Abstract
The small γ-butyrolactone A-factor is an important autoregulatory signaling molecule for the soil-inhabiting streptomycetes. Starvation is a major trigger for development, and nutrients are provided by degradation of the vegetative mycelium via a process of programmed cell death, reusing proteins, nucleic acids, and cell wall material. The A-factor regulon includes many extracellular hydrolases. Here we show via proteomics analysis that many nutrient-scavenging and stress-related proteins were overexpressed in an A-factor non-producing mutant of Streptomyces griseus B-2682. Transcript analysis showed that this is primarily due to differential transcription of the target genes during early development. The targets include proteins relating to nutrient stress and environmental stress and an orthologue of the Bacillus sporulation control protein Spo0M. The enhanced expression of these proteins underlines the stress that is generated by the absence of A-factor. Wild-type developmental gene expression was restored to the A-factor non-producing mutant by the signaling protein Factor C in line with our earlier observation that Factor C triggers A-factor production.
Bacteria of the Gram-positive filamentous Streptomyces are a well known model system for the study of prokaryotic multicellular differentiation. They have a complex mycelial life cycle starting with a vegetative mycelium that develops into aerial mycelium, which then produces chains of spores at the ends of the hyphae (1). The onset of development is triggered by nutritional signals (2) and temporally relates to the production of antibiotics and other secondary metabolites (3). Autoregulatory molecules play a key role in controlling both the onset of cellular differentiation and secondary metabolism. The best studied autoregulator is A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone), a small microbial hormone-like molecule (243 Da) that induces both morphological and physiological differentiation in Streptomyces griseus (4, 5). The γ-butyrolactone regulatory system is widespread in streptomycetes. Virginiae butanolides control virginiamycin production in Streptomyces virginiae (6), and SCB1 plays an important role in the control of actinorhodin and undecylprodigiosin biosynthesis and a cryptic, type I polyketide synthase (cpk) gene cluster in Streptomyces coelicolor (7, 8). In S. griseus binding of A-factor to its cellular receptor ArpA derepresses expression of the transcriptional activator AdpA. Although initially identified as the activator of streptomycin production through strR, this protein acts as a central switch, and the AdpA regulon includes several important positive regulators of development (ssgA, amfR, and adsA (bldN)) and secreted proteases (for reviews, see Refs. 9 and 10). A-factor-deficient mutants are neither able to sporulate nor able to produce antibiotics (streptomycin).
Another interesting autoregulator is the secreted signaling protein Factor C (molecular mass 34.555 kDa) originally isolated from the culture fluid of “S. griseus 45H” (11), which was recently shown to be identical to a laboratory strain known as Streptomyces flavofungini, itself a member of the Streptomyces albidoflavus species group (12). The Factor C producer strain like S. griseus readily sporulates in submerged culture (13). Similarly to A-factor, Factor C also plays a key role in cellular communication and cytodifferentiation.
A-factor mutants fail to develop aerial hyphae and spores and are therefore classified as bald mutants. Expression of facC from a low copy plasmid in a spontaneous A-factor-deficient bald mutant of S. griseus NRRL B-2682 restored its A-factor production as well as aerial mycelium and spore formation on solid media (14). The wild-type strain itself does not produce Factor C as shown by immunoblotting (15) and by DNA hybridization studies (16). Our previous results (14) indicate a connection between two highly divergent types of signaling molecules and possible interplay between their regulatory networks. In preliminary experiments we observed characteristic differences between the extracellular proteomes of the strains that prompted detailed further analysis facilitated by the currently available DNA sequence of the genome of S. griseus IFO 13350 (17). Here we show that the bald A-factor non-producing mutant (AFN)1 overexpressed several ABC transporter solute-binding proteins and stress response proteins compared with the wild-type S. griseus B-2682 strain or with the facC transformant of the AFN in an effort to supply the cells with nutrients.
EXPERIMENTAL PROCEDURES
Strains and Preparation of Extracellular Protein Fractions
Strains of S. griseus were grown on R2YE agar plates (18, p. 408) covered with a polycarbonate track etch membrane (Poretics 0.2-μm pore size). The strains were S. griseus NRRL B-2682 (parental strain; in short B-2682), its bald S. griseus NRRL B-2682 AFN, and a transformant of AFN (designated AFN/pSGF4) that harbors facC on the pHJL401-based low copy number plasmid pSGF4 (16). Protein extracts were prepared from spent agar of surface-grown cultures by crumbling the solid medium and passing it through a syringe with frits at 4 °C by centrifugation. Samples of ∼300 μg of protein (measured using the Coomassie Protein Assay Reagent, Pierce) were purified using the ReadyPrepTM 2-D Cleanup kit (Bio-Rad) according to the instructions manual, and dissolved in Rehydration Buffer (8 m urea, 2% CHAPS, 50 mm DTT, 0.2% 100× Bio-Lyte 3/10 (or 4/7) ampholyte, 0.002% bromphenol blue).
