Abstract
There is a mounting evidence of the existence of autoantibodies associated to cancer progression. Antibodies are the target of choice for serum screening because of their stability and suitability for sensitive immunoassays. By using commercial protein microarrays containing 8000 human proteins, we examined 20 sera from colorectal cancer (CRC) patients and healthy subjects to identify autoantibody patterns and associated antigens. Forty-three proteins were differentially recognized by tumoral and reference sera (p value <0.04) in the protein microarrays. Five immunoreactive antigens, PIM1, MAPKAPK3, STK4, SRC, and FGFR4, showed the highest prevalence in cancer samples, whereas ACVR2B was more abundant in normal sera. Three of them, PIM1, MAPKAPK3, and ACVR2B, were used for further validation. A significant increase in the expression level of these antigens on CRC cell lines and colonic mucosa was confirmed by immunoblotting and immunohistochemistry on tissue microarrays. A diagnostic ELISA based on the combination of MAPKAPK3 and ACVR2B proteins yielded specificity and sensitivity values of 73.9 and 83.3% (area under the curve, 0.85), respectively, for CRC discrimination after using an independent sample set containing 94 sera representative of different stages of progression and control subjects. In summary, these studies confirmed the presence of specific autoantibodies for CRC and revealed new individual markers of disease (PIM1, MAPKAPK3, and ACVR2B) with the potential to diagnose CRC with higher specificity and sensitivity than previously reported serum biomarkers.
Colorectal cancer (CRC)1 is the second most prevalent cancer in the western world. The development of this disease takes decades and involves multiple genetic events. CRC remains a major cause of mortality in developed countries because most of the patients are diagnosed at advanced stages because of the reluctance to use highly invasive diagnostic tools like colonoscopy. Actually only a few proteins have been described as biomarkers in CRC (carcinoembryonic antigen (CEA), CA19.9, and CA125 (1–3)), although none of them is recommended for clinical screening (4). Proteomics analysis is actively used for the identification of new biomarkers. In previous studies, the use of two-dimensional DIGE and antibody microarrays allowed the identification of differentially expressed proteins in CRC tissue, including isoforms and post-translational modifications responsible for modifications in signaling pathways (5–8). Both approaches resulted in the identification of a collection of potential tumoral tissue biomarkers that is currently being investigated.
However, the implementation of simpler, non-invasive methods for the early detection of CRC should be based on the identification of proteins or antibodies in serum or plasma (9–13). There is ample evidence of the existence of an immune response to cancer in humans as demonstrated by the presence of autoantibodies in cancer sera. Self-proteins (autoantigens) altered before or during tumor formation can elicit an immune response (13–19). These tumor-specific autoantibodies can be detected at early cancer stages and prior to cancer diagnosis revealing a great potential as biomarkers (14, 15, 20). Tumor proteins can be affected by specific point mutations, misfolding, overexpression, aberrant glycosylation, truncation, or aberrant degradation (e.g. p53, HER2, NY-ESO1, or MUC1 (16, 21–25)). In fact, a number of tumor-associated autoantigens (TAAs) were identified previously in different studies involving autoantibody screening in CRC (26–28).
Several approaches have been used to identify TAAs in cancer, including natural protein arrays prepared with fractions obtained from two-dimensional LC separations of tumoral samples (29, 30) or protein extracts from cancer cells or tissue (9, 31) probed by Western blot with patient sera, cancer tissue peptide libraries expressed as cDNA expression libraries for serological screening (serological analysis of recombinant cDNA expression libraries (SEREX)) (22, 32), or peptides expressed on the surface of phages in combination with microarrays (17, 18, 33, 34). However, these approaches suffer from several drawbacks. In some cases TAAs have to be isolated and identified from the reactive protein lysate by LC-MS techniques, or in the phage display approach, the reactive TAA could be a mimotope without a corresponding linear amino acid sequence. Moreover, cDNA libraries might not be representative of the protein expression levels in tumors as there is a poor correspondence between mRNA and protein levels.
Protein arrays provide a novel platform for the identification of both autoantibodies and their respective TAAs for diagnostic purposes in cancer serum patients. They present some advantages. Proteins printed on the microarray are known “a priori,” avoiding the need for later identifications and the discovery of mimotopes. There is no bias in protein selection as the proteins are printed at a similar concentration. This should result in a higher sensitivity for biomarker identification (13, 35, 36).
In this study, we used commercially available high density protein microarrays for the identification of autoantibody signatures and tumor-associated antigens in colorectal cancer. We screened 20 CRC patient and control sera with protein microarrays containing 8000 human proteins to identify the CRC-associated autoantibody repertoire and the corresponding TAAs. Autoantibody profiles that discriminated the different types of CRC metastasis were identified. Moreover a set of TAAs showing increased or decreased expression in cancer cell lines and paired tumoral tissues was found. Finally an ELISA was set up to test the ability of the most immunoreactive proteins to detect colorectal adenocarcinoma. On the basis of the antibody response, combinations of three antigens, PIM1, MAPKAPK3, and ACVR2B, showed a great potential for diagnosis.
EXPERIMENTAL PROCEDURES
Clinical Information and Serum Collection
For microarray screening, serum samples from 12 individuals were collected after CRC diagnosis (Hospital Universitario de Salamanca). Those samples were selected for having CRC in advanced stages as well as for developing metastasis to liver (seven patients), liver and lung (four patients), or liver and bone marrow (one patient). The median age for the CRC patients was 64.5 years (range, 41–84 years). Eight control serum samples were obtained from healthy subjects and were selected to match both the median age of the CRC population and the same proportion of male and female subjects. Clinical data from the patients are provided in Table I. For ELISA validations, another set of 52 serum samples from CRC patients representative of the different Dukes stages (A–D) and 42 control serum samples from healthy subjects was used for the validation screening (supplemental Table S1).
Table I.
Clinical information of the CRC patients tested in the high density human protein microarrays
| Serum | Agea | Genderb | Outcomec | Survival timed | Metastasis |
|---|---|---|---|---|---|
| VH1 | 84 | F | Alive | — | Liver |
| MH1 | 60 | F | Dead | 15 | Liver |
| MHP1 | 65 | M | Dead | 64 | Liver-lung |
| MHP2 | 41 | M | Dead | 62 | Liver-lung |
| MH2 | 55 | M | Dead | 14 | Liver |
| MHP3 | 62 | M | Dead | 51 | Liver-lung |
| VP1 | 71 | F | Alive | — | Lung-bone |
| VH2 | 75 | M | Alive | — | Liver |
| MH3 | 76 | M | Dead | 31 | Liver |
| MH4 | 64 | M | Dead | 28 | Liver |
| VHP1 | 51 | M | Alive | — | Liver-lung |
| VH3 | 74 | M | Alive | — | Liver |
a In years.
b M, male; F, female.
c Outcome of the CRC patients after collecting the serum.
d Survival time in months after collecting the serum samples. —, survival time was longer than five years.
All sera were processed using identical procedures. Blood samples were left at room temperature for a minimum of 30 min (and a maximum of 60 min) to allow clot formation and then centrifuged at 3000 × g for 10 min at 4 °C. The serum was frozen and stored at −80 °C until use.
Protein Arrays
Twenty serum samples (12 from the CRC tumor group and eight from the control group; Table I) were probed in the Human ProtoArrayTM v4.0 (Invitrogen). These microarrays contained 8000 human GST-tagged proteins expressed in Sf9 insect cells and spotted in duplicate. ProtoArrays were used according to the recommendations of the manufacturer. Briefly the slides were equilibrated at 4 °C for 15 min and then incubated with blocking buffer (1% BSA in 0.1% Tween 20, PBS) for 1 h at 4 °C with gentle agitation. Then 150 μl of human serum (diluted 1:50 in blocking buffer) was overlaid on the arrays, covered with cover glass (Corning), and incubated for 90 min at 4 °C. The slides were washed three times for 10 min with probe buffer (1% BSA, 0.5 mm DTT, 5% glycerol, 0.05% Triton X-100 in PBS). Human bound antibodies were detected after incubation with Alexa Fluor 647-labeled goat anti-human IgG (Invitrogen; diluted 1:2000 in probe buffer) for 90 min at 4 °C. The arrays were washed and dried by centrifugation at 1000 rpm for 1 min at room temperature. As a first control, ProtoArrays v4.0 were probed with goat anti-GST antibody to check the uniformity of the proteins spotted in the array followed by incubation with Alexa Fluor 555-labeled anti-goat IgG. The other control array was only incubated with the secondary antibody Alexa Fluor 647-labeled anti-human IgG for background determination. Finally the slides were scanned on a ScanArrayTM 5000 (Packard BioChip Technologies) to produce red (Alexa Fluor 647) or green images (Alexa Fluor 555). The Genepix Pro 5.1 (Axon Laboratories) image analysis software was used for the quantification.
