Abstract
SUMMARY
The mTORC1 and mTORC2 pathways regulate cell growth, proliferation, and survival. We identify DEPTOR, also called DEPDC6, as an mTOR-interacting protein whose expression is negatively regulated by mTORC1 and mTORC2. Loss of DEPTOR activates S6K1, Akt, and SGK1; promotes cell growth and survival; and activates mTORC1 and mTORC2 kinase activities. DEPTOR overexpression suppresses S6K1 but, by relieving feedback inhibition from mTORC1 to PI3K signaling, activates Akt. Consistent with many human cancers having activated mTORC1 and mTORC2 pathways, DEPTOR expression is low in most cancers. Surprisingly, DEPTOR is highly overexpressed in a subset of Multiple Myelomas harboring Cyclin D1/D3 or c-MAF/MAFB translocations. In these cells, high DEPTOR expression is necessary to maintain PI3K and Akt activation and a reduction in DEPTOR levels leads to apoptosis. Thus, we identify a novel mTOR-interacting protein whose deregulated overexpression in Multiple Myeloma cells represents a new mechanism for activating PI3K/Akt signaling and promoting cell survival.
INTRODUCTION
Mammalian TOR (mTOR) is an evolutionarily conserved serine/threonine kinase that integrates signals from growth factors, nutrients, and stresses to regulate multiple processes, including mRNA translation, cell cycle progression, autophagy, and cell survival (reviewed in (Sarbassov et al., 2005a)). It is increasingly apparent that deregulation of the mTOR pathway occurs in common diseases, including cancer and diabetes, emphasizing the importance of identifying and understanding the function of the components of the mTOR signaling network. mTOR resides in two distinct multiprotein complexes referred to as mTOR complex 1 (mTORC1) and 2 (mTORC2) (reviewed in (Guertin and Sabatini, 2007)). mTORC1 is composed of the mTOR catalytic subunit and three associated proteins, raptor, PRAS40, and mLST8/GβL. mTORC2 also contains mTOR and mLST8/GβL, but instead of raptor and PRAS40, contains the proteins rictor, mSin1, and protor.
mTORC1 controls cell growth in part by phosphorylating S6 Kinase 1 (S6K1) and the eIF-4E-binding protein 1 (4E-BP1), key regulators of protein synthesis. mTORC2 modulates cell survival in response to growth factors by phosphorylating its downstream effectors Akt/PKB and Serum/Glucocorticoid Regulated Kinase 1 (SGK1) (reviewed in (Guertin and Sabatini, 2007)).
In addition to directly activating Akt as part of mTORC2, mTOR, as part of mTORC1, also negatively regulates Akt by suppressing the growth factor-driven pathways upstream of it. Specifically, mTORC1 impairs PI3K activation in response to growth factors by downregulating the expression of Insulin Receptor Substrate 1 and 2 (IRS-1/2) and Platelet-Derived Growth Factor Receptor-Beta (PDGFR-β) (reviewed in (Sabatini, 2006)). The activation of Akt that results from treating cells with the mTORC1 inhibitor rapamycin may contribute to the limited success to date of this drug and its analogs as cancer therapies.
While most information concerning the involvement of the mTOR pathway in human cancers is consistent with a role for mTOR in directly promoting tumor growth, there are also indications in the literature that mTOR possesses tumor suppressor-like properties. Thus, the tumors that develop in patients with Tuberous Sclerosis Complex (TSC), a syndrome characterized by mTORC1 hyperactivation, are thought to have a limited growth potential due to the PI3K inactivation caused by the aforementioned feedback loop (Manning et al., 2005; Zhang et al., 2007). In addition, partial loss of function alleles of mTOR confer susceptibility to plasmacytomas in mice, though the mechanism for this effect has not been clarified (Bliskovsky et al., 2003).
Here, we identify DEPTOR as an mTOR binding protein that normally functions to inhibit the mTORC1 and mTORC2 pathways. When greatly overexpressed, DEPTOR inhibits mTORC1, and, unexpectedly, this leads to the activation of the PI3K/mTORC2/Akt pathway. This indirect mode of PI3K activation is important for the viability of a subset of Multiple Myeloma cells which otherwise lack PI3K-activating mutations. We propose that DEPTOR is an endogenous inhibitor of mTOR whose deregulated overexpression promotes cell survival in a subset of Multiple Myelomas.
RESULTS
DEPTOR is an mTOR Interacting Protein
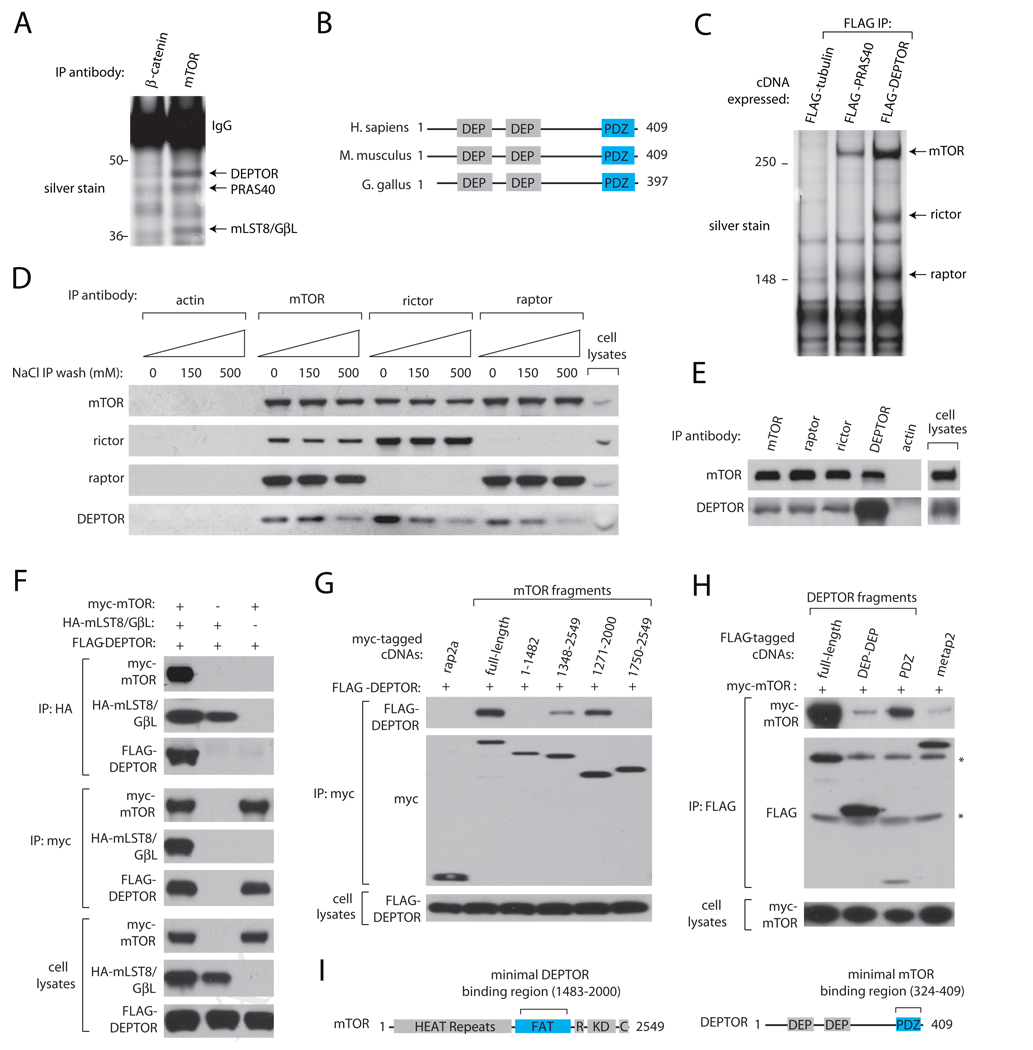
Using low-salt purification conditions designed to isolate PRAS40 (Sancak et al., 2007), we identified within mTOR immunoprecipitates a 48 kDa protein assigned the NCBI Gene Symbol DEPDC6 (NCBI Gene ID: 64798) (Figure 1A). The gene for DEPDC6 is found only in vertebrates, and encodes a protein with tandem N-terminal DEP (Dishevelled, Egl-10, Pleckstrin) domains and a C-terminal PDZ (Postsynaptic density 95, Discs large, Zonula occludens-1) domain (reviewed in (Chen and Hamm, 2006; Jemth and Gianni, 2007) (Figure 1B). Because no previous studies refer to the function of the DEPDC6 gene product, we named it DEPTOR in reference to its DEP domains and its specific interaction with mTOR (see below). In purified preparations of recombinant DEPTOR stably expressed in HEK-293E cells, we detected via mass spectrometry endogenous mTOR, as well as raptor and rictor, mTORC1 and mTORC2-specific components, respectively. Analogous preparations of recombinant PRAS40, a raptor binding protein, contained only mTORC1 (Figure 1C). Consistent with DEPTOR being part of mTORC1 and mTORC2, endogenous DEPTOR was also detected in immunoprecipitates prepared from HEK-293E (Figure 1D) or HeLa cells (Figure 1E) using antibodies that recognize endogenous mTOR, raptor, or rictor, but not actin. As was the case with PRAS40 (Sancak et al., 2007), buffers with increased salt concentrations reduced the amounts of DEPTOR recovered in the immunoprecipitates (Figure 1D). Reciprocal experiments confirmed that endogenous mTOR specifically co-immunoprecipitates with DEPTOR (Figure 1E).
Figure 1. DEPTOR is an mTOR-Interacting Protein.
(A) Silver stain of SDS-PAGE analysis of mTOR immunoprecipitates prepared from HEK-293E cells.
(B) Schematic representation of structural features of human DEPTOR and its mouse and chicken orthologues.
(C) Endogenous mTOR, raptor, and rictor co-immunoprecipitate with epitope-tagged DEPTOR. FLAG immunoprecipitates from HEK-293E cells expressing FLAG-DEPTOR, FLAG-PRAS40, or FLAG-tubulin were analyzed by SDS-PAGE followed by silver staining.
(D) Interaction of endogenous DEPTOR with endogenous mTORC1 and mTORC2 is sensitive to high salt-containing buffers. mTOR, raptor, rictor, or actin immunoprecipitates were prepared from HEK-293E cells, washed with buffers containing indicated amounts of NaCl and analyzed by SDS-PAGE followed by immunoblotting for indicated proteins.
(E) Endogenous DEPTOR co-immunoprecipitates endogenous mTOR. DEPTOR immunoprecipitates were prepared from HeLa cells, washed in a buffer containing 150 mM NaCl, and analyzed as in (D).
(F) DEPTOR interacts with mTOR and not mLST8/GβL. Indicated cDNAs in expression vectors were co-expressed in HEK-293T cells, cell lysates prepared, and used to prepare anti-HA immunoprecipitates and anti-myc immunoprecipitates. Both immunoprecipitates and cell lysates were analyzed as in (D).
