Abstract
The performances of 10 different normalization methods on data of endogenous brain peptides produced with label-free nano-LC-MS were evaluated. Data sets originating from three different species (mouse, rat, and Japanese quail), each consisting of 35–45 individual LC-MS analyses, were used in the study. Each sample set contained both technical and biological replicates, and the LC-MS analyses were performed in a randomized block fashion. Peptides in all three data sets were found to display LC-MS analysis order-dependent bias. Global normalization methods will only to some extent correct this type of bias. Only the novel normalization procedure RegrRun (linear regression followed by analysis order normalization) corrected for this type of bias. The RegrRun procedure performed the best of the normalization methods tested and decreased the median S.D. by 43% on average compared with raw data. This method also produced the smallest fraction of peptides with interblock differences while producing the largest fraction of differentially expressed peaks between treatment groups in all three data sets. Linear regression normalization (Regr) performed second best and decreased median S.D. by 38% on average compared with raw data. All other examined methods reduced median S.D. by 20–30% on average compared with raw data.
Peptidomics is defined as the analysis of the peptide content within an organism, tissue, or cell (1–3). The proteome and peptidome have common features, but there are also prominent differences. Proteomics generally identifies proteins by using the information of biologically inactive peptides derived from tryptic digestion, whereas peptidomics tries to identify endogenous peptides using single peptide sequence information only (4). Endogenous neuropeptides are peptides used for intracellular signaling that can act as neurotransmitters or neuromodulators in the nervous system. These polypeptides of 3–100 amino acids can be abundantly produced in large neural populations or in trace levels from single neurons (5) and are often generated through the cleavage of precursor proteins. However, unwanted peptides can also be created through post-mortem induced proteolysis (6). The later aspect complicates the technical analysis of neuropeptides as post-mortem conditions increase the number of degradation peptides. The possibility to detect, identify, and quantify lowly expressed neuropeptides using label-free LC-MS techniques has improved with the development of new sample preparation techniques including rapid heating of the tissue, which prevents protein degradation and inhibition of post-mortem proteolytic activity (7, 8).
It has been suggested by us (4, 5) and others (9) that comparing the peptidome between samples of e.g. diseased and normal tissue may lead to the discovery of biologically relevant peptides of certain pathological or pharmacological events. However, differences in relative peptide abundance measurements may not only originate from biological differences but also from systematic bias and noise. To reduce the effects of experimentally induced variability it is common to normalize the raw data. This is a concept well known in the area of genomics studies using gene expression microarrays (10–12). As a consequence, many methods developed for microarray data have also been adapted for normalizing peptide data produced with LC-MS techniques (10–16). Normally the underlying assumption for applying these techniques is that the total or mean/median peak abundances should be equal across different experiments, in this case between LC-MS analyses. Global normalization methods refer to cases where all peak abundances are used to determine a single normalization factor between experiments (13, 15, 16), a subset of peaks assumed to be similarly abundant between experiments (16) is used, or spiked-in peptides are used as internal standards. In a study by Callister et al. (14), normalization methods for tryptic LC-FTICR-MS peptide data were compared. The authors concluded that global or iterative linear regression works best in most cases but also recommended that the best procedure should be selected for each data set individually. Methods used for normalizing LC-MS data have been reviewed previously (14, 17, 18), but to our knowledge only Callister et al. (14) have used small data sets to systematically evaluate such methods. None of these studies have targeted data of endogenous peptides.
In this study, the effects of 10 different normalization methods were evaluated on data produced by a nano-LC system coupled to an electrospray Q-TOF or linear trap quadrupole (LTQ)1 mass spectrometer. Normalization methods that originally were developed for gene expression data were used, and one novel method, linear regression followed by analysis order normalization (RegrRun), is presented. The normalization methods were evaluated using three data sets of endogenous brain peptides originating from three different species (mouse, rat, and Japanese quail), each consisting of 35–45 individual LC-MS analyses. Each data set contained both technical and biological replicates.
EXPERIMENTAL PROCEDURES
This study on normalization methods for label-free relative quantification of endogenous peptides from brain tissue was based on data from three different data sets with different experimental end points. The data set named “Mouse” originates from striatal tissue samples of a Parkinsonian model of unilaterally lesioned mice (Mus musculus) with and without l-3,4-dihydroxyphenylalanine treatment (19). The data set named “Rat” originates from nucleus accumbens tissue samples from rats of two different strains (Sprague-Dawley and Wistar) with or without naloxone-precipitated withdrawal during morphine tolerance (20). The data set named “Quail” originates from embryonic diencephalon tissue samples of Japanese quail (Coturnix japonica) from males and females at two different embryonic stages with or without ethinylestradiol exposure.2 More detailed information about the different data sets can be found in supplemental Data 1.
Tissue Extraction and Sample Preparation
The animals were either euthanized by focused microwave irradiation (Mouse), or the dissected frozen tissue samples were denaturized by rapid heating at 95 °C using a heat transfer inactivation instrument (Rat and Quail) (Stabilizor, Denator AB, Gothenburg, Sweden) to minimize post-mortem protein/peptide degradation (7). The tissue samples were extracted as described previously (19, 20).2
Experimental Setup
All three data sets were analyzed in a randomized block design fashion. In the Mouse experimental procedure a reference sample of pooled material (all 25 samples) was analyzed first followed by four samples, one from each treatment group selected by random. The procedure was repeated until all individual samples had been analyzed. The biological samples in block 8–10 were technical replications of the samples in block 5–7 except for a single sample that failed in block 10. In the Rat experimental procedure a reference sample (pool of 10 samples) was analyzed first followed by four samples, one from each treatment group selected by random. The procedure was repeated until all individual samples had been analyzed. In the Quail experimental procedure a sample from each treatment group was selected by random, and a technical replicate of a single sample was analyzed at the end of each block. The procedure was repeated until all individual samples had been analyzed. In the Mouse and Rat data sets each group of four to five samples (six in the last block in the Rat data set) and in the Quail data set six to nine samples were considered a block in the statistical analysis. Some blocks were incomplete because the number of available samples in each treatment group was uneven (Table I and supplemental Fig. 1).
Table I.
Experimental setup
See also supplemental Fig. 1.
| Data set | Biological groups | Biological samples in each group | Technical replicate (termed Pool) | Number of LC-MS experiments of the Pool | Total number of LC-MS experiments | Additional information |
|---|---|---|---|---|---|---|
| Mouse | 4 | 6–7 | Pool of all 25 samples | 11 | 45 | Some biological samples were analyzed twice |
| Rat | 4 | 6–7 | Pool of 10 samples | 8 | 35 | |
| Quail | 8 | 4–5 | A single sample | 6 | 41 |
Peptide Profiling
The profiling analyses of the peptide samples were performed on a nano-LC system (Ettan MDLC, GE Healthcare) coupled to either a Q-TOF mass spectrometer (Waters; Mouse and Rat) or a linear ion trap mass spectrometer (LTQ, Thermo Electron, San José, CA; Quail). Data were collected during a 40-min gradient. More detailed information about the nano-LC-MS conditions can be found in supplemental Data 1.