2D Gel Electrophoresis and Image Analysis
Separation of protein extracts (∼300 μg) in the first dimension was performed by isoelectric focusing using 17-cm-long Immobiline DryStrip Gels (IPG) in the pH range of 3–10 or 4–7 (Bio-Rad) on a Protean IEF cell (Bio-Rad). Samples were focused at 250 V for 15 min followed by an increase to 8000 V over 2.5 h and kept at this voltage for 45,000 V-h. Focused strips were separated on the basis of relative molecular weight in the second dimension on 13% SDS-polyacrylamide gels in a Protean II XL vertical gel system (Bio-Rad). For quantitative comparison of extracellular protein profiles gels were stained with colloidal Coomassie G-250 (19). Gels were scanned using a GS-800 Imaging Densitometer (Bio-Rad), and images were analyzed with PDQuestTM software (Bio-Rad). Histograms comparing spot quantity were generated with this software. 2-fold differences between the mutant and the parental B-2682 were considered as significant changes. The data below are from a single representative experiment, but at least two additional biological replicas were performed, and they showed similar results.
In-gel Digestion
Gel slices containing 2D PAGE-separated proteins were cut, diced, and then washed with 25 mm NH4HCO3 in 50% (v/v) acetonitrile, water. After reduction with 20 μl of 10 mm DTT (30 min at 56 °C) and alkylation with 25 μl of 55 mm iodoacetamide (30 min at room temperature in the dark) the proteins were digested with side chain-protected porcine trypsin (Promega, Madison, WI) at 37 °C for 4 h. Tryptic digests were extracted into 1% formic acid, 50% ACN, H2O solution and desalted on C18 ZipTips (Millipore, Bedford, MA). Mass spectrometric analysis of the unfractionated tryptic digests was performed in positive ion, reflectron mode on a Reflex III MALDI-TOF mass spectrometer (Bruker, Karlsruhe, Germany) using 2,5-dihydroxybenzoic acid as the matrix. Two-point external calibration was applied; this guarantees a mass accuracy within 200 ppm. The peak lists were generated with X-Tof (version 5.1.5) software. Mascot database search software (Matrix Science) was used to identify proteins in the full NCBI database (November 16, 2007, 5,633,163 sequences). Searches also were performed using an in-house Mascot server (version 2.2.04) and a S. griseus database (2008, 7138 sequences) downloaded from the Streptomyces griseus Genome Project Server. Protein scores greater than 80 (full NCBI database search) or 51 (S. griseus database) were considered significant (p < 0.05).
Search parameters were as follows: mass accuracy, 200 ppm; only tryptic cleavages permitted; and two missed cleavages considered. Carbamidomethylation of Cys residues was a fixed modification; methionine oxidation, protein N-acetylation, pyroglutamic acid formation from amino-terminal Gln (and Glu) residues, and methyl esterification of Asp and Glu residues (Coomassie Brilliant Blue-staining side reaction (20) proven by PSD) were the considered variable modifications. Esterified peptides were only accepted when the peptide was also detected without the modification. Protein identification was confirmed by sequence information obtained from MS/MS (postsource decay) spectra acquired in 10–12 steps, lowering the reflector voltage by 25% in each step and then stitching the data together. Some MS/MS experiments were performed on an Agilent XCT Plus 3D ion trap equipped with an atmospheric pressure MALDI source using 4-OH-α-CN-cinnamic acid as the matrix. Search parameters for MS/MS data were as follows: mass accuracy 200 ppm for the precursor ion and 1 Da for the fragment ions. Cleavage specificity and covalent modifications were considered as described above. MS/MS spectra are shown in supplemental Figs. S1–S10. Instead of the accession number the SGR locus number is listed in Table I except for protein ID 14, which is not present in the sequenced S. griseus IFO 13350 strain.
Table I.
Proteins with enhanced expression in S. griseus B-2682 AFN
Match, peptides identified/masses searched; Seq. cov., sequence coverage; Sec. no., presence of a secretion signal; SCO, S. coelicolor database; SAV, S. avermitilis database; SSP, standard spot number generated by PDQuest; SGR, Streptomyces griseus database. The percentage in parentheses refers to amino acid identity between the predicted SGR and SCO proteins.
| ID | SSP | Accession no. | Protein name | Match | Seq. cov. | Molecular mass | Sequence confirmed by PSD or CID | Sec. no. | ADP-ribosylation consensus | Orthologue in SCO/SAV |
|---|---|---|---|---|---|---|---|---|---|---|
| % | kDa | |||||||||
| 1 | 3002 | SGR1460 | OsmC-like ATP/GTP-binding protein | 4/9 | 24 | 14.5 | 6VAHTNWEGNLIEGK19, 20GVVTFDSSGIGDHPVSWPAR39 | No | No | SCO6531 (83%), SAV1862 (84%) |
| 2 | 0004 | SGR2245 | Superoxide dismutase (SodN) | 11/15 | 32 | 15 | 85YPELHQLINDTLK97 | No | No | SCO5254 (87%), SAV2988 (86%) |
| 3 | 4603 | SGR3109 | Putative aminotransferase | 31/43 | 51 | 39.4 | 2ADIQIPADIKPADGR16, 226HIPEFFSLPTAIDNSLK242 | No | No | SCO4366 (87%), SAV3883 (87%) |