Proteins, Antibodies, and Cell Lines
A cDNA encoding the full-length human PIM1 was introduced into the pET28a expression vector (Novagen). The His6-PIM1 fusion protein was then expressed in Escherichia coli strain BL21(DE3) (Invitrogen) and purified by affinity chromatography on a HiTrap chelating column (GE Healthcare) followed by gel filtration on a Superdex 200 column (GE Healthcare). Human MAPKAPK3 protein was purchased from GenWay (San Diego, CA). Human ACVR2B cDNA was cloned into the pDEST527 (a gift from Dr. J. L. Hartley, National Institutes of Health), expressed, and purified as mentioned above. Human Annexin IV cDNA was cloned into pTT3 expression vector (kindly provided by Dr. Y. Durochet, Biotechnology Research Institute, Montreal, Canada) and expressed in HEK293-EBNA cells. The recombinant protein was produced by the transiently transfected cells and purified by affinity chromatography on a nickel-chelating resin (GE Healthcare). CEA and human seroalbumin were purchased from Sigma. Antibodies against MAPKAPK3 and PIM1 used in ELISA were purchased from Abnova. Antibodies against MAPKAPK3, PIM1, and ACVR2B used for immunoblotting and tissue microarray were purchased from Abcam.
CRC cell lines (RKO, HCT116, HCT15, SW48, SW480, and Colo205) and control BxPc3 (pancreatic adenocarcinoma) and Molt4 (lymphoblastoid) cells were grown according to established protocols. Neutrophiles (Neut) and lymphocytes (lymph) were isolated from peripheral blood cells from a healthy individual. Murine embryo fibroblasts were immortalized by infecting a primary culture with the Epstein-Barr virus and grown according to established protocols.
Western Blot Analysis
Protein extracts from paired tissues from CRC patients were prepared as described previously (6). Briefly protein extracts were obtained after lysis with 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate in 150 mm NaCl, 5 mm EDTA, 10 mm Tris-HCl (pH 7.2) containing protease inhibitor mixture (Roche Applied Science). After clarifying by centrifugation at 12,000 × g for 15 min, protein concentrations were determined with the 2-D Quant kit (GE Healthcare). For Western blot, 50 μg of protein extracts were separated by 10% SDS-PAGE and transferred to nitrocellulose membranes (Hybond-C Extra) according to standard procedures (37). After blocking, membranes were incubated overnight at 4 °C with PIM1 (dilution, 1:100), MAPKAPK3 (dilution, 1: 500), and ACVR2B (dilution, 1:200) antibodies. Immunodetection on the membranes was achieved by using either peroxidase-labeled, anti-goat IgG (Dako Cytomation) at a dilution of 1:5000 for ACVR2B and 1:20,000 for PIM1 or peroxidase-labeled anti-chicken IgY (Jackson ImmunoResearch Laboratories) at 1:20,000 for MAPKAPK3. The signal was developed by ECL (GE Healthcare).
Immunohistochemistry
Tissue microarrays (TMAs) specific for colorectal cancer with 45 different paired samples (tumoral and normal) were prepared as described previously (7). Sections were cut at a thickness of 3 μm and dried for 16 h at 56 °C before being dewaxed in xylene and rehydrated through graded ethanol series to water. A heat-induced epitope retrieval step was performed in 0.01 m trisodium citrate solution with heating for 2 min in a conventional pressure cooker. After heating, slides were rinsed in cool running water for 5 min and quickly washed in TBS (pH 7.4). TMAs were incubated with mouse monoclonal anti-PIM1 (dilution, 1:50) and goat polyclonal anti-ACVR2B (dilution, 1:10). Specific binding was followed by anti-IgG conjugated with biotin. Visualization of specific interaction was monitored by using the EnVision FLEX system (Dako Cytomation) or Bond (Vision BioSystems). Diaminobenzidine (DAB+) was used as substrate chromogen after the sections were counterstained with hematoxylin. A positive control was included within each staining experiment to ensure consistency between consecutive runs. The evaluation of the TMA was performed by two independent pathologists according to the following scale: 0, no staining; 1, weak staining; 2, normal staining; and 3, strong staining of the tissue cylinder.
ELISA
An ELISA was developed to test the ability of target proteins to screen for CRC status in sera. Briefly, microtiter plates (Maxisorp, Nunc) were coated overnight with 0.3 μg of the purified recombinant proteins, including human seroalbumin as negative control in 50 μl of PBS. After washing three times with PBS, plates were blocked with 3% skimmed milk in PBS (MPBS) for 2 h at room temperature. Then serum samples (dilution, 1:50 in 3% MPBS) were incubated for 2 h at room temperature. After washing, peroxidase-labeled anti-human IgG (Dako) (dilution, 1:3000 in 3% MPBS) was added for 2 h at room temperature. Then the signal was developed with 3,3′,5,5′-tetramethylbenzidine substrate for 10 min (Sigma). The reaction was stopped with 1 m H2SO4, and absorption was measured at 450 nm.
Statistical Analysis
Microarrays were analyzed with the ProtoArray Prospector Analyzer 4.0 (Invitrogen), which relies on Chebyshev's inequality principle (13). After applying quantile normalization, the algorithm compares the signal from each protein with the signal from the negative control features among the array and assigns a CI-p value for each protein. Then the software identifies the significant signals (those that can be determined as different from the background) and calculates the Z-scores, which reflect the signal strength relative to all protein features. Finally the program compares the two groups and identifies those proteins that exhibit increased signal values in both groups. In addition, a p value is calculated for each protein across the hypothesis that there is no signal increase in one group compared with another. Clusters were performed using Multi Experiment Viewer (Dana-Farber Cancer Institute, Boston, MA) based on Pearson's correlation distant metrics using normalized values in log10 to visualize the discrimination between the two groups analyzed.
For the analysis of ELISA data sets, a one-tailed Student's t test was performed assuming unequal variances to assess whether the means of normal and tumoral groups were statistically different from each other. Each individual marker was evaluated by a receiver operating characteristic (ROC) curve, and the corresponding area under the curve (AUC) was calculated using JMP® 7 (SAS, Cary, NC). Finally, we fitted logistic regressions using the bootstrap applied to the complete procedure (i.e. including the variable selection step) to determine the effect of the combination of the biomarkers (38). All models were fitted using Harrell's design library (39) with the R statistical computing system (40).
RESULTS
Profiling of Colorectal Cancer-specific Autoantibodies
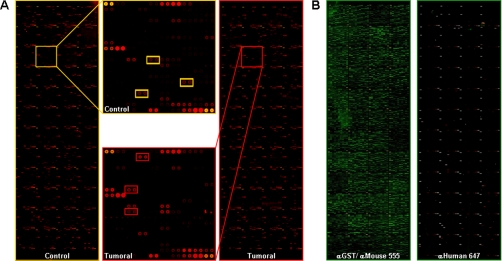
Twenty serum samples (12 CRC patients and 8 controls) were probed in the protein microarrays. Clinical data are shown in Table I. A representative image of the autoantibody response in patients and controls is shown in Fig. 1A. A different pattern of immunoreactivity was observed between the CRC patients and the reference sera. After quantifying the spot intensities, the data were normalized with the method of the quantiles and processed with ProtoArray Prospector Analyzer. Control slides indicated a good performance of the ProtoArrays with a low background and specific reactivity (Fig. 1, B and C).
Fig. 1.
Identification of proteins reactive to autoantibodies. A, ProtoArrays with over 8000 human proteins were probed with 12 sera from CRC patients and eight sera from control subjects (Table I). A representative array from each group is shown. The same subarray is highlighted in both images, and the reactive proteins differentially recognized by the autoantibodies of the patients and controls are identified by colored squares: red, proteins reacting with CRC patient serum; yellow, proteins reacting with control serum. B, control protein microarrays probed with anti-GST monoclonal antibody and Alexa Fluor 555-labeled goat anti-mouse IgG as protein printing control and with Alexa Fluor 647-labeled goat anti-human IgG as a negative control to detect false positive spots.