Peptide Identification
Details about peptide identification for each data set can be found in the studies addressing the biological questions of the data sets (19, 20).2
Peak Matching and Filtering
The DeCyder MS2.0 software (22) was used for matching of peaks (23). Peaks were detected using a signal-to-noise cutoff of 5 and background subtracted quantification (smooth surface). For all data sets the peak matching was manually checked to minimize the number of missing values and mismatched peaks across analyses. Prior to applying all normalization methods except “DeCyder” and “Spike” (see below) the data were filtered so that only peaks that were successfully matched across ≥50% analyses in each data set were retained. For evaluation of normalization methods, peaks were included based on different criteria for each data set (Table II).
Table II.
Peak detection and filtering
| Data set | Peaks matched across >50% of all analyses (n) | Peaks matched across all LC-MS experiments (n) | Filtering criteria | Peaks remaining after filtering (n) |
|---|---|---|---|---|
| Mouse | 935 | 240 | A maximum of three missing observation allowed in each biological group | 575 |
| Rat | 930 | 309 | At least five observations in all biological groups | 545 |
| Quail | 534 | 160 | A maximum of one missing observation allowed in each biological group | 268 |
Normalization Procedures
DeCyder MS Normalization (DeCyder and Spike)
The DeCyder software provides two types of normalization methods, global normalization (here named DeCyder) and normalization based on internal standards (here named Spike). For global normalization the intensity distributions of the peaks detected in each sample were used for normalization (23). In the Mouse and Rat experiments, the extraction buffer was spiked with deuterated Met-enkephalin (YGGFM where F is Phe-d8), neurotensin (pyr(Q)LYENKPRRPYIL where L is Leu-d3), and substance P (RPKPQQFFGLM-amide where F is Phe-d8) at a concentration of 40 fmol/μl. These peptides were used for normalization purposes. The shift of data for each analysis was calculated by fitting a linear regression to the spiked-in peptides in each analysis and then subtracting the average regression coefficient across all analyses from each run. The normalized values produced with the DeCyder MS2.0 software were exported for both normalization procedures.
Local Regression Normalization of MA-transformed Data (LoessMA)
This method assumes systematic bias to be nonlinearly dependent on the magnitude of peak intensities. The approach is based upon the idea of an M versus A plot where M is the difference in log intensity values and A is the average log intensity values. Lowess smoothing is used to estimate intensity-dependent differences in pairs of analyses, and these differences are then removed by shifting the Lowess line to 0. Here the scatter plot of a pair of analyses was constructed instead of a single two-color microarray, which was used when the method was originally developed (12). The procedure was iterated until intensity-dependent differences were removed from all experimental analyses. This procedure, known as cyclic Lowess, was first implemented for the use in Affymetrix gene expression microarray data (10). It can be computationally very demanding for large data sets. The method has been used previously for normalizing peptide data (14) produced with LC-FTICR-MS. The fraction of peaks used for fitting the nonlinear regression line at each point was set at 0.4 (the span), and the iteration process continued until the difference between the mean of all abundance ratios from the previous iteration and current iteration was <0.005. This was the same criteria as used by Callister et al. (14).
Linear Regression Normalization of MA-transformed Data (LinRegMA)
This method assumes systematic bias to be linearly dependent on the magnitude of peak intensities and was also evaluated by Callister et al. (14). Linear regression normalization is performed by applying least squares regression to the M versus A plot, and normalized values are calculated by subtracting predicted peak ratios calculated from the regression equation. As for method LoessMA, the procedure was iterated until intensity-dependent differences were removed from all experimental analyses. The iteration process continued until the difference between the mean of all intensity ratios from the previous iteration and current iteration was <0.005. For normalization methods LoessMA and LinRegMA two iterations were sufficient to reach the stopping criteria.
Median Scale Normalization (MedScale)
Median normalization scales the log intensity values for one analysis using the global median value. Median or mean normalization has often been used for normalizing gene expression data across microarrays (12) and has been applied to peptide data produced with LC-FTICR-MS (14).
Quantile Normalization (Quantile)
This non-parametric method was first developed for the normalization between Affymetrix high density gene expression arrays (10). The method is frequently used with this type of data, and it has also been used for normalizing peptide data (14, 24). The aim of the method is to make the distribution of intensities the same across all analyses.
Reference Run Normalization (RefRun)
This method is a type of global intensity normalization where one analysis is chosen as a reference, and then all other runs are normalized to the chosen one. A single normalization constant for each analysis is obtained from the median of the ratios of intensities for all matched peaks between the analyses in question and the reference analysis (13, 15, 16). Herein the analysis in each data set with the most peaks matched after filtering was chosen as reference.
Variance Stabilization Normalization (Vsn)
This method was originally developed for normalizing single and two-channel gene expression microarray data. The function calibrates sample-to-sample variations through shifting and scaling of intensities and transforms the intensities to a scale where the variance is approximately independent of the mean intensity (11).
Linear Regression Normalization (Regr)
This method also assumes systematic bias to be linearly dependent on the magnitude of peak intensities, but instead of constructing MA plots of pairs of analyses as for method LinRegMA, a single reference analysis was constructed by taking the median peak intensity for all matched peaks. Linear regression normalization was then performed on this constructed reference by applying least squares regression to each individual analysis. Based on the linear regression equation new values were predicted for each analysis, taking both intercept and slope of the regression line into account.
RegrRun
Indications that the analysis order of the LC-MS experiments contributes to bias in the data were found. Normalization based only on analysis order will not correct for global differences, such as differences in amount/concentration of one sample compared with the rest of the samples. Regr was combined with analysis order normalization to address the different types of biases. A locally weighted polynomial regression (Lowess (25)), using function loessFit () in the R package limma (26, 27) (Version 2.12.0), was fitted for each matched peak versus the analysis order, and the mean value across all analyses for each peak was then added to retain the native intensity dimension. For each matched peak a certain proportion of neighbors (analyses), weighted by their distance to the measurement, was used for controlling the smoothness of the fit, the span. A high span value gives more smoothness, and a value of 1 returns values similar to those of linear regression. Equal weight was given for all types of samples, and default settings were used for the loessFit () function (span of 0.3).