| 4 | 4507 | SGR3109 | Putative aminotransferase | 13/29 | 37 | 39.4 | 2ADIQIPADIKPADGR16, 226HIPEFFSLPTAIDNSLK242 | No | No | SCO4366 (87%), SAV3883 (87%) |
| 5 | 1502 | SGR1498 | Xylose ABC transporter solute-binding protein | 32/34 | 56 | 38.8 | 1–27 | 21LAACG25 | SCO6009 (78%), SAV2247 (77%) | |
| 6 | 2402 | SGR1498 | Xylose ABC transporter solute-binding protein | 22/27 | 39 | 38.8 | 47IGLLLPENQTAR58, 125AKDAGIPVVAFDR137, 254GAGISPLPPVTGQDAELAGVQR275 | 1–27 | 21LAACG25 | SCO6009 (78%), SAV2247 (77%) |
| 7 | 2405 | SGR1498 | Xylose ABC transporter solute-binding protein | 41/49 | 56 | 38.8 | 47IGLLLPENQTAR58, 127DAGIPVVAFDR137, 254GAGISPLPPVTGQDAELAGVQR275 | 1–27 | 21LAACG25 | SCO6009 (78%), SAV2247 (77%) |
| 8 | 2404 | SGR1498 | Xylose ABC transporter solute-binding protein | 31/41 | 44 | 38.8 | 47IGLLLPENQTAR58, 127DAGIPVVAFDR137, 254GAGISPLPPVTGQDAELAGVQR275 | 1–27 | 21LAACG25 | SCO6009 (78%), SAV2247 (77%) |
| 9 | 8202 | SGR5704 | Putative Spo0M-type sporulation control protein | 16/24 | 29 | 28.5 | 38IQGGSVAQQIEGLNVGLQAR57, 192GLHQVELTFVADDR205 | 1–17 | No | SCO1793 (81%), SAV6485 (83%) |
| 10 | 4305 | SGR2237 | Putative arginine/ornithine-binding protein | 34/47 | 69 | 33.5 | 178SIEDLCGKPAAVQR191, 231SGGSVAGVNDYPVAVDLAR249, 286EAVDAIIADGSYQK299 | 1–25 | 29LTACG33 | SCO5260 (58%), SAV2982 (58%) |
| 11 | 6701 | SGR5275 | Maltose-binding protein MalE | 21/45 | 43 | 44.8 | 73YVNVPFGDAQNK84, 87NAAQAGDGAPDVIR100 | 1–29 | 18ATACG22 | SCO2231 (48%), SAV5977 (48%) |
| 12 | 1307 | SGR1737 | Glutamate ABC transporter substrate-binding protein | 33/35 | 55 | 33 | 77ADQPYLGFEDQATK90, 149QVGFAGPYYK158, 197KPEYGASVVELAK209 | 1–22 | 19ATACG23 | SCO5776 (40%), SAV2485 (55%) |
| 13 | 1303 | SGR1737 | Glutamate ABC transporter substrate-binding protein | 35/38 | 56 | 33 | 77ADQPYLGFEDQATK90, 131GQVDYYVGTYTINDER146, 149QVGFAGPYYK158, 197KPEYGASVVELAK209 | 1–22 | 19ATACG23 | SCO5776 (40%), SAV2485 (55%) |
| 14 | 4204 | Manganese-superoxide dismutase SodA | 9/14 | 56 | 22.4 | 115FKSDFAAAAAGR126 | No | No | ||
| 15 | 2702 | SGR5280 | Putative α-amylase | 21/25 | 27 | 60.7 | 295SAVFVDNHDTER306, 406AYVAINHEGSALTR419, 103AAFAAMVNTCHAAGVK118, 229LTNPNVYWK237 | 1–35 | No | SCO7020 (78%), SAV5981 (73%), |
| 16 | 1803 | SGR5280 | Putative α-amylase | 25/46 | 44 | 60.7 | 295SAVFVDNHDTER306, 406AYVAINHEGSALTR419 | 1–35 | No | SCO7020 (78%), SAV5981 (73%) |
RNA Isolation and RT-PCR
For transcript analysis, RNA was isolated from surface-grown cultures of S. griseus B-2682 or its derivative AFN. Mycelium was grown on R2YE agar plates and harvested after 36 h (onset of aerial growth) or 54 h (sporulation). RNA isolation and semiquantitative RT-PCR was carried out twice as described previously (21). 200 ng of RNA was used for each reaction (concentration was assessed using a Nanodrop® spectrophotometer). Oligonucleotides used for RT-PCR are presented in supplemental Table S1.
RESULTS AND DISCUSSION
Extracellular Proteome Analysis
The extracellular proteomes of S. griseus B-2682, of its AFN, and of AFN complemented with a plasmid expressing Factor C (called AFN/pSGF4 (16)) were studied. AFN has a non-sporulating (bald) phenotype, and introduction of Factor C-expressing plasmid pSGF4 restores wild-type levels of sporulation to this strain (16). The choice for the extracellular proteome is a logical consequence of our focus on extracellular signaling mechanisms with many of the proteins that are part of the A-factor/AdpA response regulon extracellularly localized.
To obtain an initial assessment of the protein expression profiles in the different strains, SDS-PAGE was performed; it revealed that several protein bands varied strongly in intensity. Considering the low separative capacity of one-dimensional gels the samples were analyzed further by 2D proteome analysis, and initial analysis with a pI range of 3–10 revealed that the vast majority of the proteins appeared in the pI range of 4–7, which was then used for all experiments. Over 200 detectable protein spots were identified on each gel by colloidal Coomassie staining (Fig. 1). In total 42 spots differed significantly between the parent and the mutants, and these were analyzed by MALDI-TOF mass spectrometry. In a previous study (14) we showed that Factor C acts by restoring A-factor production and normal sporulation to S. griseus AFN. Restoration of A-factor production also resulted in an increase in the expression of several A-factor-responsive secreted proteases to wild-type levels. However, at the time detailed analysis of the proteomes of AFN and AFN/pSGF4 was severely hampered by the lack of adequate genome information of S. griseus. Although our data now indicate (see below) that most proteins studied are highly conserved in S. coelicolor and Streptomyces avermitilis a single amino acid substitution in the peptides will prevent protein identification. This significant hurdle was overcome once the full genome sequence of S. griseus became publicly available (17).
