Abstract
Verification of candidate biomarkers requires specific assays to selectively detect and quantify target proteins in accessible biofluids. The primary objective of verification is to screen potential biomarkers to ensure that only the highest quality candidates from the discovery phase are taken forward into preclinical validation. Because antibody reagents for a clinical grade immunoassay often exist for a small number of candidates, alternative methodologies are required to credential new and unproven candidates in a statistically viable number of serum or plasma samples. Using multiple reaction monitoring coupled with stable isotope dilution MS, we developed quantitative, multiplexed assays in plasma for six proteins of clinical relevance to cardiac injury. The process described does not require antibodies for immunoaffinity enrichment of either proteins or peptides. Limits of detection and quantitation for each signature peptide used as surrogates for the target proteins were determined by the method of standard addition using synthetic peptides and plasma from a healthy donor. Limits of quantitation ranged from 2 to 15 ng/ml for most of the target proteins. Quantitative measurements were obtained for one to two signature peptides derived from each target protein, including low abundance protein markers of cardiac injury in the nanogram/milliliter range such as the cardiac troponins. Intra- and interassay coefficients of variation were predominantly <10 and 25%, respectively. The configured multiplex assay was then used to measure levels of these proteins across three time points in six patients undergoing alcohol septal ablation for hypertrophic obstructive cardiomyopathy. These results are the first demonstration of a multiplexed, MS-based assay for detection and quantification of changes in concentration of proteins associated with cardiac injury in the low nanogram/milliliter range. Our results also demonstrate that these assays retain the necessary precision, reproducibility, and sensitivity to be applied to novel and uncharacterized candidate biomarkers for verification of proteins in blood.
Discovery of disease-specific biomarkers with diagnostic and prognostic utility has become an important challenge in clinical proteomics. In general, unbiased discovery experiments often result in the confident identification of thousands of proteins, hundreds of which may vary significantly between case and control samples in small discovery studies. However, because of the stochastic sampling of proteomes in discovery “omics” experiments, a large fraction of the protein biomarkers “discovered” in these experiments are false positives arising from biological or technical variability. Clearly discovery omics experiments do not lead to biomarkers of immediate clinical utility but rather produce candidates that must be qualified and verified in larger sample sets than were used for discovery (1).
Traditional, clinical validation of biomarkers has relied primarily on immunoassays because of their specificity and sensitivity for the target analyte and high throughput capability. However, antibody reagents for a clinical grade immunoassay often only exist for a short list of candidates. The development of a reliable sandwich immunoassay for one target protein is expensive, has a long development time, and is dependent upon the generation of high quality protein antibodies. For the large majority of new, unproven candidate biomarkers, an intermediate verification technology is required that has shorter assay development time lines, lower assay cost, and effective multiplexing of dozens of candidates in low sample volumes. Ideally the approach should be capable of analyzing hundreds of samples of serum or plasma with good precision. The desired outcome of verification is a small number of highly credentialed candidates suitable for traditional preclinical and clinical validation studies.
Multiple reaction monitoring (MRM)1 coupled with stable isotope dilution (SID) MS has recently been shown to be well suited for direct quantification of proteins in plasma (2–4) and has emerged as the core technology for candidate biomarker verification. MRM assays can be highly multiplexed such that a moderate number of candidate proteins (in the range of 10–50) can be simultaneously targeted and measured in the statistically viable number of patient samples required for verification (hundreds of serum samples). However, sensitivity for unambiguous detection and quantification of proteins by MS-based assays is often constrained by sample complexity, particularly when the measurements are being made in complex fluids such as plasma.
Many biomarkers of current clinical importance, such as prostate-specific antigen and the cardiac troponins, reside in the low nanogram/milliliter range in plasma and, until recently, have been inaccessible by non-antibody approaches. Our laboratory has recently shown for the first time that a combination of abundant protein depletion with limited fractionation at the peptide level prior to SID-MRM-MS provides robust limits of quantitation (LOQs) in the 1–20 ng/ml range with coefficient of variation (CV) of 10–20% at the LOQ for proteins in plasma (3).
Here we demonstrate that this work flow can be extended to configure assays for a number of known markers of cardiovascular disease and, more importantly, can be deployed to measure their concentrations in clinical samples. We modeled a verification study comprising six patients undergoing alcohol septal ablation treatment for hypertrophic obstructive cardiomyopathy, a human model of “planned” myocardial infarction (PMI), and obtained targeted, quantitative measurements for moderate to low concentrations of cardiac biomarkers in plasma. This work provides additional evidence that MS-based assays can be configured and applied to verification of new protein targets for which high quality antibody reagents are not available.
MATERIALS AND METHODS
Blood Collection and Processing
A total of six patients undergoing PMI using alcohol septal ablation for the treatment of symptomatic hypertrophic obstructive cardiomyopathy were included in this study. The protocol for obtaining blood from patients was approved by the Massachusetts General Hospital Institutional Review Board, and all subjects gave written informed consent. Inclusion criteria for patients to receive alcohol ablation treatment were as described previously (5). The most proximal accessible septal branch was instrumented using standard angioplasty guiding catheters and guide wires and 1.5 or 2.0-mm × 9-mm MaverickTM balloon catheters. Radiographic and echocardiographic contrast injections confirmed proper selection of the septal branch and balloon catheter position. Ethanol was infused through the balloon catheter at 1 ml/min. Additional injections in the same or other septal branches were administered as needed, causing cessation of blood flow to the isolated myocardium, and to reduce the gradient in the left ventricular outflow tract caused by the excessive heart muscle to <20 mm Hg (6).
Blood was drawn from femoral venous catheters at base line (just prior to the onset of the ablation) and at 10 min, 1 h, 2 h, 4 h, and 24 h following the onset of injury. Samples were collected in K2EDTA-treated tubes (BD Biosciences). All blood samples were centrifuged at 2000 × g for 10 min to pellet cellular elements within 10 min of sample collection. The supernatant plasma was then aliquoted and immediately frozen at −80 °C to minimize freeze-thaw degradation. Additional blood samples were sent to the clinical chemistry laboratory for evaluation of the standard cardiac markers creatine kinase, creatine kinase MB, and troponin T (Roche Diagnostics). For this study, only plasma sampled at base line and 4 and 24 h postinjury was analyzed by the targeted LC-MS/MS-based approach.