(G) Myc-tagged mTOR, its indicated fragments, or rap2a were co-expressed in HEK-293T cells with FLAG-DEPTOR, and anti-myc immunoprecipitates were analyzed as in (D).
(H) The PDZ domain of DEPTOR interacts with mTOR. FLAG-tagged DEPTOR, its fragments, or metap2 were co-expressed in HEK-293T cells with myc-mTOR, and anti-FLAG immunoprecipitates were analyzed as in (D). Asterisks (*) indicate non-specific bands.
(I) Schematic indicating the regions of mTOR and DEPTOR that mediate their interaction. The abbreviations for and residues that define known mTOR domains are: HEAT Repeats, 1–1382; FAT, 1383–2014; R (FKBP12-Rapamycin Binding), 2015–2114; KD (PI3K-like Kinase Domain), 2115–2431; C (FATC), 2432–2549.
As the above results indicate that DEPTOR is part of mTORC1 and mTORC2, we expected DEPTOR to interact with mTOR and/or mLST8/GβL, the only proteins common to both mTORC1 and mTORC2. To test this, we co-overexpressed DEPTOR with epitope-tagged mTOR or mLST8/GβL and analyzed mTOR or mLST8/GβL immunoprecipitates for the presence of DEPTOR. The results indicate that DEPTOR is an mTOR interacting protein: DEPTOR co-immunoprecipitated with mTOR irrespective of whether or not mLST8/GβL was co-expressed with it, while DEPTOR co-immunoprecipitated with mLST8/GβL only when it and mTOR were expressed together (Figure 1F). DEPTOR interacts with a C-terminal portion of mTOR (a.a. 1483–2000) that is upstream of its kinase domain and does not encompass the mLST8/GβL binding site (a.a. 2001–2549) (Figure 1G, 1I), providing further evidence that, within mTORC1 and mTORC2, DEPTOR interacts specifically with mTOR and not with mLST8/GβL. Finally, reciprocal experiments showed that the PDZ domain of DEPTOR mediates its interaction with mTOR (Figure 1H, 1I).
DEPTOR Depletion Activates mTORC1 and mTORC2 signaling in Intact Cells and Enhances the In Vitro Kinase Activities of the Complexes
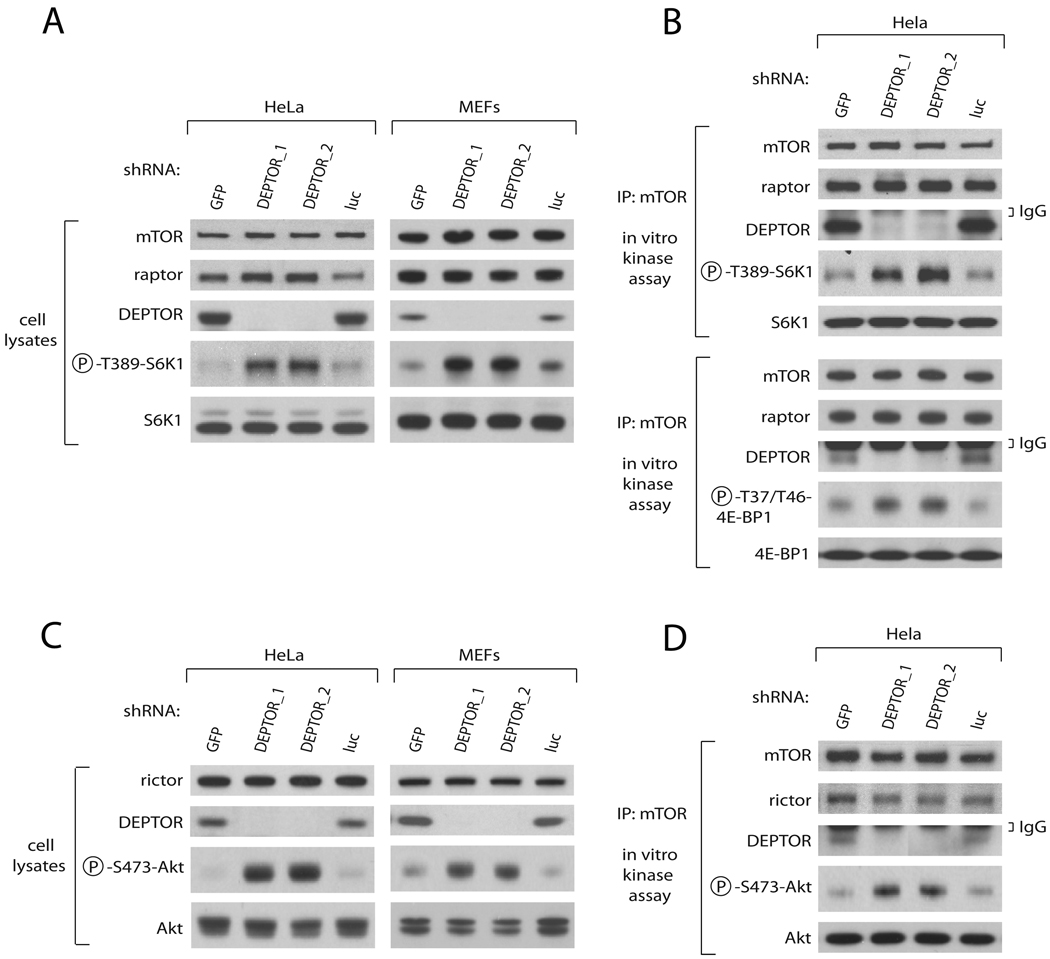
To test the idea that DEPTOR regulates mTOR function, we measured the activity of the mTOR pathway in cells with RNAi-mediated reductions in DEPTOR expression. Human or mouse cells expressing either of two shRNAs targeting DEPTOR, but not control proteins, had increased mTORC1 and mTORC2 signaling as judged by the phosphorylation states of the mTORC1 and mTORC2 substrates, S6K1 and Akt, respectively (Figure 2A, 2C). Concomitant with the increase in mTORC1 signaling, mTORC1 immunopurified from cells depleted of DEPTOR had increased in vitro kinase activity towards two known substrates, S6K1 and 4E-BP1 (Figure 2B). Likewise, mTORC2 isolated from DEPTOR-depleted cells had increased in vitro kinase activity towards its substrate Akt1 (Figure 2D). Taken together, the loss of function data indicate that within cells DEPTOR is an inhibitor of mTORC1 and mTORC2 activities.
Figure 2. DEPTOR Depletion in Cells Activates mTORC1 and mTORC2 signaling and In Vitro Kinase Activities.
(A) Knockdown of DEPTOR activates the mTORC1-S6K1 pathway. HeLa cells or p53−/− MEFs were infected with lentiviruses expressing shRNAs targeting the indicated genes. Cell lysates were analyzed by immunoblotting for the levels of the indicated proteins and phosphorylation states.
(B) Knockdown of DEPTOR activates mTORC1 kinase activity towards S6K1 and 4E-BP1. HeLa cells were infected with lentiviruses expressing shRNAs targeting the indicated genes. mTOR immunoprecipitates were prepared from cell lysates (1 mg total protein) and analyzed for mTORC1 kinase activity toward S6K1 and 4E-BP1 and for levels of mTOR, raptor, and DEPTOR.
(C) Knockdown of DEPTOR activates the mTORC2-Akt pathway. The experiment was performed as in (A) except that Akt phosphorylation and levels were measured.
(D) Knockdown of DEPTOR activates mTORC2 kinase activity towards Akt1. The experiment was performed as in (B) except that mTORC2 activity towards Akt was measured.
DEPTOR Depletion Increases Cell Size and Protects Cells from Apoptosis
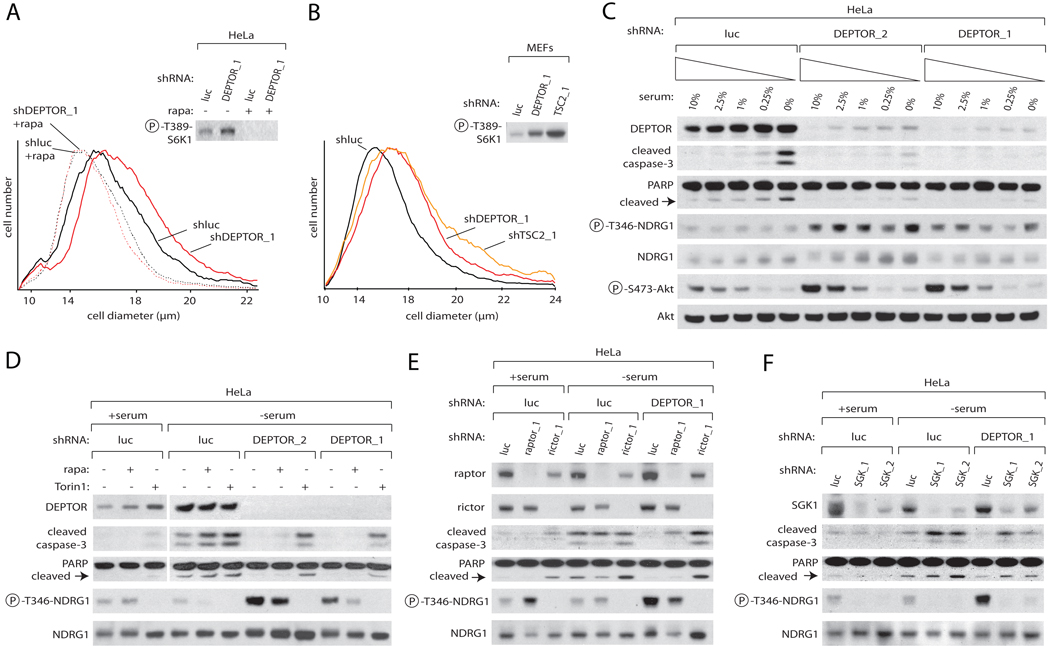
To determine if the effects of DEPTOR loss on mTORC1/2 signaling are physiologically significant, we measured in DEPTOR-deficient cells key outputs of mTORC1 and mTORC2 function, namely cell growth and survival, respectively. Cells with reduced DEPTOR expression were larger than control cells and rapamycin treatment reversed this phenotype, consistent with DEPTOR acting upstream of mTORC1 (Figure 3A). The size of the DEPTOR-deficient cells was comparable in magnitude to, although slightly smaller than, those with reduced expression of TSC2, a well-established negative regulator of mTORC1 (reviewed in (Sabatini, 2006)) (Figure 3B).
Figure 3. DEPTOR Depletion Increases Cell Size and Protects Cells from Apoptosis Induced by Serum Deprivation.
(A) Knockdown of DEPTOR increases cell size in an mTORC1-dependent fashion. Cell size distributions (graphs) and S6K1 phosphorylation (immunoblot) are shown for HeLa cells stably expressing shRNAs targeting DEPTOR or luciferase. Where indicated, cells were pre-treated with 100 nM rapamycin or vehicle before infection. 72 hours after infection, cell size was measured using a Coulter counter.