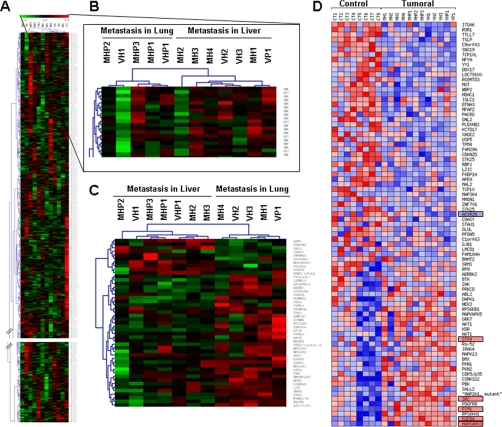
We investigated the use of autoantibody profiles to discriminate among cancer and normal sera and between different metastatic progression patterns. An unsupervised clustering analysis using the Multi Experiment Viewer software failed to discriminate between tumoral and normal sera (data not shown) probably due to the heterogeneity in the natural antibody response in healthy subjects. However, a good discrimination was obtained in metastatic samples, splitting them into two main branches corresponding to patients with metastasis in liver or lung; only two samples were misclassified (Fig. 2, A and B). Supervised analysis offered a correct separation (Fig. 2C). Prospector analysis of the 12 cancer sera identified a collection of TAAs with a prevalence higher than 60% associated to either liver (22 proteins) or lung (15 proteins) metastasis (Table II).
Fig. 2.
Hierarchical cluster analysis of serum samples. A, a panel representing the unsupervised hierarchical cluster analysis comprising the complete set of 8000 proteins, when incubated with the sera of CRC patients, shows a good separation between the metastasis in liver and metastasis in lung CRC patients. Red, immunoreactive protein; green, non-reactive protein. B, the top of the cluster was enlarged to show the separation between metastasis in liver and metastasis in lung groups. C, supervised hierarchical cluster analysis of the most significant 37 CRC metastasis-associated reactive proteins shows a perfect separation in two branches of the CRC patients according to their metastasis status. D, heat map representation of the autoantibody signature that distinguishes sera from CRC and healthy individuals (p value <0.1). Red, higher reactivity in CRC; blue, lower reactivity in CRC.
Table II.
Proteins reacting with autoantibodies associated to metastasis
| Name | Liver prevalence | Lung prevalence | p value | Function |
|---|---|---|---|---|
| % | % | |||
| Increased reactivity in liver metastasis | ||||
| PHLDB1 | 88 | 14 | 0.0022 | Unknown |
| AKT3 | 75 | 14 | 0.0130 | Signal transduction |
| PRKCH | 75 | 14 | 0.0130 | Signal transduction |
| MAPKAPK3 | 88 | 29 | 0.0152 | Ras protein signal transduction |
| C9orf43 | 88 | 29 | 0.0152 | Hypothetical protein |
| EGFR | 88 | 29 | 0.0152 | Signal transduction |
| CAMKV | 63 | 14 | 0.0455 | Kinase, cell signaling |
| THAP3 | 63 | 14 | 0.0455 | Apoptosis |
| C15orf38 | 63 | 14 | 0.0455 | Hypothetical protein |
| EPB41L5 | 63 | 14 | 0.0455 | Cell-cell adhesion |
| PGAM1 | 63 | 14 | 0.0455 | Metabolism, energy pathways |
| PADI4 | 63 | 14 | 0.0455 | Metabolism, energy pathways |
| UBE2T | 63 | 14 | 0.0455 | Protein metabolism |
| C9orf78 | 63 | 14 | 0.0455 | Hypothetical protein |
| WDR61 | 63 | 14 | 0.0455 | Transcriptional regulation |
| PRKCB1 | 63 | 14 | 0.0455 | Signal transduction |
| PRKCD | 63 | 14 | 0.0455 | Signal transduction |
| ZAP70 | 63 | 14 | 0.0455 | T cell development and activation |
| ABL2 | 63 | 14 | 0.0455 | Signal transduction |
| WEE1 | 63 | 14 | 0.0455 | Cell cycle |
| DCAMKL2 | 63 | 14 | 0.0455 | Unknown |
| TRIM21 | 63 | 14 | 0.0455 | Transcriptional regulation |
| Lung prevalence | Liver prevalence | |||
|---|---|---|---|---|
| % | % | |||
| Increased reactivity in lung metastasis | ||||
| PAK1 | 86 | 13 | 0.00216 | Cell motility and morphology |
| HOMER2 | 86 | 25 | 0.01299 | Cell growth |
| IRAK4 | 86 | 25 | 0.01299 | Signal transduction |
| PRKD2 | 86 | 25 | 0.01299 | Signal transduction |
| AK075484 | 86 | 13 | 0.01299 | Hypothetical protein |
| C2orf13 | 71 | 13 | 0.01515 | Hypothetical protein |
| PSCD3 | 71 | 13 | 0.01515 | Signal transduction |
| SH3BGRL2 | 71 | 13 | 0.01515 | Unknown |
| CDK2 | 71 | 13 | 0.01515 | Cell cycle |
| DAPK2 | 71 | 13 | 0.01515 | Apoptosis |
| TRPT1 | 86 | 38 | 0.04545 | RNA-binding protein |
| PDGFRB | 86 | 38 | 0.04545 | Signal transduction |
| NEK1 | 86 | 38 | 0.04545 | DNA repair |
| SOCS3 | 86 | 25 | 0.04545 | Cytokine signaling |
| EPHA4 | 86 | 25 | 0.04545 | Signal transduction, angiogenesis |
Characterization of the Most Prevalent Tumor-associated Antigens in Colorectal Cancer
Then we searched for individual autoantibodies able to discriminate between cancer and normal samples by using the ProtoArray Prospector tool. Data were sorted according to the calculated p value and the prevalence of the autoantibodies in each group. A total of 432 proteins of the microarray were reactive with the autoantibodies present in the sera. Only autoantibodies with a prevalence higher than 50% were selected. From these, 43 proteins showed a statistically significant p value under 0.04 (Table III). Among them, 25 exhibited increased autoantibody prevalence in CRC patients, and 18 had a lower prevalence with respect to the controls. A heat map for the visualization of the autoantibody profile using all proteins with a p value lower than 0.1 is shown in Fig. 2D.
Table III.
Proteins reacting with autoantibodies associated to colorectal cancer
Proteins were classified according to the calculated p values and to the prevalence of the protein in the CRC or control groups.
| Name | Cancer prevalence | Control prevalence | p value | Function |
|---|---|---|---|---|
| % | % | |||
| Increased reactivity in colorectal cancer | ||||
| MAPKAPK3 | 71.4 | 10 | 0.0099 | Ras protein signal transduction |
| PIM1 | 71.4 | 20 | 0.0099 | Cell proliferation |
| STK4 | 71.4 | 20 | 0.0099 | Cell morphogenesis |
| FGFR4 | 71.4 | 20 | 0.0099 | Fibroblast growth factor receptor signaling pathway |
| TRIM21 | 71.4 | 20 | 0.0099 | Transcription regulation |
| SRC | 57.1 | 10 | 0.0102 | Ras protein signal transduction |
| AKT1 | 57.1 | 10 | 0.0102 | G-protein-coupled receptor protein signaling pathway |
| KDR | 57.1 | 10 | 0.0102 | Angiogenesis |
| PKN1 | 57.1 | 10 | 0.0102 | Activation of JNK activity |
| CSNK1G2 | 92.9 | 50 | 0.0144 | Wnt receptor signaling pathway |
| DAPK1 | 92.9 | 50 | 0.0144 | Antiapoptosis |
| PBK | 78.6 | 30 | 0.0154 | Mitosis |
| NEK3 | 85.7 | 30 | 0.0181 | Cell cycle |
| PRKCD | 85.7 | 40 | 0.0181 | Intracellular signaling cascade |
| SALL2 | 50.0 | 10 | 0.0238 | Regulation of transcription, DNA-dependent |
| GRK7 | 50.0 | 10 | 0.0238 | G-protein-coupled receptor kinase 1 activity |
| IRAK4 | 50.0 | 10 | 0.0238 | I-κB kinase/NF-κB cascade |
| MAPKAPK5 | 50.0 | 10 | 0.0238 | Ras protein signal transduction |
| PKN2 | 50.0 | 10 | 0.0238 | Signal transduction |
| ABL2 | 50.0 | 10 | 0.0238 | Cell adhesion |
| RPS6KA1 | 64.3 | 10 | 0.0249 | Signal transduction |