Labeled Peptide Internal Standards
Supplemental Table 1 lists the protein targets and their “signature peptides” for which final MRM assays were configured. Signature peptides have both high responses in electrospray LC-MS/MS and are sequence-unique when searched against a non-redundant human protein database (NCBInr). Signature peptides were identified for each protein by tryptic digest of the protein standards and analysis of the resulting peptides on a LTQ ion trap mass spectrometer as described previously (Fig. 1A) (3). C-reactive protein, sCD40L, myeloperoxidase, cardiac troponin T, and cardiac troponin I were purchased from EMD Chemicals Inc. (San Diego, CA); BNP and NT-proBNP were purchased from United States Biological (Swampscott, MA); and IL-33 was a kind gift from Dr. Richard Lee (Brigham and Women's Hospital, Boston, MA). Peptides for MRP14 were chosen based on previous knowledge. The detailed criteria for selection of peptides for synthesis and ultimately for assay configuration were described previously (3).
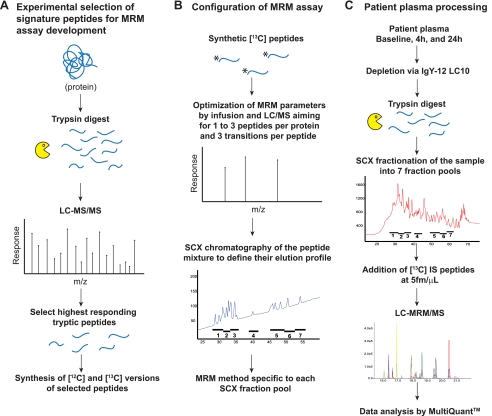
Fig. 1.
Assay configuration and sample preparation work flow. Work flow A represents the method used to select signature peptides for proteins associated with cardiac injury. Work flow B represents assay configuration conducted in parallel for MS instrument optimization and peptide separation by SCX. Work flow C represents the plasma processing and limited fractionation/MRM assay used for all six patients and time points (baseline and 4 and 24 h post injury). Three technical replicates and two process replicates for all samples were performed. IS, internal standard.
Stable isotope-labeled amino acids, [13C6]leucine (98.4 atom % isotopic enrichment), [13C6]isoleucine, and [13C5]valine (98 atom % isotopic enrichment) were purchased from Cambridge Isotope Laboratories (Andover, MA). Twenty-seven peptides derived from the target proteins were synthesized with a single, uniformly labeled [13C6]leucine, [13C6]isoleucine, or [13C5]valine using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry (Massachusetts Institute of Technology, Cambridge, MA). Unlabeled 12C forms of each peptide were synthesized by GL Biochem. For methionine-containing peptides derived from cardiac troponin T, NT-proBNP, and BNP-32, the sulfoxide forms of the same sequence were synthesized to account for this modification during sample processing. Synthetic peptides were purified to >90% purity and analyzed by amino acid analysis (Dana-Farber Cancer Institute, Boston, MA). Calculations of concentration were based upon the amino acid analysis.
Protein Depletion and Enzymatic Digestion
Plasma from all patients and time points was processed as described previously (3) (Fig. 1C). Briefly volumes of 0.8–1.2 ml of plasma per time point per patient were depleted of 12 high abundance proteins using an IgY-12 high capacity LC10 column (12.7 × 79 mm; GenWay Biotech, San Diego, CA) according to the manufacturer's instructions. Depleted plasma was concentrated to the original starting volume via Vivaspin 15R concentrators (5000 molecular weight cutoff; Vivascience, Hannover, Germany). Protein concentration of depleted, concentrated plasma was determined by Coomassie Plus Bradford assay (Pierce) and was 4.5 ± 1.1 mg/ml.
100 μl of depleted patient plasma was denatured with 6 m urea, 10 mm Tris, pH 8.0; reduced with 20 mm dithiothreitol at 37 °C for 30 min; and alkylated with 50 mm iodoacetamide at room temperature in the dark for 30 min. Urea concentration was diluted 10-fold with water prior to overnight digestion at 37 °C with porcine trypsin (sequencing grade modified; Promega, Madison, WI) using a 1:50 (w/w) enzyme to substrate ratio. Digests were terminated with formic acid to a final concentration of 1% and desalted using Oasis HLB 1cc (30 mg) reversed phase cartridges (Waters) as described previously (3). Eluates were frozen, dried to dryness via vacuum centrifugation, and stored at −80 °C.
Strong Cation Exchange Chromatography (SCX)
Digested plasma samples from each patient and time point were normalized to total protein amount such that ∼350 μg of total protein was injected per patient per time point. Each sample was processed in duplicate. Detailed methods for SCX fractionation can be found in Keshishian et al. (3). Briefly the fractionation was done on an Agilent 1100 capillary LC system (Agilent Technologies, Palo Alto, CA) using a BioBasic 1 × 250-mm column (ThermoFisher, San Jose, CA) at a flow rate of 50 μl/min and mobile phases that consisted of 5 mm potassium phosphate in 25% acetonitrile, pH 3.0 (A) and 500 mm potassium chloride in 5 mm potassium phosphate, 25% acetonitrile, pH 3.0 (B). After loading the sample onto the column, the mobile phase was held at 1% B for 15 min. Peptides were then separated with a linear gradient of 1–20% B in 20 min, 20–40% B in 10 min, and 40–100% B in 10 min. Fractions were collected every minute, and acetonitrile was removed from collected fractions by vacuum centrifugation. The elution profile of the peptide internal standards was predefined and used to generate seven pools of SCX fractions for MRM analysis per patient per time point (Fig. 1B). Each pool was desalted using Oasis HLB 1cc (10 mg) reversed phase cartridges as described previously (3) and stored at −80 °C until LC-MRM/MS analysis. To ensure detection of peptides that eluted in or near the void volume of the SCX column (e.g. fractions 1–2), the flow-through was desalted and analyzed by LC-MRM/MS.
LC-MRM/MS
Pooled SCX fractions were reconstituted in 25 μl of 5% formic acid, 3% acetonitrile and spiked with a mixture of 13C-labeled peptides for a final concentration of 5 fmol/μl. Nano-LC-MRM/MS/MS was performed on a 4000 Q Trap hybrid triple quadrupole/linear ion trap mass spectrometer coupled to a Tempo LC system (Applied Biosystems, Foster City, CA). Chromatography was performed with Solvent A (0.1% formic acid) and Solvent B (90% acetonitrile in 0.1% formic acid). Full loop injection of 1 μl of each sample was done in triplicate on PicoFrit columns (75-μm inner diameter, 10-μm tip opening; New Objective, Woburn, MA) packed in house with 11–12 cm of ReproSil-Pur C18-AQ 3-μm reversed phase resin (Dr. Maisch, GmbH). Sample was eluted at 300 nl/min with a gradient of 3–20% Solvent B for 3 min, 20–55% solvent B for 35 min, and 55–80% solvent B for 3 min. Data were acquired with an ion spray voltage of 2200 V, curtain gas of 20 p.s.i., nebulizer gas of 3 p.s.i., and an interface heater temperature of 150 °C. Collision energy (7), declustering potential and collision cell exit potential were optimized for maximum transmission and sensitivity of each MRM transition with infusion of each peptide standard and the Quantification Optimization function provided in Analyst (Fig. 1B). Identical declustering potential, collision energy, and collision cell exit potential values were used for each 12C/13C pair. A dwell time of 10–75 ms was used for different SCX fraction pools based upon the number of peptides and transitions monitored in that pool. In all experiments cycle times did not exceed 1 s, and a minimum of five to six data points were collected per peak. Three MRM transitions per peptide (supplemental Table 1) were monitored and acquired at unit resolution both in the first and third quadrupoles (Q1 and Q3) to maximize specificity. In general, transitions were chosen based upon relative abundance and m/z greater than the precursor m/z in the full-scan MS/MS spectrum recorded on the 4000 Q Trap mass spectrometer. The final MRM method included 162 optimized MRMs for nine target proteins. These MRMs were distributed among seven SCX fractions in accordance with the elution profile of the synthetic peptides.