(B) A knockdown of DEPTOR has similar effects on cells size as a TSC2 knockdown. p53 null MEFs were infected with shRNAs and cell size measured as in (A).
(C) DEPTOR knockdown protects against apoptosis induced by serum withdrawal. HeLa cells expressing shRNAs targeting luciferase or DEPTOR were grown in media containing the indicated concentrations of serum for 30 hours. Cell lysates were then analyzed by immunoblotting for the levels of the indicated proteins and phosphorylation states.
(D) mTOR inhibition re-sensitizes DEPTOR knockdown cells to apoptosis induced by serum withdrawal. HeLa cells expressing shRNAs targeting luciferase or DEPTOR were serum deprived for 30 hours in the presence of 100 nM rapamycin, 250 nM Torin1, or vehicle. Cell lysates were then analyzed as in (C).
(E) Rictor but not raptor inhibition re-sensitizes DEPTOR knockdown cells to apoptosis induced by serum withdrawal. HeLa cells co-expressing shRNAs targeting luciferase or DEPTOR along with shRNAs targeting luciferase or rictor or raptor were serum deprived for 30 hours. Cell lysates were then analyzed as in (C).
(F) SGK1 inhibition re-sensitizes DEPTOR knockdown cells to apoptosis induced by serum withdrawal. HeLa cells co-expressing shRNAs targeting luciferase or DEPTOR along with shRNAs targeting luciferase or SGK1 were analyzed as in (C).
Because Akt promotes cell survival (Dudek et al., 1997) and DEPTOR suppression within cells causes an increase in Akt phosphorylation (Figure 2C), we tested whether DEPTOR-deficient cells are resistant to apoptosis induction. In HeLa cells expressing a control shRNA, reductions in the serum concentration of the media caused a dose-dependent decrease in Akt phosphorylation and a concomitant increase in the cleaved forms of caspase-3 and PARP, two hallmarks of apoptosis (Datta et al., 1999; Tewari et al., 1995) (Figure 3C). In contrast, DEPTOR-deficient cells were resistant to PARP and caspase-3 cleavage in a manner that correlated with the efficiency of DEPTOR suppression (Figure 3C). However, in contrast to what is observed in the presence of serum, in the absence of serum DEPTOR-deficient HeLa cells did not have higher levels of Akt phosphorylation than control cells (Figure 3C). This suggests that, despite the capacity of DEPTOR suppression to protect cells from apoptosis, the effect was unlikely to be mediated by Akt. As Akt shares some of its pro-survival functions with the related family of SGK kinases (reviewed in (Tessier and Woodgett, 2006)) and recent evidence indicates that mTOR activates SGK1 by directly phosphorylating it (Garcia-Martinez and Alessi, 2008; Hong et al., 2008), we reasoned that DEPTOR suppression might protect HeLa cells from apoptosis by promoting SGK1 activity. Consistent with this, the DEPTOR-deficient HeLa cells had, at all serum concentrations, increased phosphorylation levels of NDRG1, an established SGK1 substrate (Murray et al., 2004) (Figure 3C). Because it is still controversial if SGK1 is an mTORC2 and/or mTORC1 substrate (Garcia-Martinez and Alessi, 2008; Hong et al., 2008), we determined if DEPTOR-deficient cells could be re-sensitized to pro-apoptotic stimuli by inhibiting mTORC1 with rapamycin or by inhibiting both mTORC1 and mTORC2 with Torin1, a highly selective and potent ATP-competitive inhibitor of mTOR (Thoreen et al., 2009). Consistent with mTORC2 mediating the pro-survival effects of DEPTOR suppression, treatment of the serum-deprived and DEPTOR-deficient HeLa cells with Torin1, but not rapamycin, restored caspase-3 and PARP cleavage and reversed the hyperphosphorylation of NDRG1 caused by DEPTOR loss (Figure 3D). In accord with the results obtained with Torin1, a knockdown of rictor, but not raptor, abolished NDRG1 hyperphosphorylation and re-activated apoptosis in DEPTOR knockdown cells deprived of serum (Figure 3E). Lastly, a reduction in SGK1 levels in the DEPTOR knockdown HeLa cells reduced NDRG1 phosphorylation and re-sensitized the cells to apoptosis induction upon serum deprivation (Figure 3F). DEPTOR suppression also prevented caspase-3 cleavage in serum-deprived HT-29 cells, though in this cell type the anti-apoptotic effects of DEPTOR loss correlated with a partial rescue of Akt phosphorylation (Figure S1). Other perturbations that promote Akt activation, such as suppression of raptor expression (Skeen et al., 2006), also protected HT-29 cells from apoptosis induced by serum withdrawal (Figure S1). Thus, analyses of DEPTOR-deficient cells indicate that DEPTOR physiologically controls cellular processes regulated by mTOR signaling.
mTOR Negatively Regulates DEPTOR Expression at the Transcriptional and Post-Translational levels
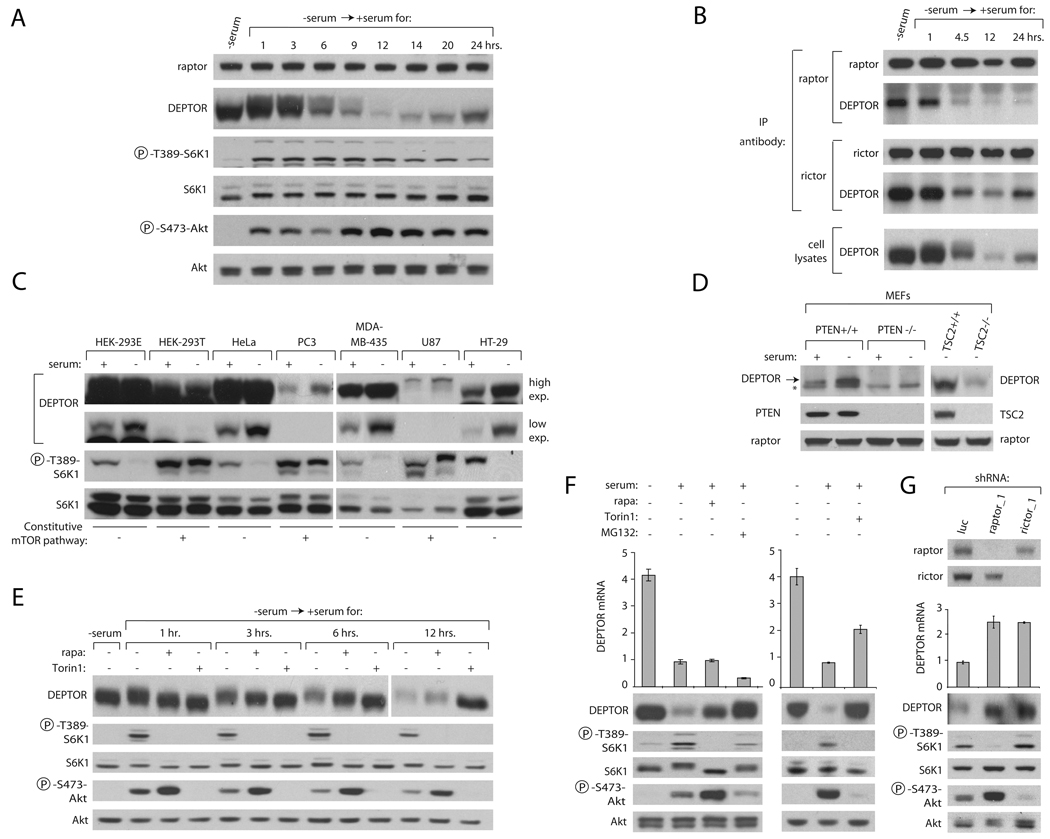
In our previous experiments, we noted that prolonged serum withdrawal caused an increase in DEPTOR protein expression (Figure 3C). To gain insight into the temporal regulation of DEPTOR expression, we starved and stimulated HeLa cells with serum for various time intervals. Serum stimulation led to a time-dependent retardation in DEPTOR migration in SDS-PAGE until about 6 hours after serum addition, at which point DEPTOR expression began to fall, and by 12 hours, almost disappeared (Figure 4A). Interestingly, these qualitative and quantitative changes in DEPTOR correlated with inflections in mTORC1 and mTORC2 pathway activities as measured by the phosphorylations of S6K1 and Akt, respectively. As expected, the amount of DEPTOR bound to mTORC1 and mTORC2 (Figure 4B) reflected the serum-induced changes in DEPTOR expression detected in cell lysates (Figures 4A and 4B). Serum starvation and stimulation also regulated DEPTOR expression in a variety of other cancer cell lines in addition to HeLa cells (Figure 4C). We noted that cell lines known to have serum-independent mTORC1 and mTORC2 signaling, such as HEK-293T, PC3, and U87 cells (Sarbassov et al., 2006) have markedly reduced levels of DEPTOR that are largely insensitive to serum (Figure 4C).
Figure 4. mTOR Negatively Regulates DEPTOR Protein and mRNA Expression.
(A) Serum starvation and stimulation regulates DEPTOR protein expression. HeLa cells were serum starved for 30 hours and stimulated with serum (10% final concentration) for the indicated times. Cell lysates were analyzed by immunoblotting for the levels of indicated proteins and phosphorylation states.
(B) DEPTOR amounts in mTORC1 and mTORC2 correlate with its expression within cells. HeLa cells were serum starved for 30 hours and stimulated with serum (10% final concentration) for the indicated times. Cell lysates and raptor and rictor immunoprecipitates were analyzed by immunoblotting for indicated proteins.
(C) DEPTOR protein expression is downregulated in cancer cell lines with constitutive mTOR signaling. Cell lines were seeded at equal density and grown in the presence or absence of serum for 30 hours and cell lysates analyzed as in (A).
(D) DEPTOR protein expression is downregulated in PTEN null or TSC2 null MEFs. Indicated MEF lines were seeded at equal density and grown in the presence or absence of serum for 30 hours and cell lysates analyzed for levels of DEPTOR, PTEN, TSC2, and raptor. An asterisk (*) indicates a non-specific band.
(E) Rapamycin treatment partially and Torin1 treatment fully rescues the serum-induced decrease in DEPTOR protein. HeLa cells were serum-deprived for 30 hours, and then stimulated with serum in the presence of 100 nM rapamycin, 250 nM Torin1, or vehicle for the specified times. Cell lysates were analyzed as in (A).
(F) Regulation of DEPTOR protein and mRNA expression. HeLa cells were serum-deprived for 30 hours, and then stimulated with serum in the presence of 100 nM rapamycin, 10 uM of MG132, 250 nM Torin1, or vehicle for the specified times. Cell lysates were analyzed by immunoblotting for the levels of indicated proteins and phosphorylation states. DEPTOR mRNA was measured by qRT-PCR and normalized to GADPH mRNA levels. Error bars indicate standard deviation for n=3.
