Summary
GTPases of the Rab1 subclass are essential for membrane traffic between the endoplasmic reticulum (ER) and Golgi complex in animals, fungi and plants. Rab1-related proteins in higher plants are unusual because sequence comparisons divide them into two putative subclasses, Rab-D1 and Rab-D2, that are conserved in monocots and dicots. We tested the hypothesis that the Rab-D1 and Rab-D2 proteins of Arabidopsis represent functionally distinct groups. RAB-D1 and RAB-D2a each targeted fluorescent proteins to the same punctate structures associated with the Golgi stacks and trans-Golgi-network. Dominant-inhibitory N121I mutants of each protein inhibited traffic of diverse cargo proteins at the ER but they appeared to act via distinct biochemical pathways as biosynthetic traffic in cells expressing either of the N121I mutants could be restored by coexpressing the wild-type form of the same subclass but not the other subclass. The same interaction was observed in transgenic seedlings expressing RAB-D1 [N121I]. Insertional mutants confirmed that the three Arabidopsis Rab-D2 genes were extensively redundant and collectively performed an essential function that could not be provided by RAB-D1, which was non-essential. However, plants lacking RAB-D1, RAB-D2b and RAB-D2c were short and bushy with low fertility, indicating that the Rab-D1 and Rab-D2 subclasses have overlapping functions.
Keywords: AtRab1b, ARA5, Ypt1, Rab GTPases, pOp6/LhGR
Introduction
Membrane traffic in eukaryotes is dependent on accurate targeting of transport vesicles between the diverse membrane-bound compartments of the biosynthetic and endocytic pathways. One of the key regulatory protein families is the Rab family of small GTPases, which contribute variously to the specification of membrane identity, the accuracy of vesicle targeting and the recruitment of molecular motors to membranes (Olkkonen and Stenmark, 1997; Behnia and Munro, 2005; Grosshans et al., 2006; Markgraf et al., 2007; Woollard and Moore, 2008). Rab GTPases undergo a regulated cycle between GDP- and GTP-bound forms and in so doing they cycle on and off particular endomembranes where they participate in specific membrane trafficking events (Olkkonen and Stenmark, 1997; Markgraf et al., 2007; Woollard and Moore, 2008).
Genomic data have shown that yeasts, animals and plants have each elaborated distinct sets of Rab proteins. Arabidopsis and mammals each have roughly 60 Rab GTPases compared to the six to ten that are found in yeasts, but it is striking that 80% of the predicted mammalian Rab GTPase subclasses are missing in Arabidopsis (Pereira-Leal and Seabra, 2001; Rutherford and Moore, 2002; Vernoud et al., 2003; Woollard and Moore, 2008). The diversity of the Rab GTPases family in mammals relative to that of yeasts appears to reflect the greater complexity and diversity of membrane trafficking events that occur in the diverse cell types of mammals (Bock et al., 2001; Pereira-Leal and Seabra, 2001). The situation is less clear in angiosperms, where phylogenetic analysis shows that all 57 Arabidopsis RAB sequences fall into just eight clades (RAB-A to RAB-H). Each of these clades is clearly related to one of the six Rab subclasses that are common to all three kingdoms or to mammalian Rab2 and Rab18 (Rutherford and Moore, 2002; Vernoud et al., 2003). Nevertheless, analysis of specificity-determining regions of Rab GTPases (Moore et al., 1995; Pereira-Leal and Seabra, 2000) predicts that several of the eight plant Rab clades might contain functionally distinct proteins (Pereira-Leal and Seabra, 2001; Rutherford and Moore, 2002), leading to the proposal that Arabidopsis could have as many as 18 distinct functional Rab subclasses. If so, it would mean that the Rab GTPase family has undergone distinct adaptive radiations in mammalian and angiosperm lineages and that many Rab subclasses in Arabidopsis are plant-specific (Rutherford and Moore, 2002). So far, however, there has been little experimental testing of these phylogenetic predictions. It has been shown that proteins of the Rab5-related Rab-F1 (ARA6) and Rab-F2 subclasses (ARA7 and RHA1) localise to different but overlapping populations of prevacuolar or endosomal compartments (Ueda et al., 2004) even though they share an exchange factor (Goh et al., 2007).
