Abstract
MS-based quantitative proteomics is widely used for large scale identification of proteins. However, an integrated approach that offers comprehensive proteome coverage, a tool for the quick categorization of the identified proteins, and a standardized biological study method is needed for helping the researcher focus on investigating the proteins with biologically important functions. In this study, we utilized isobaric tagging for relative and absolute quantification (iTRAQ)-based quantitative differential LC/MS/MS, functional annotation with a proprietary gene ontology tool (Molecular Annotation by Gene Ontology (MANGO)), and standard biochemical methods to identify proteins related to neuronal differentiation in nerve growth factor-treated rat pheochromocytoma (PC12) cells, which serve as a representative model system for studying neuronal biological processes. We performed MS analysis by using both nano-LC-MALDI-MS/MS and nano-LC-ESI-MS/MS for maximal proteome coverage. Of 1,482 non-redundant proteins semiquantitatively identified, 72 were differentially expressed with 39 up- and 33 down-regulated, including 64 novel nerve growth factor-responsive PC12 proteins. Gene ontology analysis of the differentially expressed proteins by MANGO indicated with statistical significance that the up-regulated proteins were mostly related to the biological processes of cell morphogenesis, apoptosis/survival, and cell differentiation. Some of the up-regulated proteins of unknown function, such as PAIRBP1, translationally controlled tumor protein, prothymosin α, and MAGED1, were further analyzed to validate their significant functions in neuronal differentiation by immunoblotting and immunocytochemistry using each antibody combined with a specific short interfering RNA technique. Knockdown of these proteins caused abnormal cell morphological changes, inhibition of neurite formation, and cell death during each course of the differentiation, confirming their important roles in neurite formation and survival of PC12 cells. These results show that our iTRAQ-MANGO-biological analysis framework, which integrates a number of standard proteomics strategies, is effective for targeting and elucidating the functions of proteins involved in the cellular biological process being studied.
MS-based quantitative proteomics strategies such as iTRAQ1 (1) and stable isotope labeling with amino acids in cell culture (2) are powerfully effective for the comprehensive characterization of biological phenomena (1–5). Although these methods have been applied for cancer biomarker (6, 7) and drug target (8) discovery, their use in the elucidation of biological and functional processes has been limited because of certain technical problems that arise when attempting to meaningfully process the immense amount of data obtained from such experiments. The following four main issues are typically the sources of such difficulties. 1) Quantitative identification by one type of MS system may fail to cover the total proteome because of ionization efficiency differences, such as those between ESI and MALDI, for certain peptides, leading to theoretical limitations in proteome coverage. 2) The public protein databases are often insufficient for searching non-human species because of the limited available genomic information. 3) The identification of the functions and biological processes of thousands of proteins is a formidable task because of the lack of simple and user-friendly software to automate gene ontology (GO) annotation. Furthermore it is difficult to convert large lists of taxonomically diverse proteins into their human orthologs to obtain the richest GO information available. 4) Lastly biological validation strategies for identified proteins have not been standardized. Therefore, we believe an analysis framework that provides (a) comprehensive proteome data; (b) a simple and quick tool for organizing, enriching, and sorting those data to reveal candidate molecules for relation to certain processes; and (c) a standardized biological validation technique would greatly benefit this field. We therefore designed a concise, three-step, sequential proteomics strategy that addresses the above concerns and utilized it successfully in studying the mechanism of neuronal differentiation in PC12 cells.
PC12 cells (9) have been widely used as a model of neurons because of their unique advantages, such as stability, homogeneity, strong nerve growth factor (NGF) responsiveness, high differentiation potential, and a wealth of accessible background material, which help to facilitate their manipulation (10). This cell line has also been used for studying the mechanisms of neuronal disorders such as Alzheimer (11), Huntington (12), and Parkinson diseases (13) and neurofibromatosis type 1 (14–16). Here we used PC12 cells as a model for characterizing the mechanisms of neuronal differentiation and neurodegenerative disorders by means of MS-based quantitative proteomics.
NGF is one member of a family of structurally and functionally related dimeric polypeptides, neurotrophins, that are essential for the development and maintenance of distinct neuronal populations in the central and peripheral nervous systems (17). The initial signaling cascades in the neuronal cells right after NGF stimulation have been subjected to thorough investigation and characterization by using PC12 cells. After binding of extracellular NGF to the cell membrane-localized tropomyosin-related kinase A (TrkA) receptor, TrkA receptors dimerize and subsequently autophosphorylate each other. Then the phosphorylated receptors recruit a complex of signaling molecules and induce a number of intracellular signaling cascades involving the signaling molecules, such as phosphoinositide 3-kinase, phospholipase C-γ, and Ras (18). The posttranslational modifications, such as phosphorylation cascades, triggered by NGF stimulation play important roles in PC12 cell differentiation. However, knowledge of the precise dynamic molecular events of protein expression in response to NGF signaling in PC12 cells after an interval that allows the stimulation to take full effect and produce morphological changes remains far from complete.
Several reported studies have applied such methods as expressed sequence tag (19), restriction landmark cDNA scanning (20), targeted display (21), serial analysis of gene expression (22), and cDNA microarray (23) to survey the global change of differentially expressed genes in PC12 cells before and after NGF treatment (19–23). However, the genes and underlying mechanisms associated with the acquisition of a neuronal phenotype in these cells have not been clarified. Also a few proteomics approaches have been used for identifying the proteins related to NGF-inducible neurite formation in PC12 cells. For example, 2-D electrophoresis was applied in whole-cell extract separation to study the NGF modulation of protein synthesis (24); however, only two peptides were identified (25). Even currently available PC12 cell 2-D databases include merely a few proteins related to NGF stimulation (26–29). There is thus a paucity of functional proteomic information related to PC12 cell biological processes that may be attributed to technical limitations such as those listed above.
In this study, we performed the first proteomics survey of proteins differentially expressed in PC12 cells during NGF treatment by using a semiquantitative differential LC shotgun method, namely isobaric tagging for relative and absolute quantitation (iTRAQ) coupled with concurrent use of two tandem MS/MS systems, namely nano-LC-MALDI-TOF-TOF and nano-LC-ESI-Quadrupole/quadrupole/time-of-flight mass spectrometers. The total list of proteins identified was converted into a new file linked to the GO database by our proprietary GO analysis tool for proteomes (MANGO) and categorized by biological process and function using specific classification methods. Thereafter we classified the subset of proteins that were up- or down-regulated during neurite formation into specific molecular categories by combining the differential data obtained by iTRAQ with the proteomic GO analysis results. We then attempted to characterize the functional mechanism of NGF-induced PC12 cell neuronal differentiation. Interestingly the specific up-regulated groups classified in this study were related to apoptosis/cell survival in addition to cell motility, differentiation, stress stimulation, and morphogenesis. To investigate the molecular functions of the up-regulated proteins in relation to both PC12 cell differentiation and apoptosis/survival during neurite formation, some of them were further analyzed with a biochemical and cellular biological strategy using a combined antibody and siRNA technique. Lastly we demonstrated the advantages that our concise, sequential proteomics strategy offers for studying the molecular mechanisms of cellular biological events such as cell differentiation and survival/apoptosis.
EXPERIMENTAL PROCEDURES
Cell Culture, NGF Treatment, and Preparation of Cell Lysate
PC12 cells were cultured under 5% CO2 at 37 °C in Dulbecco's modified Eagle's medium supplemented with 10% horse serum and 5% fetal bovine serum. We performed four independent cell cultures for a fourplex iTRAQ analysis. Two of them were used as duplicated samples for controls, and the other two samples were used as NGF-treated cells. For NGF stimulation, the cells were cultured onto collagen-coated culture dishes (Iwaki) in the same medium and stimulated with 50 ng/ml 2.5 S NGF (Wako) at 48 h. For preparation of cell lysate, cells were solubilized with the lysis buffer containing 8 m urea, 2% CHAPS, 2 mm Na2VO4, 10 mm NaF, 1 μm okadaic acid, and 1% (v/v) protease inhibitor mixture (Sigma) and passed through a 25-gauge syringe 15 times. Lysates were centrifuged at 13,000 × g for 20 min at 4 °C, and the protein concentration of the supernatants was determined using the Bio-Rad protein assay.
iTRAQ Sample Labeling
One hundred micrograms of each protein sample was precipitated using a 2-D Clean-Up kit (Amersham Biosciences), and the precipitants were dissolved in 10 μl of 6 m urea. iTRAQ sample labeling was performed according to the manufacturer's protocol with minimum modification. For the fourplex iTRAQ labeling, the four lysates of PC12 cells separately cultured were treated with iTRAQ reagents in parallel. Twenty microliters of dissolution buffer and 1 μl of denaturant reagent were added to the samples. The samples were reduced by addition of 2 μl of reducing reagent and incubation at 60 °C for 1 h. Reduced cysteine residues were then blocked by addition of 1 μl of cysteine blocking reagent and incubated at room temperature for a further 10 min. Tryptic digestion was initiated by the addition of 12.5 μl of trypsin solution (Promega; prepared as 1 μg/μl in water solution) and incubated at 37 °C for 16 h. To label the peptides with iTRAQ reagents, one vial of labeling reagent was thawed and reconstituted in 80 μl of ethanol. The reagents 114 and 115 for two samples from untreated cells and the reagents 116 and 117 for two samples from NGF-stimulated cells were added to the digests and incubated for 1 h at room temperature. The labeled samples were then mixed together before fractionation using a cation exchange column.
Sample Fractionation and Desalting
To remove excess, unbound iTRAQ reagent and to simplify the peptide mixture, the labeled peptide mixture was purified and fractionated using a GE Healthcare AKTA system. The mixed sample was diluted in loading buffer (20% (v/v) ACN and 10 mm potassium phosphate, pH 3.0) and loaded onto a Mono S column (GE Healthcare) equilibrated with loading buffer. Peptides were eluted with a gradient of solvent B (10 mm potassium phosphate, pH 3.0, and 1 m KCl in 20% (v/v) ACN) as follows: 0–2 min, 0–7% B; at 6 min, to 14% B; at 8 min, to 32% B; at 13 min, to 70% B; and at 21 min, to 100% B. Twenty-five fractions that included the iTRAQ-labeled peptides were used for analysis. The fractions were dried in a vacuum centrifuge and rehydrated with solution containing 2% ACN and 0.1% TFA. The samples were desalted with ZipTipTM μ-C18 pipette tips (Millipore). The desalted peptides were divided into two fractions to analyze the same samples by using nano-LC-MALDI-TOF-TOF and nano-LC-ESI-QqTOF systems.