Data Analysis
Data analysis was performed using MultiQuantTM software (AB/MDS Sciex, Foster City, CA). The relative ratios of the three transitions selected and optimized for the final MRM assay were predefined in the absence of plasma (i.e. in buffer) for each peptide using the 13C internal standards (3). The most abundant transition for each pair was used for quantification unless interference from the matrix was observed. In these cases, another transition free of interference was chosen for quantification.
12C/13C peak area ratios were used to calculate concentrations of target proteins in plasma by the following equation: Measured concentration = (Peak area ratio)(5 fmol/μl internal standard)(Protein molecular weight)(Analysis volume)/(Process volume)/1000 (see above). Intra-assay imprecision (CV) for each peptide was based on the calculated average protein concentration for a set of triplicate injections. Interassay CV was calculated for each peptide from two process replicates per time point per patient. All data are summarized in supplemental Table 2.
Standard Addition Experiments
The limit of detection (LOD) and LOQ for each signature peptide derived from the target proteins were determined in SCX matrix by the method of standard addition. Female plasma from a healthy donor was immunoaffinity-depleted, digested, and SCX-fractionated to generate SCX-fractionated plasma pools. Response curves ranging from 0.1 to 10 fmol/μl were generated for each 12C-peptide by spiking them into corresponding SCX pools. Labeled internal standard peptides were spiked in at a constant concentration of 5 fmol/μl prior to LC-MRM/MS. This concentration range is equivalent to 5–800 ng/ml of corresponding target proteins. For this study a modification of the Linnet and Kondratovich (8) method was used for calculating LOD. The LOD was based on the variance of the blank sample and the variance of the lowest level spike-in sample. Assuming a type I error rate α = 0.05 for deciding that the analyte is present when it is not and a type II error rate β = 0.05 for not detecting the analyte when it is present, the LOD was derived as follows.
The limit of blank (LOB) was defined as the 95th percentile of the blank samples. This was estimated as meanb + t1 − α × S.D.b where meanb and S.D.b are the mean and standard deviation of the blank sample, and S.D.s is the standard deviation of the low level spike. For a relatively small number of repeated measurements for the blank, cβ was approximated as t1 − β. t1 − α and t1 − β are the (1 − α) and (1 − β) percentile, respectively, of the standard t distribution on (n − 1) degrees of freedom where the blank and low level spike samples have n = 3 replicate measurements. In this study, the LOD for the sample is obtained from the three replications as measured by the best transition. When three values are averaged to obtain the final measurement, the LOD calculation requires the S.D. estimates to be divided by [rad]3[/rad] = 1.73. Because α = β = 0.05, the LOD equation becomes Equation 2.
Once the LOD was determined separately for each transition of each peptide, the LOQ was calculated using the customary relation: LOQ = 3 × LOD (9).
Depending on the LC-MS matrix in which the analyte is measured, an endogenous non-zero analyte level may exist even in the blank sample for some of the peptides under consideration. This endogenous level of an analyte was estimated by using the linear part of the concentration curve. A robust linear fit using least median squares regression (10) was performed. The 99% confidence interval of the regression line intercept was calculated using bootstrap estimation (11). If the lower limit of the confidence interval is positive, then the analyte is deemed to have an endogenous level equal to the regression intercept. If the lower 99% confidence interval is zero or negative, there is no expected endogenous level for that analyte.
ELISA
Plasma concentrations of CRP, MPO, MRP14, NT-proBNP, and cTnT for patients 1–3 before and after immunoaffinity depletion were determined by immunoassay at the Department of Laboratory Medicine at Children's Hospital (Boston, MA). Samples were analyzed by the following kits or systems: CRP, DiaSorin; MPO, Alpco; MRP14, Peninsula; NT-proBNP, Cobas, Roche Diagnostics; and cTnT, Elecsys TnT.
RESULTS
Fig. 1 illustrates the strategy used for sample processing and assay configuration. Peptides derived from the target proteins (supplemental Table 1) were selected based upon experimental data obtained by tryptic digestion of standard protein and subsequent analysis by LC-MS/MS (Fig. 1A). Two to five peptides per protein were selected based upon criteria described previously (3) and synthesized both in 12C and 13C forms. Assay configuration was conducted to optimize the SCX peptide separation and MS instrument parameters to maximize detection of the target peptides (Fig. 1B). For peptide fractionation via SCX, the elution profile of all signature peptides was evaluated to determine the pooling strategy for patient samples. For MRM acquisition, three transitions per peptide were selected and monitored to achieve maximum selectivity and sensitivity in the MRM assay. Peptides that were synthesized but that did not make the final MRM assay after optimization were dropped because of poor electrospray response or poor chromatographic behavior by reversed phase. The final MRM assay included two to four peptides per protein (supplemental Table 1). Fig. 1C shows the final limited fractionation/MRM assay in which patient plasma was depleted of high abundance plasma proteins before reduction, alkylation, and tryptic digestion. Peptides were minimally fractioned by SCX, pooled according to a predefined elution profile, and analyzed by LC-MRM/MS following addition of heavy labeled internal peptide standards. Three technical replicates were performed for each fraction, and two process replicates were performed across all patients and time points.