Here, we have investigated the Rab1-related proteins of Arabidopsis. The Rab1 subclass is one of the most highly conserved in eukaryotes and members of this subclass have been shown to act in early stages of biosynthetic traffic to or through the Golgi complex in yeasts, animals and plants (Plutner et al., 1991; Tisdale et al., 1992; Pind et al., 1994; Jedd et al., 1995; Batoko et al., 2000; Alvarez et al., 2003; Zheng et al., 2005). Animal Rab1 and its budding-yeast homologue, Ypt1p, are known to interact with similar regulatory and effector proteins (such as Uso1p/p115 and the TRAPP complex) to promote the targeting of COPII vesicles to the cis-Golgi (Beard et al., 2005; Grosshans et al., 2006; Markgraf et al., 2007; Cai et al., 2008). Rab1/Ypt1 proteins of higher plants are unusual, however, because sequence comparisons suggest that they can be divided into two distinct clades, Rab-D1 and Rab-D2, that could represent distinct functional subclasses (Pereira-Leal and Seabra, 2001; Rutherford and Moore, 2002; Vernoud et al., 2003) (supplementary material Fig. S1).
In Arabidopsis, the Rab-D1 subclass is represented by a single gene, RAB-D1 (also known as RABD1, At3g11730, ATFP8), whereas the Rab-D2 subclass is represented by three genes, RAB-D2a, RAB-D2b and RAB-D2c (RABD2A, At1g02130, ARA5, Rab1b; RABD2B, At5g47200; and RABD2C, At4g17530, respectively) which are predicted to be extensively redundant (Biermann et al., 1996; Rutherford and Moore, 2002) (supplementary material Fig. S1). Several reports have shown that a dominant-negative mutant of Arabidopsis RAB-D2a inhibits traffic between the ER and Golgi in plant cells (Batoko et al., 2000; Saint-Jore et al., 2002; Park et al., 2004; Zheng et al., 2005; Samalova et al., 2006). By contrast, the higher plant Rab-D1 subclass has not been investigated. Here, by use of protein tagging, dominant-negative mutants and loss of function mutants we provide evidence that the Arabidopsis Rab-D1 and Rab-D2 proteins do indeed represent distinct functional groups that act in transport between the endoplasmic reticulum (ER) and Golgi complex via distinct interactions.
Results
Tagged Arabidopsis RAB-D1 and RAB-D2a colocalise on Golgi and TGN
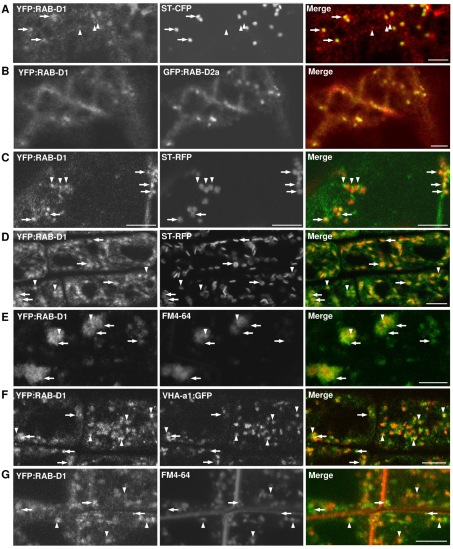
We first investigated the membrane targeting signals in RAB-D proteins in tobacco leaf epidermal cells where yellow fluorescent protein (YFP)-tagged RAB-D2a (ARA5; AtRab1b) has been localised to Golgi-associated punctate structures (Zheng et al., 2005). Similarly, in cells that exhibited the lowest detectable accumulation of YFP:RAB-D1, the fusion protein was observed in the cytoplasm and on mobile punctate structures that were often closely associated with the mobile Golgi stacks (Fig. 1A; supplementary material Fig. S2A,B). In cells exhibiting higher expression levels, YFP:RAB-D1 accumulated in the cytoplasm, indicating that targeting to the punctate structures was saturable (not shown). No labelling of the ER network was observed (supplementary material Fig. S2C). GFP:RAB-D2a colocalised with YFP:RAB-D2a and YFP:RAB-D1, indicating that both proteins target the same mobile punctate structures (Fig. 1B; supplementary material Fig. S2D,E).
Fig. 1.
Localisation of YFP:RAB-D1. Confocal laser scanning microscope (CLSM) images of tobacco (A,B) or Arabidopsis (C-G) cells coexpressing YFP:RAB-D1 (green) with various markers (red). (A,B) YFP:RAB-D1 in tobacco leaf abaxial epidermis coexpressing either the Golgi marker ST-CFP (A) or GFP:RAB-D2a (B). (C) Arabidopsis leaf abaxial epidermis coexpressing the Golgi marker ST-RFP. (D-G) Localisation of YFP:RAB-D1 in Arabidopsis root tips together with the Golgi marker ST-RFP (D), the TGN marker VHA-a1:GFP (F), or the endosomal dye FM4-64 (E,G). Root tips in E were treated with brefeldin A. Arrows indicate YFP:RAB-D1-labelled punctate structures that associate with Golgi stacks. Arrowheads show structures that associate with the TGN. Scale bars: 5 μm.