| BMX | 64.3 | 20 | 0.0249 | Intracellular signaling cascade |
| PDGFRB | 64.3 | 20 | 0.0249 | Platelet-derived growth factor receptor signaling pathway |
| CDK5/p35 | 64.3 | 20 | 0.0249 | Acetylcholine receptor signaling, muscarinic pathway |
| RPS6KA2 | 71.4 | 30 | 0.0399 | Signal transduction |
| Control prevalence | Cancer prevalence | |||
|---|---|---|---|---|
| % | % | |||
| Lower reactivity in colorectal cancer | ||||
| RBPJ | 60 | 7.1 | 0.0036 | DNA recombination |
| ITGA6 | 80 | 28.6 | 0.0099 | Cell adhesion |
| ACVR2B | 70 | 21.4 | 0.0144 | BMP signaling pathway |
| NFYA | 50 | 7.1 | 0.0144 | Transcriptional regulation |
| TTLL7 | 50 | 7.1 | 0.0144 | Cell differentiation |
| C9orf43 | 50 | 7.1 | 0.0144 | Unknown |
| ZNF706 | 50 | 7.1 | 0.0144 | Unknown |
| HDAC1 | 50 | 7.1 | 0.0144 | Antiapoptosis |
| TPM4 | 50 | 7.1 | 0.0144 | Cell motility |
| TSLP | 70 | 21.4 | 0.0154 | Cytokine, cell signaling |
| WBP2 | 70 | 21.4 | 0.0154 | Unknown |
| STAU1 | 60 | 14.3 | 0.0181 | RNA-binding protein |
| PFDN5 | 60 | 14.3 | 0.0181 | Protein folding |
| COASY | 60 | 14.3 | 0.0181 | Biosynthesis of coenzyme A |
| IGLC1 | 80 | 35.7 | 0.0249 | tRNA aminoacylation for protein translation |
| MFAP2 | 70 | 21.4 | 0.0399 | Cytoskeleton |
| BHMT2 | 70 | 21.4 | 0.0399 | Methyltransferase |
| EFNA3 | 70 | 28.6 | 0.0399 | Cell-cell signaling |
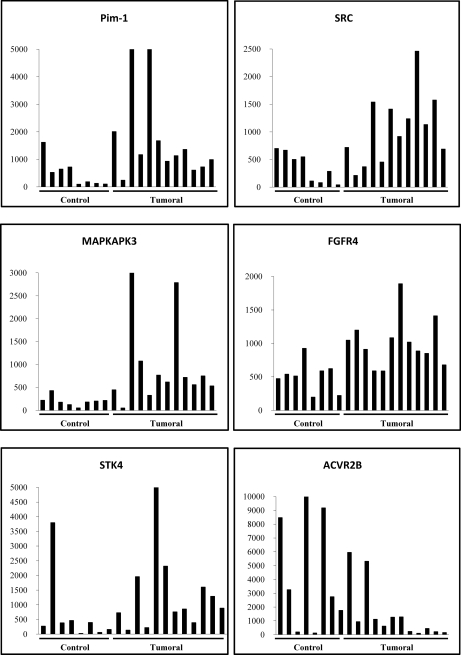
Six proteins (MAPKAPK3, PIM1, STK4, SRC, FGFR4, and ACVR2B) showed the highest differential prevalence and signal intensity obtained from the microarrays between both groups (Fig. 3A). Five of them were among the most prevalent in cancer sera according to the Prospector analysis, showing between 50 and 70% cancer prevalence and less than 20% prevalence in the reference subjects. Although there were significant variations in the individual response, MAPKAPK3, PIM1, STK4, SRC, and FGFR4 were significantly recognized by the cancer sera. In contrast, ACVR2B was preferentially recognized by the control sera.
Fig. 3.
Autoantibody response to six reactive proteins. Signal intensity for PIM1, SRC, MAPKAPK3, FGFR4, STK4, and ACVR2B obtained from the microarray analysis for each individual serum sample after quantile normalization is shown.
Tumor-associated Antigen Expression in Colorectal Cell Lines and Tumoral Tissues
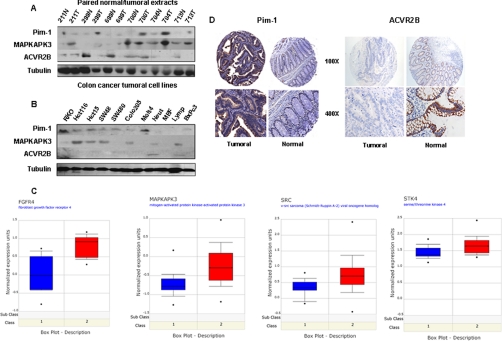
We hypothesized that stronger autoantibody reactivity would be associated to overexpression of those proteins in CRC tissues, whereas a weaker recognition would indicate a repression of those proteins in the tumor. Three autoantigens (PIM1, MAPKAPK3, and ACVR2B) were selected for initial validation. First we analyzed by immunoblotting the expression in paired normal/tumoral extracts (Fig. 4A). PIM1 and MAPKAPK3 showed an intense staining in tumoral tissues with more abundant expression in late stages. ACVR2B exhibited a weaker expression in the tumoral tissues, with more abundant expression in earlier stages. Then extracts from six different colon cancer cell lines together with five reference cell lines were analyzed (Fig. 4B). PIM1 and MAPKAPK3 expression was detected in most CRC cell lines, except MAPKAPK3 in SW480 cells. ACVR2B was not significantly expressed in colon cancer cells, whereas it was detected in control cells, including neutrophiles and lymphocytes.
Fig. 4.
Analysis of the expression of ACVR2B, PIM1, and MAPKAPK3 in cell lines and tumoral tissue. A, Western blot analysis was carried out with commercial antibodies against ACRV2B, PIM1, and MAPKAPK3 using tubulin as a control. 50 μg of cell extracts from six different CRC cell lines and five controls were separated by 10% SDS-PAGE and transferred to nitrocellulose membranes. B, 50 μg of protein extracts from paired normal (N) and tumoral (T) tissues from six CRC patients (Dukes stages A, 211 and 299; B, 699, 700, and 704; and C, 713) were resolved and transferred onto nitrocellulose membranes and incubated with the specific antibodies. Staining was developed using ECL (Amersham Biosciences) or SuperSignal Femto (Pierce). C, relative levels of gene expression for FGFR4 (58), MAPKAPK3 (59), SRC (59), and STK4 (60) were assessed with the use of publicly available DNA microarray data taken from the Oncomine database. The modified box plots represent the interquartile range, the bars represent the 10–90% range, the dots indicate the minimum and maximum expression. Red, tumoral tissue; blue, normal tissue. D, analysis of the expression of PIM1 and ACVR2B in CRC tissue using specific TMAs. Images were taken at different magnifications (100× and 400×). PIM1 expression was observed in the epithelial cells surrounding the crypts of the tumoral tissues, giving a cytoplasmic staining. ACVR2B staining was mainly localized at the membrane level of the epithelial cells in the normal tissues with a clear down-regulation in tumoral epithelia.
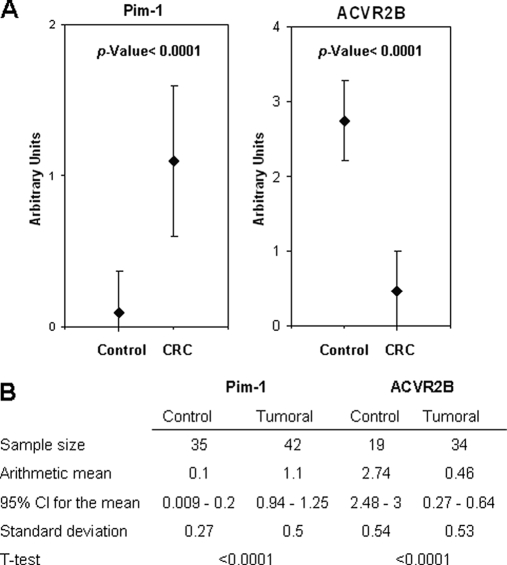
To determine the mRNA expression levels for the six selected targets, we investigated Oncomine (41), a Web-based cancer microarray database, for reported gene expression data. Data were available for FGFR4, MAPKAPK3, SRC, and STK4 (Fig. 4C). All of them showed an increased expression level in CRC. No data were available for PIM1 and ACVR2B gene expression in CRC. Then, to corroborate the differential expression of ACVR2B and PIM1, we used a CRC-specific tissue microarray (Fig. 4D). No antibody was available for MAPKAPK3 immunohistochemistry analysis. Enhanced expression of PIM1 was observed in the epithelial cells surrounding the crypts of the tumoral tissues. The staining was mainly cytoplasmic. Infiltrating lymphocytes and macrophages were also strongly stained. For PIM1, 36 of 42 CRC tumoral tissues showed weak to strong staining; 30 of 35 normal adjacent tissues showed no staining. According to the staining scale applied for the evaluation of the TMA, we found a mean value of 1.1 (95% CI = 0.94–1.25) and 0.1 (95% CI = 0.009–0.2) for tumoral and normal tissue, respectively, giving a p value <0.0001, which confirms a statistically significant higher expression of PIM1 in tumoral tissue (Fig. 5). In contrast, ACVR2B expression was down-regulated in cancer patients (six of 34 patients showed positive staining; mean = 0.46, 95% CI = 0.27–0.64, p value <0.0001), whereas normal tissues showed significant levels of ACVRB2 expression (18 of 19 patients showed positive staining; mean = 2.74, 95% CI = 2.48–3). In this case, staining was mainly localized at the membrane level of the epithelial cells in accordance with the receptor characteristics of ACVR2B.