Determination of LOD/LOQ by Method of Standard Addition
12C-Peptides derived from each of the target proteins were added to SCX-fractionated plasma from a healthy donor to determine the LOD and LOQ for each peptide as described under “Materials and Methods.” Response curves for all peptides are shown in supplemental Fig. 1. For seven of nine proteins, at least one signature peptide derived from the target protein had an LOD in plasma that ranged from 0.052 (cTnT) to 0.287 fmol/μl (MPO) and an LOQ in plasma that ranged from 0.157 to 0.862 fmol/μl (Table I). These molar concentrations are equivalent to 0.9–24 and 5–72 ng/ml target protein for LOD and LOQ, respectively. Values for LOD and LOQ listed in Table I for CRP and MRP14 reflect endogenous levels of these proteins detected in the background plasma (equivalent to ∼50 and 3 ng/ml for CRP and MRP14, respectively) as determined by SID-MRM-MS. The concentration of CRP in the background plasma was also determined by clinical immunoassay and found to be 250 ng/ml. In general, the values measured for CRP by clinical immunoassay for all samples assayed (including patient plasma) were higher then those obtained by MRM (see below). This trend emphasizes that MRM assays provide relative quantitation between cases and controls and not absolute quantitation.
Table I.
LOD/LOQ for target peptides in healthy, female plasma
LOD/LOQ were determined by standard addition performed in depleted, digested, and SCX-fractionated plasma from a healthy donor. Mox, methionine sulfoxide; Camc, carbamidomethyl cysteine.
| Protein | Molecular mass | Signature peptide | LOD |
LOQ |
||
|---|---|---|---|---|---|---|
| fmol of peptide | ng/ml proteina | fmol of peptide | ng/ml proteina | |||
| kDa | ||||||
| NT-proBNP | 11 | EVATEGIR | 0.334b | 3.77 | 1.003b | 11.31 |
| MVLYTLR | 0.082c | 0.97 | 0.246c | 2.78 | ||
| MoxVLYTLR | 0.098c | 1.11 | 0.294c | 3.32 | ||
| MRP14 | 13 | LTWASHEK | 0.397b,d | 5.25 | 1.190b,d | 15.76 |
| LGHPDTLNQGEFK | 0.475c,d | 6.29 | 1.425c,d | 18.87 | ||
| BNP-32 | 15 | ISSSSGLGCamcK | 0.074c | 0.256 | 0.223c | 0.768 |
| MVQGSGCamcFGR | 0.081b | 0.281 | 0.244b | 0.843 | ||
| MoxVQGSGCamcFGR | 0.216b | 0.743 | 0.647b | 2.23 | ||
| Troponin I | 24 | TLLLQIAK | 0.088b | 2.11 | 0.265b | 6.33 |
| NITEIADLTQK | 0.141c | 3.38 | 0.424c | 10.13 | ||
| NIDALSGMEGR | 0.282b | 6.74 | 0.847b | 20.22 | ||
| CRP | 25 | ESDTSYVSLK | 1.454c,d | 36.40 | 4.361c,d | 109.19 |
| GYSIFSYATK | 1.949b,d | 48.80 | 5.847b,d | 146.41 | ||
| sCD40L | 29 | TTSVLQWAEK | 0.202b | 5.92 | 0.606b | 17.75 |
| EASSQAPFIASLCamcLK | 0.286c | 8.38 | 0.859c | 25.15 | ||
| SQFEGFVK | 0.203c | 5.94 | 0.609c | 17.83 | ||
| SLSLLNCEEIK | 0.313b | 9.16 | 0.939b | 27.49 | ||
| IL-33 | 31 | DNHLALIK | 0.606c | 18.64 | 1.818c | 55.93 |
| TDPGVFIGVK | 0.172b | 5.28 | 0.515b | 15.84 | ||
| DFWLHANNK | 0.271c | 8.32 | 0.812c | 24.98 | ||
| VLLSYYESQHPSNESGDGVDGK | 0.308b | 9.46 | 0.923b | 28.38 | ||
| Troponin T | 34 | VLAIDHLNEDQLR | 0.154e | 5.30 | 0.461e | 15.89 |
| SFMPNLVPPK | 0.126e | 4.35 | 0.378e | 13.04 | ||
| SFMoxPNLVPPK | 0.196e | 6.76 | 0.588e | 20.28 | ||
| YEINVLR | 0.052c | 1.80 | 0.157c | 5.40 | ||
| MPO | 84 | IPCamcFLAGDTR | 0.287c | 24.09 | 0.862c | 72.27 |
| IANVFTNAFR | 0.329b | 27.63 | 0.988b | 82.90 | ||
a Equivalent ng of protein/1 ml of plasma.
b LOD and LOQ values are reported for the third transition of the peptide.
c LOD and LOQ values are reported for the second transition of the peptide.
d LOD and LOQ values reflect endogenous levels detected in the background plasma from a healthy donor.
e LOD and LOQ values are reported for the first transition of the peptide.
Quantification of Target Proteins in Planned Myocardial Infarction Patients
Plasma from six patients undergoing septal ablation was sampled at base line (just prior to the ablation), 4 h, and 24 h after onset of injury. The septal ablation recapitulated important clinical features of spontaneous myocardial infarction, including substernal chest pain and electrocardiographic changes, as well as the development of echocardiographic evidence of septal wall motion abnormalities as described previously (7, 12, 13). The standard biochemical metrics of myocardial injury, creatine kinase MB and troponin T, were within normal limits prior to septal ablation for the six patients used in this study and increased to 140 ± 67 and 5.2 ± 4.4 ng/ml 4 h after the onset of the injury, respectively, as assessed in the hospital clinical laboratory on a Roche Diagnostics Elecsys platform.
Reproducibility of the plasma digestions and the SCX separations is shown in supplemental Fig. 2. Elution profiles for duplicate SCX separations of the base-line, 4-h, and 24-h samples for a single patient (A) and one replicate SCX separation of the base-line sample across four of six patients (B) are comparable. This indicates that the digestion efficiency of the plasma and subsequent SCX peptide fractionation was consistent across all samples and replicates.
Quantitative measurements were successfully obtained for eight of the 27 peptides in all six patients for which MRM assays were configured (supplemental Table 1). These eight peptides represent six of the nine target proteins and are derived from CRP, MRP14, MPO, cTnT, cTnI, and NT-proBNP. No peptides derived from IL-33, BNP-32, and sCD40L were detected most likely because their concentrations in plasma were below the LOD of the MRM assays. MRM analysis of recombinant IL-33 spiked into plasma from a healthy donor indicated that IL-33 had a recovery of >80% through all processing steps including depletion, ultrafiltration, and digestion (supplemental Table 3). Therefore, we can conclude that in the case of IL-33 the plasma concentration in the PMI patients was below 4.7 ng/ml, our LOD for IL-33 by SID-MRM-MS.