The identity of the structures that did not associate with Golgi stacks was unclear but it has been suggested that they might represent ER-export sites (Zheng et al., 2005). To investigate their identity in Arabidopsis, we used YFP-tagged forms of RAB-D1 and RAB-D2a from the Wave collection (Wave 25y and Wave 29y respectively) (Geldner et al., 2009). As in tobacco leaf epidermis, in leaf and root tip epidermis of Arabidopsis, YFP:RAB-D1 and YFP:RAB-D2a labelled punctate structures, some of which colocalised with Golgi stacks while others were distinct from, but often close to, the Golgi (Fig. 1C,D; supplementary material Fig. S3A,B). Surprisingly, treatment of root tips with the drug brefeldin A (BFA) caused a portion of the YFP:RAB-D1 and YFP:RAB-D2a signal to accumulate in the core of the BFA-bodies (Fig. 1E; supplementary material Fig. S3C). These bodies are aggregations of trans-Golgi-network (TGN) and endosomal compartments with Golgi stacks clustered at their periphery (Grebe et al., 2003; Dettmer et al., 2006; Richter et al., 2007; Teh and Moore, 2007). This suggested that a portion of the YFP:RAB-D1 and YFP:RAB-D2a signal localised to the TGN or endosomes in addition to Golgi stacks.
Introduction of VHA-a1:GFP, a marker of the TGN in Arabidopsis (Dettmer et al., 2006), confirmed that some of the structures labelled by YFP:RAB-D1 and YFP:RAB-D2a colocalised with the TGN (Fig. 1F; supplementary material Fig. S3D). The TGN also acts as an early endosome in Arabidopsis, accumulating the endocytic dye FM4-64 shortly after it is taken up into root-tip cells (Dettmer et al., 2006; Chow et al., 2008). Consistent with this, FM4-64 also accumulated in a subset of the compartments labelled with YFP:RAB-D1 and YFP:RAB-D2a (Fig. 1G; supplementary material Fig. S3E). Similar observations have been made with RAB-D2b using independent markers of the Golgi and the TGN in Arabidopsis (N.G. and J.C., unpublished). We concluded that proteins of the Arabidopsis Rab-D1 and Rab-D2 subclasses target YFP to both the Golgi stack and the TGN.
Dominant-negative RAB-D1 inhibits traffic between the ER and Golgi complex
To investigate the pathways in which the Rab-D1 subclass acts, we adopted a dominant mutant approach based on well-characterised amino acid substitutions in small GTPases (Olkkonen and Stenmark, 1997). We generated RAB-D1 [S22N], which is expected to promote the inactive GDP-bound state, and RAB-D1 [N121I], which is expected to promote the nucleotide free state. Each of these substitutions in Rab GTPases can generate a dominant-negative mutant owing to the titration of interactors (such as nucleotide-exchange factors) in a non-functional complex (Tisdale et al., 1992; Jedd et al., 1995; Jones et al., 1995; Olkkonnen and Stenmark, 1997; Goh et al., 2007). We also generated RAB-D1 [Q67L], which is predicted to be GTPase-deficient and thus to promote the active state (Olkkonnen and Stenmark, 1997; Richardson et al., 1998).