Mixed Linear Models
A linear mixed model was applied to each data set using restricted maximum likelihood estimation (28). Because samples were analyzed in a randomized block order there should ideally be no differences or at least only minor interblock differences. To calculate the fraction of peaks with significant block differences in each data set, blocks were considered fixed effects, whereas biological and technical variations were random effects. To calculate the fraction of peaks with statistically significant LC-MS run order differences in the Mouse data set, the Pool replicates were divided into two groups, the first five and the last five, leaving one intermediate replicate. The model was fitted with and without including the block factor. To calculate the fraction of peaks with biologically differential expression, biological effects were considered fixed effects, whereas block and technical replicates were random effects. Both models were fitted with and without including the block factor. Analysis of variance for mixed linear models in the R (29) (Version 2.6.0)/maanova (30) (Version 1.5.1) Bioconductor (31) package (Version 2.1) was used to obtain tabulated p values, and missing values were imputed using k-nearest neighbors (k = 10). A p value of p < 0.05 was considered statistically significant. Some bias may originate from the fact that some blocks were incomplete; however, this probably only had a minor impact on the overall result. For displaying trends in matching success of peaks and for k-means cluster analysis the median peak intensity value in block 1 was subtracted from all analyses. For k-means cluster analysis the function kmeans () with default settings (k = 4) available in the stats (29) (Version 2.6.0) R package was used. Three of the four largest clusters were displayed.
Ranking of Normalization Methods
For the variation between technical replicates (Pool) and the variation including both technical and biological variation, biological samples belonging to the treatment group (BioRep) were ranked based on their average percent reduction in median log2 S.D. and reduction of log2 pooled estimate of variance (PEV) compared with raw data. In the Rat and Quail data sets the BioRep only consisted of different biological samples, whereas the Mouse data set included one to three technical replicates of biological samples. For the Quail data set, which lacked spiked-in peptides, only nine methods were evaluated, resulting in a slight bias in the average rank across the different data sets, but this will only have a minor effect on the final result. To get a more general overview of how much the different normalization methods decreased median S.D. and PEV compared with the raw data, the average percent reduction for the three data sets was calculated.
RESULTS
The performances of 10 different normalization methods on three different data sets of endogenous peptide data produced with label-free nano-LC mass spectrometry were investigated. The samples in all three data sets were derived from brain tissue (mouse, rat, and quail), and each data set consisted of 35–45 individual LC-MS analyses. Before analyzing a new set of samples, the analytical LC column was replaced. The samples in the Mouse, Rat, and Quail experiments were analyzed in a randomized block design fashion, and each data set was produced within a time period of 50–70 h. After matching and filtering different numbers of peaks were retained for further analysis in the Mouse, Rat, and Quail data sets, respectively (Table II).
Matching Success of Peaks
Successful matching of peaks across samples is important to keep the number of missing values at an absolute minimum. For peaks matched across >50% of all analyses in each data set there was a decrease at the end of the analysis in matching success for the Mouse data set, whereas the opposite was found for the Quail data set (Fig. 1). No clear analysis order-dependent peak matching pattern was found for the Rat data set.
Fig. 1.
The fraction of successfully matched peaks across LC-MS analyses. Open spots are different biological samples, and gray spots are technical replicates. In the Mouse data set there was a weak trend of lower peak matching success at the end of the experiment, whereas the opposite was found in the Quail data set. Analyses with overall lower matching success were often accompanied with overall low intensity values, resulting in lack of detection of lowly expressed peaks in these analyses.
Global Intensity Distribution
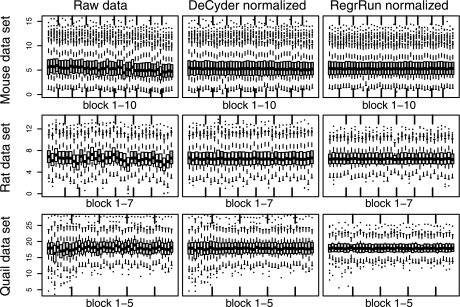
After matching and filtering there were global differences in total peak intensity between LC-MS analyses (Fig. 2). The basic assumption for normalizing this type of data is that the biological variability and different treatments only affect a relatively small fraction of the measured peaks so that after normalization all analyses should have approximately the same intensity distribution. All methods investigated except for the Spike method successfully removed global intensity bias. The remaining methods not shown in Fig. 2 displayed a pattern similar to that of the DeCyder- and RegrRun-normalized data. In the Rat data set, lower mean intensity for a particular analysis (raw data) was accompanied with a decreased matching success for that particular analysis. The reason for this is that lowly expressed peaks will have an intensity value close to the background intensity. A similar but not as clear relationship was also found in the Mouse and Quail data sets.
Fig. 2.
Box plots of the log2 intensity values (y axis) for the individual LC-MS analyses in each data set before and after normalization using the DeCyder and RegrRun methods. The data are displayed in the analysis order of the samples. Open boxes are different biological samples, and gray box plots are technical replicates. Both normalization methods successfully assured that the intensity values have approximately the same average and empirical distribution across all analyses for each data set. The box plots display the median, first and third quartiles. The whiskers extend to the extreme values or at most 1.5 times the interquartile range from the box. Observations outside the whiskers are plotted as dots.
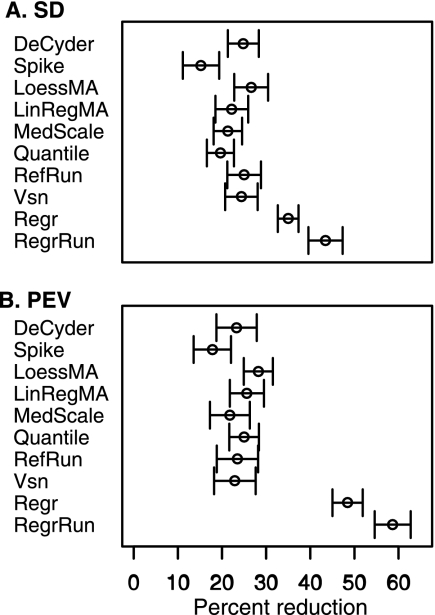
Technical Variability
Median S.D. and PEV of raw data for the Pool ranged between 0.25 and 0.65 and between 0.1 and 1.44, respectively, across the three data sets. Across all three data sets the largest improvement was found using the Regr and RegrRun methods with an average decreased median S.D. of 38 and 43% and on average a 48 and 59% decrease in PEV, respectively. For all other examined methods the reduction was generally lower and ranged between 20 and 30% and between 20 and 28% on average in median S.D. and PEV, respectively, across all data sets (Table III). For the Pool replicates, quantified using the Q-TOF (Mouse and Rat) the S.D. and PEV were found to be lower compared with quantification using the LTQ (Quail). After normalization using the RegrRun method the median S.D. for the Pool replicates was 0.17 for both the Mouse and Rat data sets, and in the Quail data set it was 0.42. After data normalization the relationship between technical replicates should ideally be a straight line with a slope of 1. By using the RegrRun method the regression coefficient was close to 1 for the technical replicates (for the Mouse data set the technical replicates of the different biological samples were also included). The raw and DeCyder-normalized data produced regression coefficients deviating from 1, indicating that bias still remains (Fig. 3).
Table III.