Fig. 5.
Immunohistochemistry analysis of PIM1 and ACVR2B. A, immunohistochemistry results for PIM1 and ACVR2B in CRC tissue and the normal adjacent mucosa of 45 CRC patients were quantified by two independent investigators on different days according to the following criteria: 0, no staining; 1, weak staining; 2, normal staining; 3, strong staining. Error bars represent the S.D. of the assay. B, statistical analysis of the TMA results. Sample size, mean, S.D., 95% CI for the mean, and t test results are shown.
PIM1, MAPKAPK3, and ACVR2B as Biomarkers for Colorectal Cancer Screening
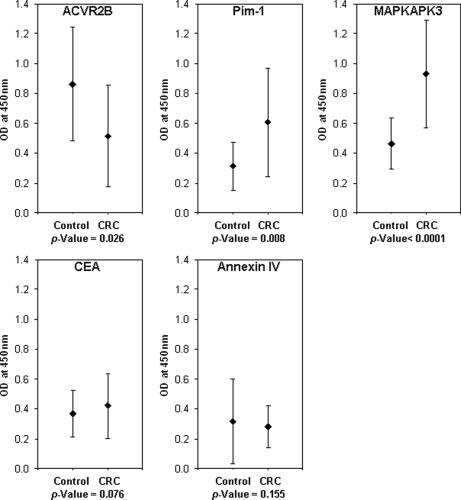
We next developed an ELISA test to study the ability of PIM1, MAPKAPK3, and ACVR2B to discriminate between diseased individuals and reference subjects using a different and wider serum cohort. We used recombinant PIM1, MAPKAPK3, and ACVR2B purified from E. coli. CEA and recombinant Annexin IV purified from mammalian cells were used as controls. CEA is the most widely used marker in colorectal malignancy (42), and Annexin IV was selected as a representative of the proteins overexpressed in colorectal cancer tissue (6). This direct ELISA was used to test 94 serum samples different from those tested previously in the discovery phase; 52 were from patients with colorectal cancer in different stages of progression, and 42 were from control subjects (supplemental Table S1). In total, 20 of 52 cancer sera corresponded to early stages A and B. Autoantibodies to PIM1, MAPKAPK3, and ACVR2B were able to discriminate between colorectal cancer and control samples (Fig. 6). In the case of ACVR2B, immunoreactivity was higher in sera from healthy individuals (mean = 0.86, 95% CI = 0.74–0.98, p < 0.026) than in sera from patients with CRC (mean = 0.67, 95% CI = 0.55–0.79). PIM1 immunoreactivity was higher for sera from patients with colorectal cancer (mean = 0.61, 95% CI = 0.50–0.71, p < 0.008) than in control subjects (mean = 0.44, 95% CI = 0.37–0.51). Better results were found for MAPKAPK3 in cancer (mean = 0.93, 95% CI = 0.83–1.03, p < 0.0001) and controls (mean = 0.65, 95% CI = 0.57–0.72). CEA only showed a weak discriminating capacity (mean cancer = 0.79, 95% CI = 0.67 to 0.90, p < 0.1; mean control = 0.66, 95% CI = 0.58–0.74). Annexin IV did not exhibit any difference between tumoral (mean = 0.42, 95% CI = 0.36–0.48, p < 0.16) and control samples (mean = 0.36, 95% CI = 0.31–0.41).
Fig. 6.
ELISA-based analysis of serum samples using the selected TAAs. A total of 94 serum samples (52 from patients with CRC cancer and 42 from controls) were used for the implementation and testing of an ELISA based on the recombinant TAAs. CEA and Annexin IV were used as controls. Results are displayed as the mean of the absorbance values obtained with the cancer and reference sera populations for MAPKAPK3, PIM1, ACVR2B, CEA, and Annexin IV. Error bars represent the S.D. of the assay.
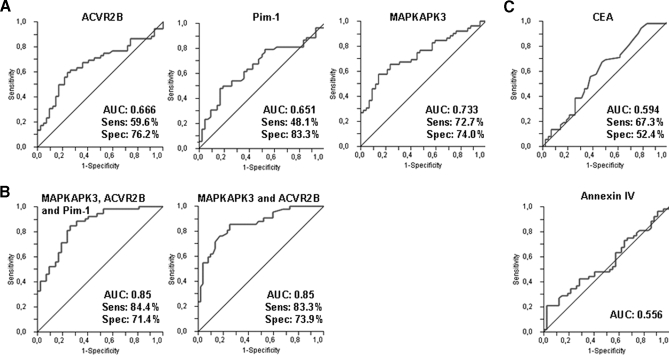
Then, we investigated this humoral response as a predictor to detect CRC. ROC curves were generated from the antibody response against ACVR2B, PIM1, and MAPKAPK3 (Fig. 7A). We found for ACVR2B a specificity and a sensitivity of 76.2 and 60%, respectively (using a cutoff of 0.55) and an area under the curve of 0.66 (95% CI = 0.56–0.76). For PIM1, the specificity was 83.3%, the sensitivity was 48.1% (using a cutoff of 0.53), and the area under the curve was 0.65 (95% CI = 0.54–0.74). In the case of MAPKAPK3, the specificity was 74% and the sensitivity 72.7% (for a cutoff of 0.76) with an area under the curve of 0.73 (95% CI = 0.63–0.82). Moreover, we examined whether different combinations of these proteins would improve the diagnostic capacity of the candidate proteins. By fitting the data to a logistic curve, we performed logistic regressions and produced different models using different combinations of the potential biomarkers (Fig. 7B). The initial model included four proteins (PIM1, MAPKAPK3, ACVR2B, and CEA). CEA and PIM1 did not improve the fit over MAPKAPK3 and ACVR2B. This was confirmed by comparing the complete model including the four predictors (AUC = 0.85) with a model including only MAPKAPK3 and ACVR2B (AUC = 0.85). Then, the combination of MAPKAPK3 and ACVR2B yielded a specificity and sensitivity of 73.9 and 83.3%, respectively, with an AUC of 0.85. CEA autoantibodies gave a lower specificity (59.5%) and sensitivity (63.5%) (using a cutoff of 0.61) with an area under the curve of 0.61 (95% CI = 051–0.72). Annexin IV showed an AUC of 0.55 (95% CI = 0.45–0.66), confirming the absence of specific autoantibodies for this protein (Fig. 7C).
Fig. 7.
Validation of the selected TAAs in colorectal cancer. A, performance of MAPKAPK3, PIM1, and ACVR2B in discriminating serum of CRC patients from reference subjects independently in a validation set with a total of 94 samples (52 from patients with CRC cancer and 42 from controls). B, specificity and sensitivity obtained from ROC curve analysis using a combination of the three proteins or a combination of MAPKAPK3 and PIM1. C, specificity (Spec) and sensitivity (Sens) obtained using the controls, CEA and Annexin IV, to discriminate CRC patients from reference subjects independently.
Bootstrap Analysis of the Predictor Model
To assess the predictive ability, we computed the AUC using the bootstrap (43) with 1000 bootstrap samples. The initial model included linear terms for all four predictors (ACVR2B, MAPKAPK3, PIM1, and CEA). The bias-corrected AUC for this model was 0.82. If we allowed for variable selection (backward selection using Akaike's information criterion as the stopping rule), the final model retained only ACVR2B and MAPKAPK3. We again assessed predictive ability of the models with variable selection, bootstrapping the complete process; the bias-corrected AUC was 0.83. Among the bootstrapped models, the vast majority (776 of 1000) included two variables, 170 included three variables, and 50 models included the complete set of four variables. Among all models MAPKAPK3 and ACVR2B were selected simultaneously in 984 cases. In models with three variables, PIM1 appeared in 66% of the cases, ACVR2B appeared in 100% of the cases, and MAPKAPK3 appeared in 99% of the cases.
However, partial residual plots of the original models suggested that the relationship between ACVR2B and tumor status could be better modeled by including non-linear terms. We explored this possibility using restricted cubic splines with three knots (there was no evidence that more knots were warranted). The model with the non-linear term fitted the data significantly better (χ2 test for differences in deviances; p = 2.596e−06). The bias-corrected bootstrapped estimate (using 1000 bootstrap samples and the 0.632 rule) of AUC was 0.89. If we allowed for variable selection, it was also 0.89. Of the 600 models with two variables, 599 were made of MAPKAPK3 and ACVR2B; of the 312 bootstrapped models with three variables, MAPKAPK3 and ACVR2B were present in 100%, and PIM1 was present in 87%.