(G) Reductions in raptor or rictor expression increase DEPTOR protein and mRNA levels. Five days after transductions with raptor or rictor shRNA-expressing lentiviruses, HeLa cells were lysed and analyzed as in (A). DEPTOR mRNA was prepared and analyzed as in (F).
As both PC3 and U87 cells lack PTEN (SANGER COSMIC database), a negative regulator of PI3K signaling that controls the responsiveness of the mTORC1 and mTORC2 pathways to growth factors (reviewed in (Carracedo and Pandolfi, 2008)), it seemed likely that PTEN loss suppresses DEPTOR expression. Consistent with this, in PTEN-null MEFs DEPTOR was virtually absent in the presence or absence of serum, while serum still regulated DEPTOR expression in MEFs that express PTEN (Figure 4D). As PTEN loss activates both mTORC1 and mTORC2 signaling we determined if DEPTOR expression is also sensitive to perturbations that only activate mTORC1. Indeed, MEFs lacking TSC2, a tumor suppressor whose loss leads to activation of only mTORC1, had reduced levels of DEPTOR even when cultured in the absence of serum (Figure 4D).
The above findings suggested that both mTORC1 and mTORC2 negatively regulate DEPTOR expression (Figures 4A–D). Supporting this idea, treatment of cells with the mTORC1 and mTORC2 inhibitor Torin1 fully blocked the mobility shift and decrease in expression of DEPTOR caused by serum stimulation, while treatment with the mTORC1 inhibitor rapamycin had only partial effects on DEPTOR mobility and expression (Figure 4E). To determine at which regulatory level(s) cells control DEPTOR expression we measured the amounts of DEPTOR protein and mRNA in HeLa cells treated in various ways (Figure 4F and 4G). The drop in DEPTOR protein caused by serum stimulation correlated with a significant decrease in the mRNA for DEPTOR (Figure 4F), suggesting that transcriptional regulation contributes to the control of DEPTOR expression. However, it was clear that post-transcriptional mechanisms are also in play because the partial rescue by rapamycin treatment of the serum-induced decrease in DEPTOR protein amounts occurred in the absence of any change in DEPTOR mRNA levels (Figure 4F). Moreover, treatment of cells with the proteasome inhibitor MG132 (Tsubuki et al., 1993) blocked the serum-induced decrease in DEPTOR protein without increasing the DEPTOR mRNA, indicating that the degradation of DEPTOR is a key step in the regulation of its expression (Figure 4F). Treatment of cells with Torin1 blocked the serum-induced drop in DEPTOR protein levels and, in contrast to rapamycin, also partially prevented the decrease in the DEPTOR mRNA (Figure 4F).
A comparison of the effects of rapamycin with those of Torin1 on DEPTOR expression suggested that both mTORC1 and mTORC2 negatively regulate DEPTOR protein expression, but that only mTORC2 negatively regulates DEPTOR mRNA expression. To further test this, we depleted mTORC1 or mTORC2 using shRNAs targeting raptor or rictor, respectively, and examined DEPTOR protein and mRNA levels. As expected, both the raptor and rictor knockdowns increased DEPTOR protein levels in serum-replete cells (Figure 4G). However, both knockdowns also increased DEPTOR mRNA (Figure 4G), indicating that the regulation of DEPTOR mRNA expression by mTORC1 is rapamycin resistant, consistent with the increasing evidence that only some mTORC1 functions are sensitive to rapamycin (Choo et al., 2008; Feldman et al., 2009; Thoreen et al., 2009). Thus, both mTOR complexes negatively regulate DEPTOR mRNA expression, explaining why cell lines with high mTORC1 and mTORC2 activity, such as the PTEN-null PC3 or U87 cells, have low levels of DEPTOR mRNA (Figure S2A). In aggregate, these results suggest that the regulation of DEPTOR expression is complex and involves post-translational and transcriptional mechanisms mediated by both mTORC1 and mTORC2.
DEPTOR is Phosphorylated in an mTOR-dependent Fashion
Because the mTOR inhibitor Torin 1 prevents the serum-induced degradation and SDS-PAGE mobility shift of DEPTOR, we reasoned that a mechanism through which mTORC1 and mTORC2 might regulate DEPTOR is by controlling its phosphorylation. To investigate this, we purified DEPTOR from serum-stimulated cells and analyzed it by mass spectrometry for the presence of phosphorylated residues. This led to the identification of 13 serine and threonine phosphorylation sites, all of which are located in the linker between the C-terminal DEP domain and the PDZ domain (Figure S3A). Interestingly, many of the phosphorylation sites fall within “proline-directed” motifs like those in 4E-BP1 (Gingras et al., 2001) which are known to be phosphorylated by mTOR (Brunn et al., 1997; Burnett et al., 1998; Gingras et al., 1999). Extensive characterization of DEPTOR from serum-stimulated cells supported the notion that DEPTOR is heavily phosphorylated in an mTOR-dependent manner (Figures S3B–D). We changed all 13 phosphorylation sites to alanine to generate a non-phosphorylatable mutant protein that we call DEPTOR 13×S/T->A. In serum-stimulated cells the mutant protein did not undergo a mobility shift on SDS-PAGE (Figure S3E). When overexpressed, neither wild-type nor 13×S/T->A DEPTOR decreased in expression upon serum stimulation of cells (compare Figures 4B and S3E), suggesting that the machinery that degrades DEPTOR is readily saturated. Consistent with this, stable and modest 2–5 fold overexpression of epitope-tagged wild-type DEPTOR severely inhibited the loss of endogenous DEPTOR that normally occurs upon serum stimulation (Figure S3F). Because overexpression of DEPTOR impaired the regulation its expression (Figure S3E–F), we focused on other functions for DEPTOR phosphorylation that could be relevant to DEPTOR turnover and, in particular, examined the potential role of DEPTOR phosphorylation on regulating the DEPTOR-mTOR interaction. Compared to wild-type DEPTOR, the 13×S/T->A DEPTOR mutant bound better to mTOR within mTORC1 and mTORC2 (Figure 5A). These results suggest that the phosphorylation of DEPTOR promotes its release from mTOR and correlates with its loss. In addition, experiments described below indicate that DEPTOR phosphorylation is necessary to reverse the inhibitory effects of DEPTOR on mTORC2 activity.
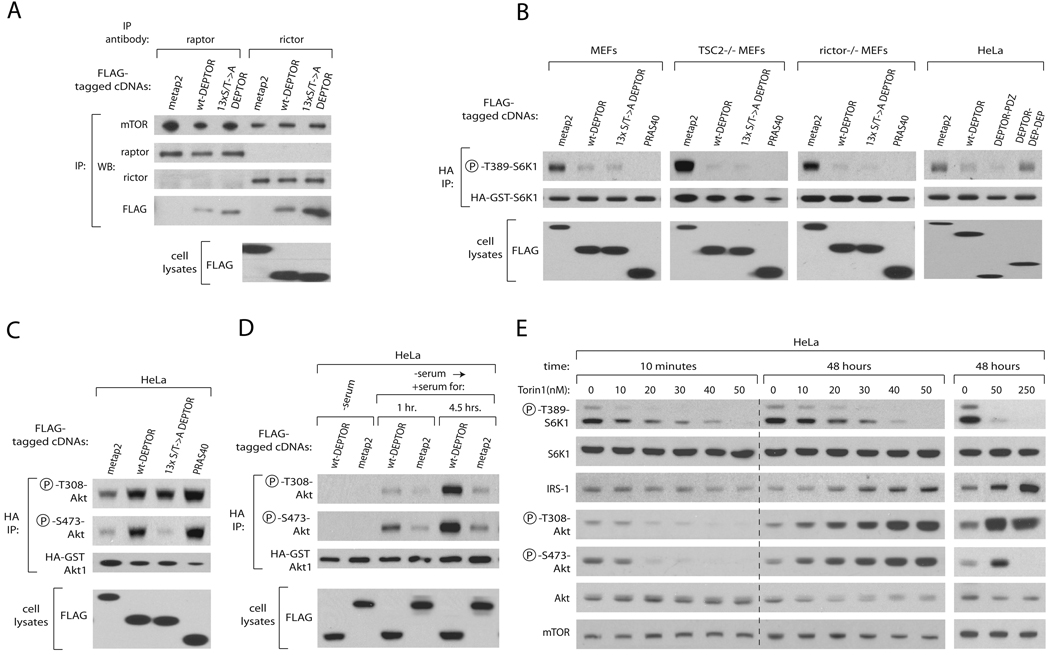
Figure 5. DEPTOR Overexpression Inhibits mTORC1 but Activates PI3K/Akt Signaling.
(A) The 13×S/T->A DEPTOR mutant binds mTORC1 and mTORC2 more tightly than wild-type DEPTOR. HEK-293T cells were transfected with 500 ng of expression vectors encoding the indicated FLAG-tagged proteins for 48 hours. Raptor or rictor immunoprecipitates were prepared from cell lysates, washed with buffers containing 150 mM NaCl, and analyzed along with cell lysates by immunoblotting for indicated proteins.
(B) Overexpression of DEPTOR inhibits T389 phosphorylation of S6K1. HeLa cells or the indicated p53−/− MEFS were cotransfected with expression plasmids encoding HA-GST-S6K1 as well as the indicated FLAG-tagged proteins. Cell lysates were prepared 24 hours after transfection and were analyzed by immunoblotting for the levels of the indicated proteins and phosphorylation states.
(C) Overexpression of DEPTOR activates T308 and S473 Akt1 phosphorylation. HeLa cells were cotransfected with expression plasmids encoding HA-GST-Akt1 as well as the indicated FLAG-tagged proteins. Cell lysates were analyzed as in (B).
(D) Overexpression of DEPTOR activates T308 and S473 Akt1 phosphorylation only in serum-replete cells. HeLa cells were transfected as in (B), serum starved for 24 hours and stimulated for the indicated times and cell lysates were analyzed as in (B).
(E) Partial inhibition of mTOR with Torin1 inhibits S6K1 phosphorylation but activates PI3K/mTORC2/Akt signaling. HeLa cells were treated with the specified concentrations of Torin1 or vehicle for either 10 minutes or 48 hours and cell lysates were analyzed as in (B).
DEPTOR Overexpression Inhibits mTORC1 but Activates PI3K/Akt Signaling
Because our loss-of-function experiments indicate that DEPTOR is an mTOR inhibitor, we hypothesized that DEPTOR overexpression should suppress both the mTORC1 and mTORC2 pathways. Indeed, transient overexpression of wild-type DEPTOR, but not a control protein, in MEFs reduced the phosphorylation of T389 of co-expressed S6K1, an established mTORC1 substrate, to a similar degree as overexpression of PRAS40, a known mTORC1 inhibitor (Figure 5B). DEPTOR overexpression also strongly inhibited mTORC1 in MEFs lacking TSC2 or rictor (Figure 5B). Overexpression of just the PDZ domain of DEPTOR, the region of DEPTOR that binds mTOR (Figure 1H), but not of the tandem DEP domains of DEPTOR, also inhibited mTORC1 signaling (Figure 5B).