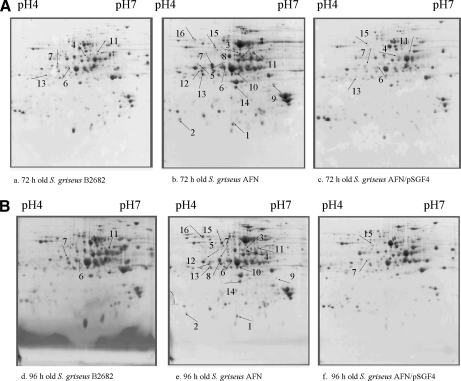
Fig. 1.
2D gel electrophoresis maps of secreted proteins extracted from 72-h-old surface-grown cultures of B-2682 (a), AFN (b), AFN/pSGF4 (c) and from 96-h-old surface-grown cultures of B-2682 (d), AFN (e), AFN/pSGF4 (f). Numbers correspond to the IDs of proteins in Table I.
The main question we sought to address this time is what proteins are expressed or overexpressed in the AFN to compensate for the lack of extracellular hydrolases that normally supply the developing colonies with nutrients. 16 of 42 spots fulfilled the selection criteria (for details see “Experimental Procedures”), and they were unambiguously identified (Table I). The expression profiles of all 16 protein spots were very similar in the 72- and 96-h cultures with the majority completely absent in wild-type cultures and in the facC-complemented AFN strain. A few proteins were visible in B-2682 and AFN/pSGF4 but with strongly reduced intensity (Fig. 1, A–F, and Fig. 2, A and B). The 16 spots represented 10 different proteins, namely SGR1460 (ID 1), SGR2245 (ID 2), SGR3109 (ID 3–4), SGR1498 (ID 5–8), SGR5704 (ID 9), SGR2237 (ID 10), SGR5275 (ID 11) SGR1737 (ID 12–13), SGR5280 (ID 15–16), and SodA (ID 14). Interestingly SodA or manganese-superoxide dismutase is absent in the sequenced S. griseus strain IFO 13350. All proteins were overexpressed in both 72- and 96-h surface-grown cultures (Fig. 2, A and B). Detailed information regarding the identification is provided in Table I.
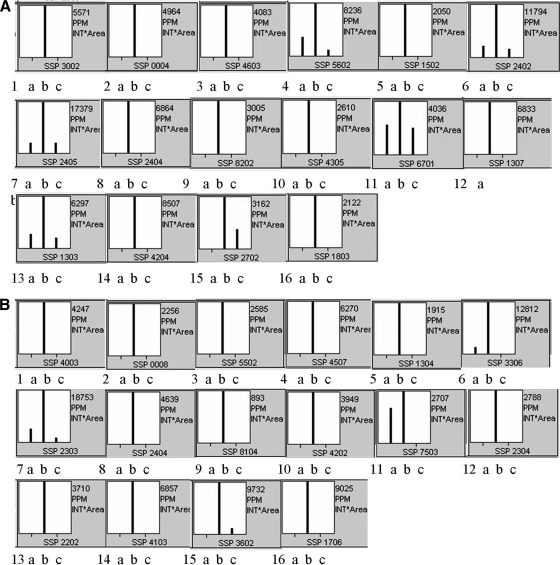
Fig. 2.
Quantitative image analysis of the proteins performed by PDQuest 2-D Analysis Software (Bio-Rad) in 72-h-old (A) and 96-h-old (B) cultures. Numbers underneath each plot correspond to the protein ID in Fig. 1 and Table I. The bars represent the relative amount of the different proteins in the strains in the order of B-2682 (a), AFN (b), and AFN/pSGF4 (c).
Interestingly all proteins with the exception of SodA are well conserved in S. avermitilis and S. coelicolor (Table I), suggesting that these proteins play a universal role in the Streptomyces life cycle. In fact, the proteins are conserved (more than 50% amino acid identity for the gene products) in the sequenced genomes of Streptomyces species Streptomyces clavuligerus ATCC 27064, Streptomyces sp. Mg1, Streptomyces sviceus ATCC 29083, Streptomyces pristinaespiralis ATCC 25486, Streptomyces sp. SPB74, and Streptomyces scabies (data not shown). All identified proteins relate to stress and may primarily be subdivided into environmental stress and nutrient stress (i.e. starvation) proteins. One exception is perhaps SGR5704, an orthologue of the Bacillus sporulation control protein Spo0M. However, sporulation results in the autolytic breakdown of the vegetative mycelium, and in fact it is a major stress mechanism in bacteria (22, 23).
The environmental stress control proteins are SGR1460, SGR2245, and SodA. SGR1460 is a putative ATP/GTP-binding protein and a homologue of the Escherichia coli osmotically inducible OsmC protein that is involved in the osmotic stress response. Superoxide dismutases SodN (SGR2245) and SodA are involved in the control of oxidative stress. Although these are predicted cytoplasmic proteins, SodF (SGR4906) is known to be autotransported as a leaderless protein (14, 24), and our experiments suggest that the same is true for SGR1460, SodN, and SodA.