In summary, the modeling showed that it was enough to use only MAPKAPK3 and ACVR2B in predictive models. Slightly better results could be achieved if ACVR2B was modeled including a non-linear term.
DISCUSSION
Blood is the optimal source for the diagnostic screening of large human populations for non-invasive markers. Serum and plasma are easily obtained, and moreover blood circulation facilitates the contact with every body tissue, including representative tumor antigens. However, tumor leakage antigens are probably present at the very low range of concentration in plasma, and they probably suffer from extensive proteolysis in a relatively short period (44). Therefore, the search for tumor-specific antigens in blood is a complicated task. Approaches based on a peptide search (“peptidomics”), such as SELDI, are prone to artifacts due to variability issues and require ultraextensive standardization for every step of the procedure, rendering them unsuitable for routine clinical use.
Antibodies are very stable serum molecules with a long tradition of use in immunoassays, which facilitates their standardization. Autoantibodies present in the serum of patients appear to be a promising alternative for biomarker identification. The use of autoantibodies profits from the immune system amplification of the humoral response to low abundance antigens, making quantification easier. Although antibodies against reactive autoantigens are not present in every cancer patient, the use of proteomics platforms has been shown to be a powerful discovery tool of new TAAs (12–15, 19, 20). A critical advantage of the protein arrays is the possibility to customize the content, testing for biologically relevant molecules, without many of the biases present in other proteomics approaches. Furthermore, they use a sensitive technique, such as ELISA, for the detection and relative quantification of proteins printed in the proper conformation. ELISA is more sensitive than other techniques such as Western blot or immunohistochemistry. This high sensitivity would explain the significant increase in the number of identified proteins in this study, why the prevalence of autoantibodies in cancer patients was higher than observed previously (28), and why some reactivity in control patients was detected. We believe that the diagnostic assay should be based on a combination of autoantibodies with the highest possible prevalence and complementarity as we did not find autoantigens with exclusive reactivity to cancer sera.
Although the final goal of this study was the identification of autoantibodies specific for early cancer stages, we decided to start with sera from late and advanced stages as a proof of concept. We hypothesized that advanced cancer stages are more likely to contain a higher number of protein mutations, which would cause an increase in the levels of autoantibodies, facilitating their recognition. Moreover, the identified autoantibodies might be particularly relevant for the detection of recurrence in CRC, a critical aspect in disease survival and patient outcome. Subsequent validation studies with an independent set of samples, representing the different Dukes stages, confirmed the validity of this assertion.
Protein microarrays facilitated the detection of tumor-associated autoantigens. Most of the detected antigens were novel. They were involved mainly in cell signaling processes, transcriptional regulation, and cell growth and differentiation. In fact, many of the identified biomarkers were kinases, which are usual targets of therapeutic intervention. From a total of 43 cancer-discriminating proteins, we selected three protein kinases, PIM1, MAPKAPK3, and ACVR2B, to evaluate the performance of an ad hoc designed ELISA test. This ELISA was developed for further screening of larger cohorts of patients and potential clinical implementation. We found that the combination of MAPKAPK3 with ACVR2B had the best discriminatory power. Interestingly, we calculated the correlation of these three proteins and found that the correlation between PIM1 and MAPKAPK3 was 0.64 indicating that their reactivity pattern was similar and that the presence of both markers was redundant in the model. On the other hand, the correlation between MAPKAPK3 and ACVR2B was 0.34. Both markers induced a different immunoreactivity pattern among the patients and when used in combination were more informative and permitted a better discrimination between groups. Indeed, the final model with these two markers gave the highest specificity and sensitivity. MAPKAPK3 and ACVR2B specificity (73.9%) and sensitivity (83.3%) show promising values for the screening of a widespread disease such as colorectal cancer. However, we consider that the source of the recombinant proteins (baculovirus versus E. coli), the purity level of the protein, the presence of different tags (GST versus His6), or the solid support of the assay (nitrocellulose versus polyvinyl) might all be relevant for assay optimization as described previously (19). So there should be further room for specificity and sensitivity improvements in this diagnostic ELISA based on recombinant proteins.
PIM1, MAPKAPK3, and ACVR2B participate in the control of cell growth and differentiation. PIM1 and MAPKAPK3 are oncogene-like proteins, whereas ACVR2B has shown a suppressor-like activity. There were no previous reports of association between PIM1 and MAPKAPK3 with colorectal cancer. The proto-oncogene PIM1 is a constitutively active serine/threonine kinase involved in the control of cell growth and differentiation (45) that has been described to inhibit apoptosis (46). PIM1 has been reported to be overexpressed in prostate cancer (47), lymphomas (48), and head and neck carcinomas (49). PIM1 was recently identified as a target of aberrant somatic hypermutation in non-Hodgkin lymphoma and B cell lymphoma (50). MAPKAPK3 is a relatively unknown enzyme that is the target of different mitogen-activated protein kinases, such as p38α/β24 (51). In vitro studies demonstrated that ERK, p38 mitogen-activated protein kinase, and Jun N-terminal kinase were all able to phosphorylate and activate MAPKAPK3. In a previous report, we found ERK, MAPK, and Jun to be overexpressed in colorectal cancer (7), confirming the well known activation of the ERK1/2 MAPK pathway. There are no previous reports of MAPKAPK3 up-regulation in other tumors. Regarding ACVR2B, activins are dimeric growth and differentiation factors that belong to the transforming growth factor-β superfamily. Activins signal through a heteromeric complex of receptor serine kinases that include at least two type I (I and IB) and two type II (II and IIB) receptors. ACVR2B and its ligand activin can regulate cell differentiation, proliferation, and apoptosis in a variety of cancer cell types (52–54). Indeed, the expression of ACVR2B seems to be down-regulated in CRC cells, CRC-induced xenografts, and primary colon cancer specimens as a consequence of the mutation status associated with high frequency microsatellite instability (55). After restoring its expression, there was a decrease in the growth of colon cancer cells, which may suggest a role for ACVR2B as a tumor suppressor (53, 55).
Although Annexin IV has been reported to be overexpressed in colorectal cancer (6, 8), our results showed a complete absence of autoantibodies against Annexin IV. Therefore, overexpression is not sufficient for the induction of an immune response. Other factors such as the presence of mutations should play a more relevant role. Different types of mutations have been reported for PIM1 and ACVR2B. Because MAPKAPK3 is hardly characterized, no mutations have been described yet. Thus, we can speculate that aberrant mutations probably also occur in MAPKAPK3. We can hypothesize that the occurrence of autoantibodies requires the presence of mutations in tumor-associated proteins in combination with overexpression. Further research will be necessary to correlate the presence of mutations in tumor proteins with the induction of autoantibodies.
One of the findings of our study was the identification of several targets for natural autoantibodies in reference subjects. Interestingly autoantibodies against ACVR2B were diminished in cancer patients. The presence of natural autoantibodies is common in all individuals (56), and they are characterized by a low affinity and polyreactivity (57), which could explain the significant reactivity against many proteins. It is tempting to speculate that the lower autoantibody reactivity observed in cancer patients against these antigens could be due to the appearance of deleterious mutations, truncations, or a significant repression in the protein expression levels that would abolish or decrease this natural reactivity. In fact, TMA data support this repression in the expression of ACVR2B in tumors.
Proteins identified in this study were different from previously identified TAAs in colon cancer probably because among the 8000 human proteins of our high density microarrays there were many low abundance proteins, which increased considerably the chances of finding more CRC-specific TAAs. Although these potential CRC biomarkers were initially identified with sera from patients with metastasis, further ELISA screening with sera representing different stages of CRC demonstrated the applicability of these TAAs for a global diagnosis of CRC. Furthermore, the identified TAAs are not only good candidates for the diagnosis of CRC but interesting new targets for therapeutic intervention because they are altered, and probably mutated, kinase proteins.
Supplementary Material
Acknowledgments
Tissue samples were provided by the Tissue Bank Network of the CNIO. Serum samples were provided by F. Bonilla (Hospital Puerta de Hierro), M. Urioste (CNIO), and J. Casinello (Hospital de Guadalajara). We thank the Histology and Immunohistochemistry Unit of the CNIO for technical assistance with the TMA staining and Dr. Marta Cañamero for the TMA analysis.
Footnotes
* This work was supported in part by Spanish Ministry of Education and Science Grant BIO2006-07689 and grants from the Colomics program of the regional government of Madrid and from the Fundación Médica Mutua Madrileña.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- CRC
- colorectal cancer
- CEA
- carcinoembryonic antigen
- TAA
- tumor-associated autoantigen
- TMA
- tissue microarray
- CI
- confidence interval
- ROC
- receiver operating characteristic
- AUC
- area under the curve
- MAPKAPK
- mitogen-activated protein kinase-activated protein kinase
- ERK
- extracellular signal-regulated kinase.