Median S.D. and PEV
The median S.D. and PEV for the three data sets when omitting (Pool) and including biological variability (BioRep) are shown. For each data set, normalization method and subset, the median S.D., and PEV are given followed by the ranking score and percent reduction in median S.D. and PEV compared with raw data (within parentheses). The method resulting in the lowest median S.D. and PEV was given the rank 1 and so forth. For each normalization method and across the different data sets the average rank scores are given (±S.E.). n.a., not applicable.
| Method | Data set Mouse |
Data set Rat |
Data set Quail |
Mean rank (±S.E.) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEV |
Median S.D. |
PEV |
Median S.D. |
PEV |
Median S.D. |
||||||||
| Pool | BioRep | Pool | BioRep | Pool | BioRep | Pool | BioRep | Pool | BioRep | Pool | BioRep | ||
| Raw | 0.32 | 0.51 | 0.42 | 0.54 | 0.10 | 0.70 | 0.25 | 0.67 | 1.44 | 1.50 | 0.65 | 0.75 | 10. ± 0.1 |
| 11 | 11 | 11 | 11 | 11 | 11 | 11 | 11 | 10 | 10 | 10 | 10 | ||
| DeCyder | 0.22 | 0.35 | 0.29 | 0.40 | 0.08 | 0.43 | 0.19 | 0.48 | 1.25 | 1.35 | 0.45 | 0.68 | 5.5 ± 0.3 |
| 5 | 7 | 5 | 5 | 5 | 6 | 3 | 6 | 7 | 7 | 6 | 4 | ||
| (31%) | (29%) | (30%) | (27%) | (22%) | (37%) | (25%) | (28%) | (13%) | (8%) | (31%) | (8%) | ||
| Spike | 0.24 | 0.39 | 0.31 | 0.45 | 0.08 | 0.63 | 0.21 | 0.63 | n.a. | n.a. | n.a. | n.a. | 9.9 ± 0.1 |
| 9 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | ||||||
| (26%) | (21%) | (25%) | (16%) | (19%) | (6%) | (15%) | (5%) | ||||||
| LoessMA | 0.22 | 0.34 | 0.28 | 0.39 | 0.08 | 0.42 | 0.20 | 0.46 | 1.02 | 1.18 | 0.41 | 0.66 | 3.5 ± 0.2 |
| 3 | 3 | 4 | 3 | 6 | 4 | 5 | 3 | 3 | 3 | 2 | 3 | ||
| (32%) | (31%) | (32%) | (28%) | (22%) | (39%) | (22%) | (31%) | (29%) | (16%) | (36%) | (10%) | ||
| LinRegMA | 0.23 | 0.35 | 0.29 | 0.39 | 0.08 | 0.42 | 0.20 | 0.48 | 1.15 | 1.24 | 0.49 | 0.70 | 6.1 ± 0.3 |
| 6 | 4 | 6 | 4 | 7 | 5 | 7 | 7 | 5 | 5 | 8 | 9 | ||
| (31%) | (31%) | (30%) | (27%) | (21%) | (39%) | (20%) | (27%) | (20%) | (13%) | (24%) | (5%) | ||
| MedScale | 0.23 | 0.36 | 0.31 | 0.41 | 0.08 | 0.45 | 0.21 | 0.51 | 1.28 | 1.38 | 0.47 | 0.69 | 7.9 ± 0.3 |
| 7 | 9 | 9 | 9 | 4 | 9 | 8 | 8 | 9 | 9 | 7 | 7 | ||
| (28%) | (28%) | (26%) | (25%) | (22%) | (35%) | (19%) | (24%) | (11%) | (6%) | (28%) | (7%) | ||
| Quantile | 0.24 | 0.35 | 0.31 | 0.40 | 0.08 | 0.43 | 0.21 | 0.51 | 1.08 | 1.22 | 0.53 | 0.69 | 7.4 ± 0.4 |
| 10 | 5 | 8 | 7 | 8 | 8 | 9 | 9 | 4 | 4 | 9 | 8 | ||
| (25%) | (30%) | (26%) | (26%) | (20%) | (37%) | (18%) | (23%) | (25%) | (13%) | (18%) | (6%) | ||
| RefRun | 0.22 | 0.35 | 0.28 | 0.40 | 0.08 | 0.43 | 0.19 | 0.47 | 1.25 | 1.36 | 0.45 | 0.69 | 5.4 ± 0.4 |
| 4 | 6 | 3 | 6 | 3 | 7 | 4 | 5 | 8 | 8 | 5 | 6 | ||
| (32%) | (29%) | (33%) | (26%) | (23%) | (37%) | (24%) | (29%) | (13%) | (7%) | (32%) | (7%) | ||
| Vsn | 0.23 | 0.36 | 030 | 0.40 | 0.08 | 0.41 | 0.20 | 0.46 | 1.25 | 1.33 | 0.44 | 0.68 | 6.2 ± 0.4 |
| 8 | 8 | 7 | 8 | 9 | 3 | 6 | 4 | 6 | 6 | 4 | 5 | ||
| (28%) | (28%) | (29%) | (26%) | (19%) | (40%) | (21%) | (30%) | (13%) | (9%) | (33%) | (8%) | ||
| Regr | 0.20 | 0.30 | 0.28 | 0.37 | 0.05 | 0.31 | 0.16 | 0.43 | 0.58 | 0.67 | 0.36 | 0.53 | 1.8 ± 0.1 |
| 2 | 2 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 1 | 2 | ||
| (37%) | (41%) | (34%) | (32%) | (49%) | (54%) | (37%) | (35%) | (59%) | (51%) | (45%) | (28%) | ||
| RegrRun | 0.11 | 0.21 | 0.17 | 0.28 | 0.06 | 0.24 | 0.17 | 0.36 | 0.55 | 0.54 | 0.42 | 0.45 | 1.3 ± 0.1 |
| 1 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | ||
| (66%) | (58%) | (59%) | (48%) | (39%) | (66%) | (33%) | (46%) | (62%) | (61%) | (36%) | (39%) | ||
Fig. 3.
Box plots of the regression coefficient between technical replicates for raw, DeCyder-, and RegrRun-normalized data. For the RegrRun normalized data the regression coefficient was close to 1 in all three data sets. The box plots display the median, first and third quartiles. The whiskers extend to the extreme values or at most 1.5 times the interquartile range from the box. Observations outside the whiskers are plotted as open circles.
Technical and Biological Variability
In general the variability including biological variability (BioRep) was higher compared with the Pool. The median S.D. and PEV of the raw data ranged between 0.55 and 0.75 and between 0.51 and 1.50, respectively. Across all three data sets the largest improvement was found using the Regr and RegrRun methods with an average decrease in median S.D. of 38 and 42%, respectively. The largest decrease in PEV was also found for the Regr and RegrRun methods, 48 and 62% on average, respectively. In the Quail data set only methods Regr and RegrRun resulted in a significant reduction of median S.D. and PEV. As previously found for the Pool, all other examined methods generally produced lower reductions of median S.D. and PEV, 10–23 and 13–29%, respectively (Table III).