Quantification of Moderately Abundant Cardiovascular Biomarkers in Plasma of PMI Patients
In this study, moderately abundant proteins were defined as proteins with a measured concentration of >50 ng/ml in the MRM assay (e.g. CRP and MRP14). Each protein was represented by two distinct signature peptides. Overall 74% of the quantitative measurements for CRP and MRP14 in patient plasma across the time course had an intra-assay CV of <10, and 24% of the MRM measurements had a CV between 10 and 20% (supplemental Table 2). Only 2% of the measurements had an intra-assay CV of >20%. Interassay CV of the process replicates was <25% for the majority of MRM measurements (78%) with the remaining 20% of the measurements having an interassay CV between 25 and 46% and 2% having interassay CVs >70%. Overall greater variability was observed for patient 1 likely due to the fact that optimization of the work flow was still ongoing during processing of the first replicate.
Overall CRP concentrations in patient plasma ranged from a median value of 130 ng/ml in base-line samples to 2.4 μg/ml at 24 h for both surrogate peptides. Although the determined concentrations of the two CRP peptides differed from one another, the trend for both peptides clearly showed very significant increases in concentration 24 h after cardiac injury (Fig. 2A) as reported previously (14). The difference is most likely because of variation in digestion and recovery efficiency for each peptide and is consistent with previous findings for CRP measured by MRM in plasma (2). Supplemental Fig. 3A shows the median concentration for each peptide across all six patients and time points. Although the concentration of CRP in patient 4 as determined by the ESDTSYVSLK peptide showed 10–30 times less CRP than the other five patients, the trend of increasing concentration across the time course was consistent (supplemental Table 2).
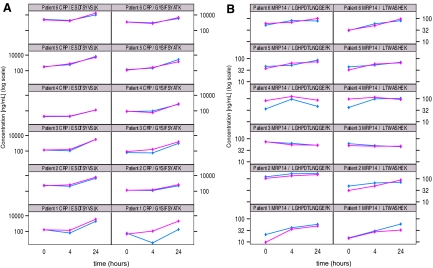
Fig. 2.
Quantitation of moderately abundant proteins in patient plasma samples. Line plots of log(concentration) versus time of CRP (A) and MRP14 (B) at baseline and 4 and 24 h post injury as measured by MRM assay with two different signature peptides for each protein across six patients. Pink and blue traces indicate two different process replicates for each patient and time point.
The concentration of MRP14 in patient plasma was also measured using two different signature peptides, and the median values for each peptide across all six patients and time points are shown in supplemental Fig. 6B. The concentration of MRP14 as measured in the MRM assay generally increased from base line to 4 h and continued to rise at 24 h for four of six patients. In patients 3 and 4, the measured concentrations of MRP14 decreased over time, peaking at 4 h. These trends were consistent for both peptides derived from MRP14 across both process replicates (Fig. 2B).
Quantification of Low Abundance Cardiovascular Biomarkers in Plasma of PMI Patients
In this study, low abundance proteins were defined as proteins with a measured concentration of <50 ng/ml in the MRM assay. MPO, cTnI, cTnT, and NT-proBNP reside in this category, and we confirmed these levels for MPO, cTnT, and NT-proBNP by immunoassay (supplemental Table 4). Each of these proteins was represented by one distinct peptide. Overall 46% of the quantitative measurements for these four proteins in patient plasma across the time course had an intra-assay CV of <10%, and ∼40% of the MRM measurements had an intra-assay CV between 10 and 20% (supplemental Table 2). The remaining 14% of the measurements had an intra-assay CV of >20%. Because the measured concentrations were predominantly <5 ng/ml, signal to noise ratios for peptides derived from MPO, cTnI, cTnT, and NT-proBNP were generally lower than for peptides derived from CRP and MRP14, resulting in increased CV (supplemental Fig. 4). Interassay CV of the process replicates was <25% for approximately two-thirds of the MRM measurements with another 25% of the measurements having interassay CVs between 25 and 40%. Approximately one-tenth of the measurements had CVs greater than 40% where detection of peptides was below 1 ng/ml.
For MPO, the plasma concentration in PMI patients peaked at 4 h and then returned to base-line concentration (Fig. 3A and supplemental Fig. 3C). This trend was consistent for four of six patients. The highest level of MPO was detected in patient 2 (96 ng/ml). Patients 5 and 6 had the lowest concentrations of MPO ranging from 1.5 to 3.3 ng/ml and showed minimal change over time.
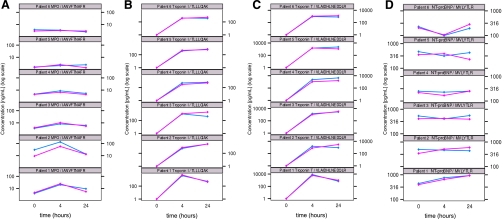
Fig. 3.
Quantitation of low abundance proteins in patient plasma samples. Line plots of log(concentration) versus time of MPO (A), cTnI (B), cTnT (C), and NT-proBNP (D) at baseline and 4 and 24 h post injury as measured by MRM assay with one representative signature peptide for each protein across six patients. Pink and blue traces indicate two different process replicates for each patient and time point.
cTnI was determined to be quantified at high pg/ml concentrations up to low ng/ml concentrations in plasma samples from all six patients using the peptide TLLLQIAK (Fig. 3B). Median concentrations of 0.5 and 0.6 ng/ml were measured in patient plasma sampled at 4 and 24 h, respectively, after cardiac injury (supplemental Fig. 3D). cTnI concentrations were slightly elevated from 4 to 24 h in all patients except patient 1 where the concentration decreased at 24 h with respect to 4 h. No peptides derived from cTnI or cTnT were observed in plasma sampled at base line from any of the PMI patients (Fig. 3, B and C).
For cTnT, peptide VLAIDHLNEDQLR was consistently detected in plasma sampled at 4 and 24 h post-PMI in all six patients (Fig. 3C). Median concentrations of 1.4 and 1.8 ng/ml were measured in patient plasma sampled at 4 and 24 h, respectively, after cardiac injury (supplemental Fig. 3E). Plasma concentrations of cTnT were slightly elevated in four of six patients from 4 to 24 h and generally resided in the 1–8 ng/ml range. In patient 1 the concentration of cTnT was elevated to an average of 7.12 ng/ml at 4 h across two process replicates; however, the concentration decreased at 24 h in this same patient consistent with the ELISA measurements for this patient (see below). No change in cTnT was observed in patient 6. A second peptide derived from cTnT (YEINVLR) was detected in four of six patients, and calculated concentrations of cTnT based upon this peptide followed the same trend as that for peptide VLAIDHLNEDQLR (data not shown).