LC-MALDI-MS/MS Analysis
Samples were separated by C18 nano-LC using DiNa Map (KYA Tech Corp.) equipped with a device spotting eluted fractions on a MALDI plate. Sample was injected onto a C18 column (0.5-mm inner diameter × 1-mm length, KYA Tech Corp.) equilibrated with solvent A (2% ACN and 0.1% TFA) and resolved on a C18 nanocolumn (0.15-mm inner diameter × 100-mm length; KYA Tech Corp.) at a flow rate of 300 nl/min with a 90-min gradient of solvent B (70% ACN and 0.1% TFA) as follows: 0–20% B from 0 to 10 min, to 50% B at 65 min, and to 100% B at 75 min. Column effluent was mixed with matrix (2 mg/ml α-cyano-4-hydroxycinnamic acid in 50% ACN and 0.1% TFA) at a flow rate of 1.4 μl/min. Fractions were spotted at 30-s intervals onto a stainless steel MALDI target plate (192 wells/plate; Applied Biosystems). Mass spectra of the peptides were acquired on a 4700 Proteomics Analyzer (Applied Biosystems) using 4000 Series Explorer software (Version 3.6). Mass spectra from m/z 800 to 4,000 were acquired for each fraction with 1,500 laser shots. To analyze the less abundant peptides, all of the peaks with a signal to noise ratio threshold from 50 to 75 and from 75 to 100 in each MS spectrum were selected for MS/MS analysis with 5,000 and 4,000 laser shots, respectively. Next all of the peaks above a signal to noise ratio threshold of 100 were selected for MS/MS analysis with 3,000 laser shots. Fragmentation of the labeled peptides was induced by the use of atmosphere as a collision gas with a pressure of 1 × 10−6 Torr and a collision energy of 1 kV.
LC-ESI-MS/MS Analysis
Samples were analyzed by nano-LC-ESI-MS/MS using the LC Packings Ultimate instrument fitted with a 20-μl sample loop. Samples were loaded onto a 5-mm RP C18 precolumn (LC Packings) at 30 μl/min and washed for 10 min before switching the precolumn in line with the separation column. The separation column used was a 75-μm internal diameter × 150-mm length PepMap RP column from LC Packings packed with 3-μm C18 beads with 100-Å pores. The flow rate used for separation on the RP column was 200 nl/min with a 90-min gradient of solvent B (85% ACN and 0.1% formic acid) as follows: 0–40% B from 0–60 min to 100% B at 70 min. The samples were divided into two fractions beforehand, and the first analysis was performed on a QSTAR Pulsar i mass spectrometer (Applied Biosystems/MDS Sciex), and the software used for data acquisition was Analyst QS 1.1 (Applied Biosystems/MDS Sciex) with the scan cycles set up to perform a 1-s MS scan followed by three MS/MS scans of the three most abundant peaks for 3 s each. Data acquisition was performed with an exclusion of 60 s for previous target ions. To analyze the less abundant peptides, the second analysis was performed under the same condition except for input of the m/z list to exclude the analyses of peptide ions already analyzed in the first run. The labeled peptides were fragmented under CID conditions designed to give iTRAQ reporter ions.
Data Analysis
Data from MALDI or ESI analysis were analyzed using the ParagonTM algorithm (30) of ProteinPilot Version 2.0 (Applied Biosystems), and the database searched was the Swiss-Prot database with all taxonomy (Revision number 53, 269,293 sequence entries, updated on May 29, 2007). Identified proteins were grouped by the Paragon algorithm of the software to minimize redundancy. This software has a function of automatic grouping of identified proteins according to the identified peptide sequence. The identified proteins were automatically grouping by the Paragon algorithm. Peptides used for the quantification of proteins were chosen by this algorithm of ProteinPilot software. All peptides used for the calculation of protein ratios were unique to the given protein or proteins within the group; peptides that were common to other isoforms or proteins of the same family that were reported separately were ignored. The ProteinPilot cutoff score used was 1.3, which corresponds to a confidence limit of 95%. The six user-defined options used included (i) cysteine alkylation, methyl methane thiosulfate; (ii) digestion, trypsin digestion; (iii) special factors, none; (iv) species, all species; (v) identification focus, biological modifications; and (vi) search effort, thorough identification search. For quantification of each protein identified in the MALDI or ESI analysis, a -fold change of each protein expression was calculated by comparing the average iTRAQ ratio of 116 and 117 as NGF-treated groups with the average ratio of 114 and 115 as control groups. Proteins quantified with a -fold change of more than 20% (average iTRAQ ratio >1.20 or <0.83) and a p value less than 0.05 (Student's t test) in the MALDI or ESI analysis were identified as differentially expressed proteins.
Proteomics GO Analysis by MANGO Method
To automate both the conversion of all identified proteins of multiple taxonomies into their human orthologs and annotation with GO information, a tool for taxonomy conversion/GO annotation, Molecular Annotation by Gene Ontology (MANGO), was designed as a Web-based application using the mySQL 4.0 database management system and scripts written in Java. Ensembl Mart and UniProt GOA (GOA UniProt Version 49.0, June 2007 update) files were integrated into the database and were used as the external reference data for the taxonomy conversion and the GO annotation, respectively. For the proteins that could not be automatically converted to the human orthologs using this conversion/annotation tool, we searched the human orthologs by using National Center for Biotechnology Information basic local alignment search tool (NCBI BLAST) programs. A list of 1,404 human GO-annotated proteins was compiled and was designated as the PC12 proteome reference set for analysis with GeneSpring GX (supplemental Table 4). GeneSpring GX Version 7.3.1 was used to determine the GO categories that were statistically overrepresented in the iTRAQ data set. GO categories, by which at least three proteins were annotated, were accepted at the significance level of p < 0.05 (Fisher's exact test). The annotation data of the 72 differentially expressed proteins were used to determine the overrepresented biological processes related to NGF-induced PC12 cell differentiation. A GO tree view composed of enriched biological processes in the up-regulated proteins was built according to the tree view in AmiGO, a tool for searching and browsing the Gene Ontology database (31).
Western Blotting
Cell lysate samples containing 20 μg of total protein were electrophoresed on SDS-polyacrylamide gels, transferred onto a PVDF membrane by electroblotting, and subjected to immunoblotting with the indicated antibody. In the case of prothymosin α (ProTα), cell lysate samples were transferred onto a membrane by electroblotting with acidic buffer (20 mm sodium acetate buffer, pH 5.2) followed by fixation with 0.5% glutaraldehyde (32, 33). Membranes were probed with different primary antibodies followed by horseradish peroxidase-conjugated mouse, rabbit, and goat secondary antibodies (GE Healthcare). The images were visualized with ECL (GE Healthcare). The following primary antibodies were used: neurosecretory protein VGF, CRMP-4, galectin-1, ProTα, MAGED1, and PCNA (Santa Cruz Biotechnology); plasminogen activator inhibitor 1 RNA-binding protein (PAIRBP1) (Abnova Co.); translationally controlled tumor protein (TCTP) (MBL International Corp.); β-actin (Sigma); peroxiredoxin 6 (Lab Frontier); and protein-disulfide isomerase (Stressgen). For the quantitative analysis, the ECL patterns were scanned using LabScan 5.0 (GE Healthcare) with transparent mode and a resolution of 300 dpi. The intensities were measured using ProGenesis Workstation Version 2005 (PerkinElmer Life Sciences).
Transfection of PC12 Cells with siRNA
Transfection of PC12 cells with siRNA was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Four target sequences for rat, PAIRBP1, TCTP, ProTα, and MAGED1 siRNA, were designed as follows. A 21-oligonucleotide siRNA duplex was designed as recommended elsewhere and was synthesized by Gene Link to target the PAIRBP1 sequence (5′-1125AAGUGCUUCUGCUCCUGACTT-3′), the TCTP sequence (5′-357AAGCACATCCTTGCTAATTTT-3′), the ProTα sequence (5′-52AAGGAGAAGAAGGAAGUUGTT-3′), and the MAGED1 sequence (5′-1928AAGUGCUGAGAUUCAUUGCTT-3′). A Silencer Negative Control siRNA 1 (Ambion) was used as a control siRNA for the analysis.
Immunofluorescence Analysis
PC12 cells cultured on 35-mm collagen-coated culture dishes were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature and then permeabilized with 0.1% Triton X-100 in PBS on ice for 15 min. After being washed with PBS, cells were incubated in primary antibodies diluted in PBS containing 5% bovine serum albumin followed by anti-mouse or -rabbit Alexa Fluor® 488-conjugated IgG (Invitrogen) for 60 min at room temperature. After being washed with PBS, the cells were incubated for 10 min at room temperature with rhodamine-phalloidin (Invitrogen) to stain cellular F-actin. After being washed with PBS, the cells were incubated for 10 min at room temperature with 20 μg/ml Hoechst33342 (Invitrogen) to stain nuclear actin. Analysis was performed with a fluorescence microscope (with 20 × 1.6 Olympus IX71) (DPController, DPManager).
Quantification of Neurite Outgrowth
For quantification of the neurite outgrowth of PC12 cells, the cells transfected with siRNAs were cultured onto collagen-coated culture dishes (Iwaki) under the condition of 1% horse serum and stimulated with 50 ng/ml 2.5 S NGF (Wako) at 48 h. Total neurite length of NGF-stimulated PC12 cells was measured using MetaMorph software (Molecular Devices). The total number of tip ends was manually counted to represent the number of neurites from individual cells. For each measurement, at least 50 cells per dish were analyzed from randomly selected fields. Each experiment was repeated three times.
Evaluation of PC12 Cell Death
Propidium iodide (PI) was used for the evaluation of PC12 cell death. Cells transfected with siRNAs were cultured onto 35-mm collagen-coated culture dishes (Iwaki) under the condition of 10% horse serum and 5% fetal bovine serum and stimulated with 50 ng/ml 2.5 S NGF (Wako) at 48 h. The cells were fixed with 4% paraformaldehyde in PBS and incubated with 0.2 μg/ml PI for apoptotic cells and 20 μg/ml Hoechst33342 (Invitrogen) for total cells for 10 min at room temperature. PI-positive cells were counted using a fluorescence microscope (with 20 × 1.6 Olympus IX71) (DPController, DPManager). For each counting, at least 500 cells per dish were counted from randomly selected fields. Each experiment was repeated three times.
Time Lapse Video Analysis
Cells were cultured on a collagen-coated glass bottom plate with 6 wells (Iwaki). The plate was maintained at 37 °C under the air condition of 5% CO2 in the chamber set under the camera during the observation. Images were obtained by using a 20× UPlan SApo objective (Olympus IX81). The camera, shutters, and filter wheel were controlled by MetaMorph imaging software (Molecular Devices), and the images were collected every 10 min with an exposure time of 50 ms.
RESULTS
Identification of Differentially Expressed Proteins in Response to NGF in PC12 Cells
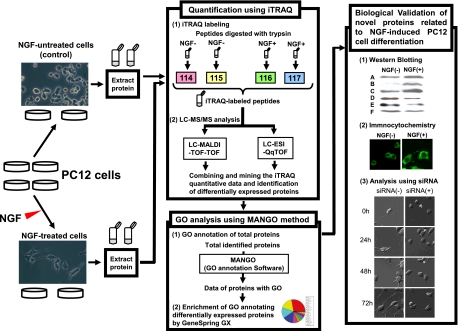
For the fourplex iTRAQ labeling, four independent, separately cultured cell lysates were prepared in parallel. The tryptic peptides from untreated cells were labeled with mass 114 and 115 isobaric iTRAQ tags, and those from NGF-treated cells were labeled with mass 116 and 117 isobaric iTRAQ tags. The iTRAQ-labeled peptides were then fractionated by cation exchange column chromatography and analyzed with LC-MS/MS using both MALDI and ESI (Fig. 1).