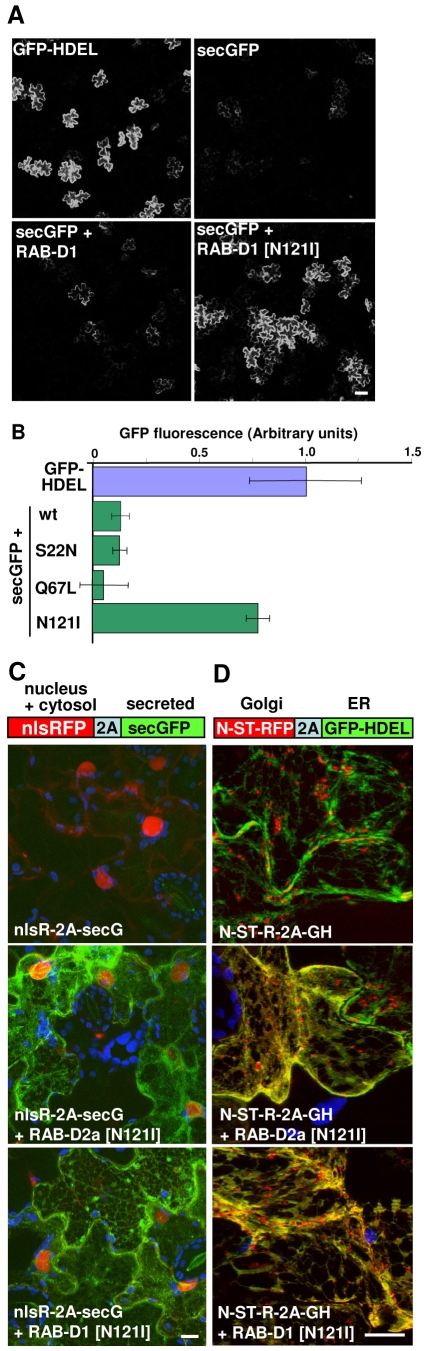
To establish whether the RAB-D1 mutants interfered with secretory traffic in plant cells, they were coexpressed with the secreted GFP marker (secGFP) using Agrobacterium-mediated transient expression in tobacco leaf epidermis, in which the function of RAB-D2a in biosynthetic traffic has been extensively characterised (Batoko et al., 2000; Saint-Jore et al., 2002; Zheng et al., 2005; Samalova et al., 2006; Duarte et al., 2008). In this assay, inhibiting the traffic of secGFP to the apoplast (where GFP fluorescence is not favoured) results in the accumulation of fluorescent GFP molecules within endomembrane compartments and this increased fluorescence signal can be imaged and quantified (Zheng et al., 2004; Zheng et al., 2005; Samalova et al., 2006). Quantitative confocal microscopy of transfected epidermal cells revealed that expression of RAB-D1 [N121I] but not wild-type RAB-D1 [S22N] or RAB-D1 [Q67L] resulted in marked intracellular accumulation of secGFP (Fig. 2A,B). In this respect RAB-D1 behaved like yeast Ypt1p and mammalian RAB1A or RAB1B where the equivalent N-to-I mutants are also more inhibitory than the corresponding S-to-N mutants and the Q-to-L mutant forms are not inhibitory (Tisdale et al., 1992; Pind et al., 1994; Richardson et al., 1998; Yoo et al., 1999; Alvarez et al., 2003). RAB-D1 [N121I] was selected for further work and compared with the corresponding mutant of RAB-D2a, which has been used to characterise RAB-D2a-dependent trafficking (Batoko et al., 2000; Saint-Jore et al., 2002; Park et al., 2004; Zheng et al., 2005; Duarte et al., 2008).
Fig. 2.
RAB-D1 [N121I] inhibits secretory traffic between the ER and Golgi. (A) Confocal images of tobacco leaf epidermal cells expressing the ER luminal marker GFP-HDEL or the secreted GFP marker secGFP either alone or together with wild-type or the N121I mutant forms of RAB-D1. (B) Quantification of the effect of RAB-D1 and the indicated mutants on the intracellular accumulation of secGFP (green bars). The fluorescence signal from the ER-lumenal marker GFP-HDEL (blue bar) was set to 1 and that for secGFP alone to 0. Error bars represent standard deviation of each sample. (C,D) Use of ratiometric fluorescent polyproteins to localise secreted and Golgi markers in the presence of RAB-D1 [N121I] and RAB-D2a [N121I]. (C) nlsR-2A-secG expresses secGFP (green) and a nuclear-targeted RFP (red) in fixed stoichiometry as a polyprotein linked by the self-cleaving 2A polypeptide (Samalova et al., 2006). Transfected cells are marked by red nuclei but do not accumulate GFP, which is secreted to the cell wall (top). In the presence of RAB-D1 [N121I] (bottom) or RAB-D2a [N121I] (centre), cells expressing similar quantities of the marker have similarly bright RFP signals in their nuclei, but now secGFP accumulates in a polygonal and planar ER network. (D) N-ST-R-2A-GH expresses the Golgi marker N-ST-RFP (red) in fixed stochiometry with the ER-marker GFP-HDEL (green), which indicates the relative expression level in each cell. In the presence of RAB-D1 [N121I] and RAB-D2a [N121I], the Golgi marker accumulates in the ER where it colocalises with GFP. In C and D, chlorophyll is blue. Scale bars:100 μm in A; 10 μm in C and D.