Several of the proteins with enhanced expression in AFN are involved in supplying nutrients to the growing colonies, and the strongly enhanced expression in AFN suggests that this strain suffers from nutrient deprivation. SGR3109 is an orthologue of the phosphoserine aminotransferase SerC of Mycobacterium tuberculosis (NCB1 Entrez Protein CAA97391; 66% amino acid identity), an aminotransferase that plays an important role in amino acid and vitamin biosynthesis notably of glycine, serine, threonine, and vitamin B6. SGR1498 is an ABC transporter solute-binding protein that is known to be involved in xylose uptake (25). SGR2237 is a predicted arginine/ornithine-binding protein and likely involved in the transport of polar amino acids. SGR5275 is MalE or maltose-binding protein, which is the solute-binding protein belonging to the maltose uptake system in streptomycetes (26, 27). SGR1737 is a glutamate ABC transporter substrate-binding protein and likely involved in sensing and processing of environmental information. SGR1498, SGR1737, SGR2237, and SGR5275 all have an amino-terminal secretion signal sequence and a conserved prokaryotic membrane lipoprotein lipid attachment site with a cysteine residue that is most likely a site for ADP-ribosylation.
Finally SGR5704 is an orthologue of the well studied sporulation control protein Spo0M of Bacillus subtilis and most likely in an operon with the phospholipid-binding protein SGR5703. Mutation/deletion in spo0M or overexpression of spo0M in B. subtilis results in impaired sporulation (28).
The identified proteins are all overexpressed in the AFN, which is deficient in A-factor production. A-factor as a pleiotropic autoregulator in S. griseus during normal development induces the biosynthesis of several secreted hydrolases, which function to digest biopolymers and supply the differentiating colony with nutrients (29–31). In the AFN these hydrolases are not secreted (14); therefore the supply of small molecule nutrients is probably scarce.
Many of the proteins overexpressed in AFN are specific ligand-binding proteins associated to the transmembrane translocator component of the ABC transporters (31). It is likely that the overproduction of these nutrient-scavenging proteins is a response to compensate for the reduced availability of nutrients. All of the ABC transporter substrate-binding proteins have an amino-terminal secretion signal sequence and a properly positioned conserved prokaryotic membrane lipoprotein lipid attachment site with a cysteine residue. Secretion of these proteins is regulated by ADP-ribosylation. When the cysteine is ADP-ribosylated these proteins cannot be exported to the surface and are therefore nonfunctional in transport (32, 33). ADP-ribosylation-mediated inactivation of substrate-binding proteins most likely takes place particularly during normal growth and development when the supply of nutrients is plentiful.
Transcriptional Analysis of the Overexpressed Proteins
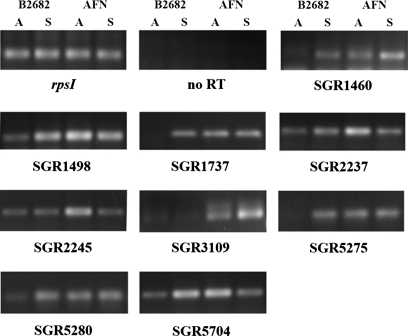
To our surprise, many of the overexpressed proteins detected extracellularly are predicted lipoproteins whose final destination is the cell surface, and their amount in the secretome therefore does not necessarily reflect their gene expression. To analyze whether the enhanced accumulation of proteins in the AFN was at least in part due to enhanced transcription, RNA was isolated from surface-grown cultures of S. griseus B-2682 and its AFN derivative. For this, mycelium was grown on R2YE agar plates and harvested at the time when aerial mycelium was formed as well as during sporulation. We used rpsI (SGR2801, encoding ribosomal protein S9) as a positive control, and reactions without the reverse transcription step were performed to confirm the lack of DNA contamination in the samples. As shown in Fig. 3, although rpsI transcript levels were the same in wild type and mutant and did not vary in time, transcription of SGR1460, -1498, -1737, -2237, -2245, -3109, -5275, -5280, and -5704 was strongly up-regulated in the AFN during early aerial growth. This corresponds to the critical phase in the Streptomyces life cycle when programmed cell death is initiated to provide the cells with nutrients. The transcriptional analysis showed that the enhanced protein levels observed in the proteome of AFN (Figs. 1 and 2) is at least in part due to enhanced gene expression in the absence of A-factor. Interestingly differences in transcript levels were far less pronounced during sporulation with only SRG1460 and SGR3109 up-regulated at this time point, whereas SGR5704 was lower in the mutant than in the parent. This strongly suggests that transcriptional up-regulation is specific for the time corresponding to programmed cell death.
Fig. 3.
Transcriptional analysis by semi-quantitative RT-PCR. Samples were harvested from R2YE agar-grown mycelium of S. griseus B-2682 and its AFN. Time points were: A, aerial growth (36 h); S, sporulation (54 h). No RT, the same as for RT-PCR with rpsI but without cDNA synthesis (control for absence of DNA contamination). Transcripts are indicated below the panels.
Interestingly we also observe elevated protein levels for the stress-related proteins in the AFN during sporulation, which is not explained by differences at the transcriptional level alone. A likely explanation is provided by the fact that A-factor activates a multitude of extracellular proteases (10, 14), and hence secreted proteins are expected to be much more stable in the AFN, further contributing to the accumulation of proteins in the mutant.