REFERENCES
- 1.Crawford N. P., Colliver D. W., Galandiuk S. (2003) Tumor markers and colorectal cancer: utility in management. J. Surg. Oncol 84, 239–248 [DOI] [PubMed] [Google Scholar]
- 2.Sidransky D. (2002) Emerging molecular markers of cancer. Nat. Rev. Cancer 2, 210–219 [DOI] [PubMed] [Google Scholar]
- 3.Duffy M. J., van Dalen A., Haglund C., Hansson L., Holinski-Feder E., Klapdor R., Lamerz R., Peltomaki P., Sturgeon C., Topolcan O. (2007) Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur. J. Cancer 43, 1348–1360 [DOI] [PubMed] [Google Scholar]
- 4.Locker G. Y., Hamilton S., Harris J., Jessup J. M., Kemeny N., Macdonald J. S., Somerfield M. R., Hayes D. F., Bast R. C., Jr. (2006) ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J. Clin. Oncol 24, 5313–5327 [DOI] [PubMed] [Google Scholar]
- 5.Kopf E., Shnitzer D., Zharhary D. (2005) Panorama Ab Microarray Cell Signaling kit: a unique tool for protein expression analysis. Proteomics 5, 2412–2416 [DOI] [PubMed] [Google Scholar]
- 6.Alfonso P., Núñez A., Madoz-Gurpide J., Lombardia L., Sánchez L., Casal J. I. (2005) Proteomic expression analysis of colorectal cancer by two-dimensional differential gel electrophoresis. Proteomics 5, 2602–2611 [DOI] [PubMed] [Google Scholar]
- 7.Madoz-Gúrpide J., Cañamero M., Sanchez L., Solano J., Alfonso P., Casal J. I. (2007) A proteomics analysis of cell signaling alterations in colorectal cancer. Mol. Cell. Proteomics 6, 2150–2164 [DOI] [PubMed] [Google Scholar]
- 8.Alfonso P., Cañamero M., Fernández-Carbonié F., Núñez A., Casal J. I. (2008) Proteome analysis of membrane fractions in colorectal carcinomas by using 2D-DIGE saturation labeling. J. Proteome Res 7, 4247–4255 [DOI] [PubMed] [Google Scholar]
- 9.Hanash S. M., Pitteri S. J., Faca V. M. (2008) Mining the plasma proteome for cancer biomarkers. Nature 452, 571–579 [DOI] [PubMed] [Google Scholar]
- 10.Pitteri S. J., Faca V. M., Kelly-Spratt K. S., Kasarda A. E., Wang H., Zhang Q., Newcomb L., Krasnoselsky A., Paczesny S., Choi G., Fitzgibbon M., McIntosh M. W., Kemp C. J., Hanash S. M. (2008) Plasma proteome profiling of a mouse model of breast cancer identifies a set of up-regulated proteins in common with human breast cancer cells. J. Proteome Res 7, 1481–1489 [DOI] [PubMed] [Google Scholar]
- 11.Pitteri S. J., Hanash S. M. (2007) Proteomic approaches for cancer biomarker discovery in plasma. Expert Rev. Proteomics 4, 589–590 [DOI] [PubMed] [Google Scholar]
- 12.Anderson K. S., Ramachandran N., Wong J., Raphael J. V., Hainsworth E., Demirkan G., Cramer D., Aronzon D., Hodi F. S., Harris L., Logvinenko T., LaBaer J. (2008) Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. J. Proteome Res 7, 1490–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hudson M. E., Pozdnyakova I., Haines K., Mor G., Snyder M. (2007) Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc. Natl. Acad. Sci. U.S.A 104, 17494–17499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman C., Murray A., Chakrabarti J., Thorpe A., Woolston C., Sahin U., Barnes A., Robertson J. (2007) Autoantibodies in breast cancer: their use as an aid to early diagnosis. Ann. Oncol 18, 868–873 [DOI] [PubMed] [Google Scholar]
- 15.Chapman C. J., Murray A., McElveen J. E., Sahin U., Luxemburger U., Türeci O., Wiewrodt R., Barnes A. C., Robertson J. F. (2008) Autoantibodies in lung cancer: possibilities for early detection and subsequent cure. Thorax 63, 228–233 [DOI] [PubMed] [Google Scholar]
- 16.Soussi T. (2000) p53 Antibodies in the sera of patients with various types of cancer: a review. Cancer Res 60, 1777–1788 [PubMed] [Google Scholar]
- 17.Chatterjee M., Mohapatra S., Ionan A., Bawa G., Ali-Fehmi R., Wang X., Nowak J., Ye B., Nahhas F. A., Lu K., Witkin S. S., Fishman D., Munkarah A., Morris R., Levin N. K., Shirley N. N., Tromp G., Abrams J., Draghici S., Tainsky M. A. (2006) Diagnostic markers of ovarian cancer by high-throughput antigen cloning and detection on arrays. Cancer Res 66, 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X., Yu J., Sreekumar A., Varambally S., Shen R., Giacherio D., Mehra R., Montie J. E., Pienta K. J., Sanda M. G., Kantoff P. W., Rubin M. A., Wei J. T., Ghosh D., Chinnaiyan A. M. (2005) Autoantibody signatures in prostate cancer. N. Engl. J. Med 353, 1224–1235 [DOI] [PubMed] [Google Scholar]
- 19.Sreekumar A., Laxman B., Rhodes D. R., Bhagavathula S., Harwood J., Giacherio D., Ghosh D., Sanda M. G., Rubin M. A., Chinnaiyan A. M. (2004) Humoral immune response to alpha-methylacyl-CoA racemase and prostate cancer. J. Natl. Cancer Inst 96, 834–843 [DOI] [PubMed] [Google Scholar]
- 20.Anderson K. S., LaBaer J. (2005) The sentinel within: exploiting the immune system for cancer biomarkers. J. Proteome Res 4, 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y., Lin P., Qiu S., Peng X. X., Looi K., Farquhar M. G., Zhang J. Y. (2007) Autoantibodies to Ca2+ binding protein Calnuc is a potential marker in colon cancer detection. Int. J. Oncol 30, 1137–1144 [PubMed] [Google Scholar]
- 22.Chen Y. T., Scanlan M. J., Sahin U., Türeci O., Gure A. O., Tsang S., Williamson B., Stockert E., Pfreundschuh M., Old L. J. (1997) A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc. Natl. Acad. Sci. U.S.A 94, 1914–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schubert U., Antón L. C., Gibbs J., Norbury C. C., Yewdell J. W., Bennink J. R. (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774 [DOI] [PubMed] [Google Scholar]
- 24.Ulanet D. B., Torbenson M., Dang C. V., Casciola-Rosen L., Rosen A. (2003) Unique conformation of cancer autoantigen B23 in hepatoma: a mechanism for specificity in the autoimmune response. Proc. Natl. Acad. Sci. U.S.A 100, 12361–12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Mensdorff-Pouilly S., Petrakou E., Kenemans P., van Uffelen K., Verstraeten A. A., Snijdewint F. G., van Kamp G. J., Schol D. J., Reis C. A., Price M. R., Livingston P. O., Hilgers J. (2000) Reactivity of natural and induced human antibodies to MUC1 mucin with MUC1 peptides and N-acetylgalactosamine (GalNAc) peptides. Int. J. Cancer 86, 702–712 [DOI] [PubMed] [Google Scholar]
- 26.Reipert B. M., Tanneberger S., Pannetta A., Bedosti M., Poell M., Zimmermann K., Stellamor M. T. (2005) Increase in autoantibodies against Fas (CD95) during carcinogenesis in the human colon: a hope for the immunoprevention of cancer? Cancer Immunol. Immunother 54, 1038–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scanlan M. J., Chen Y. T., Williamson B., Gure A. O., Stockert E., Gordan J. D., Türeci O., Sahin U., Pfreundschuh M., Old L. J. (1998) Characterization of human colon cancer antigens recognized by autologous antibodies. Int. J. Cancer 76, 652–658 [DOI] [PubMed] [Google Scholar]
- 28.Scanlan M. J., Welt S., Gordon C. M., Chen Y. T., Gure A. O., Stockert E., Jungbluth A. A., Ritter G., Jäger D., Jäger E., Knuth A., Old L. J. (2002) Cancer-related serological recognition of human colon cancer: identification of potential diagnostic and immunotherapeutic targets. Cancer Res 62, 4041–4047 [PubMed] [Google Scholar]
- 29.Bouwman K., Qiu J., Zhou H., Schotanus M., Mangold L. A., Vogt R., Erlandson E., Trenkle J., Partin A. W., Misek D., Omenn G. S., Haab B. B., Hanash S. (2003) Microarrays of tumor cell derived proteins uncover a distinct pattern of prostate cancer serum immunoreactivity. Proteomics 3, 2200–2207 [DOI] [PubMed] [Google Scholar]