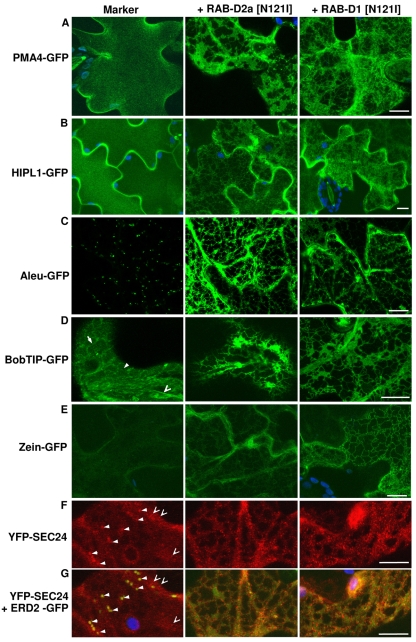
In the presence of either RAB-D1 or RAB-D2a N121I mutants, secGFP accumulated in a polygonal and planar network (Fig. 2C) typical of the ER in the presence of RAB-D2a [N121I] (Batoko et al., 2000; Samalova et al., 2006). Similarly, the Golgi marker N-ST-RFP co-accumulated with a coexpressed ER marker, GFP-HDEL, indicating that RAB-D1 [N121I] interfered with traffic between the ER and Golgi (Fig. 2D) as reported previously for RAB-D2a [N121I] (Batoko et al., 2000; Samalova et al., 2006). Both mutants also inhibited trafficking of diverse other fluorescently tagged cargo molecules at the ER (Fig. 3). These molecules included integral and glycosylphosphatidylinositol (GPI)-anchored plasma membrane proteins [the H+-ATPase PMA4-GFP and HIPL1-GFP, respectively (Lefebvre et al., 2004; Borner et al., 2005)] and markers of the vacuole lumen and tonoplast [aleu-GFP and BobTIP-GFP, respectively (Kotzer et al., 2004)] all of which accumulated in the ER (Fig. 3A-D). Similarly, both mutants promoted the accumulation of zein-GFP (Fig. 3E), which follows a recently described vacuolar pathway for degradation of proteins that misfold in the ER (Foresti et al., 2008).
Fig. 3.
Inhibition of plasma membrane and vacuolar traffic by RAB-D1 [N121I] and RAB-D2a [N121I]. Projections in z of series of confocal images from the cortical cytoplasm of transfected tobacco leaf epidermal cells expressing diverse fluorescent markers based on GFP (green) or YFP (red) in the absence (left) or presence of RAB-D1 [N121I] and RAB-D2a [N121I] (right and centre, respectively); chlorophyll, blue. (A,B) The H+-ATPase marker PMA4-GFP and the GPI-anchored protein marker HIPL1-GFP each accumulate at the plasma membrane in control cells but are trapped in the ER in the presence of RAB-D1 [N121I] or RAB-D2a [N121I]. (C) The vacuolar luminal marker aleu-GFP is usually visible only in punctate prevacuolar compartments en route to the vacuole (left) but is trapped in the ER in the presence of RAB-D1 [N121I] or RAB-D2a [N121I]. (D) In control cells (left), the tonoplast marker BobTIP-GFP is visible over the surface of the tonoplast underlying the cortical cytoplasm (solid arrowhead) and around transvacuolar strands (open arrowhead) with a faint signal from newly synthesised proteins in the ER (arrow); the marker is confined to the ER in the presence of RAB-D1 [N121I] or RAB-D2a [N121I]. (E) The misfolded ER-protein zein-GFP is faintly visible in control cells (left) but accumulates in the presence of RAB-D1 [N121I] and RAB-D2a [N121I]. (F,G) The COPII vesicle coat component YFP-SEC24 (red) accumulates on Golgi-associated ERES (solid arrowheads) and on additional independent punctate structures (open arrowheads), which are more numerous and pronounced in the presence of RAB-D1 [N121I] or RAB-D2a [N121I]. (G) The same cells as F, showing the coexpressed Golgi marker ERD2-GFP (green), which colcalises with ERES in control cells but accumulates in the ER in the presence of RAB-D1 [N121I] or RAB-D2a [N121I]; arrowheads as in F. Scale bars: 10 μm.
Finally, we investigated the effect of each mutant on the COPII-vesicle-coat component AtSEC24, which normally distributes between the cytoplasm and Golgi-associated ER-export sites (ERES) (Fig. 3F,G) (Stefano et al., 2006). Both mutants caused YFP-SEC24 to accumulate in a large number of dots, which were apparently smaller and brighter than ERES and did not associate with residual Golgi stacks (Fig. 3F,G). Thus, both RAB-D1 [N121I] and RAB-D2a [N121I] inhibit traffic of diverse biosynthetic trafficking markers at the ER.