Plasma concentrations of NT-proBNP were consistently the lowest observed in this study (Fig. 3D), ranging from 100 to 930 pg/ml across all samples and replicates. A median concentration of 0.4 ng/ml was measured in patient plasma sampled at all three time points. (supplemental Fig. 3F). There was no significant change observed across the time course for any patient. The signature peptide derived from NT-proBNP was also observed in the oxidized form (methionine sulfoxide) at subnanogram/milliliter concentrations in plasma sampled at 4 and 24 h in three of four patients. The measured concentration for the methionine sulfoxide form was one-fourth to one-half that of the non-oxidized form in two patients, whereas the measured concentrations of both forms were equivalent in the remaining patient. Oxidation of Met is a common artifact of sample handling and will always be observed to varying extents in protein digests.
Correlation of Protein Concentration Determined by MRM-MS Assay and Immunoassay in Plasma of PMI Patients
Clinical immunoassays were used to define the amounts of CRP, MRP14, MPO, cTnT, and NT-proBNP present in the plasma of patients 1–3 and to define the losses of these proteins following immunoaffinity depletion and ultrafiltration of the plasma using the GenWay IgY-12 column. The levels of cTnT, MRP14, and NT-proBNP were all in the low (<13) ng/ml level in patient plasma, whereas levels of MPO and CRP ranged from tens of ng/ml to μg/ml levels, respectively. After depletion and ultrafiltration, 50–70% of MPO, NT-proBNP, and CRP proteins were recovered (supplemental Table 4). In contrast, only ∼15% of cTnT was recovered indicating that substantial amounts of this protein are being lost either to the depletion column or to the ultrafiltration membrane used postdepletion. Despite this high loss of protein that reduced the amounts of cTnT as measured by ELISA to levels ranging from below detectable to ∼2 ng/ml, the intra- and interassay CVs for cTnT were predominantly ≤20 and ≤30%, respectively, using MRM-MS (supplemental Table 2).
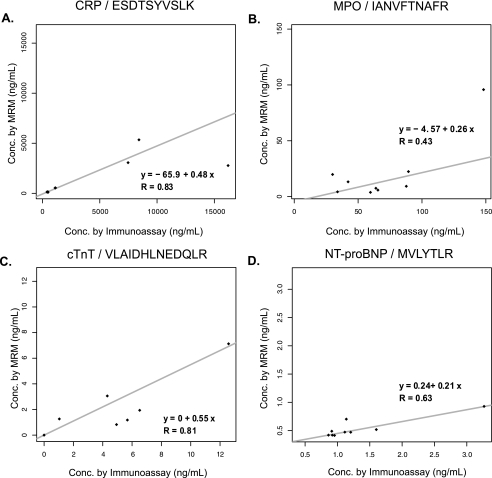
We also examined the correlation of the protein concentrations obtained by immunoassay of non-depleted plasma samples with the values obtained by depletion, fractionation, and MRM. Although absolute concentrations measured by the assays were not identical, rank regression (15) and Spearman rank correlation analysis (16) indicate that the temporal trends observed among patients were consistent. Fig. 4, A and B, show the correlation (r) between both assays for CRP (r = 0.83) and MPO (r = 0.43), respectively. Correlation for cTnT and NT-proBNP was 0.81 and 0.63, respectively (Fig. 4, C and D). The rank regression line is also shown in Fig. 4. Supplemental Fig. 5 shows the plots of concentration versus time for MRP14 across patients 1 and 2 as determined by MRM for both surrogate peptides (LGHPDTLNQGEFK (pink) and LTWASHEK (green)) and by commercial immunoassay for the intact protein (blue).
Fig. 4.
Comparison of MRM assay with clinical immunoassay. Rank regression analysis of protein concentrations obtained by immunoassay of non-depleted plasma samples and values obtained by MRM for CRP (A), MPO (B), cTnT (C), and NT-proBNP (D) is shown. Spearman rank correlation (R) is calculated for each peptide/protein.
The measured concentrations for the target proteins in patient plasma by SID-MRM-MS were generally lower then those obtained by immunoassay of both non-depleted and depleted plasma samples (supplemental Tables 2 and 4). This differential reflects sample loss incurred from plasma processing that includes depletion, ultrafiltration, and digestion of each candidate protein as well as postdigestion processing and separation of the signature peptides. Because losses resulting from the multistep processing are not accounted for by adding labeled internal standard peptides just prior to LC-MRM/MS (as was done in this study), the measured concentrations by MRM assay are an underestimate of the protein concentrations in the initial plasma sample. Addition of labeled peptides earlier in the plasma processing or the use of the identical form of intact protein labeled with heavy stable isotopes would help to account for sample loss and improve the accuracy and precision of the MRM measurements.
DISCUSSION
Proteomics-based biomarker discovery experiments can produce hundreds to thousands of candidate biomarkers that must be further credentialed before the substantial investments in time and money are committed for their clinical evaluation. This credentialing process is referred to as verification, and its primary objective is to screen potential biomarkers to ensure that only the highest quality candidates from the discovery phase are taken forward into validation. Verification has been the bottleneck in biomarker discovery primarily for two reasons. First, technologies capable of testing large numbers of protein biomarker candidates in even moderate sample sets have not been available, and second, the dynamic range of proteins in plasma makes this medium extremely challenging for detecting and reliably quantifying low abundance plasma proteins (<100 ng/ml). Our goal here was to define the analytical performance of SID-MRM-MS combined with minimal fractionation/MRM using existing and emerging protein biomarkers of cardiovascular disease present at the low ng/ml range or higher.
We successfully configured multiplexed, quantitative assays that coupled a plasma processing work flow with SID-MRM-MS for known protein biomarkers related to cardiovascular injury, including CRP, MRP14, MPO, cTnI, cTnT, and NT-proBNP. The expected plasma concentrations of these proteins in patients with cardiovascular disease span 3–4 orders of magnitude in abundance, ranging from pg/ml to μg/ml levels, and ELISAs were available for most of them. Therefore, these proteins represented an excellent opportunity to assess the precision, reproducibility, and sensitivity of SID-MRM-MS assays across a wide range of concentrations. Our results show that reliable detection and precise, relative quantification can be achieved for proteins in the high pg/ml to low ng/ml range (and higher) in patient plasma without antibody reagents or affinity enrichment. Intra- and interassay CVs for the moderately abundant proteins CRP and MPO were consistently ≤10 and ≤25%, respectively. Intra- and interassay CVs for MPO, cTnI, cTnT, and NT-proBNP were predominantly ≤25 and ≤40%, respectively. The good inter- and intra-assay performance obtained indicates that variability introduced by such sample processing steps as immunoaffinity depletion, enzymatic digestion, and SCX fractionation is small and within acceptable standards for analytical assays (17). Overall these results show, for the first time, that multiplexed MRM-MS assays can be configured and used reliably and reproducibly to measure endogenous levels of clinically relevant proteins in patient plasma without affinity enrichment when these proteins are recovered in the 1–10 ng/ml concentration range after processing.