In contrast to the expected effects we observed with regards to mTORC1, DEPTOR overexpression led to an apparent increase instead of inhibition of mTORC2 signaling as monitored by Akt S473 phosphorylation (Figures 5C and Figure S4). We suspected that because DEPTOR overexpression inhibits mTORC1, it relieves the inhibitory feedback signal normally transmitted from mTORC1 to PI3K. This would lead to hyperactive PI3K signaling, which then overcomes the direct inhibitory effects of DEPTOR on mTORC2, just as the hyperactive rheb found in TSC2 null cells can overcome the inhibitory effects of PRAS40 on mTORC1 (Sancak et al., 2007). Consistent with this interpretation, overexpression of DEPTOR, like that of PRAS40, not only caused an increase in the phosphorylation of Akt at S473 but also at T308, the PDK1-catalyzed site which serves as a read-out of PI3K activity (Alessi, 2001) (Figure 5C and Figure S4). The activation of Akt by DEPTOR overexpression was not observed in the absence of serum, as expected from the known requirement for growth factors for mTORC1 to PI3K feedback signaling (O'Reilly et al., 2006) (Figure 5D). As was the case with wild-type DEPTOR, overexpression of the non-phosphorylatable DEPTOR mutant inhibited mTORC1 signaling (Figure 5B) and triggered feedback activation of PI3K as detected by T308 phosphorylation of Akt (Figure 5C). However, unlike wild-type DEPTOR, the 13×S/T->A DEPTOR mutant did not promote Akt phosphorylation on S473, indicating that PI3K activation cannot activate mTORC2 when DEPTOR cannot be phosphorylated (Figure 5C).
We propose that the effects of DEPTOR overexpression on the mTOR pathway—inhibition of mTORC1 signaling but activation of mTORC2 signaling— result from DEPTOR-mediated inhibition of mTORC1, which then leads to the hyperactivation of PI3K and Akt. In agreement with this model, in many cell types treatment with rapamycin or overexpression of PRAS40 (Figure 5C) has similar effects on mTOR signaling as DEPTOR overexpression (O'Reilly et al., 2006; Sarbassov et al., 2006). However, both rapamycin and PRAS40 are predominantly mTORC1 inhibitors, while our loss of function data indicates that DEPTOR inhibits mTORC1 and mTORC2. To gain evidence that a dual mTORC1 and mTORC2 inhibitor can also lead to a stable cell state characterized by inhibition of mTORC1 signaling but and activation of PI3K/mTORC2/Akt signaling we explored the time- and dose-dependent effects of the mTOR inhibitor Torin1 (Thoreen et al., 2009). As expected, acute Torin1 treatment of cells for 10 minutes led to a dose-dependent inhibition of both mTORC1 and mTORC2 signaling, as monitored by S6K1 and Akt phosphorylation, respectively (Figure 5E). By 48 hours of Torin1 treatment, however, mTORC1 signaling was still inhibited but was now accompanied by increased T308 and S473 Akt phosphorylation and IRS-1 levels, molecular phenotypes consistent with the loss of the inhibitory feedback signal from mTORC1 to the PI3K pathway (Figure 5E). At a higher concentration (250 nM) of Torin1, Akt T308 phosphorylation and IRS-1 expression were very high indicating that, at this dose, PI3K signaling was still hyperactive. However, like overexpression of a non-phosphorylatable DEPTOR mutant (Figure 5C), the higher dose of Torin1 eliminated Akt S473 phosphorylation, revealing that mTORC2 was now fully inhibited. PP242, a distinct ATP-competitive inhibitor of mTOR (Feldman et al., 2009), had similar time- and dose-dependent effects on mTOR and PI3K signaling as Torin1 (Figure S5). Thus, because of the inhibitory feedback signal from mTORC1 to the PI3K pathway, partial mTOR inhibition caused by either DEPTOR overexpression or chemical inhibitors leads to an asymmetrical state of mTOR signaling characterized by an inhibition of mTORC1 but an activation of mTORC2-dependent outputs.
In a Subset of Multiple Myelomas DEPTOR Overexpression is Necessary for Akt Activation and Cell Survival
The dynamic nature of DEPTOR expression and the interesting consequences of DEPTOR overexpression on mTORC1 and PI3K/mTORC2/Akt signaling prompted us to examine the levels of DEPTOR mRNA in databases of transcriptional profiles of human tumors and cancer cell lines. Consistent with mTORC1 and mTORC2 being active in many cancers (reviewed in (Sabatini, 2006)), the levels of DEPTOR mRNA were significantly lower in most cancer types compared to the normal tissues from which they are derived (Figure 6A, Table S1). Anomalously, this was not the case for Multiple Myelomas (MM), a malignancy of antibody-producing plasma cells (reviewed in (Kuehl and Bergsagel, 2002)), in which the DEPTOR mRNA is overexpressed (Figure 6A, Table S1).
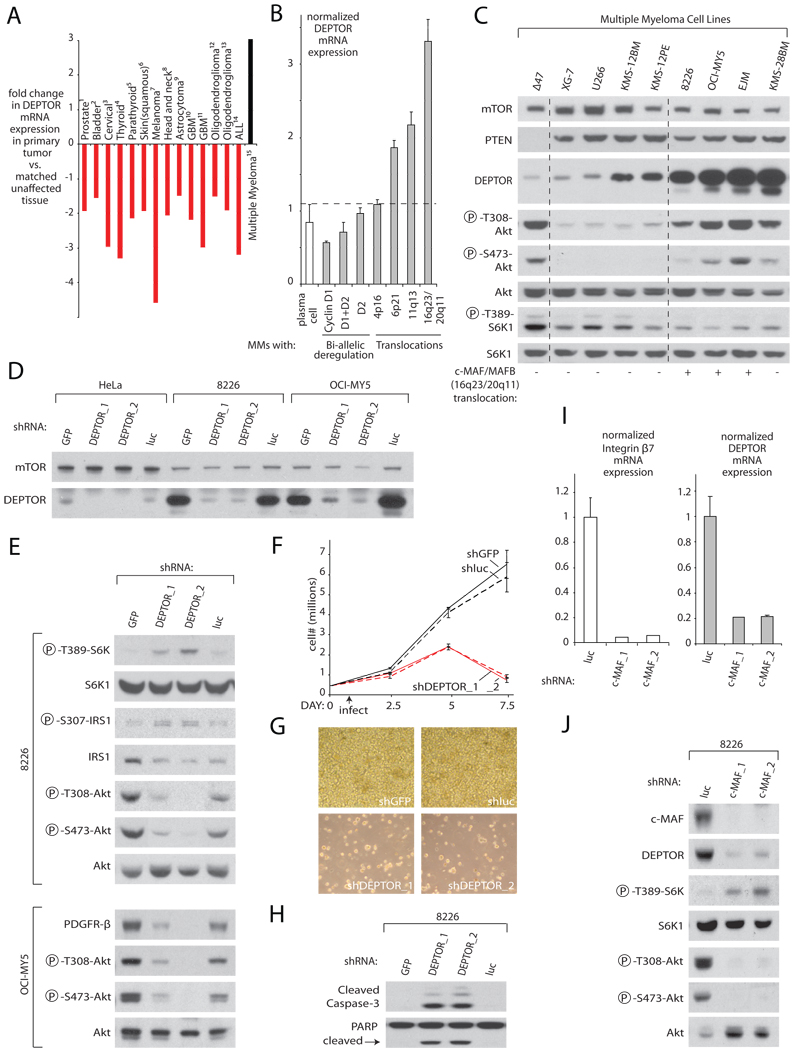
Figure 6. DEPTOR is Overexpressed in a Subset of Multiple Myeloma Cells and in these Cells Activates PI3K/Akt signaling and Promotes Cell Survival.
(A) DEPTOR mRNA expression is downregulated in most cancers, yet is up-regulated in Multiple Myelomas. Relative DEPTOR mRNA expression in various cancer types versus matched unaffected tissue was collated from publicly available microarray studies. For all studies shown, p<0.05 for DEPTOR mRNA downregulation or upregulation by one-tailed T-test.
(B) DEPTOR mRNA is mostly highly expressed in the subset of Multiple Myelomas which possess Cyclin D1/D3 or c-MAF/MAFB translocations. DEPTOR mRNA levels were normalized by the summed mean values of the housekeeping genes GAPDH, β-actin, and FIP (7 total probe-sets). Error bars indicate standard error for n=7. The differences in mean DEPTOR mRNA levels between plasma cells and MMs with Cyclin D1, Cyclin D3, or c-MAF/MAFB translocations are significant to at least p<0.005.
(C) Human Multiple Myeloma cell lines with high DEPTOR protein expression have activated PI3K and suppressed mTORC1 signaling. Cell lysates were analyzed by immunoblotting for the levels of the indicated proteins and phosphorylation states. Cell lines with known translocations of c-MAF or MAFB are indicated.
(D) RNAi-mediated knockdown of DEPTOR in 8226 and OCI-MY5 cells reduces DEPTOR expression to levels similar to those in HeLa cells. Five days after transductions with shRNA-expressing lentiviruses, cells were lysed and as in (C).
(E) DEPTOR suppression in 8226 and OCI-MY5 cells activates mTORC1 signaling, inhibits IRS-1 or PDGFR-β expression, and reduces PI3K/mTORC2/Akt signaling. Five days after transduction with shRNA-expressing lentiviruses, cells were lysed and analyzed as in (C).
(F) DEPTOR suppression markedly decreases 8226 cell number. Cell number was measured at indicated time points with a Coulter Counter. Error bars indicate mean ± standard deviation for n=3 samples per time point.
(G) Representative 20× light microscopy images of 8226 cells expressing shRNAs targeting DEPTOR, GFP, or luciferase at day 8 post-infection.
(H) DEPTOR suppression causes apoptosis. Three days after transduction with shRNA-expressing lentiviruses, 8226 cells were lysed and analyzed as in (C). (I) A reduction in c-MAF expression decreases DEPTOR mRNA levels. Five days after transduction of 8226 cells with lentiviruses expressing the indicated shRNAs, DEPTOR and Integrin β7 mRNA levels were measured by qRT-PCR and normalized to GADPH mRNA levels. Error bars indicate standard deviation for n=3.
(J) A c-MAF knockdown activates mTORC1 and downregulates PI3K signaling. Five days after transductions with shRNA-expressing lentiviruses, lysates of 8226 cells were analyzed as in (C).