Ranking of Normalization Methods and Reduction of Variability
Performance comparisons of the different normalization methods were considered by using their rank derived from the percent reduction in median S.D. and PEV. The method with the largest reduction was given the score of 1 and so forth to ranking score 11 (10 in the Quail data set). Taking both S.D. and PEV ranks into account, the RegrRun method had the best ranking score followed by Regr and LoessMA. Methods DeCyder, LinRegMA, RefRun, and Vsn all performed similarly with mean rankings in the range of 5.4–6.2, and the MedScale and Quantile had lower rankings (7.4 and 7.9, respectively). The Spike method had the least favorable ranking. To get a more general overview of how much the different normalization approaches decreased median S.D. and PEV compared with raw data, the average reductions for the three data set was calculated. The methods Regr and RegrRun resulted in a 35 and 44% average median S.D. reduction and a 48 and 59% PEV reduction, respectively. The remaining methods except for Spike, which only resulted in a 15–18% reduction, resulted in ∼20–28% average median S.D. and PEV reductions (Fig. 4).
Fig. 4.
The average median S.D. and PEV reduction after applying different normalization methods compared with raw data. The largest reductions were observed with the methods Regr and RegrRun, and the smallest reduction was observed with the Spike method. Intermediate reductions were observed with the remaining seven methods. The error bars display 1 standard error of the mean.
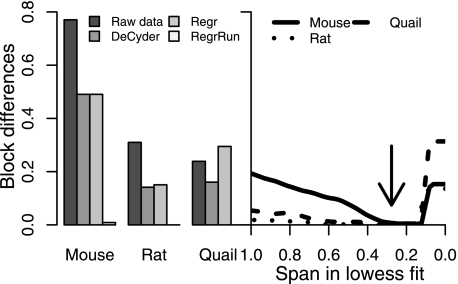
Span in Lowess Fit
Because samples were analyzed in a randomized block order there should ideally be no differences or at least small interblock differences. All normalization methods except RegrRun resulted in large (Mouse) or smaller fractions (Rat and Quail) of peaks with significant interblock differences. By fitting, in principle, a linear regression (span of 1) versus the analysis order, all three data sets displayed a substantial decrease in interblock variability. Using the default span of 0.3 in the Lowess fit resulted, in principle, in no peaks with significant interblock differences. Using too small span settings (<0.15) resulted in an increase of interblock variability again that was probably due to overfitting (Fig. 5). A similar pattern of overfitting was also found when studying the decrease in PEV within replicates belonging to the same treatment type at different span settings (supplemental Fig. 2).
Fig. 5.
The fractions of peaks with statistically significant (p < 0.05) interblock differences using raw, DeCyder-, and Regr-normalized data in the three data sets (left panel) are shown. The fractions decreased (Mouse and Rat) after normalizing the data using the DeCyder and Regr methods. Using the RegrRun method with default Lowess span setting (0.3; indicated by the arrow) resulted, in principle, in no peaks displaying significant interblock differences (left and right panels). Span settings <0.15 resulted in increased interblock differences due to overfitting.
Analysis Order Bias
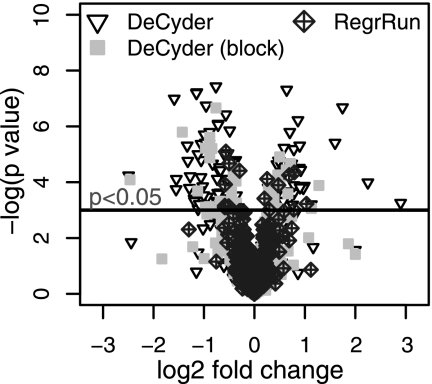
If normalization works properly there should be no or at least only minor differences in peak intensity between pool samples. To test this, the 11 pool samples in the Mouse data set were divided into two groups of five, the first five and the last five, leaving one sample intermediate. The expression differences between the two groups were then tested using DeCyder-normalized data (with or without inclusion of the block term) and RegrRun-normalized data. It is clear that both the fraction and the magnitude of the -fold difference of peaks displaying run order-dependent bias is much larger for DeCyder-normalized data (Fig. 6.). For RegrRun-normalized data only 2.6% of the peaks were statistically significant between the two groups irrespective of the magnitude of the -fold difference. For DeCyder-normalized data 19.5% of peaks displayed significant expression differences. This fraction decreased to 6.8% after including the block term in the model. To further investigate this, a k-means cluster analysis was performed on the Mouse data set to examine the global intensity distributions prior to and after normalization (supplemental Fig. 3). After the DeCyder normalization, one cluster contained data with no analysis order bias, whereas two clusters contained data that displayed decreasing or increasing peak intensities. In all three data sets, a large (∼70%; Mouse data set) or a small fraction (∼20–30%; Rat and Quail data sets) of examined peaks displaying analysis order-dependent bias was found. In all three data sets and for a majority of all matched peaks there was a trend of decreasing intensity values at the end of the LC-MS analysis. Only the RegrRun method, which combines global and analysis order normalization, successfully removed analysis order-dependent bias.
Fig. 6.
Volcano plot displaying the log2 -fold change and −log (p value) between the first five pool replicates compared with the last five replicates in the Mouse data set. For DeCyder-normalized data, 19.5% of all peaks were statistically significantly different (p < 0.05) between the two groups. Both the fraction and the magnitude of log2 -fold change decreased when including the block term in the linear model. For RegrRun-normalized data only 2.6% of the peaks were statistically significantly different, and the log2 -fold changes were much smaller compared with the DeCyder-normalized data.
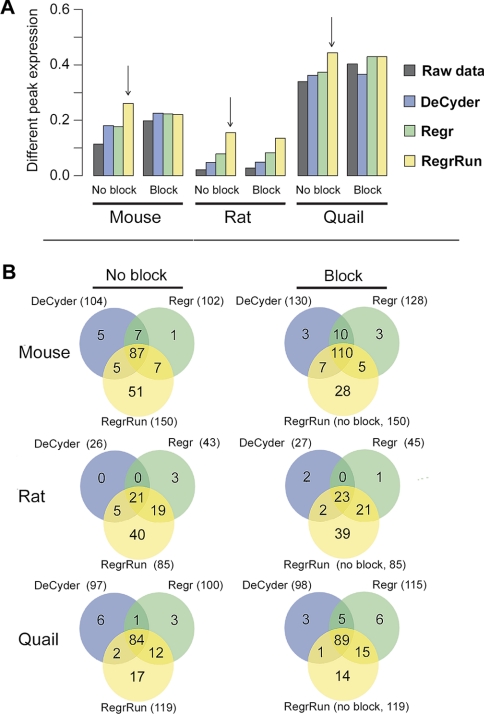
Detection of Differential Expression
To compare the performance of DeCyder-, Regr-, and RegrRun-normalized data in terms of detecting differential expression, a mixed linear model was fitted for each data set, including or omitting the block term in the model. The RegrRun method omitting the block term detected the largest fraction of differential expression in all three data sets (Fig. 7). For the DeCyder- and Regr-normalized data, including the block term in the model increased the fraction of differential expression detected. For example, in the Mouse data set there is a good agreement of the differentially expressed peaks found using the different normalization methods. By including the block term in the model for the DeCyder and Regr methods the agreement to the RegrRun method was improved. Including the block term resulted in an increase of 27% of peaks detected as differentially expressed with all methods compared with omitting the block term in the model. A similar trend was also found in the Rat and Quail data sets (Fig. 7).