These studies also showed that RAB-D1 [N121I] and RAB-D2a [N121I] each inhibited an essential cellular function because two weeks after infiltration of tobacco leaves, areas expressing either protein developed necrotic lesions (supplementary material Fig. S4). These lesions appeared earlier and were more extensive as the titre of Agrobacterium in the transfection was increased.
RAB-D1 and RAB-D2a act in ER-Golgi traffic via different interactors
Dominant-negative Rab GTPase mutants inhibit membrane trafficking by titration of an essential interacting partner such as a nucleotide exchange factor (Jones et al., 1995; Olkkonnen and Stenmark, 1997; Walch-Solimena et al., 1997; Cool et al., 1999; Goh et al., 2007). If this interactor has additional activities that are independent of its interaction with the mutated Rab GTPase, the effects of the dominant-negative mutant might not reflect the true activity of the Rab GTPase. Therefore, to establish whether inhibition of secretory traffic by RAB-D1 [N121I] can be attributed to loss of Rab-D1 function, we asked whether trafficking could be restored by coexpression of wild-type RAB-D1 to compete with the N121I mutant. This approach has been used previously with dominant-negative mutants of RAB-D2a and RAB-F2b (Batoko et al., 2000; Kotzer et al., 2004). For these experiments, we again used the quantitative secGFP accumulation assay in tobacco leaf epidermis (Zheng et al., 2005). Using this quantitative method we were able to confirm the previous observation (Batoko et al., 2000) that inhibition of ER-Golgi traffic by RAB-D2a [N121I] could be restored by coexpression of wild-type RAB-D2a but not by coexpression of RAB-E1d (At5g03520; AtRAB-E1d) which is a member of a different but closely related Arabidopsis Rab subclass that is implicated in post-Golgi secretory traffic (Zheng et al., 2005) (Fig. 4A, grey bars). Similarly, inhibition of secGFP trafficking by RAB-D1 [N121I] was substantially restored by coexpression of wild-type RAB-D1 but not RAB-E1d (Fig. 4B, grey bars). It is unlikely that the rescue phenomenon resulted from co-suppression at high expression levels because doubling the titre of bacteria expressing either of the dominant-negative mutants resulted in increased rather than reduced secGFP accumulation. Immunoblot analysis with an anti-Rab-D2 serum confirmed that Arabidopsis RAB-D2a was expressed in the rescued samples (Fig. 4C, lane c). We concluded that wild-type RAB-D1 and RAB-D2 proteins each act in membrane traffic between the ER and Golgi.