To expand upon these findings, we measured DEPTOR mRNA levels in a collection of 581 human Multiple Myelomas and found that 28% (160/581) of the MMs had mRNA levels that were 4-fold or greater than those in normal plasma cells (see methods and Table S1). Multiple Myelomas can be grouped into two broad types: (1) hyperdiploid MMs, which are associated with multiple trisomies of 8 chromosomes and bi-allelically deregulate Cyclin D1/D2 mRNA expression, and (2) non-hyperdiploid MMs, which mono-allelically deregulate Cyclin D1/D3, c-MAF/MAFB, or FGFR3/MMSET expression through translocation events involving these genes (Bergsagel et al., 2005) (In Figure 6B these two types are referred to as “Bi-allelic deregulation” and “Translocation”). We noted that many of the MMs with high levels of DEPTOR mRNA were of the non-hyperdiploid type and had translocations involving the genes for Cyclin D1 or D3, or, in particular, for the c-MAF or MAFB transcription factors (Table S1). In these MMs, the mean level of DEPTOR mRNA is significantly higher than in those without the translocations (Figure 6B).
We next examined DEPTOR mRNA and protein expression and mTORC1 and PI3K/mTORC2/Akt signaling in a set of MM cell lines. Many MM cell lines, particularly those with c-MAF/MAFB translocations, have levels of DEPTOR protein and mRNA expression that are many folds greater than those found in non-MM cancer cell lines, such as HeLa and PC3 cells (Figures 6D, S6A–B, and Table S1). Consistent with our exogenous DEPTOR overexpression studies (Figure 5), DEPTOR expression across the cell lines correlated positively with the phosphorylation of Akt on T308 and S473 (PDK1/PI3K and mTORC2 outputs, respectively), but negatively with S6K1 T389 phosphorylation (an mTORC1 output) (Figure 6C). The sole exception was the Δ47 cell line, which is one of the few MM cell lines that lack PTEN and thus has high levels of S6K1 and Akt phosphorylation (Figure 6C). Remarkably, the MM cell lines with high DEPTOR expression had levels of S473, and, particularly, T308 Akt phosphorylation comparable to those in the PTEN-null Δ47 cell line.
As PTEN inactivation and PI3K-activating mutations are rare in MM (Chang et al., 2006) (Kuehl et al., unpublished), we considered the possibility that overexpression of endogenous DEPTOR might also be a mechanism to promote activation of PI3K and Akt and cell survival in a subset of human MMs. To test this, we used RNAi to suppress the very high levels of DEPTOR expression in 8226 and OCI-MY5 Multiple Myeloma cell lines to levels comparable to those in HeLa cells (Figure 6D). DEPTOR suppression led to an increase in mTORC1 activity as detected by S6K1 T389 phosphorylation, IRS1 levels, and IRS1 phosphorylation, but to a drop in PI3K/PDK1 and mTORC2 outputs as measured by T308 and S473 Akt phosphorylation, respectively (Figure 6E). Even though the DEPTOR knockdown decreased the cellular levels of phospho-S473 Akt, it caused an increase in the in vitro kinase activity of mTORC2 isolated from the DEPTOR knockdown cells (Figure S7), a result consistent with DEPTOR being a negative regulator of mTORC2 kinase activity (Figure 2D). DEPTOR suppression had similar inhibitory effects on T308 and S473 Akt phosphorylation in OCI-MY5 cells except that in this cell line IRS-1 was not detected and decreased Akt phosphorylation instead correlated instead with a loss of PDGFR-β levels (Figure 6E, see also Figure S8). Lastly, co-knockdown of raptor with DEPTOR partially reversed the effects of the DEPTOR knockdown on T308 and S473 Akt phosphorylation, suggesting that mTORC1 activation mediates the reduction in PI3K signaling seen in the 8226 cells upon a reduction in DEPTOR expression (Figure S9).
Consistent with the pro-survival role of PI3K/Akt signaling, suppression of DEPTOR in 8226 cells abolished cell proliferation over a 7-day culture period (Figure 6F) and caused apoptosis as detected by the presence of numerous cell fragments in the culture media (Figure 6G) and increased cleaved caspase-3 and PARP (Figure 6H). Thus, in a subset of Multiple Myeloma cell lines, high expression of DEPTOR contributes to Akt activation and is an endogenous mechanism for maintaining cell survival.
Because PI3K/mTORC2/Akt signaling represses DEPTOR mRNA levels (Figures 4F–G and Figures S2A–B), we wondered how certain Multiple Myeloma cells can keep DEPTOR at elevated levels in the face of hyperactive PI3K signaling. DEPTOR mRNA levels are highest in MM tumors and cell lines in which chromosomal translocations cause the overexpression of c-MAF or MAFB. Moreover, a transcriptional profiling study shows that DEPTOR is highly induced upon forced MAFB overexpression (Hurt et al., 2004; van Stralen et al., 2009). To test if deregulated overexpression of c-MAF or MAFB drives DEPTOR expression, we used RNAi to reduce c-MAF expression in 8226 cells, a MM cell line with a translocation involving the c-MAF gene (Chesi et al., 1998). Indeed, loss of c-MAF substantially diminished DEPTOR mRNA and protein levels and, by activating mTORC1 and inhibiting mTORC2 outputs, recapitulated the effects of a DEPTOR knockdown on the mTOR pathway (Figure 6I, 6J). As expected, the knockdown of c-MAF decreased the expression of the mRNA for Integrin β7, a known c-MAF target (Figure 6I). These results are consistent with deregulated overexpression of c-MAF or MAFB driving DEPTOR expression to high levels and, thus, hyperactivating PI3K signaling.
DISCUSSION
Our loss of function data indicate that DEPTOR inhibits both the mTORC1 and mTORC2 pathways. However, by inhibiting mTORC1, DEPTOR overexpression relieves mTORC1-mediated inhibition of PI3K, causing an activation of PI3K and, paradoxically, of mTORC2-dependent outputs, like Akt.
mTOR interacts with DEPTOR via its PDZ domain and so far there is no information about the function of the tandem DEP domains the protein also contains. In other proteins DEP domains mediate protein-protein interactions (Ballon et al., 2006; Yu et al., 2007), but in numerous DEPTOR purifications we have failed to identify additional DEPTOR-interacting proteins besides the known components of mTORC1 and mTORC2 (data not shown). Therefore, based on our current evidence, DEPTOR appears dedicated to mTOR regulation, and we propose that in vertebrates it is likely to be involved in regulating other outputs of the mTOR signaling network besides the growth and survival pathways we have examined. The mTOR complexes and DEPTOR negatively regulate each other, suggesting the existence of a feedforward loop in which the loss of DEPTOR leads to an increase in mTOR activity, which then further reduces DEPTOR expression. This type of regulatory circuit should result in DEPTOR expression being tightly coupled to mTOR activity, and, interestingly, we have noted that DEPTOR mRNA levels strongly anti-correlate with cell size, a readout of mTORC1 activity (Figure S6B).
We find that about 28% of human Multiple Myelomas (MM) overexpress DEPTOR. Our results are consistent with a published survey of 67 MM tumors and 43 MM cell lines, in which 21% were shown to possess copy number gains and associated expression increases of the genes within a 6 Mb region of chromosome 8q24 that contains DEPTOR (Carrasco et al., 2006). Furthermore, it appears that deregulated overexpression of c-MAF and MAFB is an additional, perhaps even more prevalent, mechanism for increasing DEPTOR expression in MMs. The related c-MAF and MAFB transcription factors are expressed–frequently as the result of chromosomal translocations–in a large fraction of MMs, but not in the plasma cells from which they are derived (reviewed in (Eychene et al., 2008)). Consistent with c-MAF playing a key role in promoting DEPTOR expression, a knockdown of c-MAF in a MM cell line having a c-MAF translocation decreases the expression of DEPTOR and mimics the effects of a DEPTOR knockdown on mTOR and PI3K signaling. The levels of the DEPTOR and c-MAF or MAFB mRNAs highly correlate with each other (van Stralen et al., 2009) and, importantly, DEPTOR expression correlates with poor survival in patients with Multiple Myeloma (Carrasco et al., 2006).
In many Multiple Myeloma cell lines DEPTOR is massively overexpressed compared to the levels found in other cancer cell lines, such as HeLa cells. In these cells, the great overexpression of DEPTOR inhibits mTORC1 growth signaling and drives outputs dependent on PI3K. Interestingly, a reduction in DEPTOR expression to the lower levels seen in non-Multiple Myeloma cell lines causes cell death via apoptosis. This suggests that a pharmacologically induced reduction in DEPTOR expression or disruption of the DEPTOR-mTOR interaction could have therapeutic benefits for the treatment of Multiple Myeloma. There has been progress in developing small molecule inhibitors of protein-protein interactions mediated by PDZ domains (Daw et al., 2000; Fujii et al., 2007), so it is conceivable that blockers of the DEPTOR-mTOR interaction could be made.
Although a number of other cancer cell lines have high levels of DEPTOR (data not shown), as a class only Multiple Myelomas appear to consistently overexpress it. Besides activating PI3K/Akt signaling, DEPTOR overexpression in MM cells may provide these cells with benefits that are not relevant in other cancer types or perhaps even detrimental. For example, the high demand that MM cells place on the protein synthesis machinery to produce large amounts of immunoglobulins (reviewed in (Shapiro-Shelef and Calame, 2004)), causes a significant ER stress which renders these cells susceptible to apoptosis-induction via agents which induce further ER stress, such as proteasome inhibitors (Lee et al., 2003; Meister et al., 2007). DEPTOR overexpression, by partially inhibiting protein synthesis through the suppression of mTORC1, may reduce the levels of ER stress below the threshold that triggers apoptosis. In contrast, in other cancer cells in which ER stress is not a significant factor, DEPTOR overexpression may be selected against because reduced rates of protein synthesis may not be tolerated. That mTORC1-stimulated protein synthesis leads to ER stress is already appreciated as TSC1 or TSC2 null cells have increased sensitivity to ER stress-induced death (Ozcan et al., 2008).
It is curious that DEPTOR is overexpressed mostly in MMs characterized by chromosomal translocations instead of those which are hyperdiploid because of aneuploidy (Figure 6B) (Bergsagel et al., 2005). Elevated DEPTOR expression might be tolerated better in the non-hyperdiploid MMs because aneuploidy itself increases sensitivity to conditions, like mTORC1 inhibition, that interfere with protein synthesis (Torres et al., 2007). Moreover, the state of high mTORC2 and low mTORC1 signaling that our work indicates some MM cells prefer, cannot be achieved by mutations that activate PI3K signaling, perhaps explaining why Multiple Myelomas exhibit low rates of PTEN-inactivating or PI3K-activating mutations (Chang et al., 2006)(Kuehl et al., unpublished).