Real world verification studies could use the sample processing protocol described here and be completed within an acceptable amount of time. For example, using three process replicates, each run in singlicate, 100 case and 100 control samples could be analyzed in approximately 10 months using a single MS system with dual column switching. Importantly the number of signature peptides analyzed could readily be increased to 50 or more with little effect on this time line as the number of SCX fractions analyzed would not change. To effectively analyze an even higher number of patient samples in a verification study without increasing study duration, pooling strategies could also be used (18).
Our objective in these studies was not to replace current clinical grade assays that are used by clinical cardiologists for diagnosis, therapy, and/or risk stratification. Rather our goal was to generate a multiplexed assay for existing markers on which data from new proteins can be overlaid (14). As noted above, ELISAs for the troponin proteins and other cardiovascular biomarker proteins are already in routine clinical use and precisely measure these proteins in the low picogram-to-nanogram/milliliter range (19–22). However, although the SID-MRM-MS verification technology described here is clearly not yet competitive with the most sensitive clinical assays currently in use, the present studies support the feasibility of this technology to rapidly and reliably configure quantitative assays for proteins in cases where high quality sandwich immunoassays do not exist, which at present is the majority of candidate disease biomarker proteins. This is demonstrated here for IL-33 for which no reliable commercial immunoassay is currently available.
Although detection of proteins in the mid-to-low pg/ml range in plasma is not yet possible by SID-MRM-MS, there are alternative assays being explored that combine the advantages of specific immunoaffinity enrichment of either the target protein or a signature peptide derived from the target protein with the structural specificity and quantitative capabilities of MRM. For example, a recent study by Ackermann and co-workers (23) reported an MS-based assay for NT-proBNP that took advantage of the commercial availability of an ELISA grade antibody to immunoprecipitate the target protein prior to digestion and MRM analysis. A lower LOQ of 100 pg/ml was achieved. However, this approach is unlikely to be a general solution for candidate biomarker verification because the high quality antibodies required for immunoprecipitation of the intact proteins are presently not available for the majority of candidates. In Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) anti-peptide antibodies are used that are relatively easy to produce and have shown a high success rate for selective enrichment of their target peptides from complex mixtures (24, 25) SISCAPA assays for clinically relevant proteins are just beginning to be developed. Hoofnagle et al. (25) recently presented the first example of SISCAPA-MRM assays for a clinically relevant biomarker, serum thyroglobulin, where the currently available ELISAs are subject to interference and erroneous results. Good assay performance with an LOQ of ∼3 ng/ml was demonstrated. We recently reported on the development of a multiplexed SISCAPA assay to measure cTnI and IL-33 in plasma that demonstrated detection sensitivities and precision similar to those of the thyroglobulin assay (26). Although the cost of SISCAPA-MRM-MS assays are higher than for MRM-MS assays because of the price of producing and qualifying the anti-peptide antibodies, no fractionation of the plasma is required to achieve performance similar to that of MRM-MS, greatly simplifying sample processing prior to MS analysis.
Accurate quantification of proteins by SID-MRM-MS is theoretically possible using, for example, isotopically labeled versions of the candidate proteins for use as internal standards. However, such standards are generally not available, and therefore accurate quantification cannot be accomplished in the absence of knowledge of protein and peptide recovery following every sample handling step used, including depletion, membrane concentration, enzymatic digestion, peptide fractionation, and desalting. At present, the best way to achieve accurate quantitation would require synthesis of isotopically labeled versions of the candidate proteins for use as internal standards so that losses could be accounted for. Clearly this is not practically achievable for all proteins of interest. Here and previously (3) we have emphasized that the analytical goal of SID-MRM-MS is to achieve precise, relative quantitation of protein abundance between cases and controls. Accuracy, although an important goal, is not required to use SID-MRM-MS for verification of candidate biomarkers in patient samples; reproducibility of assay performance (e.g. CVs of ≤25%) and sufficient sensitivity to detect and quantify the analyte in the linear range for the assay are required. Despite loss at the depletion/ultrafiltration step of between 50 and 85% of the proteins evaluated (as determined by ELISA and MRM; supplemental Tables 3 and 4), we were still able to configure robust assays suitable for precise, relative quantification of these proteins, three of which were present in the original plasma samples at the low ng/ml range. Although the depletion/ultrafiltration process can introduce significant losses, it significantly decreases the dynamic range and complexity of plasma, reduces biological “noise” that can interfere with the MRM-MS measurements, and enables (together with limited peptide fractionation by SCX) precise measurement of signature peptides from proteins present in plasma at low ng/ml levels.
In conclusion, we used known markers of cardiac injury to demonstrate how simple sample processing and SID-MRM-MS can be used to readily configure multiplexed assays to quantify clinically relevant proteins in patient plasma with concentrations that span 4 orders of magnitude. In addition, we demonstrated that these assays have the necessary precision, reproducibility, and sensitivity to be applied to new and uncharacterized candidate biomarkers for verification studies. This study is further demonstration that SID-MRM-MS is a valuable technology in a systematic biomarker pipeline.
Supplementary Material
Acknowledgments
We thank Dick Cook, Rick Schiavoni, and Katie Tone from the Biopolymers Laboratory at the Massachusetts Institute of Technology for the synthesis and purification of the heavy labeled peptides and Will Beavers and Shannon Kerivan from the Dana-Farber Cancer Institute for amino acid analysis of all protein and peptide stocks. We also thank Nadar Rifai and Gary Bradwin from Children's Hospital Boston and Harvard Medical School for assistance in obtaining the immunoassay data for patients 1–3.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grants 1U24 CA126476-02 from the NCI as part of the NCI's Clinical Proteomic Technologies Initiative and U01-HL081341 from the NHLBI (to S. A. C., R. E. G., and M. S. S.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- MRM
- multiple reaction monitoring
- SID
- stable isotope dilution
- CV
- coefficient of variation
- PMI
- planned myocardial infarction
- CRP
- C-reactive protein
- MRP14
- myeloid related protein 14
- MPO
- myeloperoxidase
- cTnI
- cardiac troponin I
- cTnT
- cardiac troponin T
- BNP
- B-type natriuretic peptide
- NT-proBNP
- N-terminal prohormone B-type natriuretic peptide
- IL-33
- interleukin 33
- sCD40L
- soluble CD40 ligand
- SCX
- strong cation exchange chromatography
- LOQ
- limit of quantitation
- LOD
- limit of detection
- SISCAPA
- Stable Isotope Standards and Capture by Anti-Peptide Antibodies.