Fig. 7.
The number of differentially expressed peaks in the Mouse, Rat, and Quail data sets using the RegrRun method compared with raw, DeCyder-, and Regr-normalized data (p < 0.05). The largest fraction of differentially expressed peaks was found in the RegrRun-normalized data (panel A, vertical arrows). For raw, DeCyder-, and Regr-normalized data the numbers of differentially expressed peaks were calculated both with and without including the block term in the linear model. The largest number of overlapping peaks compared with the RegrRun method was found when including the block term for raw, DeCyder-, and Regr-normalized data (panel B).
DISCUSSION
Basic Normalization Assumptions
The purpose of normalizing LC-MS data is to reduce or remove bias due to differences in peptide extraction, LC separation efficiency, drifts in ionization and detector efficiencies, or other systematic sources introducing bias. In this study, the performances of 10 different normalization methods for the relative quantification of endogenous brain peptides were investigated. The underlying assumption behind most global normalization methods is that the differences in overall peak intensity of either all or a subset of peaks should be minor across experiments. However, this assumption may be invalid due to severe imbalance in missing observations in peptide data (32). In an attempt to solve this problem the data were filtered so that only peaks matched across ≥50% of the data set were included in each analysis. In some analyses high intensity peaks may be present due to unknown contamination. The normalization factor for these analyses can differ substantially whether these unmatched peaks are included in the normalization procedure or not. In agreement with what others have reported (14, 33) we also found that the log2 peak intensity was linearly dependent on the magnitude of measured intensities. This motivates the usage of linear regression for normalizing peptide data. After constructing a reference analysis, fitting a linear regression, and predicting new values to each individual analysis taking both intercept and slope of the regression line into account (method Regr), the median S.D. and PEV values decreased. Different intercepts may represent a global error such as a bias in background subtraction. However, predicting new values from regression coefficients may also introduce a bias because the linear regression is sensitive to outlier values. The 50% matching criterion used seems to handle this type of problem satisfyingly.
The endogenous peptides studied herein were not cleaved through trypsinization. However, we believe that the result of the evaluation of different normalization methods found in the present study also may be applicable to peptides derived from tryptic digestion.
Normalization Methods
Callister et al. (14) have compared the effect of different normalization procedures for relative quantification of peptides originating from trypsinized proteins from various tissues and cells. They analyzed four technical replicates in three different data sets: a simple protein mixture, a microbial system, and a mouse brain system. The LC-FTICR-MS analyses for some samples were performed over a time period of 3 months and also included replacement of the analytical LC column. The data were analyzed in a block fashion where each block of four analyses was normalized separately, whereas in our case all analyses within each data set were normalized together. Callister et al. (14) concluded that normalization by linear regression (corresponding to LinRegMA in our study) and central tendency (corresponding to MedScale in our study) performed best, but they also reported that the result may vary between different data sets. The peptides investigated in the present study are endogenous brain peptides. All LC-MS analyses for each individual experimental setup (Mouse, Rat, and Quail) were performed within a time period of 50–70 h, and the analytical LC column was replaced between experiments and not within an experimental setup. In the present study the RegrRun method performed the best in the three data sets evaluated. It appears that the basic assumption that some sort of global drift is occurring at various levels, during and between multiple MS analyses similarly affecting all peak intensities, is not perfectly true. Applying global linear regression followed by a nonlinear Lowess smoother to each peak dependent on the analysis order resulted in a clear decrease in median S.D. and PEV reduction compared with all other global normalization methods. The Regr method performed second best in reducing median S.D. and PEV, whereas the other evaluated normalization methods performed more poorly.
Using DeCyder-normalized data in the Mouse data set, almost 20% of the peaks detected in pool samples differed between the first five replicates and the last five replicates. Ideally there should be no expression differences between the pool samples. This fraction decreased (6.8%) when including the block term in the linear model. In RegrRun-normalized data only 2.6% of the peaks were statistically different in expression. Generally the use of DeCyder or the Regr method resulted in different degrees of analysis order-dependent intensity bias (including 20–70% of all detected peaks in the different data sets). This type of bias was removed when applying the RegrRun method.
Span in Lowess Fit
To correctly handle run order-dependent bias it is important to avoid confounding. The LC-MS experiments of samples belonging to different treatment groups have to be analyzed in a randomized fashion otherwise run order-dependent bias may confound with treatment effects. Proper experimental design will not be discussed further here but has thoroughly been discussed elsewhere (34–37). For all three data sets, the default settings for the loessFit () function (span of 0.3) were used and worked satisfactorily. If the span is set too small there will be a risk of overfitting. This can be investigated by estimating interblock effects as described herein. If the LC-MS experiments are performed in a completely randomized fashion an alternative approach is to calculate PEV (or median S.D.) of samples belonging to the same treatment group at different span settings (supplemental Fig. 2). For all three data sets, using a span <0.15 clearly indicated overfitting.
Spiked Peptides
The Mouse and Rat samples were spiked with deuterated peptides. However, the Spike normalization yielded the least favorable results in both data sets. It is possible that a larger number of spiked peptides would improve the performance of the method. However, based on our observations the use of spiked peptides for normalization or for evaluating the performance of normalization methods should be utilized with care because the resulting intensities may display a different type of bias compared with the rest of the peaks in the data set.
Sources of Global Drifts
The origin of analysis order-dependent intensity bias may be due to several factors such as changes of the properties in the LC column, fluctuations in the LC flow rate, or a combination of these and other unknown factors. The fact that two different detectors were used, Q-TOF in the Mouse and Rat data sets and LTQ in the Quail data set, indicates that the problem was not detector-specific. In the case of a general carryover of peaks between LC-MS analyses, it would be expected to see an increase rather than a decrease of the intensity levels. For some individual peaks there were indications of carryover, which has also been found by Daly et al. (38), whereas for other peaks there was an analysis-dependent decrease in intensity. For some peaks a clear restoration of intensity values was found after analyzing a blank sample. A blank sample was analyzed between each block of samples as a trade-off for analyzing a blank between each sample.