Fig. 1.
Work flow for the identification of the novel proteins regulating NGF-induced differentiation in PC12 cells. For the fourplex iTRAQ labeling, the four lysates of PC12 cells separately cultured were prepared in parallel. The tryptic peptides from untreated cells and NGF-treated cells were labeled using mass 114/115 and 116/117 isobaric iTRAQ tags, respectively. These labeled peptides were analyzed with LC-MALDI-TOF-TOF or LC-ESI-QqTOF, and the quantification of each protein was performed according to the obtained iTRAQ data. Further biological and functional interpretation of the differentially expressed proteins was carried out by proteomics GO analysis using the MANGO method followed by cell biological analyses. Proteins A–F are candidates for novel molecules related to NGF-induced PC12 cell differentiation.
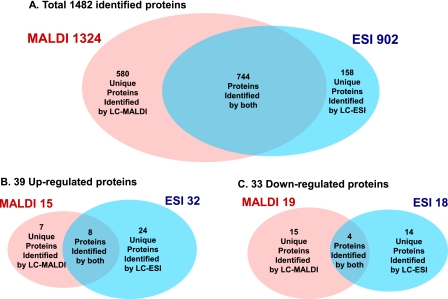
With LC-MALDI-MS/MS, 1,324 proteins were identified from 14,710 peptide sequences; in contrast, with LC-ESI-MS/MS, 902 proteins were identified from 12,769 peptide sequences. Only 744 proteins were identified by both systems, and 580 and 158 proteins were uniquely identified with LC-MALDI-MS/MS and LC-ESI-MS/MS, respectively. In total, 1,482 proteins were identified with >95% confidence (Fig. 2A and supplemental Table 1). At a -fold change of >20% (ratio >1.20 or <0.83), 15 proteins identified with LC-MALDI-MS/MS were up-regulated, and 19 were down-regulated, whereas 32 identified with LC-ESI-MS/MS were up-regulated, and 18 were down-regulated (p value <0.05) (Fig. 2, B and C). Eight up- and four down-regulated proteins were identified by both systems, and seven or 24 up- and 15 or 14 down-regulated proteins were uniquely identified with LC-MALDI-MS/MS or LC-ESI-MS/MS, respectively (Fig. 2, B and C). In total, 72 proteins (39 up- and 33 down-regulated) were differentially expressed in response to NGF. Among the NGF-responsive proteins, 64 were newly identified in this study (Table I and supplemental Tables 2 and 3).
Fig. 2.
Venn diagrams of the number of total (A), up-regulated (B), and down-regulated (C) proteins identified by iTRAQ. A, in total, 1,482 proteins of all taxonomies listed in supplemental Table 1 were identified with a confidence limit of 95%. B and C, for quantification of each protein identified by MALDI or ESI, a -fold change of each protein expression was calculated by comparing the average iTRAQ ratio of 116 and 117 as NGF-treated groups with the average ratio of 114 and 115 as control groups. Proteins quantified with a -fold change more than 20% (average iTRAQ ratio >1.20 or <0.83) and a p value less than 0.05 by MALDI or ESI were identified as differentially expressed proteins (B and C).
Table I.
List of proteins differentially-expressed in response to NGF stimulation
snRNA, small nuclear RNA; —, not applicable.
| Protein name abbreviation | Protein name | Accession no. | Theoretical molecular mass (kDa)/pI | MALDI |
ESI |
Ref.b | ||
|---|---|---|---|---|---|---|---|---|
| Average ratioa | p value | Average ratioa | p value | |||||
| Up-regulated proteins | ||||||||
| VGF | Neurosecretory protein VGF | P20156 | 68.2/4.7 | 2.935 | 0.011 | 3.770 | 0.024 | 23, 40 |
| NEUM | Neuromodulin | P07936 | 23.6/4.6 | 1.791 | 0.008 | 2.936 | 0.021 | 23, 41 |
| MOES | Moesin | O35763 | 67.7/6.2 | 1.691 | 0.035 | 1.679 | 0.030 | — |
| CMGA | Chromogranin A | P10354 | 52.0/4.7 | 1.594 | 0.003 | 1.596 | 0.003 | 42 |
| ANXA2 | Annexin A2 | Q07936 | 38.7/7.6 | 1.449 | 0.048 | 1.805 | 0.037 | 23, 43 |
| PERI | Peripherin | P21807 | 53.5/5.4 | 1.329 | 0.011 | 1.429 | 0.012 | 44 |
| AT1A1 | Sodium/potassium-transporting ATPase α-1 chain | P06685 | 11.3/5.3 | 1.271 | 0.048 | 1.341 | 0.003 | 23 |
| MAP1B | Microtubule-associated protein 1B | P15205 | 269.5/4.7 | 1.250 | 0.010 | 1.352 | 0.027 | 45 |
| SQSTM | Sequestosome-1 | O08623 | 47.7/5.1 | 1.764 | 0.042 | — | ||
| RIR2 | Ribonucleoside-diphosphate reductase M2 subunit | Q4KLN6 | 45.0/5.5 | 1.339 | 0.044 | — | ||
| SGTA | Small glutamine-rich tetratricopeptide repeat-containing protein A | O70593 | 34.2/5.1 | 1.286 | 0.035 | 1.051 | 0.787 | — |
| CROP | Cisplatin resistance-associated overexpressed protein | O95232c | 51.5/9.8 | 1.266 | 0.042 | 1.006 | 0.294 | — |
| PAIRBP1 | Plasminogen activator inhibitor 1 RNA-binding protein | Q6AXS5 | 44.8/8.6 | 1.226 | 0.013 | 1.241 | 0.113 | — |
| GSPT1 | G1 to S phase transition protein 1 homolog | P15170c | 55.8/5.5 | 1.206 | 0.042 | 0.977 | 0.841 | — |
| CRIP2 | Cysteine-rich protein 2 | P36201 | 22.7/8.9 | 1.205 | 0.020 | — | ||
| PROTA | Prothymosin α | P06302 | 12.4/3.8 | 1.642 | 0.206 | 2.173 | 0.037 | — |
| AHNAK | Neuroblast differentiation-associated protein AHNAK | Q09666c | 629.1/5.8 | 1.245 | 0.113 | 1.947 | 0.012 | 46 |
| SCG2 | Secretogranin-2 | P10362 | 71.0/4.7 | 1.339 | 0.068 | 1.633 | 0.027 | 42 |
| AT1B1 | Sodium/potassium-transporting ATPase subunit β-1 | P07340 | 35.2/8.8 | 1.423 | 0.062 | 1.630 | 0.001 | 23 |
| MAGED1 | Melanoma-associated antigen D1 | Q9ES73 | 85.8/7.1 | 1.066 | 0.654 | 1.630 | 0.020 | — |
| LEG1 | Galectin-1 | P11762 | 14.9/5.1 | 1.272 | 0.126 | 1.578 | 0.016 | 22, 23 |
| ENPL | Endoplasmin | P14625c | 92.5/4.8 | 1.052 | 0.134 | 1.556 | 0.016 | 22, 23 |
| S100A6 | Protein S100-A6 | P05964 | 10.0/5.3 | 1.362 | 0.069 | 1.552 | 0.004 | — |
| E2IG5 | E2-induced gene 5 protein homolog | Q4QQV3 | 17.8/10.0 | 1.240 | 0.103 | 1.547 | 0.023 | 23 |
| CRMP4 | Dihydropyrimidinase-related protein 3 (CRMP4) | Q62952 | 62.0/6.0 | 1.183 | 0.240 | 1.407 | 0.016 | 23 |
| SRRM2 | Serine/arginine repetitive matrix protein 2 | Q8BTI8d | 294.7/12.0 | 0.981 | 0.667 | 1.377 | 0.013 | — |
| DNJA1 | DnaJ homolog subfamily A member 1 | P63036 | 44.9/6.7 | 1.065 | 0.397 | 1.331 | 0.011 | — |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 | Q6RUV5 | 21.5/8.8 | 1.316 | 0.044 | — | ||
| TCTP | Translationally controlled tumor protein | P63029 | 19.5/4.8 | 1.359 | 0.126 | 1.291 | 0.016 | — |
| RAB2A | Ras-related protein Rab-2A | P05712 | 23.5/6.1 | 1.272 | 0.239 | 1.280 | 0.015 | — |
| OXRP | Hypoxia up-regulated protein 1 | Q63617 | 111.3/5.1 | 1.051 | 0.591 | 1.271 | 0.008 | — |
| PRDX6 | Peroxiredoxin-6 | O35244 | 24.8/5.6 | 1.128 | 0.207 | 1.268 | 0.008 | — |
| ENOA | α-Enolase | P04764 | 47.1/6.2 | 1.153 | 0.172 | 1.253 | 0.008 | 23 |
| RADI | Radixin | P35241c | 68.6/6.0 | 0.885 | 0.193 | 1.251 | 0.032 | 23 |
| ALDR | Aldose reductase | P07943 | 35.8/6.3 | 1.276 | 0.235 | 1.248 | 0.004 | — |
| PDI | Protein-disulfide isomerase | P04785 | 57.0/4.8 | 1.041 | 0.118 | 1.245 | 0.045 | 23 |
| FABPE | Fatty acid-binding protein, epidermal | P55053 | 15.1/6.73 | 1.136 | 0.268 | 1.227 | 0.049 | 23 |
| RGP1 | Ran GTPase-activating protein 1 | P46061d | 63.6/4.6 | 1.027 | 0.359 | 1.219 | 0.012 | — |
| NUCB1 | Nucleobindin-1 | Q63083 | 53.5/5.0 | 1.039 | 0.521 | 1.208 | 0.005 | — |
| Down-regulated proteins | ||||||||
| K1C18 | Keratin, type I cytoskeletal 18 | Q5BJY9 | 47.8/5.2 | 0.626 | 0.028 | 0.566 | 0.043 | — |
| K2C8 | Keratin, type II cytoskeletal 8 | Q10758 | 54.0/5.8 | 0.690 | 0.011 | 0.599 | 0.040 | — |