Fig. 4.
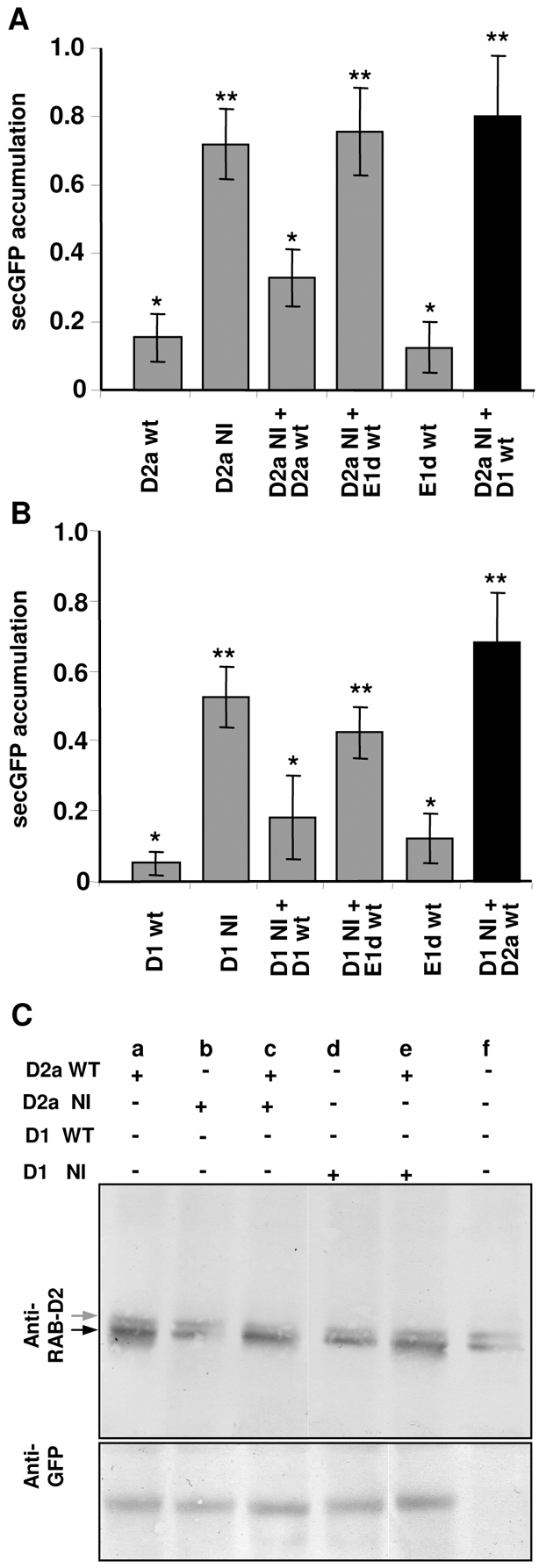
RAB-D1 and RAB-D2a act in ER-Golgi traffic via independent interactions. (A,B) Quantitation of secGFP accumulation (arbitrary units) in tobacco leaf epidermal cells expressing secGFP together with wild-type (wt) or N121I mutant forms (NI) of RAB-D1 (D1), RAB-D2a (D2a) or RAB8-E1d (E1d) alone or in combination as indicated. Error bars represent s.e.m. Values that are not significantly different (P≥0.05) are marked by an asterisk. Values with one asterisk are significantly different from values with two asterisks (P≤0.05). (A) Restoration of traffic inhibited by RAB-D2a [N121I]. (B) Restoration of traffic inhibited by RAB-D1 [N121I]. (C) Immunoblot analysis of tobacco leaves transiently expressing wild-type or mutant forms of RAB-D1 or RAB-D2a (as in A and B) using an anti-RAB-D2 antibody (see Materials and Methods). In uninfected plants (f) this antibody recognises two bands, most probably tobacco RAB-D proteins, the lower of which (black arrow) co-migrates with transiently expressed Arabidopsis RAB-D2a (lanes a, c and e). Note that the RAB-D2a [N121] mutant is not significantly overexpressed under these conditions and that it accumulates less efficiently than the wild type, consistent with previous observations that small GTPases are unstable in the nucleotide-free state when not bound to the exchange factor (Okkonen and Stenmark, 1997; Cool et al., 1999).
As RAB-D1 and RAB-D2a both act in traffic between the ER and Golgi complex we were interested to know whether they interact with the same partners. Therefore, we asked whether wild-type RAB-D1 was able to rescue membrane traffic in cells inhibited by dominant-negative RAB-D2a [N121I] and vice versa. Strikingly, in reciprocal rescue experiments we found that neither wild-type protein was able to rescue traffic inhibited by the mutant form of the other protein (Fig. 4A,B, black bars), arguing that RAB-D1 [N121I] and RAB-D2a [N121I] mutants each inhibited membrane traffic between ER and Golgi by titrating different interactors. Although Arabidopsis RAB-D2a was unable to rescue traffic inhibited by RAB-D1 [N121I], immunoblot analysis with an anti-Rab-D2 serum confirmed that it was expressed (Fig. 4C, lane e).