ABC Transporters and the Control of Streptomyces Development
Streptomycetes are soil-dwelling bacteria that are able to utilize almost all naturally occurring polymers, such as starch, cellulose, chitin, and agar. These polymers are degraded by extracellular hydrolases and subsequently imported (mainly as monomers or dimers) via specific permeases either via ABC transporters or via the phosphotransferase system (34–37). Permeases that are involved in sensing and transport therefore substantially influence their metabolism and morphogenesis. An in silico survey (25) identified 81 ABC transporters in S. coelicolor that are involved in the uptake of sugars, oligopeptides, and other nutrients. This large number of transporters and secreted hydrolases perfectly reflects the lifestyle of these microbes. In a recent analysis of the membrane-associated proteome (38) in S. coelicolor it was noted that a large proportion of these characterized proteins related to ABC transporter systems (including SCO5776, SCO2231, and SCO6009 whose orthologues we found in this study), which was considered as a snapshot of the nutritional requirement of the organism.
Interestingly recent studies revealed that in fact several sugar transporters play a crucial role in the control of development and in particular those that relate to N-acetylglucosamine transport and metabolism. These include the universal phosphotransferase system components enzyme I, enzyme IIACrr, and histidine phosphocarrier protein (2, 36) as well as components of the DasABC transporter. DasABC transports chitobiose, the dimer of N-acetylglucosamine, and DasA is a lipoprotein that functions as the chitobiose-binding protein (39). In addition to the transport of chitobiose DasA also plays a role in nutrient sensing perhaps by interacting with the chiRS two-component system (34). The dasABC operon is developmentally regulated and plays a role in the control of sporulation in an A-factor-independent manner in S. griseus (40, 41), and deletion of dasA results in a non-sporulating (bald) phenotype in S. coelicolor (34). Other oligomeric transporters that relate to the onset of development are for example the BldK oligopeptide transporter of S. coelicolor that is essential for sporulation on rich and glucose-containing media (42) and a second developmentally regulated ABC transporter, SCO7167, involved in morphogenesis of S. coelicolor (43).
Superoxide Dismutases in Streptomyces
Superoxide dismutases are a ubiquitous part of the cellular defense system against oxidative stress. They convert the toxic superoxide to hydrogen peroxide, which is further degraded by the enzyme catalase. So far in Streptomyces two classes of superoxide dismutases have been described, namely the nickel-containing SodN and the iron-zinc-containing SodF. We detected an additional superoxide dismutase, which has high similarity to the Bacillus pumilus SAFR-032 manganese-superoxide dismutase SodA (NCBI Entrez Protein YP_001487459). This protein so far has not been described in Streptomyces and may therefore represent a third class. In S. pristinaespiralis the expression of SodF is regulated by the SpbR protein, an analogue of the S. griseus ArpA and thus related to the γ-butyrolactone regulon (44). It is also hypothesized that SodF is involved in adaptation to oxidative stress generated by metabolic shifts during colony development (44). In further support of this hypothesis, a catalase is also developmentally regulated and required for aerial mycelium formation in S. coelicolor (45). Previously (14) we identified SodF in S. griseus as up-regulated in mutant strain AFN relative to the parental strain S. griseus B-2682, suggesting that SodF is negatively controlled by AdpA. In this work we identified SodN as a protein up-regulated in the AFN. Interestingly the presence of nickel in a micromolar concentration in the growth media induced the expression of SodN in S. coelicolor, whereas the expression of SodF was repressed (46). The differential expression of superoxide dismutases seems to suggest that a strict control of the level of superoxide dismutase is of great importance to cellular defenses against oxidative stress. The enhanced expression of the nutrient and environmental stress proteins in a strain that fails to produce (significant amounts of) A-factor underlines the stress that is generated by the absence of this signaling molecule.
The A-factor non-producing strain of S. griseus detailed in this work is a valuable asset in the search for genes that control the production of A-factor and the response it elicits. The afn locus does not correspond to any of the known A-factor biosynthetic genes or regulatory genes such as afsA, adpA, or arpA. Considering the fact that the afn mutant is able to produce A-factor under some conditions (such as after addition of the signaling protein Factor C) we anticipate that the mutation lies in a gene involved in the regulation of A- factor biosynthesis. The nature of the afn mutation and its role in the control of A-factor production are currently under investigation in our laboratories.
Supplementary Material
Footnotes
* This work was supported in part by Netherlands Organisation for Scientific Research Grant NWO 048.011.049 and Hungarian Scientific Research Fund (OTKA) Grant N42824 in frame of the Hungarian-Dutch Research Cooperation 2002 (to G. P. v. W. and S. B.) and by OTKA Grant T 034147 (to S. B.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- AFN
- A-factor non-producing mutant
- ABC
- ATP-binding cassette
- 2D
- two-dimensional
- NCBI
- National Center for Biotechnology Information
- SGR
- S. griseus
- ID
- identification number
- SCO
- S. coelicolor
- NRRL
- Northern Regional Research Laboratory (National Center for Agricultural Utilization Research).
