google Scholar]
- 50.Zhang Q., Wang Y. (2008) High mobility group proteins and their post-translational modifications. Biochim. Biophys. Acta 1784, 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhey K. J., Gaertig J. (2007) The tubulin code. Cell Cycle 6, 2152–2160 [DOI] [PubMed] [Google Scholar]
- 52.Appella E., Anderson C. W. (2000) Signaling to p53: breaking the posttranslational modification code. Pathol. Biol 48, 227–245 [PubMed] [Google Scholar]
- 53.Sims R. J., 3rd, Reinberg D. (2008) Is there a code embedded in proteins that is based on post-translational modifications? Nat. Rev. Mol. Cell Biol 9, 815–820 [DOI] [PubMed] [Google Scholar]
- 54.Hebbes T. R., Thorne A. W., Crane-Robinson C. (1988) A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J 7, 1395–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T. K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E. S., Bernstein B. E. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang G. G., Allis C. D., Chi P. (2007) Chromatin remodeling and cancer. Part I: Covalent histone modifications. Trends Mol. Med 13, 363–372 [DOI] [PubMed] [Google Scholar]
- 57.Wang G. G., Allis C. D., Chi P. (2007) Chromatin remodeling and cancer. Part II: ATP-dependent chromatin remodeling. Trends Mol. Med 13, 373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eckhardt F., Beck S., Gut I. G., Berlin K. (2004) Future potential of the Human Epigenome Project. Expert Rev. Mol. Diagn 4, 609–618 [DOI] [PubMed] [Google Scholar]
- 59.Jones P. A., Martienssen R. (2005) A blueprint for a Human Epigenome Project: the AACR Human Epigenome Workshop. Cancer Res 65, 11241–11246 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.