| PRPS2 | Ribose-phosphate pyrophosphokinase II | P09330 | 34.8/6.2 | 0.690 | 0.040 | 0.642 | 0.015 | — |
| LDHA | l-Lactate dehydrogenase A chain | P04642 | 36.5/8.5 | 0.768 | 0.022 | 0.792 | 0.041 | — |
| PRP19 | Pre-mRNA-processing factor 19 | Q9JMJ4 | 55.2/6.2 | 0.551 | 0.009 | 0.880 | 0.604 | — |
| VTDB | Vitamin D-binding protein | P04276 | 53.5/5.7 | 0.625 | 0.006 | — | ||
| IDHP | Isocitrate dehydrogenase (NADP), mitochondrial | P56574 | 51.0/8.9 | 0.630 | 0.005 | 0.868 | 0.068 | — |
| HNRLL | Heterogeneous nuclear ribonucleoprotein L-like | Q921F4d | 64.1/5.8 | 0.651 | 0.031 | 0.636 | 0.187 | — |
| NUCKS | Nuclear ubiquitous casein and cyclin-dependent kinase substrate | Q9EPJ0 | 27.1/5.0 | 0.747 | 0.006 | — | ||
| API5 | Apoptosis inhibitor 5 | Q9BZZ5c | 57.6/5.8 | 0.748 | 0.013 | 0.777 | 0.116 | — |
| H2B1C | Histone H2B type 1 | Q00715 | 14.0/10.4 | 0.762 | 0.040 | 0.860 | 0.003 | 23 |
| RL34 | 60 S ribosomal protein L34 | P11250 | 13.5/11.7 | 0.769 | 0.049 | — | ||
| RLA0 | 60 S acidic ribosomal protein P0 | P19945 | 34.2/5.9 | 0.772 | 0.035 | 0.962 | 0.605 | — |
| TBB4 | Tubulin β-4 chain | P04350c | 49.6/4.8 | 0.784 | 0.034 | — | ||
| SRR35 | 35-kDa SR repressor protein | Q8WXF0c | 30.5/11.7 | 0.787 | 0.001 | — | ||
| PCNA | Proliferating cell nuclear antigen | P04961 | 28.7/4.6 | 0.790 | 0.043 | 0.743 | 0.060 | — |
| UBP48 | Ubiquitin carboxyl-terminal hydrolase 48 | Q76LT8 | 118.8/5.9 | 0.793 | 0.016 | — | ||
| GSTM2 | Glutathione S-transferase Mu 2 | P08010 | 25.7/6.9 | 0.794 | 0.024 | 0.825 | 0.055 | 23 |
| CB39L | Calcium-binding protein 39-like | Q9H9S4c | 39.1/8.5 | 0.818 | 0.033 | — | ||
| LSM8 | U6 snRNA-associated Sm-like protein LSm8 | O95777c | 10.4/4.4 | 0.648 | 0.015 | — | ||
| DDC | Aromatic-l-amino-acid decarboxylase | P14173 | 54.1/6.5 | 0.669 | 0.045 | 23 | ||
| CSK21 | Casein kinase II subunit α | P19139 | 45.1/7.3 | 0.875 | 0.088 | 0.670 | 0.041 | — |
| RL6 | 60 S ribosomal protein L6 | P21533 | 33.6/10.7 | 0.904 | 0.497 | 0.681 | 0.040 | — |
| GALK1 | Galactokinase | Q9R0N0d | 42.2/5.2 | 1.222 | 0.509 | 0.730 | 0.029 | — |
| AB14B | Abhydrolase domain-containing protein 14B | Q8VCR7d | 22.5/5.8 | 0.675 | 0.292 | 0.748 | 0.030 | — |
| HNRPF | Heterogeneous nuclear ribonucleoprotein F | Q794E4 | 45.7/5.3 | 1.053 | 0.294 | 0.756 | 0.031 | — |
| APT | Adenine phosphoribosyltransferase | P36972 | 19.5/6.2 | 0.942 | 0.724 | 0.783 | 0.028 | — |
| H2A2B | Histone H2A type 2-B | Q8IUE6c | 14.0/10.9 | 0.860 | 0.157 | 0.814 | 0.024 | 23 |
| FUBP2 | Far upstream element-binding protein 2 | Q99PF5 | 74.2/6.4 | 0.949 | 0.090 | 0.818 | 0.002 | — |
| FINC | Fibronectin | P04937 | 272.5/5.5 | 0.688 | 0.172 | 0.819 | 0.000 | — |
| TIM13 | Mitochondrial import inner membrane translocase subunit Tim13 | P62076 | 10.5/8.4 | 1.003 | 0.974 | 0.820 | 0.025 | — |
| SFRS2 | Splicing factor, arginine/serine-rich 2 | Q6PDU1 | 25.5/11.9 | 0.852 | 0.249 | 0.826 | 0.005 | — |
| FEN1 | Flap endonuclease 1 | P39748c | 42.6/8.8 | 0.971 | 0.775 | 0.833 | 0.022 | 23 |
Proteomics GO Analysis of NGF-responsive Proteins
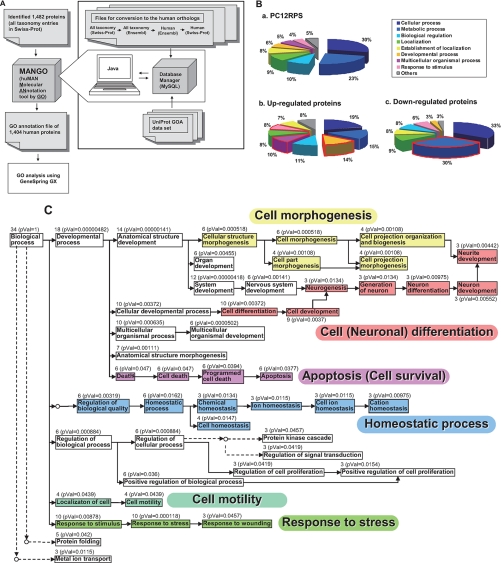
The list of differentially expressed proteins was prepared for use in GO analysis with GeneSpring using a new, in-house taxonomy conversion/GO annotation tool, MANGO (a Web application) (Fig. 3A). First the taxonomies of all 1,482 identified proteins (multiple species matches) were converted to the Homo sapiens taxonomy. 1,404 of them were assigned human orthologs and annotated with GO information from UniProt. This GO-annotated list, compiled using the MANGO tool, was designated as the PC12 proteome reference set (PC12PRS) on which two distinct proteomics GO analyses were subsequently performed (supplemental Table 4). In the first analysis, to determine the overrepresented biological processes specifically related to the NGF-induced PC12 cell differentiation, we simply compared the GO categories represented by the differentially expressed proteins in the PC12PRS with the GO categories represented by the PC12PRS overall (Fig. 3B). From this we found that the following biological processes occurred significantly more frequently in the GO annotations of the 39 up-regulated proteins than they did in the PC12PRS as a whole: “developmental process” (14 versus 6%), “multicellular organismal process” (10 versus 5%), and “response to stimulus” (8 versus 4%) (Fig. 3B, a and b). Also on the other hand, the occurrence ratio of “metabolic process” (30 versus 23%) was higher in the GO annotations of the 33 down-regulated proteins than in those of the PC12PRS as a whole (Fig. 3B, a and c). Next to obtain further statistical interpretation, a second proteomics GO analysis of the differentially expressed proteins in the PC12PRS (supplemental Table 4) was performed to determine which biological processes were enriched in the 39 up-regulated and the 33 down-regulated proteins (Table II). Using these data, a GO tree view was built to clarify the interrelationships of these biological processes (Fig. 3C). The part of the tree comprising the up-regulated proteins was mainly divided into six branches, which included “cell morphogenesis” (p value = 0.000518), “apoptosis (cell survival)” (p value = 0.0377), “homeostatic process” (p value = 0.0162), “cell motility” (p value = 0.000118), “response to stress” (p value = 0.000118), and “cell differentiation” (p value = 0.00372) (Tables II and III and Fig. 3C). On the other hand, the part of the tree comprising the down-regulated proteins included cellular metabolic processes, especially RNA and DNA metabolic processes (Tables II and III).
Fig. 3.
GO analysis of PC12 proteins differentially expressed after NGF stimulation. A, work flow for proteomics GO analysis (MANGO method). MANGO (a Web application), composed of the mySQL 4.0 and scripts written in Java, automatically executed the following steps. First, 1,482 proteins with multiple species matches were converted to human orthologs according to the conversion steps using Ensembl Mart files shown in the figure. Then the converted human orthologs were annotated with GO according to the UniProt GOA data set (described under “Experimental Procedures”). The resulting list of 1,404 GO-annotated human proteins was used as the PC12PRS as shown in supplemental Table 4. GeneSpring GX was used to determine statistically overrepresented GO categories from the iTRAQ data set. B, classification by annotated biological processes of the PC12PRS (a), 39 up-regulated (b), and 33 down-regulated (c) proteins identified with the iTRAQ method. C, a GO hierarchy tree of biological processes annotating the 39 up-regulated proteins. GO analysis was performed by using GeneSpring GX software. A GO tree view composed of enriched biological processes in the up-regulated proteins was built according to the tree view in AmiGO. Direct and indirect hierarchies are indicated by full and dotted lines with arrows, respectively. The number of molecules and p values (pVal) for biological processes annotated are shown in the tree.
Table II.