- 30.Yan F., Sreekumar A., Laxman B., Chinnaiyan A. M., Lubman D. M., Barder T. J. (2003) Protein microarrays using liquid phase fractionation of cell lysates. Proteomics 3, 1228–1235 [DOI] [PubMed] [Google Scholar]
- 31.Pereira-Faca S. R., Kuick R., Puravs E., Zhang Q., Krasnoselsky A. L., Phanstiel D., Qiu J., Misek D. E., Hinderer R., Tammemagi M., Landi M. T., Caporaso N., Pfeiffer R., Edelstein C., Goodman G., Barnett M., Thornquist M., Brenner D., Hanash S. M. (2007) Identification of 14-3-3 theta as an antigen that induces a humoral response in lung cancer. Cancer Res 67, 12000–12006 [DOI] [PubMed] [Google Scholar]
- 32.Türeci O., Sahin U., Pfreundschuh M. (1997) Serological analysis of human tumor antigens: molecular definition and implications. Mol. Med. Today 3, 342–349 [DOI] [PubMed] [Google Scholar]
- 33.Chatterjee M., Ionan A., Draghici S., Tainsky M. A. (2006) Epitomics: global profiling of immune response to disease using protein microarrays. Omics 10, 499–506 [DOI] [PubMed] [Google Scholar]
- 34.Ran Y., Hu H., Zhou Z., Yu L., Sun L., Pan J., Liu J., Yang Z. (2008) Profiling tumor-associated autoantibodies for the detection of colon cancer. Clin. Cancer Res 14, 2696–2700 [DOI] [PubMed] [Google Scholar]
- 35.Ramachandran N., Raphael J. V., Hainsworth E., Demirkan G., Fuentes M. G., Rolfs A., Hu Y., LaBaer J. (2008) Next-generation high-density self-assembling functional protein arrays. Nat. Methods 5, 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson W. H., DiGennaro C., Hueber W., Haab B. B., Kamachi M., Dean E. J., Fournel S., Fong D., Genovese M. C., de Vegvar H. E., Skriner K., Hirschberg D. L., Morris R. I., Muller S., Pruijn G. J., van Venrooij W. J., Smolen J. S., Brown P. O., Steinman L., Utz P. J. (2002) Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat. Med 8, 295–301 [DOI] [PubMed] [Google Scholar]
- 37.Barderas R., Desmet J., Timmerman P., Meloen R., Casal J. I. (2008) Affinity maturation of antibodies assisted by in silico modeling. Proc. Natl. Acad. Sci. U.S.A 105, 9029–9034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visintin I., Feng Z., Longton G., Ward D. C., Alvero A. B., Lai Y., Tenthorey J., Leiser A., Flores-Saaib R., Yu H., Azori M., Rutherford T., Schwartz P. E., Mor G. (2008) Diagnostic markers for early detection of ovarian cancer. Clin. Cancer Res 14, 1065–1072 [DOI] [PubMed] [Google Scholar]
- 39.Harrell F. (2001) Regression Modeling Strategies, Springer, New York [Google Scholar]
- 40.R Core Development Team (2009) R: a Language and Environment for Statistical Computing The R Foundation for Statistical Computing, Vienna [Google Scholar]
- 41.Rhodes D. R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A. M. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duffy M. J. (2001) Carcinoembryonic antigen as a marker for colorectal cancer: is it clinically useful? Clin. Chem 47, 624–630 [PubMed] [Google Scholar]
- 43.Efron B. (1983) Estimating the error rate of a prediction rule: improvement on cross-validation. J. Am. Stat. Assoc 78, 316–331 [Google Scholar]
- 44.Villanueva J., Shaffer D. R., Philip J., Chaparro C. A., Erdjument- Bromage H., Olshen A. B., Fleisher M., Lilja H., Brogi E., Boyd J., Sanchez-Carbayo M., Holland E. C., Cordon-Cardo C., Scher H. I., Tempst P. (2006) Differential exoprotease activities confer tumor-specific serum peptidome patterns. J. Clin. Investig 116, 271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z., Bhattacharya N., Weaver M., Petersen K., Meyer M., Gapter L., Magnuson N. S. (2001) Pim-1: a serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J. Vet. Sci 2, 167–179 [PubMed] [Google Scholar]
- 46.Lilly M., Sandholm J., Cooper J. J., Koskinen P. J., Kraft A. (1999) The PIM-1 serine kinase prolongs survival and inhibits apoptosis-related mitochondrial dysfunction in part through a bcl-2-dependent pathway. Oncogene 18, 4022–4031 [DOI] [PubMed] [Google Scholar]
- 47.Cibull T. L., Jones T. D., Li L., Eble J. N., Ann Baldridge L., Malott S. R., Luo Y., Cheng L. (2006) Overexpression of Pim-1 during progression of prostatic adenocarcinoma. J. Clin. Pathol 59, 285–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubenstein J. L., Fridlyand J., Shen A., Aldape K., Ginzinger D., Batchelor T., Treseler P., Berger M., McDermott M., Prados M., Karch J., Okada C., Hyun W., Parikh S., Haqq C., Shuman M. (2006) Gene expression and angiotropism in primary CNS lymphoma. Blood 107, 3716–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beier U. H., Weise J. B., Laudien M., Sauerwein H., Görögh T. (2007) Overexpression of Pim-1 in head and neck squamous cell carcinomas. Int. J. Oncol 30, 1381–1387 [PubMed] [Google Scholar]
- 50.Morishita D., Katayama R., Sekimizu K., Tsuruo T., Fujita N. (2008) Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res 68, 5076–5085 [DOI] [PubMed] [Google Scholar]
- 51.Gaestel M. (2006) MAPKAP kinases—MKs—two's company, three's a crowd. Nat. Rev. Mol. Cell Biol 7, 120–130 [DOI] [PubMed] [Google Scholar]
- 52.Cocolakis E., Lemay S., Ali S., Lebrun J. J. (2001) The p38 MAPK pathway is required for cell growth inhibition of human breast cancer cells in response to activin. J. Biol. Chem 276, 18430–18436 [DOI] [PubMed] [Google Scholar]
- 53.Jung B. H., Beck S. E., Cabral J., Chau E., Cabrera B. L., Fiorino A., Smith E. J., Bocanegra M., Carethers J. M. (2007) Activin type 2 receptor restoration in MSI-H colon cancer suppresses growth and enhances migration with activin. Gastroenterology 132, 633–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z., Zhao Y., Batres Y., Lin M. F., Ying S. Y. (1997) Regulation of growth and prostatic marker expression by activin A in an androgen-sensitive prostate cancer cell line LNCAP. Biochem. Biophys. Res. Commun 234, 362–365 [DOI] [PubMed] [Google Scholar]
- 55.Jung B., Doctolero R. T., Tajima A., Nguyen A. K., Keku T., Sandler R. S., Carethers J. M. (2004) Loss of activin receptor type 2 protein expression in microsatellite unstable colon cancers. Gastroenterology 126, 654–659 [DOI] [PubMed] [Google Scholar]
- 56.Avrameas S., Ternynck T. (1993) The natural autoantibodies system: between hypotheses and facts. Mol. Immunol 30, 1133–1142 [DOI] [PubMed] [Google Scholar]
- 57.Avrameas S. (1991) Natural autoantibodies: from ‘horror autotoxicus’ to ‘gnothi seauton’. Immunol Today 12, 154–159 [DOI] [PubMed] [Google Scholar]
- 58.Notterman D. A., Alon U., Sierk A. J., Levine A. J. (2001) Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res 61, 3124–3130 [PubMed] [Google Scholar]
- 59.Ki D. H., Jeung H. C., Park C. H., Kang S. H., Lee G. Y., Lee W. S., Kim N. K., Chung H. C., Rha S. Y. (2007) Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int. J. Cancer 121, 2005–2012 [DOI] [PubMed] [Google Scholar]
- 60.Watanabe T., Kobunai T., Toda E., Yamamoto Y., Kanazawa T., Kazama Y., Tanaka J., Tanaka T., Konishi T., Okayama Y., Sugimoto Y., Oka T., Sasaki S., Muto T., Nagawa H. (2006) Distal colorectal cancers with microsatellite instability (MSI) display distinct gene expression profiles that are different from proximal MSI cancers. Cancer Res 66, 9804–9808 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.