REFERENCES
- 1.Chater K. F. (1993) Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol 47, 685–713 [DOI] [PubMed] [Google Scholar]
- 2.Rigali S., Nothaft H., Noens E. E., Schlicht M., Colson S., Müller M., Joris B., Koerten H. K., Hopwood D. A., Titgemeyer F., van Wezel G. P. (2006) The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol 61, 1237–1251 [DOI] [PubMed] [Google Scholar]
- 3.Chater K. F. (2006) Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos. Trans. R. Soc. Lond. B Biol. Sci 361, 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horinouchi S., Beppu T. (1992) Autoregulatory factors and communication in actinomycetes. Annu. Rev. Microbiol 46, 377–398 [DOI] [PubMed] [Google Scholar]
- 5.Horinouchi S., Beppu T. (1994) A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol 12, 859–864 [DOI] [PubMed] [Google Scholar]
- 6.Nakano H., Takehara E., Nihira T., Yamada Y. (1998) Gene replacement analysis of the Streptomyces virginiae barA gene encoding the butyrolactone autoregulator receptor reveals that BarA acts as a repressor in virginiamycin biosynthesis. J. Bacteriol 180, 3317–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takano E., Chakraburtty R., Nihira T., Yamada Y., Bibb M. J. (2001) A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol 41, 1015–1028 [DOI] [PubMed] [Google Scholar]
- 8.Takano E., Kinoshita H., Mersinias V., Bucca G., Hotchkiss G., Nihira T., Smith C. P., Bibb M., Wohlleben W., Chater K. (2005) A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol. Microbiol 56, 465–479 [DOI] [PubMed] [Google Scholar]
- 9.Ohnishi Y., Yamazaki H., Kato J. Y., Tomono A., Horinouchi S. (2005) AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem 69, 431–439 [DOI] [PubMed] [Google Scholar]
- 10.Horinouchi S. (2007) Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci. Biotechnol. Biochem 71, 283–299 [DOI] [PubMed] [Google Scholar]
- 11.Biró S., Békési I., Vitális S., Szabó G. (1980) A substance effecting differentiation in Streptomyces griseus. Purification and properties. Eur. J. Biochem 103, 359–363 [DOI] [PubMed] [Google Scholar]
- 12.Kiss Z., Ward A. C., Birkó Z., Chater K. F., Biró S. (2008) Streptomyces griseus 45H, a producer of the extracellular autoregulator protein factor C, is a member of the species Streptomyces albidoflavus. Int. J. Syst. Evol. Microbiol 58, 1029–1031 [DOI] [PubMed] [Google Scholar]
- 13.Birkó Z., Sümegi A., Vinnai A., van Wezel G., Szeszák F., Vitális S., Szabó P. T., Kele Z., Janáky T., Biró S. (1999) Characterization of the gene for factor C, an extracellular signal protein involved in morphological differentiation of Streptomyces griseus. Microbiology 145, 2245–2253 [DOI] [PubMed] [Google Scholar]
- 14.Birkó Z., Bialek S., Buzás K., Szájli E., Traag B. A., Medzihradszky K. F., Rigali S., Vijgenboom E., Penyige A., Kele Z., van Wezel G. P., Biró S. (2007) The secreted signaling protein factor C triggers the A-factor response regulon in Streptomyces griseus: overlapping signaling routes. Mol. Cell. Proteomics 6, 1248–1256 [DOI] [PubMed] [Google Scholar]
- 15.Szeszák F., Vitális S., Tóth F., Valu G., Fachet J., Szabó G. (1990) Detection and determination of factor C—a regulatory protein—in Streptomyces strains by antiserum and monoclonal antibody. Arch. Microbiol 154, 82–84 [DOI] [PubMed] [Google Scholar]
- 16.Biró S., Birkó Z., van Wezel G. P. (2000) Transcriptional and functional analysis of the gene for factor C, an extracellular signal protein involved in cytodifferentiation of Streptomyces griseus. Antonie van Leeuwenhoek 78, 277–285 [DOI] [PubMed] [Google Scholar]
- 17.Ohnishi Y., Ishikawa J., Hara H., Suzuki H., Ikenoya M., Ikeda H., Yamashita A., Hattori M., Horinouchi S. (2008) The genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol 190, 4050–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000) Practical Streptomyces Genetics, p. 408, John Innes Foundation, Norwich, UK [Google Scholar]
- 19.Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G. M., Carnemolla B., Orecchia P., Zardi L., Righetti P. G. (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 20.Haebel S., Albrecht T., Sparbier K., Walden P., Körner R., Steup M. (1998) Electrophoresis-related protein modification: alkylation of carboxy residues revealed by mass spectrometry. Electrophoresis 19, 679–686 [DOI] [PubMed] [Google Scholar]
- 21.Noens E. E., Mersinias V., Willemse J., Traag B. A., Laing E., Chater K. F., Smith C. P., Koerten H. K., van Wezel G. P. (2007) Loss of the controlled localization of growth stage-specific cell-wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Mol Microbiol 64, 1244–1259 [DOI] [PubMed] [Google Scholar]
- 22.Rigali S., Titgemeyer F., Barends S., Mulder S., Thomae A. W., Hopwood D. A., van Wezel G. P. (2008) Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep 9, 670–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González-Pastor J. E., Hobbs E. C., Losick R. (2003) Cannibalism by sporulating bacteria. Science 301, 510–513 [DOI] [PubMed] [Google Scholar]
- 24.Henderson I. R., Cappello R., Nataro J. P. (2000) Autotransporter proteins, evolution and redefining protein secretion: response. Trends Microbiol 8, 534–535 [DOI] [PubMed] [Google Scholar]