Overrepresented biological processes by proteomics GO analysis of proteins differentially expressed in response to NGF stimulation
| Categories | All proteins in category | Percentage of all proteins in category | Up- or down-regulated proteins in category | Percentage of up- or down-regulated proteins in category | p value |
|---|---|---|---|---|---|
| Up-regulated proteins | |||||
| GO:8150: biological process | 1,245 | 100 | 34 | 100 | 1 |
| GO:32502: developmental process | 231 | 18.55 | 18 | 52.94 | 4.82e−06 |
| GO:48856: anatomical structure development | 126 | 10.12 | 14 | 41.18 | 1.41e−06 |
| GO:32501: multicellular organismal process | 186 | 14.94 | 13 | 38.24 | 0.000635 |
| GO:48731: system development | 99 | 7.952 | 12 | 35.29 | 4.18e−06 |
| GO:7275: multicellular organismal development | 125 | 10.04 | 12 | 35.29 | 5.02e−05 |
| GO:6950: response to stress | 95 | 7.631 | 10 | 29.41 | 0.000118 |
| GO:48869: cellular developmental process | 145 | 11.65 | 10 | 29.41 | 0.00372 |
| GO:30154: cell differentiation | 145 | 11.65 | 10 | 29.41 | 0.00372 |
| GO:50896: response to stimulus | 163 | 13.09 | 10 | 29.41 | 0.00878 |
| GO:48468: cell development | 121 | 9.719 | 9 | 26.47 | 0.0037 |
| GO:9653: anatomical structure morphogenesis | 63 | 5.06 | 7 | 20.59 | 0.00111 |
| GO:32989: cellular structure morphogenesis | 40 | 3.213 | 6 | 17.65 | 0.000518 |
| GO:902: cell morphogenesis | 40 | 3.213 | 6 | 17.65 | 0.000518 |
| GO:7399: nervous system development | 48 | 3.855 | 6 | 17.65 | 0.00141 |
| GO:65008: regulation of biological quality | 56 | 4.498 | 6 | 17.65 | 0.00319 |
| GO:48513: organ development | 60 | 4.819 | 6 | 17.65 | 0.00455 |
| GO:48518: positive regulation of biological process | 93 | 7.47 | 6 | 17.65 | 0.036 |
| GO:6915: apoptosis | 94 | 7.55 | 6 | 17.65 | 0.0377 |
| GO:12501: programmed cell death | 95 | 7.631 | 6 | 17.65 | 0.0394 |
| GO:8219: cell death | 99 | 7.952 | 6 | 17.65 | 0.047 |
| GO:48523: negative regulation of cellular process | 99 | 7.952 | 6 | 17.65 | 0.047 |
| GO:16265: death | 99 | 7.952 | 6 | 17.65 | 0.047 |
| GO:6457: protein folding | 72 | 5.783 | 5 | 14.71 | 0.042 |
| GO:30030: cell projection organization and biogenesis | 18 | 1.446 | 4 | 11.76 | 0.00108 |
| GO:48858: cell projection morphogenesis | 18 | 1.446 | 4 | 11.76 | 0.00108 |
| GO:32990: cell part morphogenesis | 18 | 1.446 | 4 | 11.76 | 0.00108 |
| GO:19725: cell homeostasis | 36 | 2.892 | 4 | 11.76 | 0.0147 |
| GO:42592: homeostatic process | 37 | 2.972 | 4 | 11.76 | 0.0162 |
| GO:6928: cell motility | 50 | 4.016 | 4 | 11.76 | 0.0439 |
| GO:51674: localization of cell | 50 | 4.016 | 4 | 11.76 | 0.0439 |
| GO:31175: neurite development | 13 | 1.044 | 3 | 8.824 | 0.00442 |
| GO:48666: neuron development | 14 | 1.124 | 3 | 8.824 | 0.00552 |
| GO:30003: cation homeostasis | 17 | 1.365 | 3 | 8.824 | 0.00975 |
| GO:30182: neuron differentiation | 17 | 1.365 | 3 | 8.824 | 0.00975 |
| GO:6873: cell ion homeostasis | 18 | 1.446 | 3 | 8.824 | 0.0115 |
| GO:30001: metal ion transport | 18 | 1.446 | 3 | 8.824 | 0.0115 |
| GO:50801: ion homeostasis | 18 | 1.446 | 3 | 8.824 | 0.0115 |
| GO:22008: neurogenesis | 19 | 1.526 | 3 | 8.824 | 0.0134 |
| GO:48699: generation of neurons | 19 | 1.526 | 3 | 8.824 | 0.0134 |
| GO:48878: chemical homeostasis | 19 | 1.526 | 3 | 8.824 | 0.0134 |
| GO:8284: positive regulation of cell proliferation | 20 | 1.606 | 3 | 8.824 | 0.0154 |
| GO:9966: regulation of signal transduction | 29 | 2.329 | 3 | 8.824 | 0.0419 |
| GO:42127: regulation of cell proliferation | 29 | 2.329 | 3 | 8.824 | 0.0419 |
| GO:7243: protein kinase cascade | 30 | 2.41 | 3 | 8.824 | 0.0457 |
| GO:9611: response to wounding | 30 | 2.41 | 3 | 8.824 | 0.0457 |
| Down-regulated proteins | |||||
| GO:8150: biological process | 1,245 | 100 | 29 | 100 | 1 |
| GO:44237: cellular metabolic process | 824 | 66.18 | 24 | 82.76 | 0.0386 |
| GO:43283: biopolymer metabolic process | 389 | 31.24 | 15 | 51.72 | 0.0161 |
| GO:61 nucleobase, nucleoside, nucleotide, and nucleic acid | 337 | 27.07 | 14 | 48.28 | 0.011 |
| GO:6397: mRNA processing | 86 | 6.908 | 7 | 24.14 | 0.00263 |
| GO:16071: mRNA metabolic process | 101 | 8.112 | 7 | 24.14 | 0.0066 |
| GO:6396: RNA processing | 107 | 8.594 | 7 | 24.14 | 0.00907 |
| GO:8380: RNA splicing | 86 | 6.908 | 6 | 20.69 | 0.0119 |
| GO:6259: DNA metabolic process | 70 | 5.622 | 5 | 17.24 | 0.0201 |
| GO:6281: DNA repair | 23 | 1.847 | 3 | 10.34 | 0.0147 |
| GO:6974: response to DNA damage stimulus | 25 | 2.008 | 3 | 10.34 | 0.0185 |
| GO:9719: response to endogenous stimulus | 27 | 2.169 | 3 | 10.34 | 0.0228 |
Table III.
The differentially expressed proteins related to the enriched biological processes
| GO categories | Protein name (abbreviation)a |
|---|---|
| Up-regulated proteins | |
| Cell differentiation | SQSTM, RAC1, MAP1B, ENPL, CROP, NEUM, GAL1, TCTP, SCG2, S100A6, PROTA,b PAIRBP1b |
| Response to stress | RAC1, SQSTM, ENPL, DNJA1, CROP, AT1A1, SCG2, ANXA2, OXRP, PRDX6 |
| Cell morphogenesis | RAC1, RADI, S100A6, ENOA, NEUM, MAP1B, MAGED1,b CRMP4b |
| Apoptosis (cell survival) | SQSTM, ENPL, TCTP, CROP, LEG1, SCG2, PAIRBP1,b MAGED1,b PROTAb |
| Homeostatic process | ENPL, AT1A1, PDI, TCTP, |
| Cell motility | MOES, AT1A1, SGTA, RAC1 |
| Down-regulated proteins | |
| DNA and RNA metabolic process | HNRLL, SRR35, LSM8, FUBP2, HNRPF, SFRS2, PRP19, PCNA, H2B1C, H2B2A, FEN1 |
| Other cellular metabolic process | VTDB, TBB4, GALK1, RL34, UBP48, CSK21, RLA0, IDHP, TIM13, PRPS2, DDC, RL6, APT |
a The abbreviation of the protein name listed in Table I are shown in this table.
Biological Validation of Newly Identified NGF-responsive Proteins Related to GO Categories Extracted by MANGO
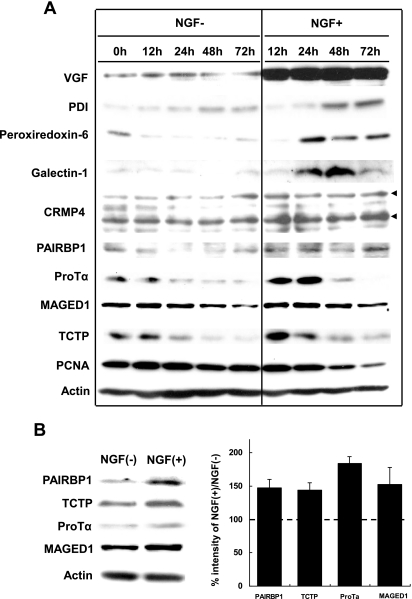
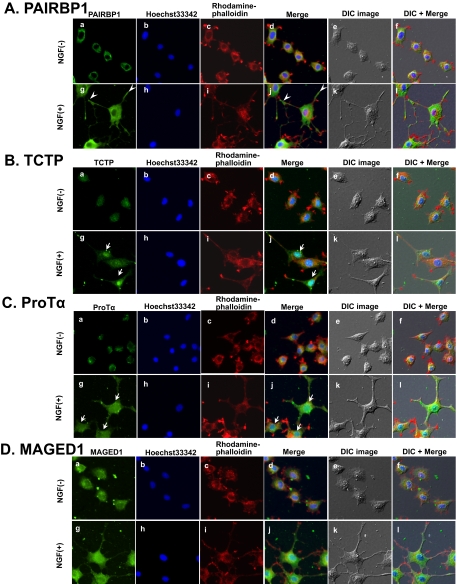
Of the six up-regulated groups listed in Table II, we focused on cell morphogenesis and apoptosis/cell survival (Fig. 3C and Table III) and used a time course analysis by Western blotting to study the expression patterns of the up-regulated proteins related to these processes (Fig. 4). Neurosecretory protein VGF, the most significantly up-regulated protein identified by both LC-MALDI-MS/MS and LC-ESI-MS/MS (Table I and supplemental Fig. 1), was used as a positive control of the time-dependently increased protein by Western blotting (Fig. 4). Seven proteins, CRMP-4, MAGED1, PAIRBP1, protein-disulfide isomerase, peroxiredoxin 6, ProTα, and TCTP, were up-regulated in response to NGF with different expression patterns in the time course study (Fig. 4, A and B, and supplemental Fig. 2). Interestingly TCTP and MAGED1 expression increased for 12 h and then gradually decreased. On the other hand, galectin-1 and ProTα expression levels peaked at 48 and 24 h, respectively. CRMP-4 and PAIRBP1 expression levels were constantly up-regulated during 12–72 h of NGF stimulation. The down-regulated protein, PCNA, was also consistently regulated in response to NGF (Fig. 4A). PAIRBP1 (34, 35), TCTP (36), ProTα (37), and MAGED1 (38) have been speculated to be functionally related to cellular differentiation and survival/apoptosis. We therefore sought to identify the roles of these proteins. Immunocytochemistry (ICC) showed that PAIRBP1 was expressed not only in the cytoplasm but also in the neurites, especially at the tips and junctions (Fig. 5A, g and i, arrowheads), whereas MAGED1 was expressed only in the cytoplasm (Fig. 5D, g and i). TCTP and ProTα were strongly expressed in the nucleus in response to NGF (Fig. 5, B and C, g and i, arrows). These results suggested that those proteins were involved in NGF-treated cellular responses.
Fig. 4.
Immunoblot images of PC12 proteins differentially expressed in response to NGF stimulation. Cell lysates were prepared at different time points as indicated. Actin expression was assessed for equal loading for all the proteins. A, the representative images are shown in four reproducible experiments. Upper and lower arrowheads show isoforms 1 and 2 of CRMP4, respectively. Some of those proteins showed a change in their expression levels even in untreated cells probably because of certain signals from collagen coated on the plates. B, Western blot images and quantification of the above four proteins PAIRBP1, TCTP, ProTα, and MAGED1 in NGF-stimulated PC12 cells. Cell lysates were prepared at 48 h after NGF stimulation. Actin expression was assessed for equal loading for all the proteins. The representative images are shown in four reproducible experiments. The ratios of percent intensities of the four proteins in NGF-stimulated versus untreated PC12 obtained from four separate identical experiments are shown in the histogram. Error bars represent S.D. The ratios were largely consistent with the corresponding iTRAQ ratios.
Fig. 5.
Analysis of NGF-responsive proteins by ICC. PC12 cells were treated with NGF for 48 h. Cells were fixed and incubated with antibodies against the indicated proteins (A, PAIRBP1; B, TCTP; C, ProTα, and D, MAGED1) followed by detection with Alexa Fluor 488-labeled secondary antibodies (a and g) and observation with a fluorescence microscope. Nuclear and F-actin were stained with Hoechst33342 (b and h) and rhodamine-phalloidin (c and i), respectively. The merged images for a, b, c and g, h, i are shown in d and j, respectively. The merged images for d, e and j, k are shown in f and l. Differential interference contrast (DIC) images of PC12 cells in the same field are shown in e and k. Arrowheads indicate the PAIRBP1 expression in the neurite junctions and tips. Arrows indicate the TCTP and ProTα expression in the nucleus.