The change in relative matching success (Fig. 1) may be associated with slight changes in flow rate. In the Rat data set there were large differences in the relative matching success between samples compared with the Mouse and Quail data sets. It is important to remember that normalization will remove bias but not improve matching success. As clearly seen in the Mouse and Quail data sets, the matching success of the technical replicates was dependent on other sources of bias (drifts) than only the content of the sample. Analyzing a single technical replicate of each sample would minimize the number of missing peaks for each biological sample. The replicates should be analyzed as far apart as possible.
Detecting Differential Expression
Mixed-effects statistical modeling has previously been used to estimate relative protein concentrations where each peptide is modeled and summarized to global relative protein abundances (38, 39). Mixed-effects modeling was used to compare the fraction of differentially expressed peaks detected using DeCyder-, Regr-, and RegrRun-normalized data.
A normalization method evaluated based on the fraction of differentially expressed peaks is approximate at best, considering that the “true” fraction of differential expression is unknown. However, in the field of gene expression data, this is a common approach for comparing the performance of different normalization methods (21, 40). The largest fraction of differentially expressed peaks for all three data set was found using the method RegrRun. For Regr- and DeCyder-normalized data the agreement in differentially expressed peaks compared with RegrRun-normalized data increased when including the block term in the linear model. Despite blocking, the fraction of differentially expressed peaks was found to be smaller for DeCyder- and Regr-normalized data compared with RegrRun-normalized data. Blocking assumes the error to be similar within each block; however, in this type of data the analysis order bias is spanning across blocks. The presence of incomplete blocks may also contribute to the differences, but at the same time this argues for using the RegrRun method because there will always be a risk of incomplete blocks when performing large LC-MS experiments.
All three different data sets evaluated in this study displayed large differences in the fraction of differential expression. Compared with the number of peaks detected, the Mouse and Rat data sets displayed moderate differences, whereas the Quail data set displayed large differences (Fig. 7, panel A). This is also what could be expected from a biological point of view. In the Quail data set two different embryonic developmental stages were compared.2 The fact that such a large fraction of all peaks differed may have an impact on the success of the normalization procedure because only a small fraction of the peaks is assumed to be differentially expressed between groups.
Conclusions
In this study the performances of 10 different normalization methods on endogenous peptide data generated with LC-MS were investigated. The methods presented and evaluated are all, except for one commercially developed method (DeCyder), publicly available and fairly straightforward. The novel method presented here named RegrRun (which corrects for both global differences between LC-MS analyses and analysis order-dependent bias) and Regr decreased the median S.D. by 42–43 and 38% compared with raw data, respectively, both when omitting and including biological variability. The intensity levels of detected peaks in a set of LC-MS analyses may display analysis order-dependent bias. Only the RegrRun method effectively removed this type of bias. Other normalization methods operating on a global level only reduced the median S.D. by 15–28%.
Supplementary Material
Acknowledgments
We thank Denator AB for providing their prototype for inactivation of protein degradation and Rolf Danielsson and Hanna Göransson for valuable discussions.
Footnotes
* This work was supported by the Swedish Research Council (Vetenskapsrådet) Grants 521-2007-3017 and 621-2007-4686 and the Knut and Alice Wallenberg Foundation (to P. E. A.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
2 B. Scholz, H. Alm, A. Mattsson, A. Nilsson, M. Savitski, M. Falth, K. Kultima, K. Skold, B. Brunstrom, P. E. Andren, and L. Dencker, manuscript in preparation.
1 The abbreviations used are:
- LTQ
- linear trap quadrupole
- A
- average log intensity
- BioRep
- biological samples belonging to the treatment group
- DeCyder
- global normalization available in the DeCyder software
- LinRegMA
- linear regression normalization of MA-transformed data
- LoessMA
- local regression normalization of MA-transformed data
- M
- difference in log intensity
- MedScale
- median scale normalization
- PEV
- pooled estimate of variance
- Pool
- technical replicates
- Quantile
- quantile normalization
- RefRun
- reference run normalization
- Regr
- linear regression normalization
- RegrRun
- linear regression followed by analysis order normalization
- Spike
- normalization based on internal standards
- Vsn
- variance stabilization normalization.
REFERENCES
- 1.Clynen E., Baggerman G., Veelaert D., Cerstiaens A., Van der Horst D., Harthoorn L., Derua R., Waelkens E., De Loof A., Schoofs L. (2001) Peptidomics of the pars intercerebralis-corpus cardiacum complex of the migratory locust, Locusta migratoria. Eur. J. Biochem 268, 1929–1939 [DOI] [PubMed] [Google Scholar]
- 2.Schulz-Knappe P., Zucht H. D., Heine G., Jürgens M., Hess R., Schrader M. (2001) Peptidomics: the comprehensive analysis of peptides in complex biological mixtures. Comb. Chem. High Throughput Screen 4, 207–217 [DOI] [PubMed] [Google Scholar]
- 3.Verhaert P., Uttenweiler-Joseph S., de Vries M., Loboda A., Ens W., Standing K. G. (2001) Matrix-assisted laser desorption/ionization quadrupole time-of-flight mass spectrometry: an elegant tool for peptidomics. Proteomics 1, 118–131 [DOI] [PubMed] [Google Scholar]
- 4.Svensson M., Sköld K., Nilsson A., Fälth M., Nydahl K., Svenningsson P., Andrén P. E. (2007) Neuropeptidomics: MS applied to the discovery of novel peptides from the brain. Anal. Chem 79, 15–16, 18–21 [DOI] [PubMed] [Google Scholar]
- 5.Svensson M., Sköld K., Nilsson A., Fälth M., Svenningsson P., Andrén P. E. (2007) Neuropeptidomics: expanding proteomics downwards. Biochem. Soc. Trans 35, 588–593 [DOI] [PubMed] [Google Scholar]
- 6.Sköld K., Svensson M., Kaplan A., Björkesten L., Aström J., Andren P. E. (2002) A neuroproteomic approach to targeting neuropeptides in the brain. Proteomics 2, 447–454 [DOI] [PubMed] [Google Scholar]
- 7.Sköld K., Svensson M., Norrman M., Sjögren B., Svenningsson P., Andrén P. E. (2007) The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: stathmin 2–20 and peptides as sample quality indicators. Proteomics 7, 4445–4456 [DOI] [PubMed] [Google Scholar]