- 25.Bertram R., Schlicht M., Mahr K., Nothaft H., Saier M. H., Jr., Titgemeyer F. (2004) In silico and transcriptional analysis of carbohydrate uptake systems of Streptomyces coelicolor A3(2). J. Bacteriol 186, 1362–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Wezel G. P., White J., Young P., Postma P. W., Bibb M. J. (1997) Substrate induction and glucose repression of maltose utilization. Mol. Microbiol 23, 537–549 [DOI] [PubMed] [Google Scholar]
- 27.van Wezel G. P., White J., Bibb M. J., Postma P. W. (1997) The malEFG gene cluster of Streptomyces coelicolor A3(2): characterization, disruption and transcriptional analysis. Mol. Gen. Genet 254, 604–608 [DOI] [PubMed] [Google Scholar]
- 28.Han W. D., Kawamoto S., Hosoya Y., Fujita M., Sadaie Y., Suzuki K., Ohashi Y., Kawamura F., Ochi K. (1998) A novel sporulation-control gene (spo0M) of Bacillus subtilis with a σH-regulated promoter. Gene 217, 31–40 [DOI] [PubMed] [Google Scholar]
- 29.Méndez C., Braña A. F., Manzanal M. B., Hardisson C. (1985) Role of substrate mycelium in colony development in Streptomyces. Can. J. Microbiol 31, 446–450 [DOI] [PubMed] [Google Scholar]
- 30.Miguélez E. M., Hardisson C., Manzanal M. B. (1999) Hyphal death during colony development in Streptomyces antibioticus: morphological evidence for the existence of a process of cell deletion in a multicellular prokaryote. J. Cell Biol 145, 515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirano S., Kato J. Y., Ohnishi Y., Horinouchi S. (2006) Control of the Streptomyces subtilisin inhibitor gene by AdpA in the A-factor regulatory cascade in Streptomyces griseus. J. Bacteriol 188, 6207–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Heide T., Poolman B. (2002) ABC transporters: one, two or four extracytoplasmic substrate-binding sites? EMBO Rep 3, 938–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugawara K., Dohmae N., Kasai K., Saido-Sakanaka H., Okamoto S., Takio K., Ochi K. (2002) Isolation and identification of novel ADP-ribosylated proteins from Streptomyces coelicolor A3(2). Biosci. Biotechnol. Biochem 66, 2292–2296 [DOI] [PubMed] [Google Scholar]
- 34.Colson S., van Wezel G. P., Craig M., Noens E. E., Nothaft H., Mommaas A. M., Titgemeyer F., Joris B., Rigali S. (2008) The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology 154, 373–382 [DOI] [PubMed] [Google Scholar]
- 35.Saito A., Schrempf H. (2004) Mutational analysis of the binding affinity and transport affinity for N-acetylglucosamine of the novel ABC transporter Ngc in the chitin-degrader Streptomyces olivaceoviridis. Mol. Genet. Genomics 271, 545–553 [DOI] [PubMed] [Google Scholar]
- 36.Nothaft H., Dresel D., Willimek A., Mahr K., Niederweis M., Titgemeyer F. (2003) The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J. Bacteriol 185, 7019–7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamionka A., Parche S., Nothaft H., Siepelmeyer J., Jahreis K., Titgemeyer F. (2002) The phosphotransferase system of Streptomyces coelicolor. Eur. J. Biochem 269, 2143–2150 [DOI] [PubMed] [Google Scholar]
- 38.Kim D. W., Chater K. F., Lee K. J., Hesketh A. (2005) Effects of growth phase and the developmentally significant bldA-specified tRNA on the membrane-associated proteome of Streptomyces coelicolor. Microbiology 151, 2707–2720 [DOI] [PubMed] [Google Scholar]
- 39.Saito A., Shinya T., Miyamoto K., Yokoyama T., Kaku H., Minami E., Shibuya N., Tsujibo H., Nagata Y., Ando A., Fujii T., Miyashita K. (2007) The dasABC gene cluster, adjacent to dasR, encodes a novel ABC transporter for the uptake of N,N′-diacetylchitobiose in Streptomyces coelicolor A3(2). Appl. Environ. Microbiol 73, 3000–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo J. W., Ohnishi Y., Hirata A., Horinouchi S. (2002) ATP-binding cassette transport system involved in regulation of morphological differentiation in response to glucose in Streptomyces griseus. J. Bacteriol 184, 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohnishi Y., Seo J. W., Horinouchi S. (2002) Deprogrammed sporulation in Streptomyces. FEMS Microbiol. Lett 216, 1–7 [DOI] [PubMed] [Google Scholar]
- 42.Nodwell J. R., McGovern K., Losick R. (1996) An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol 22, 881–893 [DOI] [PubMed] [Google Scholar]
- 43.Hillerich B., Westpheling J. (2006) A new GntR family transcriptional regulator in Streptomyces coelicolor is required for morphogenesis and antibiotic production and controls transcription of an ABC transporter in response to carbon source. J. Bacteriol 188, 7477–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Folcher M., Gaillard H., Nguyen L. T., Nguyen K. T., Lacroix P., Bamas-Jacques N., Rinkel M., Thompson C. J. (2001) Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic biosynthesis, and expression of a superoxide dismutase. J. Biol. Chem 276, 44297–44306 [DOI] [PubMed] [Google Scholar]
- 45.Cho Y. H., Lee E. J., Roe J. H. (2000) A developmentally regulated catalase required for proper differentiation and osmoprotection of Streptomyces coelicolor. Mol. Microbiol 35, 150–160 [DOI] [PubMed] [Google Scholar]
- 46.Kim F. J., Kim H. P., Hah Y. C., Roe J. H. (1996) Differential expression of superoxide dismutases containing Ni and Fe/Zn in Streptomyces coelicolor. Eur. J. Biochem 241, 178–185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.