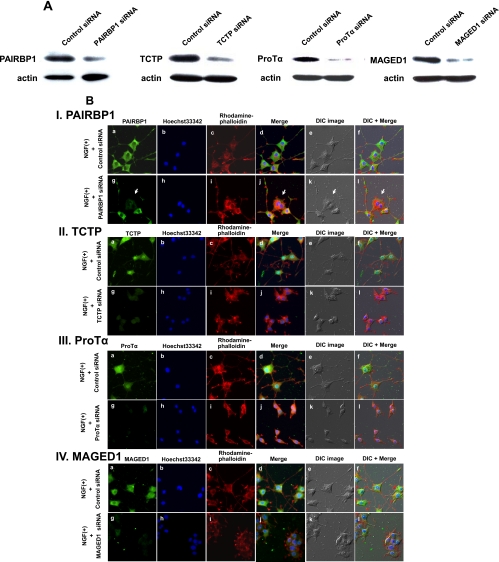
Suppression of Differentially Expressed Proteins by siRNA Treatment
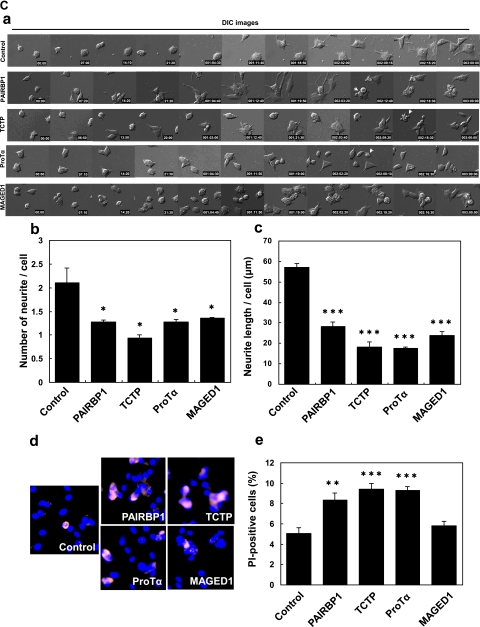
PAIRBP1, TCTP, ProTα, and MAGED1 were further analyzed using specific siRNAs to investigate their functions. siRNA-induced down-regulation was confirmed by Western blotting and ICC using their specific antibodies (Fig. 6, A and B) followed by a test of whether or not their suppression morphologically influenced the PC12 neurite formation. As expected, significant inhibition of the neurite formation was observed (Fig. 6B). Interestingly the suppression of MAGED1 caused aggregation of differentiating cells (Fig. 6, B, IV and C, a, and supplemental Movie 5). Furthermore we calculated the number and total length of neurites in differentiating cells and found that these measurements were certainly decreased by the suppression of each protein (Fig. 6C, b and c). In addition, apoptotic phenotypes were observed in NGF-stimulated cells treated with PAIRBP1, TCTP, or ProTα siRNA (Fig. 6C, a and supplemental Movies 2–4). We therefore observed the effects of the down-regulation of these proteins by siRNA on the survival of NGF-treated PC12 cells using PI staining for apoptotic cells coupled with nuclear staining by Hoechst33342. PAIRBP1, TCTP, or ProTα suppression caused a significant increase in PI-positive cells, whereas suppression of MAGED1 did not (Fig. 6C, d and e). These results suggested that, in PC12 cells, PAIRBP1, TCTP, ProTα, and MAGED1 regulate NGF-induced differentiation and that, except for MAGED1, they are involved in cell survival.
Fig. 6.
The effects caused by the suppression of the NGF-inducible proteins on PC12 cell differentiation. A, the cells were transfected with siRNA for each NGF-inducible protein or control siRNA for 24 h and then treated with NGF for 48 h. Immunoblot images were taken after treatment with each siRNA of the protein. Down-regulation of these proteins was confirmed. B, the cells were transfected with siRNA for each NGF-inducible protein or control siRNA for 24 h and then treated with NGF for 48 h. Cells were fixed and incubated with the antibodies against the indicated proteins (I, PAIRBP1; II, TCTP; III, ProTα; and IV, MAGED1) followed by detection with Alexa Fluor 488-labeled secondary antibodies and observation with a fluorescence microscope (a and g). Nuclear and F-actin were stained with Hoechst33342 (b and h) and rhodamine-phalloidin (c and i), respectively. The merged images for a, b, c and g, h, i, are shown in d and j, respectively. The merged images for d, e and j, k are shown in f and l, respectively. Differential interference contrast (DIC) images of PC12 cells in the same field were shown in e and k. The suppression of expression of each protein led to significant morphological changes, such as inhibition of neurite formation and induction of cellular aggregation in differentiating PC12 cells. Arrowheads indicate the PAIRBP1-suppressed cells. C, effects on neurite outgrowth and survival of PC12 cells of the siRNA treatment of the NGF-inducible proteins. a, representative time lapse images of NGF-stimulated PC12 cells treated with each siRNA for NGF-inducible proteins. The cells were transfected with siRNA for each NGF-inducible protein or control siRNA and stimulated with NGF for 72 h. A significant inhibition of neurite formation was observed due to each siRNA treatment. Arrows show the apoptotic phenotypes of PC12 cells. b and c, the cells were transfected with siRNA for each NGF-inducible protein or control siRNA and stimulated with NGF for 48 h under the condition of low serum (1% horse serum). The average of the number and total length of neurites of the PC12 cells are shown on the y axis (b and c). The data are expressed as means and S.D. of the three independent experiments (n = 3). For each experiment, more than 50 cells were counted. d and e, effects on PC12 cell survival of the siRNA treatment of the NGF-inducible proteins. The cells were transfected with siRNA for each NGF-inducible protein or control siRNA, stimulated with NGF for 48 h, fixed, and stained with PI and Hoechst33342 dye. Representative images of PI and Hoechst staining of PC12 cells treated with each siRNA and control and stimulated with NGF are shown (d). The percentage of PI-positive PC12 cells in total Hoechst-stained PC12 cells is shown on the y axis (e). The data and error bars are expressed as means and S.D. of the three independent experiments (n = 3), respectively. For each experiment, more than 500 cells were counted. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus NGF + control siRNA treatment (Student's t test).
DISCUSSION
MS-based techniques targeting functional proteins in biological specimens have been developed to improve sensitivity, quantitation, and throughput. However, their application to biological study has not been satisfactory because their independent development has not met the needs of cell biologists and because of the lack of a sequential protocol and associated user-friendly analysis tools. Here we present a concise strategy consisting of sequential MS-based, in silico, and cell biological analyses to study the mechanisms of neuronal differentiation using PC12 cells.
Using MALDI-TOF-TOF and ESI-QqTOF in parallel, not only were certain proteins identified by both methods, but also unique proteins were independently identified by either of the methods. These data were combined to increase the number of identified proteins. It seemed important to know what kind of differences there were between the unique peptides identified by either of the systems. Previously Stapels and Barofsky (39) reported that the ESI system favored the identification of hydrophobic peptides whereas the MALDI system tended to lead to the identification of basic and aromatic peptides in the analysis of E. coli DNA-binding proteins. We noticed that the peptides identified by ESI-QqTOF had relatively higher theoretical pI values and molecular weights (pI = 6.46 and Mr = 1,694.4 on average) compared with those identified by MALDI-TOF-TOF (pI = 5.81 and Mr = 1,651.6 on average). The discrepancy, which seems contradictory, between our results and those of Stapels and Barofsky (39) may be explained by differences in sample, type of proteins analyzed, and the presence or absence of modification such as iTRAQ labeling. These results suggest that both MS systems may be complementary due to the biases of the two ionization systems and that analyses should be performed using both instruments to obtain more comprehensive information out of a given set of samples in a proteomics study.
In total, we analyzed 1,482 proteins with iTRAQ quantitation data of 4,899 proteins from 40,037 peptides identified by using the Mascot search engine (data not shown) in this study. To understand how our identification strategy covers the proteins expressed in PC12 cells, we analyzed the cellular component locations of all the identified proteins by the MANGO method and compared them with those of all human proteins listed in the Swiss-Prot database (20,332 entries, Release 57.2 of May 5, 2009) (supplemental Table 5). Of the 1,404 total identified human orthologs (PC12RPS), 1,231 were annotated with cellular component data. Of the latter, 758 (62%), 166 (13%), and 118 (10%) proteins were annotated with “cytoplasm,” “mitochondrion,” and “plasma membrane,” respectively. On the other hand, among 16,530 human proteins from the Swiss-Prot database annotated with cellular component data, 7,106 (43%), 1,033 (6%), and 3,632 (22%) proteins were annotated with cytoplasm, mitochondrion, and plasma membrane, respectively. In the PC12RPS, the occurrence ratios of cytoplasm (62%) and mitochondrion (13%) were higher than those in the Swiss-Prot human proteins (43 and 6%, respectively), whereas the ratio of plasma membrane (10%) in PC12RPS was lower than that in the Swiss-Prot human proteins (22%). By combining this ratio of plasma membrane with the ratio of “organelle membrane” proteins (10.97%), the percentage of membrane proteins in the total proteins identified in our study becomes more than 20%. From these results, it is suggested that although cytoplasmic and mitochondrial proteins are largely favored in our method other components such as membrane and nuclear proteins can be also identified in significant numbers (supplemental Table 5).
From 1,482 proteins identified in iTRAQ quantitation, 72 differentially expressed proteins, including 39 up-regulated and 33 down-regulated, were extracted. The up-regulation of neurosecretory protein VGF (40), neuromodulin (41), chromogranin A (42), annexin A2 (43), peripherin (44), MAP1B (45), AHNAK (46), and secretogranin 2 (42) was observed as reported previously (Table II). Eighteen proteins in our results also corresponded to NGF-responsive genes in cDNA microarray data of Dijkmans et al. (23) (Table II) that coincided with our results, showing the confidence and reliability of our methods. Sixty-four proteins were heretofore unreported (Table II). The majority were cytoskeletal organizing components, such as moesin, radixin, MAP1B, annexin A2, peripherin, CRMP-4, RAC1, keratin-8, and keratin-18, whose changes in expression suggested roles for them in the cytoskeletal reorganization for cellular motility and morphogenesis. Interestingly PCNA and flap endonuclease 1 are both required for DNA replication, and both histones H2A and H2B are nucleosome components, all of which were down-regulated by NGF treatment. These results suggested that the down-regulation of these proteins was a factor in the arrest of cell division and the initiation of PC12 neuronal differentiation.
Further biological and functional interpretation of the above proteins was performed by proteomics GO analysis using the GO annotation and analysis tool. Here we encountered two problems. First, unlike microarray data, proteomics data consisting of thousands of proteins cannot be easily annotated by GO because of the general lack of either an automated method of proteomics GO annotation or GO analysis software that directly links to the proteomics database. Second, the genome information (including GO) of rat is much poorer than that of human or mouse. To overcome those difficulties, we therefore created a simple method, namely the MANGO method, that consists of the annotation tool (MANGO) and our PC12 cell proteome reference set (PC12RPS) with all non-human proteins converted into corresponding human orthologs. This method enabled us to analyze the GO of the differentially expressed proteins using the human PC12RPS very quickly.
The MANGO method helped us to narrow the focus of our study when the results suggested that NGF-induced PC12 cell differentiation involves functional up-regulation of protein groups related to six processes from which we selected cell morphogenesis and apoptosis/cell survival for deeper investigation. This led to biological validation by Western blotting, which confirmed that all of proteins in these groups were in fact up-regulated in PC12 cells by the NGF treatment (Fig. 4). Because their neuronal functions have not been reported in detail, of the proteins shown in Fig. 4, we focused on PAIRBP1 (34, 35), TCTP (36), ProTα (37), and MAGED1 (38). The indispensable roles of these proteins were further indicated by our subsequent ICC and siRNA-based studies. In this way, our MANGO method allowed us to quickly determine high probability candidate proteins whose important roles in NGF-induced PC12 cell differentiation are elaborated below.