EXPERIMENTAL PROCEDURES
Materials
Reagents were obtained from the following sources: rabbit polyclonal antibodies to DEPTOR (09–463), raptor (09–217), Akt (05–591), and phospho-S877 raptor (09–107) from Upstate/Millipore; mouse monoclonal antibody to DEPTOR from Novus Biologicals; mouse monoclonal antibodies to rictor and raptor from Assay Designs; antibodies to mTOR, β-catenin, actin, S6K1, c-MAF, as well as HRP-labeled anti-mouse, anti-goat, and anti-rabbit secondary antibodies from Santa Cruz Biotechnology; antibodies to phospho-T389 S6K1, phospho-T37/T46 4E-BP1, phospho-S473 Akt/PKB, phospho-T308-Akt, phospho-T346-NDRG1, phospho-S307-IRS-1, IRS-1, PTEN, TSC2, 4E-BP1, cleaved caspase-3, PARP, PDGFR- β, and the c-MYC epitope from Cell Signaling Technology; antibodies to rictor, HA, c-MYC from Bethyl Laboratories; Flag M2 affinity gel, Flag M2 antibody and SGK1 antibodies, ATP, and SYBR Green JumpStart Taq ReadyMix from Sigma Aldrich; mouse monoclonal antibody to mTOR and recombinant IL-6 from BD Pharmingen; protein G Sepharose and anti-sheep secondary antibody from Pierce; DMEM from SAFC Biosciences; rapamycin from LC Labs; MG-132 from Biomol; PreScission protease from Amersham Biosciences; pTREQ Tet-On vector from Clontech; Adenoviral CRE and GFP from University of Iowa Gene Transfer Vector Core; FuGENE 6 and Complete Protease Cocktail from Roche; 4E-BP1 from A.G. Scientific; SuperScript II Reverse Transcriptase, Platinum Pfx Polymerase, SimplyBlue Coomassie G, Silverquest Staining kit, and inactivated fetal calf serum (IFS) from Invitrogen. An antibody to NDRG1 was kindly provided by Dario Alessi (University of Dundee, UK). We have found that the phospho-S877 raptor antibody also recognizes immunoprecipitated phosphorylated DEPTOR and can be used to read out the DEPTOR phosphorylation state.
Cell Lines and Cell Culture
HEK-293E cells were kindly provided by John Blenis (Harvard Medical School). HeLa, HEK-293E, HEK-293T, HT-29, U87, PC3, MD-MBA-435, and MEFs were cultured in DMEM with 10% Inactivated Fetal Bovine Serum (IFS). The Human Multiple Myeloma cell lines: FR4, XG-7, U266, KMS-12BM, KMS-12PE, PE2, 8226, OCI-MY5, KMS-28BM were provided by the Kuehl lab. The EJM, MM-1S, JJN-3, and Δ47 Human Multiple Myeloma cell lines were kindly provided by Ken Anderson (Dana Farber Cancer Institute). Human Multiple Myeloma cell lines were cultured in RPMI-1640 with 10% Fetal Bovine Serum (FBS) supplemented with 2 mM glutamine. XG-7 cells were additionally supplemented with 2 ng/ml IL-6. TSC2−/−, p53−/− and TSC2+/+, p53−/− MEFs were kindly provided by David Kwitakowski (Harvard Medical School). PTEN LoxP/LoxP MEFs were generated from PTEN LoxP/LoxP mice kindly provided by Hong Wu (UCLA). To produce PTEN −/− and PTEN +/+ MEFs, 1 µl of Adenoviral CRE or Adenoviral GFP at a titer of 1×1010 PFU/mL was added to 500,000 PTEN LoxP/LoxP MEFs. Cell lysates were generated 5 days post-infection. The HeLa cell line with doxycycline-inducible DEPTOR expression was generated by retroviral transduction of HeLa that were previously modified to express rtTA with an inducible DEPTOR cDNA.
cDNA Manipulations, Mutagenesis, and Sequence Alignments
The cDNA for DEPTOR (NCBI gene symbol: DEPDC6) in the pCMV6-XL4 vector was obtained from Origene. The DEPTOR cDNA was amplified by PCR and the product subcloned into the Sal 1 and Not 1 sites of pRK5, the Xho I and Not I sites of pMSCV, or the BsiWI and BstB I sites of pTREQ. All constructs were verified by DNA sequencing. The DEPTOR cDNA in pRK5 was mutagenized using the QuikChange XLII mutagenesis kit (Stratagene) with oliogonucleotides obtained from Integrated DNA Technologies. NCBI Blosum62 Blast searches were used to identify blocks of similar sequence between DEPTOR orthologues. Amino acid sequence alignment of the phosphorylated region of DEPTOR was performed using ClustalX v1.81.
Mass Spectrometric Analysis
mTOR or FLAG immunoprecipitates prepared from 30 million HEK-293E cells were resolved by SDS-PAGE, Coomassie G-stained, and gel bands were excised and processed as described in (Sancak et al., 2007). In 4 independent experiments, a total of 12 peptides corresponding to DEPTOR were identified while no DEPTOR peptides were identified in control purifications.
DEPTOR phosphorylation sites were identified by mass spectrometry of trypsin digested FLAG-DEPTOR purified from HEK-293E or HEK-293T cells stably or transiently over-expressing FLAG-DEPTOR. All putative phosphorylated residues on DEPTOR (highlighted in BOLD) were detected on the following peptides (amino acid position according to NCBI DEPDC6 Protein Sequence NP_073620):
234 KSPSSQETHDSPFCLR 249
257 KSTSFMSVSPSK 268
278 RSSMSSCGSSGYFSSSPTLSSSPPVLCNPK 311
Label-free quantification of DEPTOR phosphorylation sites were performed using BioWorks Rev3.3 software following methodology utilized in (Stokes et al., 2007).
Mammalian Lentiviral shRNAs
Lentiviral shRNAs to human raptor, rictor, and mTOR were previously described (Sarbassov et al., 2005b). All other shRNAs were obtained from the collection of The RNAi Consortium (TRC) at the Broad Institute (Moffat et al., 2006). These shRNAs are named with the numbers found at the TRC public website:
(http://www.broad.mit.edu/genome_bio/trc/publicSearchForHairpinsForm.php)
Human DEPTOR_1 shRNA: TRC candidate; NM_022783.1-877s1c1
Human DEPTOR_2 shRNA: TRC candidate; NM_022783.1-1101s1c1
Human TSC2_1 shRNA: TRCN0000040178; NM_000548.2-4551s1c1
Human PTEN_1 shRNA: TRCN0000002746; NM_000314.×-1320s1c1
Mouse DEPTOR_1 shRNA: TRCN0000110157; NM_145470.1-1164s1c1
Mouse DEPTOR_2 shRNA: TRCN0000110159; NM_145470.1-1165s1c1
Mouse TSC2_1 shRNA: TRCN0000042727; NM_011647.1-1843s1c1
Human SGK1_1 shRNA: TRCN0000040175; NM_005627.2-964s1c1
Human SGK1_2 shRNA: TRCN0000040176; NM_005627.2-252s1c1
Human c-MAF_1 shRNA: TRCN0000000255; NM_005360.×-1839s1c1
Human c-MAF_2 shRNA: TRCN0000000257; NM_005360.×-1067s1c1
shRNA-encoding plasmids were co-transfected with the Delta VPR envelope and CMV VSV-G packaging plasmids into actively growing HEK-293T using FuGENE 6 transfection reagent as previously described (Sarbassov et al., 2005b). Virus containing supernatants were collected at 48 hours after transfection, filtered to eliminate cells, and target cells (e.g., 300,000 HeLa cells or 500,000 8226 cells) infected in the presence of 8 µg/ml polybrene. For 8226 cells, infected cells were spun at 300g for 1.5 hours before incubating at 37°C for 24 hours. For all cell types, 24 hours after infection, the cells were split into fresh media (e.g., DMEM/10%IFS for HeLa/MEFs; RPMI/10%FBS for 8226/OCI-MY5), selected with 1 µg/ml puromycin. Five days post-infection, shRNA-expressing cells were analyzed or split again and analyzed 2–3 days later. For adherent cell lines, shRNA-expressing cells were analyzed at 50–75% confluence.
Cell Lysis and Immunoprecipitations
All cells were rinsed with ice-cold PBS before lysis. All cells, with the exception of those used to isolate mTOR-containing complexes, were lysed with Triton-X 100 containing lysis buffer (40 mM HEPES [pH 7.4], 2 mM EDTA, 10 mM sodium pyrophosphate, 10 mM sodium glycerophosphate, 150 mM NaCl, 50 mM NaF, 1% Triton-X 100, and one tablet of EDTA-free protease inhibitors [Roche] per 25 ml). The soluble fractions of cell lysates were isolated by centrifugation at 13,000 arpm for 10 min in a microcentrifuge. For immunoprecipitations, primary antibodies were added to the lysates and incubated with rotation for 1.5 hr at 4°C. A 50% slurry of protein G Sepharose (60 µl) was then added, and the incubation continued for an additional 1 hr. Immunoprecipitated proteins were denatured by the addition of 20 µl of sample buffer and boiling for 5 min, resolved by 4%–12% SDS-PAGE, and analyzed by immunoblotting as described (Kim et al., 2002). To observe gel mobility shifting in DEPTOR, 8% Tris Glycine gels (Invitrogen) were used. For all other applications, 4–12% Bis-Tris gels (Invitrogen) were used. For immunoprecipitations of mTOR containing complexes, cells were lysed in ice-cold CHAPS-containing lysis buffer lacking added NaCl (40 mM HEPES [pH 7.4], 2 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 0.3% CHAPS, and one tablet of EDTA-free protease inhibitors [Roche] per 25 ml). Immunoprecipitates were washed once each in the CHAPS lysis buffer and twice with CHAPS lysis buffer containing 150 mM NaCl such that the immunopurified material would be rinsed in a solution with a phyisiologically-relevant salt concentration. When specified, the latter two washes contained 0 mM or 500 mM NaCl.
In Vitro Kinase Assay For mTORC1 and mTORC2 Activities
For kinase assays, immunoprecipitates were washed once in CHAPS lysis buffer followed by two additional washes in CHAPS lysis buffer containing 150 mM NaCl. Immunoprecipitates were then washed twice in 25 mM HEPES (pH 7.4), 20 mM KCl. Kinase assays were performed as described (Sancak et al., 2007).
cDNA Transfection-based Experiments
To examine the effects on mTOR signaling of DEPTOR overexpression, 500,000 HeLa; 300,000 p53−/− or rictor −/−, p53−/−; or 200,000 TSC2−/−, p53−/− regularly passaged (every 2–3 days) cells were plated in 6 cm culture dishes in DMEM/10%IFS. 12 hours later, cells were transfected with the pRK5-based cDNA expression plasmids indicated in the figures in the following amounts: 2 µg for all FLAG tagged cDNAs and 200 ng of HA-GST S6k1 or Akt1. All cells were lysed at 50–75% confluence 24 hours after transfection.