- 8.Svensson M., Sköld K., Svenningsson P., Andren P. E. (2003) Peptidomics-based discovery of novel neuropeptides. J. Proteome Res 2, 213–219 [DOI] [PubMed] [Google Scholar]
- 9.Boonen K., Landuyt B., Baggerman G., Husson S. J., Huybrechts J., Schoofs L. (2008) Peptidomics: the integrated approach of MS, hyphenated techniques and bioinformatics for neuropeptide analysis. J. Sep. Sci 31, 427–445 [DOI] [PubMed] [Google Scholar]
- 10.Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 11.Huber W., von Heydebreck A., Sültmann H., Poustka A., Vingron M. (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18, Suppl. 1, S96–S104 [DOI] [PubMed] [Google Scholar]
- 12.Yang Y. H., Dudoit S., Luu P., Lin D. M., Peng V., Ngai J., Speed T. P. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderle M., Roy S., Lin H., Becker C., Joho K. (2004) Quantifying reproducibility for differential proteomics: noise analysis for protein liquid chromatography-mass spectrometry of human serum. Bioinformatics 20, 3575–3582 [DOI] [PubMed] [Google Scholar]
- 14.Callister S. J., Barry R. C., Adkins J. N., Johnson E. T., Qian W. J., Webb-Robertson B. J., Smith R. D., Lipton M. S. (2006) Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J. Proteome Res 5, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy S. M., Becker C. H. (2007) Quantification of proteins and metabolites by mass spectrometry without isotopic labeling. Methods Mol. Biol 359, 87–105 [DOI] [PubMed] [Google Scholar]
- 16.Wang W., Zhou H., Lin H., Roy S., Shaler T. A., Hill L. R., Norton S., Kumar P., Anderle M., Becker C. H. (2003) Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal. Chem 75, 4818–4826 [DOI] [PubMed] [Google Scholar]
- 17.America A. H., Cordewener J. H. (2008) Comparative LC-MS: a landscape of peaks and valleys. Proteomics 8, 731–749 [DOI] [PubMed] [Google Scholar]
- 18.Listgarten J., Emili A. (2005) Statistical and computational methods for comparative proteomic profiling using liquid chromatography-tandem mass spectrometry. Mol. Cell. Proteomics 4, 419–434 [DOI] [PubMed] [Google Scholar]
- 19.Nilsson A., Fälth M., Zhang X., Kultima K., Sköld K., Svenningsson P., Andrén P. E. (2009) Striatal alterations of secretogranin-1, somatostatin, prodynorphin, and cholecystokinin peptides in an experimental mouse model of Parkinson disease. Mol. Cell. Proteomics 8, 1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossbach U., Nilsson A., Fälth M., Kultima K., Zhou Q., Hallberg M., Gordh T., Andren P. E., Nyberg F. (2009) A quantitative peptidomic analysis of peptides related to the endogenous opioid and tachykinin systems in nucleus accumbens of rats following naloxone-precipitated morphine withdrawal. J. Proteome Res 8, 1091–1098 [DOI] [PubMed] [Google Scholar]
- 21.Stafford P., Brun M. (2007) Three methods for optimization of cross-laboratory and cross-platform microarray expression data. Nucleic Acids Res 35, e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan A., Söderström M., Fenyö D., Nilsson A., Fälth M., Sköld K., Svensson M., Pettersen H., Lindqvist S., Svenningsson P., Andrén P. E., Björkesten L. (2007) An automated method for scanning LC-MS data sets for significant peptides and proteins, including quantitative profiling and interactive confirmation. J. Proteome Res 6, 2888–2895 [DOI] [PubMed] [Google Scholar]
- 23.Johansson C., Samskog J., Sundström L., Wadensten H., Björkesten L., Flensburg J. (2006) Differential expression analysis of Escherichia coli proteins using a novel software for relative quantitation of LC-MS/MS data. Proteomics 6, 4475–4485 [DOI] [PubMed] [Google Scholar]
- 24.Higgs R. E., Knierman M. D., Gelfanova V., Butler J. P., Hale J. E. (2005) Comprehensive label-free method for the relative quantification of proteins from biological samples. J. Proteome Res 4, 1442–1450 [DOI] [PubMed] [Google Scholar]
- 25.Cleveland W. S. (1979) Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc 74, 829–836 [Google Scholar]
- 26.Smyth G. K. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol 3, Article3. [DOI] [PubMed] [Google Scholar]
- 27.Smyth G. K., Speed T. (2003) Normalization of cDNA microarray data. Methods 31, 265–273 [DOI] [PubMed] [Google Scholar]
- 28.Patterson H. D., Thompson R. (1971) Recovery of inter-block information when block sizes are unequal. Biometrika 58, 545–554 [Google Scholar]
- 29.The R Development Core Team (2008) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Wien, Austria [Google Scholar]
- 30.Hao W. (2007) maanova: Tools for Analyzing MicroArray Experiments, R package Version 1.8.0, Bioconductor, Seattle, WA [Google Scholar]
- 31.Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P., Tang H., Zhang H., Whiteaker J., Paulovich A. G., Mcintosh M. (2006) Normalization regarding non-random missing values in high-throughput mass spectrometry data. Pac. Symp. Biocomput 2006, 315–326 [PubMed] [Google Scholar]
- 33.Sauve A., Speed T. (2004) Normalization, baseline correction and alignment of high-throughput spectrometry data, in Proceedings of the Workshop on Genomic Signal Processing and Statistics (GENSIPS), Baltimore, May 26–27, 2004, Institution of Electrical Engineers (IEEE), Washington, D. C. [Google Scholar]
- 34.Churchill G. A. (2002) Fundamentals of experimental design for cDNA microarrays. Nat. Genet 32, (suppl.) 490–495 [DOI] [PubMed] [Google Scholar]
- 35.Hu J., Coombes K. R., Morris J. S., Baggerly K. A. (2005) The importance of experimental design in proteomic mass spectrometry experiments: some cautionary tales. Brief. Funct. Genomic. Proteomic 3, 322–331 [DOI] [PubMed] [Google Scholar]
- 36.Rocke D. M. (2004) Design and analysis of experiments with high throughput biological assay data. Semin. Cell Dev. Biol 15, 703–713 [DOI] [PubMed] [Google Scholar]
- 37.Karp N. A., Lilley K. S. (2007) Design and analysis issues in quantitative proteomics studies. Proteomics 7, Suppl. 1, 42–50 [DOI] [PubMed] [Google Scholar]
- 38.Daly D. S., Anderson K. K., Panisko E. A., Purvine S. O., Fang R., Monroe M. E., Baker S. E. (2008) Mixed-effects statistical model for comparative LC-MS proteomics studies. J. Proteome Res 7, 1209–1217 [DOI] [PubMed] [Google Scholar]
- 39.Oberg A. L., Mahoney D. W., Eckel-Passow J. E., Malone C. J., Wolfinger R. D., Hill E. G., Cooper L. T., Onuma O. K., Spiro C., Therneau T. M., Bergen H. R., 3rd (2008) Statistical analysis of relative labeled mass spectrometry data from complex samples using ANOVA. J. Proteome Res 7, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma J., Qin Z. S. (2007) Different normalization strategies for microarray gene expression traits affect the heritability estimation. BMC Proc 1, Suppl. 1, S154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.