PAIRBP1 has been identified as a protein involved in the regulation of PAI-1 mRNA stability via its binding to the cyclic nucleotide-responsive sequence of the PAI-1 mRNA (34). PAI-1 is the primary inhibitor of both urokinase- and tissue-type plasminogen activator (47). Soeda et al. (35) have shown that PAI-1 promotes the neurite outgrowth and survival of PC12 cells. This neuroprotective activity is correlated with enhanced activation of both extracellular signal-regulated kinase following a direct phosphorylation of NGF receptor Trk A and c-Jun, suggesting that PAI-1 can act as a neurotrophic factor (35). Based on those findings, we hypothesized that PAIRBP1 would be a positive regulator of PC12 differentiation and survival via PAI-1 mRNA stability. In our study, we observed that its suppression caused the inhibition of neurite formation and cell death. Interestingly PAIRBP1 was expressed not only in the cytoplasm but also in the neurites, especially in the tips and junctions (Fig. 5A, g and i, arrowheads), suggesting that it may have some other essential roles such as in cytoskeletal reorganization for the neurite formation that are independent of PAI-1.
TCTP has been identified in many eukaryotes and is widely expressed. It is a growth-related protein whose expression is highly regulated at the transcriptional and translational levels and by a wide range of extracellular signals (48). TCTP is involved in calcium binding and microtubule stabilization and is implicated in the regulation of cell growth and the cell cycle (48). It is also postulated to have antiapoptotic activity (49). Previous reports have revealed that TCTP protein expression decreased in differentiating mouse embryonic stem cells (mESCs) (50) and in dopaminergic motor neurons having differentiated from mouse E14 cells (36), indicating that TCTP has an important role in mESC differentiation. In contrast, in our iTRAQ data, TCTP was up-regulated after 48 h of NGF stimulation. Our Western blotting analysis showed the time-dependent changes of the TCTP intensity after NGF treatment; namely the intensities had increased up to 12 h and then gradually decreased. This TCTP expression pattern in NGF-treated PC12 cells seems to be similar to that of the developing mESC. Compared with the TCTP intensities without NGF, those with NGF were higher throughout the same analysis (12–72 h) (Fig. 4 and supplemental Fig. 2). We also showed in this study that TCTP knockdown caused inhibition of neurite formation and apoptosis (Fig. 6C), suggesting that NGF-induced up-regulation of TCTP could be a trigger of neurite outgrowth and that its down-regulation after this triggering role might have another function in the neurite formation or cell survival of NGF-induced PC12 cells.
ProTα is a highly acidic protein widely expressed in mammalian cells. ProTα gene expression is correlated with cell proliferation in a wide variety of cells, providing evidence for its involvement in cell growth (51). ProTα overexpression has been shown to both accelerate proliferation and retard differentiation in several cell types, such as HL60, K562, U937, and PC12 (37). Because ProTα was up-regulated in our proteomics analysis, we evaluated the time-dependent change in ProTα protein levels after NGF treatment by Western blotting (Fig. 4 and supplemental Fig. 2). ProTα expression peaked at 24 h and was followed by immediate down-regulation, suggesting that there might be some strict feedback mechanisms for ProTα expression in NGF-induced PC12 differentiation. Also ProTα was primarily localized in the nucleus after NGF stimulation (Fig. 5C, g and i, arrows). Nuclear ProTα reportedly interacts with histone H1 to promote chromatin decondensation and facilitate transcription or replication, the mechanism of which is regulated by casein kinase II phosphorylation of ProTα (52). Interestingly we observed that the expression of the casein kinase II catalytic subunit was down-regulated by NGF (Table I). The up-regulation of ProTα with accompanying casein kinase II down-regulation could therefore lead to transcriptional activation of genes related to PC12 cell differentiation and survival.
MAGED1, neurotrophin receptor-interacting MAGE homolog, was identified as a binding partner for p75 neurotrophin receptor (p75NTR) by yeast two-hybrid assay (38). MAGED1 reportedly binds p75NTR, antagonizes p75NTR to TrkA association, inhibits cell cycle progression, and facilitates p75NTR-mediated apoptosis (38). However, there is little information about p75NTR-mediated signaling by MAGED1. Because the localization of MAGED1 is not consistent with that of p75NTR (53), MAGED1 is likely to have the potential for signaling that is independent of p75NTR. Human MAGED1 reportedly regulates homotypic cell-cell adhesion by disrupting the E-cadherin·β-catenin complex (54). We observed the aggregation of differentiating PC12 cells triggered by the siRNA-induced MAGED1 down-regulation (Fig. 6, B, IV and C, a, and supplemental Movie 5), suggesting that MAGED1 suppression may have a significant effect on cell morphogenesis rather than cell survival during NGF-induced differentiation in PC12 cells.
In this study, we constructed the proteome database for PC12 cells containing the maximum number of the peptides/proteins so far identified and successfully identified new proteins related to cellular differentiation and apoptosis/survival. Our sequential proteomics strategy demonstrated here is simple and effective for identifying the proteins most likely involved in the physiological phenomena of interest because it enables one to quickly but confidently identify an appropriate focus on biological processes when interpreting extensive proteomics data. We expect that this proteomics strategy can become a standard method to target and elucidate the functions of proteins involved in cellular biological processes, to study the onset and pathogenesis of various diseases, and to discover new drug candidates in the near future.
Supplementary Material
Acknowledgments
We thank Prof. Hideyuki Saya, Keio University, School of Medicine, for kind advice and encouragement throughout this study. We also thank the entire staff of the Department of Tumor Genetics and Biology in Kumamoto University for helpful support, especially N. Tsubota, U. Midorikawa, and M. Nagayama for collaborative endeavors and M. Morikawa, M. Shimono, and C. Midorikawa for secretarial assistance. We are also grateful to staff members of the Proteomic Analysis Core-system on General Research Core Laboratory, Kumamoto University Medical School, for important contributions to the experiments.
Footnotes
* This work was supported by grants from Cancer Research (to N. A.), Kiban Research (to N. A.), and Houga Research (to N. A.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; from The Ministry of Health and Welfare of Japan (to N. A.); from the Japan Health Sciences Foundation (to D. K. and N. A.); and from the Center of Excellence project B of Kumamoto University for proteomics research and education (to N. A.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- iTRAQ
- isobaric tagging for relative and absolute quantitation
- NGF
- nerve growth factor
- GO
- gene ontology
- PC12PRS
- PC12 proteome reference set
- ICC
- immunocytochemistry
- siRNA
- short interfering RNA
- TCTP
- translationally controlled tumor protein
- ProTα
- prothymosin α
- TrkA
- tropomyosin-related kinase A
- 2-D
- two-dimensional
- RP
- reverse phase
- GOA
- gene ontology annotation
- PCNA
- proliferating cell nuclear antigen
- PI
- propidium iodide
- PAIRBP1
- plasminogen activator inhibitor 1 RNA-binding protein
- PAI
- plasminogen activator inhibitor
- mESC
- mouse embryonic stem cell
- MAGE
- melanoma antigen
- p75NTR
- p75 neurotrophin receptor
- QqTOF
- quadrupole/quadrupole/time-of-flight mass spectrometers.
REFERENCES
- 1.Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 2.Mann M. (2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol 7, 952–958 [DOI] [PubMed] [Google Scholar]
- 3.Keller M., Rüegg A., Werner S., Beer H. D. (2008) Active caspase-1 is a regulator of unconventional protein secretion. Cell 132, 818–831 [DOI] [PubMed] [Google Scholar]
- 4.Wang Z., Gucek M., Hart G. W. (2008) Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl. Acad. Sci. U.S.A 105, 13793–13798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graumann J., Hubner N. C., Kim J. B., Ko K., Moser M., Kumar C., Cox J., Schöler H., Mann M. (2008) Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 7, 672–68318045802 [Google Scholar]
- 6.DeSouza L. V., Grigull J., Ghanny S., Dubé V., Romaschin A. D., Colgan T. J., Siu K. W. (2007) Endometrial carcinoma biomarker discovery and verification using differentially tagged clinical samples with multidimensional liquid chromatography and tandem mass spectrometry. Mol. Cell. Proteomics 6, 1170–1182 [DOI] [PubMed] [Google Scholar]
- 7.Ralhan R., Desouza L. V., Matta A., Chandra Tripathi S., Ghanny S., Datta Gupta S., Bahadur S., Siu K. W. (2008) Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol. Cell. Proteomics 7, 1162–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S., Mathieson T., Perrin J., Raida M., Rau C., Reader V., Sweetman G., Bauer A., Bouwmeester T., Hopf C., Kruse U., Neubauer G., Ramsden N., Rick J., Kuster B., Drewes G. (2007) Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol 25, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 9.Greene L. A., Tischler A. S. (1976) Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. U.S.A 73, 2424–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adler E. M. (2006) Teaching resources. Cell culture as a model system for teaching: using PC12 cells. Sci. STKE 2006, tr5. [DOI] [PubMed] [Google Scholar]
- 11.Yankner B. A., Dawes L. R., Fisher S., Villa-Komaroff L., Oster-Granite M. L., Neve R. L. (1989) Neurotoxicity of a fragment of the amyloid precursor associated with Alzheimer's disease. Science 245, 417–420 [DOI] [PubMed] [Google Scholar]