REFERENCES
- 1.Rifai N., Gillette M. A., Carr S. A. (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol 24, 971–983 [DOI] [PubMed] [Google Scholar]
- 2.Kuhn E., Wu J., Karl J., Liao H., Zolg W., Guild B. (2004) Quantification of C-reactive protein in the serum of patients with rheumatoid arthritis using multiple reaction monitoring mass spectrometry and 13C-labeled peptide standards. Proteomics 4, 1175–1186 [DOI] [PubMed] [Google Scholar]
- 3.Keshishian H., Addona T., Burgess M., Kuhn E., Carr S. A. (2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bondar O. P., Barnidge D. R., Klee E. W., Davis B. J., Klee G. G. (2007) LC-MS/MS quantification of Zn-alpha 2 glycoprotein: A potential serum biomarker for prostate cancer. Clin. Chem 53, 673–678 [DOI] [PubMed] [Google Scholar]
- 5.Lewis G. D., Wei R., Liu E., Yang E., Shi X., Martinovic M., Farrell L., Asnani A., Cyrille M., Ramanathan A., Shaham O., Berriz G., Lowry P. A., Palacios I. F., Ta°an M., Roth F. P., Min J., Baumgartner C., Keshishian H., Addona T., Mootha V. K., Rosenzweig A., Carr S. A., Fifer M. A., Sabatine M. S., Gerszten R. E. (2008) Metabolite profiling of blood from individuals undergoing planned myocardial infarction reveals early markers of myocardial injury. J. Clin. Investig 118, 3503–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baggish A. L., Smith R. N., Palacios I., Vlahakes G. J., Yoerger D. M., Picard M. H., Lowry P. A., Jang I. K., Fifer M. A. (2006) Pathological effects of alcohol septal ablation for hypertrophic obstructive cardiomyopathy. Heart 92, 1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakkis N. M., Nagueh S. F., Kleiman N. S., Killip D., He Z. X., Verani M. S., Roberts R., Spencer W. H., 3rd (1998) Echocardiography-guided ethanol septal reduction for hypertrophic obstructive cardiomyopathy. Circulation 98, 1750–1755 [DOI] [PubMed] [Google Scholar]
- 8.Linnet K., Kondratovich M. (2004) Partly nonparametric approach for determining the limit of detection. Clin. Chem 50, 732–740 [DOI] [PubMed] [Google Scholar]
- 9.Currie L. A. (1968) Limits for qualitative detection and quantitative determination. Anal. Chem 40, 586–593 [Google Scholar]
- 10.Venables W. N., Ripley B. D., Venables W. N. (2002) Modern Applied Statistics with S, 4th Ed., Springer, New York [Google Scholar]
- 11.Davison A. C., Hinkley D. V. (1997) Bootstrap Methods and Their Application, Cambridge University Press, Cambridge, UK [Google Scholar]
- 12.Lakkis N. (2000) New treatment methods for patients with hypertrophic obstructive cardiomyopathy. Curr. Opin. Cardiol 15, 172–177 [DOI] [PubMed] [Google Scholar]
- 13.Yoerger D. M., Picard M. H., Palacios I. F., Vlahakes G. J., Lowry P. A., Fifer M. A. (2006) Time course of pressure gradient response after first alcohol septal ablation for obstructive hypertrophic cardiomyopathy. Am. J. Cardiol 97, 1511–1514 [DOI] [PubMed] [Google Scholar]
- 14.Sabatine M. S., Morrow D. A., de Lemos J. A., Gibson C. M., Murphy S. A., Rifai N., McCabe C., Antman E. M., Cannon C. P., Braunwald E. (2002) Multimarker approach to risk stratification in non-ST elevation acute coronary syndromes: simultaneous assessment of troponin I, C-reactive protein, and B-type natriuretic peptide. Circulation 105, 1760–1763 [DOI] [PubMed] [Google Scholar]
- 15.Passing H., Bablok W. (1983) A new biometrical procedure for testing the equality of measurements from 2 different analytical methods—application of linear-regression procedures for method comparison studies in clinical-chemistry. 1. J. Clin. Chem. Clin. Biochem 21, 709–720 [DOI] [PubMed] [Google Scholar]
- 16.Conover W. J. (1999) Practical Nonparametric Statistics, 3rd Ed., Wiley, New York [Google Scholar]
- 17.Paulovich A. G., Whiteaker J. R., Hoofnagle A. N., Wang P. (2008) The interface between biomarker discovery and clinical validation: the tar pit of the protein biomarker pipeline. Proteomics Clin. Appl 2, 1386–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oberg A. L., Vitek O. (2009) Statistical design of quantitative mass spectrometry-based proteomic experiments. J. Proteome Res 8, 2144–2156 [DOI] [PubMed] [Google Scholar]
- 19.Sabatine M. S., Morrow D. A., de Lemos J. A., Jarolim P., Braunwald E. (2009) Detection of acute changes in circulating troponin in the setting of transient stress test-induced myocardial ischaemia using an ultrasensitive assay: results from TIMI 35. Eur. Heart J 30, 162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrow D. A., Sabatine M. S., Brennan M. L., de Lemos J. A., Murphy S. A., Ruff C. T., Rifai N., Cannon C. P., Hazen S. L. (2008) Concurrent evaluation of novel cardiac biomarkers in acute coronary syndrome: myeloperoxidase and soluble CD40 ligand and the risk of recurrent ischaemic events in TACTICS-TIMI 18. Eur. Heart J 29, 1096–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowie M. R., Jourdain P., Maisel A., Dahlstrom U., Follath F., Isnard R., Luchner A., McDonagh T., Mair J., Nieminen M., Francis G. (2003) Clinical applications of B-type natriuretic peptide (BNP) testing. Eur. Heart J 24, 1710–1718 [DOI] [PubMed] [Google Scholar]
- 22.Melanson S. E., Tanasijevic M. J., Jarolim P. (2007) Cardiac troponin assays: a view from the clinical chemistry laboratory. Circulation 116, e501–e504 [DOI] [PubMed] [Google Scholar]
- 23.Berna M., Ott L., Engle S., Watson D., Solter P., Ackermann B. (2008) Quantification of NTproBNP in rat serum using immunoprecipitation and LC/MS/MS: a biomarker of drug-induced cardiac hypertrophy. Anal. Chem 80, 561–566 [DOI] [PubMed] [Google Scholar]
- 24.Anderson N. L., Anderson N. G., Haines L. R., Hardie D. B., Olafson R. W., Pearson T. W. (2004) Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J. Proteome Res 3, 235–244 [DOI] [PubMed] [Google Scholar]
- 25.Hoofnagle A. N., Becker J. O., Wener M. H., Heinecke J. W. (2008) Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin. Chem 54, 1796–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn E., Addona T., Keshishian H., Burgess M., Mani D. R., Lee R. T., Sabatine M. S., Gerszten R. E., Carr S. A. (2009) Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin. Chem 55, 1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.