Gene Expression and Mutation Analysis in Human Cancers and Cancer Cell Lines
For quantification of DEPTOR, Integrin β7, and GAPDH mRNA expression in HeLa, PC3, or 8226 cell lines, total RNA was isolated from cells grown in the indicated conditions and reverse-transcription was performed. The resulting cDNA was diluted in DNase-free water (1:25) before quantification by real-time PCR. mRNA transcript levels were measured using Applied Biosystems 7900HT Sequence Detection System v2.3 software. Data are expressed as the ratio between the expression of DEPTOR or Integrin β7 and the housekeeping gene GAPDH. The following primers were used for quantitative real-time PCR:
DEPTOR (H. sapiens):
Forward: TTTGTGGTGCGAGGAAGTAA
Reverse: CATTGCTTTGTGTCATTCTGG
GAPDH (H. sapiens):
Forward: CTCTCTGCTCCTCCTGTTCGAC
Reverse: TGAGCGATGTGGCTCGGCT
Integrin β7 (H. sapiens):
Forward: TGGAGCGCTGCCAGTCACCATT
Reverse: CGTCTGAAGTGAACACCAGCAGC
For meta-analysis of DEPTOR mRNA expression in human cancers, “DEPDC6” was searched in NCBI GEO and Oncomine gene expression data repositories. Only those studies where data from primary human tumors could be compared with matched unaffected tissue were considered further. Fold change in DEPTOR mRNA was measured by taking the quotient of the mean level of DEPTOR mRNA in unaffected tissue versus that of the tumor sample. Statistical significance was measured by one-tailed, unequal variance T test. Only those studies with p<0.05 were included in the final analysis.
RNA isolation from primary Multiple Myelomas has been described (Zhan et al., 2002). Normalized DEPTOR mRNA expression was clustered according to the translocation/Cyclin D groups classified in (Bergsagel et al., 2005). The Multiple Myeloma gene expression data used in this study was generated on an Affymetrix U133_Plus_2 platform and can be found in its entirety in the NCBI GEO database with the following identifiers: GSE2658 for 559 newly diagnosed, untreated tumors and GSE5900 for 22 normal plasma cells, 12 smoldering myeloma, and 44 MGUS. Human Myeloma cell line mRNA data have been deposited in an MMRC genomics portal website that is sponsored by the MMRF (www.broad.mit.edu/mmgp).
PTEN mutation status in human cancer cell lines can be found at the following URL: http://www.sanger.ac.uk/genetics/CGP/CellLines/.
Live Cell Imaging
8226 cells grown in 12 well dishes were imaged at 20× magnification using a Canon Powershot 5 Megapixel digital camera.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Thijn Brummelkamp for reviewing the manuscript and the following for technical assistance: Leslie Brents, Stephen Carr, Jake Jaffe, Xana Frias, Heather Keys, Stephanie Kinkel, Doug McMillin, Jan Reiling, Eric Spooner, Ed Van Veen, and Marcel Van Vugt. We thank members of the Sabatini lab for helpful discussions. This work was supported by grants from the National Institutes of Health (R01 AI47389 and R01 CA103866) to D.M.S.; awards from the Keck Foundation and LAM Foundation to D.M.S.; a fellowship from the American Diabetes Association to T.R.P.; a fellowship from the Canadian Institutes of Health Research to M.L.; and a fellowship from the American Cancer Society to S.A.K. N.S.G. is funded in part by the Dana Farber Cancer Institute High-Tech Research Fund. D.M.S. is an investigator of the Howard Hughes Medical Institute. None of the authors have a conflict of interest related to the work reported in this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alessi DR. Discovery of PDK1, one of the missing links in insulin signal transduction. Colworth Medal Lecture. Biochem Soc Trans. 2001;29:1–14. doi: 10.1042/0300-5127:0290001. [DOI] [PubMed] [Google Scholar]
- Ballon DR, Flanary PL, Gladue DP, Konopka JB, Dohlman HG, Thorner J. DEP-domain-mediated regulation of GPCR signaling responses. Cell. 2006;126:1079–1093. doi: 10.1016/j.cell.2006.07.030. [DOI] [PubMed] [Google Scholar]
- Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J., Jr Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106:296–303. doi: 10.1182/blood-2005-01-0034. Epub 2005 Mar 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliskovsky V, Ramsay ES, Scott J, DuBois W, Shi W, Zhang S, Qian X, Lowy DR, Mock BA. Frap, FKBP12 rapamycin-associated protein, is a candidate gene for the plasmacytoma resistance locus Pctr2 and can act as a tumor suppressor gene. Proc Natl Acad Sci U S A. 2003;100:14982–14987. doi: 10.1073/pnas.2431627100. Epub 12003 Nov 14921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. PNAS. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27:5527–5541. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, Stewart JP, Zhan F, Khatry D, Protopopova M, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–325. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Chang H, Qi XY, Claudio J, Zhuang L, Patterson B, Stewart AK. Analysis of PTEN deletions and mutations in multiple myeloma. Leuk Res. 2006;30:262–265. doi: 10.1016/j.leukres.2005.07.008. Epub 2005 Aug 2019. [DOI] [PubMed] [Google Scholar]
- Chen S, Hamm HE. DEP domains: More than just membrane anchors. Dev Cell. 2006;11:436–438. doi: 10.1016/j.devcel.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Chesi M, Bergsagel PL, Shonukan OO, Martelli ML, Brents LA, Chen T, Schrock E, Ried T, Kuehl WM. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood. 1998;91:4457–4463. [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. Epub 12008 Oct 17427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Brunet A, Greenberg ME. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- Daw MI, Chittajallu R, Bortolotto ZA, Dev KK, Duprat F, Henley JM, Collingridge GL, Isaac JT. PDZ proteins interacting with C-terminal GluR2/3 are involved in a PKC-dependent regulation of AMPA receptors at hippocampal synapses. Neuron. 2000;28:873–886. doi: 10.1016/s0896-6273(00)00160-4. [DOI] [PubMed] [Google Scholar]
- Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, Greenberg ME. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- Eychene A, Rocques N, Pouponnot C. A new MAFia in cancer. Nat Rev Cancer. 2008;14:14. doi: 10.1038/nrc2460. [DOI] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active-Site Inhibitors of mTOR Target Rapamycin-Resistant Outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii N, You L, Xu Z, Uematsu K, Shan J, He B, Mikami I, Edmondson LR, Neale G, Zheng J, et al. An antagonist of dishevelled protein-protein interaction suppresses beta-catenin-dependent tumor cell growth. Cancer Res. 2007;67:573–579. doi: 10.1158/0008-5472.CAN-06-2726. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Alessi DR. mTOR complex-2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum and glucocorticoid induced protein kinase-1 (SGK1) Biochem J. 2008;17:17. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Hong F, Larrea MD, Doughty C, Kwiatkowski DJ, Squillace R, Slingerland JM. mTOR-raptor binds and activates SGK1 to regulate p27 phosphorylation. Mol Cell. 2008;30:701–711. doi: 10.1016/j.molcel.2008.04.027. [DOI] [PubMed] [Google Scholar]
- Hurt EM, Wiestner A, Rosenwald A, Shaffer AL, Campo E, Grogan T, Bergsagel PL, Kuehl WM, Staudt LM. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell. 2004;5:191–199. doi: 10.1016/s1535-6108(04)00019-4. [DOI] [PubMed] [Google Scholar]
- Jemth P, Gianni S. PDZ domains: folding and binding. Biochemistry. 2007;46:8701–8708. doi: 10.1021/bi7008618. Epub 2007 Jul 8710. [DOI] [PubMed] [Google Scholar]
- Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. 2002;2:175–187. doi: 10.1038/nrc746. [DOI] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci U S A. 2003;100:9946–9951. doi: 10.1073/pnas.1334037100. Epub 2003 Aug 9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Logsdon MN, Lipovsky AI, Abbott D, Kwiatkowski DJ, Cantley LC. Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2. Genes Dev. 2005;19:1773–1778. doi: 10.1101/gad.1314605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister S, Schubert U, Neubert K, Herrmann K, Burger R, Gramatzki M, Hahn S, Schreiber S, Wilhelm S, Herrmann M, et al. Extensive immunoglobulin production sensitizes myeloma cells for proteasome inhibition. Cancer Res. 2007;67:1783–1792. doi: 10.1158/0008-5472.CAN-06-2258. [DOI] [PubMed] [Google Scholar]
- Murray JT, Campbell DG, Morrice N, Auld GC, Shpiro N, Marquez R, Peggie M, Bain J, Bloomberg GB, Grahammer F, et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, Hotamisligil GS. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell. 2008;29:541–551. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005a;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005b;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Calame K. Plasma cell differentiation and multiple myeloma. Curr Opin Immunol. 2004;16:226–234. doi: 10.1016/j.coi.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Skeen JE, Bhaskar PT, Chen CC, Chen WS, Peng XD, Nogueira V, HahnWindgassen A, Kiyokawa H, Hay N. Akt deficiency impairs normal cell proliferation and suppresses oncogenesis in a p53-independent and mTORC1-dependent manner. Cancer Cell. 2006;10:269–280. doi: 10.1016/j.ccr.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. Epub 12007 Dec 19856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier M, Woodgett JR. Serum and glucocorticoid-regulated protein kinases: variations on a theme. J Cell Biochem. 2006;98:1391–1407. doi: 10.1002/jcb.20894. [DOI] [PubMed] [Google Scholar]
- Tewari M, Quan LT, O'Rourke K, Desnoyers S, Zeng Z, Beidler DR, Poirier GG, Salvesen GS, Dixit VM. Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mTOR inhibitor reveals rapamycin-insensitive functions of mTORC1. J Biol Chem. 2009;15:15. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Tsubuki S, Kawasaki H, Saito Y, Miyashita N, Inomata M, Kawashima S. Purification and characterization of a Z-Leu-Leu-Leu-MCA degrading protease expected to regulate neurite formation: a novel catalytic activity in proteasome. Biochem Biophys Res Commun. 1993;196:1195–1201. doi: 10.1006/bbrc.1993.2378. [DOI] [PubMed] [Google Scholar]
- van Stralen E, van de Wetering M, Agnelli L, Neri A, Clevers HC, Bast BJ. Identification of primary MAFB target genes in multiple myeloma. Exp Hematol. 2009;37:78–86. doi: 10.1016/j.exphem.2008.08.006. Epub 2008 Nov 2013. [DOI] [PubMed] [Google Scholar]
- Yu A, Rual JF, Tamai K, Harada Y, Vidal M, He X, Kirchhausen T. Association of Dishevelled with the clathrin AP-2 adaptor is required for Frizzled endocytosis and planar cell polarity signaling. Dev Cell. 2007;12:129–141. doi: 10.1016/j.devcel.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan F, Hardin J, Kordsmeier B, Bumm K, Zheng M, Tian E, Sanderson R, Yang Y, Wilson C, Zangari M, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–1757. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- Zhang H, Bajraszewski N, Wu E, Wang H, Moseman AP, Dabora SL, Griffin JD, Kwiatkowski DJ. PDGFRs are critical for PI3K/Akt activation and negatively regulated by mTOR. J Clin Invest. 2007;117:730–738. doi: 10.1172/JCI28984. Epub 2007 Feb 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.