- 12.Apostol B. L., Kazantsev A., Raffioni S., Illes K., Pallos J., Bodai L., Slepko N., Bear J. E., Gertler F. B., Hersch S., Housman D. E., Marsh J. L., Thompson L. M. (2003) A cell-based assay for aggregation inhibitors as therapeutics of polyglutamine-repeat disease and validation in Drosophila. Proc. Natl. Acad. Sci. U.S.A 100, 5950–5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryu E. J., Harding H. P., Angelastro J. M., Vitolo O. V., Ron D., Greene L. A. (2002) Endoplasmic reticulum stress and the unfolded protein response in cellular models of Parkinson's disease. J. Neurosci 22, 10690–10698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yunoue S., Tokuo H., Fukunaga K., Feng L., Ozawa T., Nishi T., Kikuchi A., Hattori S., Kuratsu J., Saya H., Araki N. (2003) Neurofibromatosis type I tumor suppressor neurofibromin regulates neuronal differentiation via its GTPase-activating protein function toward Ras. J. Biol. Chem 278, 26958–26969 [DOI] [PubMed] [Google Scholar]
- 15.Feng L., Yunoue S., Tokuo H., Ozawa T., Zhang D., Patrakitkomjorn S., Ichimura T., Saya H., Araki N. (2004) PKA phosphorylation and 14-3-3 interaction regulate the function of neurofibromatosis type I tumor suppressor, neurofibromin. FEBS Lett 557, 275–282 [DOI] [PubMed] [Google Scholar]
- 16.Patrakitkomjorn S., Kobayashi D., Morikawa T., Wilson M. M., Tsubota N., Irie A., Ozawa T., Aoki M., Arimura N., Kaibuchi K., Saya H., Araki N. (2008) Neurofibromatosis type 1 (NF1) tumor suppressor, neurofibromin, regulates the neuronal differentiation of PC12 cells via its associating protein, CRMP-2. J. Biol. Chem 283, 9399–9413 [DOI] [PubMed] [Google Scholar]
- 17.Davies A. M. (1994) The role of neurotrophins in the developing nervous system. J. Neurobiol 25, 1334–1348 [DOI] [PubMed] [Google Scholar]
- 18.Chao M. V. (2003) Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat. Rev. Neurosci 4, 299–309 [DOI] [PubMed] [Google Scholar]
- 19.Lee N. H., Weinstock K. G., Kirkness E. F., Earle-Hughes J. A., Fuldner R. A., Marmaros S., Glodek A., Gocayne J. D., Adams M. D., Kerlavage A. R., Fraser C. M., Venter J. C. (1995) Comparative expressed-sequence-tag analysis of differential gene expression profiles in PC-12 cells before and after nerve growth factor treatment. Proc. Natl. Acad. Sci. U.S.A 92, 8303–8307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayumi K., Yaoi T., Kawai J., Kojima S., Watanabe S., Suzuki H. (1998) Improved restriction landmark cDNA scanning and its application to global analysis of genes regulated by nerve growth factor in PC12 cells. Biochim. Biophys. Acta 1399, 10–18 [DOI] [PubMed] [Google Scholar]
- 21.Brown A. J., Hutchings C., Burke J. F., Mayne L. V. (1999) Application of a rapid method (targeted display) for the identification of differentially expressed mRNAs following NGF-induced neuronal differentiation in PC12 cells. Mol. Cell. Neurosci 13, 119–130 [DOI] [PubMed] [Google Scholar]
- 22.Angelastro J. M., Klimaschewski L., Tang S., Vitolo O. V., Weissman T. A., Donlin L. T., Shelanski M. L., Greene L. A. (2000) Identification of diverse nerve growth factor-regulated genes by serial analysis of gene expression (SAGE) profiling. Proc. Natl. Acad. Sci. U.S.A 97, 10424–10429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dijkmans T. F., van Hooijdonk L. W., Schouten T. G., Kamphorst J. T., Vellinga A. C., Meerman J. H., Fitzsimons C. P., de Kloet E. R., Vreugdenhil E. (2008) Temporal and functional dynamics of the transcriptome during nerve growth factor-induced differentiation. J. Neurochem 105, 2388–2403 [DOI] [PubMed] [Google Scholar]
- 24.Garrels J. I., Schubert D. (1979) Modulation of protein synthesis by nerve growth factor. J. Biol. Chem 254, 7978–7985 [PubMed] [Google Scholar]
- 25.Sussman M. A., Battenberg E., Bloom F. E., Fowler V. M. (1990) Identification of two nerve growth factor-induced polypeptides in PC12 cells. J. Mol. Neurosci 2, 163–174 [DOI] [PubMed] [Google Scholar]
- 26.Huang C. M., Shui H. A., Wu Y. T., Chu P. W., Lin K. G., Kao L. S., Chen S. T. (2001) Proteomic analysis of proteins in PC12 cells before and after treatment with nerve growth factor: increased levels of a 43-kDa chromogranin B-derived fragment during neuronal differentiation. Brain Res. Mol. Brain Res 92, 181–192 [DOI] [PubMed] [Google Scholar]
- 27.Huang Y. H., Chang A. Y., Huang C. M., Huang S. W., Chan S. H. (2002) Proteomic analysis of lipopolysaccharide-induced apoptosis in PC12 cells. Proteomics 2, 1220–1228 [DOI] [PubMed] [Google Scholar]
- 28.Zhou B., Yang W., Ji J. G., Ru B. G. (2004) Differential display proteome analysis of PC-12 cells transiently transfected with metallothionein-3 gene. J. Proteome Res 3, 126–131 [DOI] [PubMed] [Google Scholar]
- 29.Yang W., Liu P., Liu Y., Wang Q., Tong Y., Ji J. (2006) Proteomic analysis of rat pheochromocytoma PC12 cells. Proteomics 6, 2982–2990 [DOI] [PubMed] [Google Scholar]
- 30.Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 31.Carbon S., Ireland A., Mungall C. J., Shu S., Marshall B., Lewis S. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kondili K., Tsolas O., Papamarcaki T. (1996) Selective interaction between parathymosin and histone H1. Eur. J. Biochem 242, 67–74 [DOI] [PubMed] [Google Scholar]
- 33.Ueda H., Fujita R., Yoshida A., Matsunaga H., Ueda M. (2007) Identification of prothymosin-alpha1, the necrosis-apoptosis switch molecule in cortical neuronal cultures. J. Cell Biol 176, 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heaton J. H., Dlakic W. M., Dlakic M., Gelehrter T. D. (2001) Identification and cDNA cloning of a novel RNA-binding protein that interacts with the cyclic nucleotide-responsive sequence in the Type-1 plasminogen activator inhibitor mRNA. J. Biol. Chem 276, 3341–3347 [DOI] [PubMed] [Google Scholar]
- 35.Soeda S., Shinomiya K., Ochiai T., Koyanagi S., Toda A., Eyanagi R., Shimeno H. (2006) Plasminogen activator inhibitor-1 aids nerve growth factor-induced differentiation and survival of pheochromocytoma cells by activating both the extracellular signal-regulated kinase and c-Jun pathways. Neuroscience 141, 101–108 [DOI] [PubMed] [Google Scholar]
- 36.Wang D., Gao L. (2005) Proteomic analysis of neural differentiation of mouse embryonic stem cells. Proteomics 5, 4414–4426 [DOI] [PubMed] [Google Scholar]
- 37.Rodríguez P., Viñuela J. E., Alvarez-Fernández L., Buceta M., Vidal A., Domínguez F., Gómez-Márquez J. (1998) Overexpression of prothymosin alpha accelerates proliferation and retards differentiation in HL-60 cells. Biochem. J 331, 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salehi A. H., Roux P. P., Kubu C. J., Zeindler C., Bhakar A., Tannis L. L., Verdi J. M., Barker P. A. (2000) NRAGE, a novel MAGE protein, interacts with the p75 neurotrophin receptor and facilitates nerve growth factor-dependent apoptosis. Neuron 27, 279–288 [DOI] [PubMed] [Google Scholar]
- 39.Stapels M. D., Barofsky D. F. (2004) Complementary use of MALDI and ESI for the HPLC-MS/MS analysis of DNA-binding proteins. Anal. Chem 76, 5423–5430 [DOI] [PubMed] [Google Scholar]
- 40.Possenti R., Eldridge J. D., Paterson B. M., Grasso A., Levi A. (1989) A protein induced by NGF in PC12 cells is stored in secretory vesicles and released through the regulated pathway. EMBO J 8, 2217–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mobarak C. D., Anderson K. D., Morin M., Beckel-Mitchener A., Rogers S. L., Furneaux H., King P., Perrone-Bizzozero N. I. (2000) The RNA-binding protein HuD is required for GAP-43 mRNA stability, GAP-43 gene expression, and PKC-dependent neurite outgrowth in PC12 cells. Mol. Biol. Cell 11, 3191–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laslop A., Tschernitz C. (1992) Effects of nerve growth factor on the biosynthesis of chromogranin A and B, secretogranin II and carboxypeptidase H in rat PC12 cells. Neuroscience 49, 443–450 [DOI] [PubMed] [Google Scholar]
- 43.Jacovina A. T., Zhong F., Khazanova E., Lev E., Deora A. B., Hajjar K. A. (2001) Neuritogenesis and the nerve growth factor-induced differentiation of PC-12 cells requires annexin II-mediated plasmin generation. J. Biol. Chem 276, 49350–49358 [DOI] [PubMed] [Google Scholar]
- 44.Aletta J. M., Angeletti R., Liem R. K., Purcell C., Shelanski M. L., Greene L. A. (1988) Relationship between the nerve growth factor-regulated clone 73 gene product and the 58-kilodalton neuronal intermediate filament protein (peripherin). J. Neurochem 51, 1317–1320 [DOI] [PubMed] [Google Scholar]
- 45.Stroth U., Meffert S., Gallinat S., Unger T. (1998) Angiotensin II and NGF differentially influence microtubule proteins in PC12W cells: role of the AT2 receptor. Brain Res. Mol. Brain Res 53, 187–195 [DOI] [PubMed] [Google Scholar]
- 46.Lorusso A., Covino C., Priori G., Bachi A., Meldolesi J., Chieregatti E. (2006) Annexin2 coating the surface of enlargeosomes is needed for their regulated exocytosis. EMBO J 25, 5443–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrence D. A., Strandberg L., Ericson J., Ny T. (1990) Structure-function studies of the SERPIN plasminogen activator inhibitor type 1. Analysis of chimeric strained loop mutants. J. Biol. Chem 265, 20293–20301 [PubMed] [Google Scholar]
- 48.Bommer U. A., Thiele B. J. (2004) The translationally controlled tumour protein (TCTP). Int. J. Biochem. Cell Biol 36, 379–385 [DOI] [PubMed] [Google Scholar]
- 49.Yang Y., Yang F., Xiong Z., Yan Y., Wang X., Nishino M., Mirkovic D., Nguyen J., Wang H., Yang X. F. (2005) An N-terminal region of translationally controlled tumor protein is required for its antiapoptotic activity. Oncogene 24, 4778–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baharvand H., Fathi A., Gourabi H., Mollamohammadi S., Salekdeh G. H. (2008) Identification of mouse embryonic stem cell-associated proteins. J. Proteome Res 7, 412–423 [DOI] [PubMed] [Google Scholar]
- 51.Segade F., Gómez-Márquez J. (1999) Prothymosin alpha. Int. J. Biochem. Cell Biol 31, 1243–1248 [DOI] [PubMed] [Google Scholar]
- 52.Gómez-Márquez J. (2007) Function of prothymosin alpha in chromatin decondensation and expression of thymosin beta-4 linked to angiogenesis and synaptic plasticity. Ann. N.Y. Acad. Sci 1112, 201–209 [DOI] [PubMed] [Google Scholar]
- 53.Barrett G. L., Greferath U., Barker P. A., Trieu J., Bennie A. (2005) Co-expression of the P75 neurotrophin receptor and neurotrophin receptor-interacting melanoma antigen homolog in the mature rat brain. Neuroscience 133, 381–392 [DOI] [PubMed] [Google Scholar]
- 54.Xue B., Wen C., Shi Y., Zhao D., Li C. (2005) Human NRAGE disrupts E-cadherin/beta-catenin regulated homotypic cell-cell adhesion. Biochem. Biophys. Res. Commun 336, 247–251 [DOI] [PubMed] [Google Scholar]
- 55.Quinn C. C., Chen E., Kinjo T. G., Kelly G., Bell A. W., Elliott R. C., McPherson P. S., Hockfield S. (2003) TUC-4b, a novel TUC family variant, regulates neurite outgrowth and associates with vesicles in the growth cone. J. Neurosci 23, 2815–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.