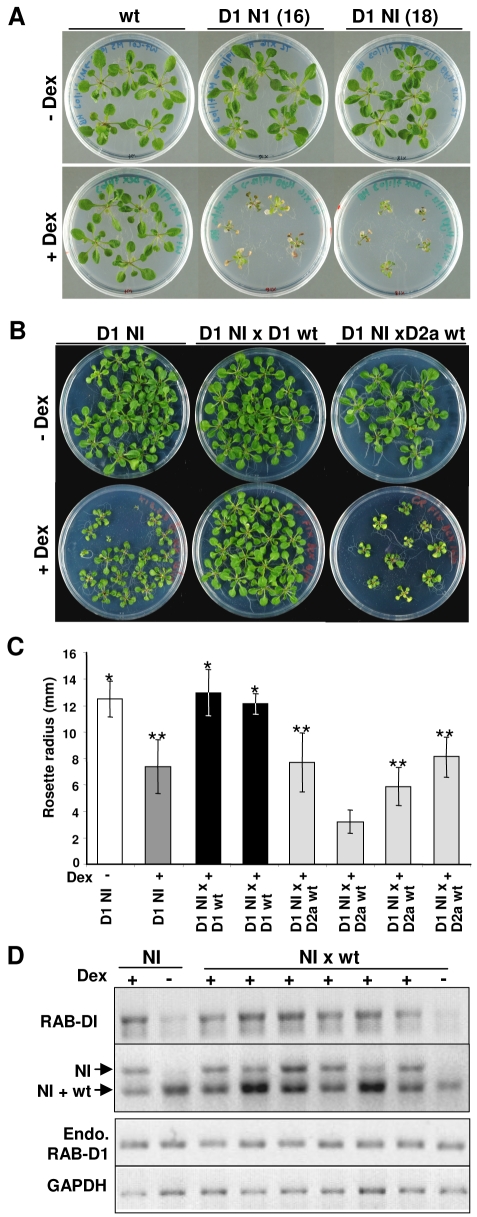
To establish whether dominant-negative mutants of the Rab-D1 subclass exhibit the same selective interactions in whole Arabidopsis plants, we expressed RAB-D1 and RAB-D1 [N121I] in seedlings using the dexamethasone-inducible pOp6/LhGR promoter system (Craft et al., 2005). When germinated on dexamethasone-containing medium, seedlings expressing RAB-D1 [N121I], but not those expressing wild-type RAB-D1, were severely dwarfed and died after prolonged exposure to dexamethasone (Fig. 5A). To test the ability of Rab-D1 and Rab-D2 proteins to suppress the dwarfing caused by RAB-D1 [N121I], plants from a homozygous RAB-D1 [N121I] line with an intermediate dwarf phenotype were crossed with plants expressing wild-type RAB-D1 or RAB-D2a from the same inducible promoter system. F1 progeny were germinated on medium with or without dexamethasone. As shown in Fig. 5B,C, seedlings from the crosses with the inducible RAB-D1 lines exhibited a wild-type growth phenotype, whereas those from four separate crosses with inducible RAB-D2a lines continued to show the dwarf phenotype.
Fig. 5.
RAB-D1 [N121I] inhibits Arabidopsis growth and cannot be rescued by RAB-D2a. (A) Seedlings exhibiting inducible expression of RAB-D1 [N121I] (D1 NI) but not wild type (wt) grown in the presence (+Dex) or absence (–Dex) of the inducer dexamethasone. Seedlings are shown from one wild-type and two N121I lines in which inducible expression of the transgene was confirmed by RNA gel blot (not shown). (B) As in A, but with seedlings expressing RAB-D1 [N121I] alone (D1 NI) compared with the F1 progeny of a cross to lines expressing wild-type RAB-D1 or RAB-D2a from the same inducible promoter system. (C) Quantitative analysis of rosette diameter in 18-day-old plants expressing RAB-D1 [N121I] only (D1 NI) in the presence (+) or absence (–) of dexamethasone (dark grey and white bars, respectively) or in the F1 progeny of crosses to lines expressing wild-type RAB-D1 (black bars) or RAB-D2a (pale grey bars). Each black and pale grey bar represents F1 progeny from an independent cross. Sets of data whose means are not significantly different (P<0.001) from D1 NI in either the absence or presence of dexamethasone are marked by single and double asterisks respectively. (D) RT-PCR analysis of transcript accumulation from the RAB-D1 wild-type and N121I mutant transgenes in the parental N121I line (NI) and the F1 progeny from crosses between lines carrying each transgene (NI×wt) in the presence (+) or absence (–) of dexamethasone (Dex). The upper panel shows total RAB-D1 transcript accumulation. The second panel shows RT-PCR products digested with HindIII to distinguish transcripts from the transgenes expressing the N121I and wild-type sequences; the upper band is characteristic for the mutant sequence and shows that the mutant is transcribed in the rescued plants that exhibit normal growth rates. The third panel shows that transcripts from the endogenous (Endo.) RAB-D1 locus are unaffected by expression of the transgenes. The lower panel shows transcripts for GAPDH, which served as a loading control. RNA was extracted after 18 days of growth.
To rule out the possibility that the apparent restoration of growth by RAB-D1 resulted from co-suppression of the RAB-D1 [N121I] transgene in the F1 seedlings, we performed reverse-transcriptase-PCR (RT-PCR) on the rescued plants and confirmed that mRNA from the RAB-D1 [N121I] transgene was still abundant (Fig. 5D). This also confirmed that the endogenous RAB-D1 transcript was expressed under all conditions, thus excluding the possibility that the dwarf phenotype associated with expression of RAB-D1 [N121I] resulted from co-suppression of the endogenous transcript. Finally, RT-PCR analysis of seedlings from a cross with an inducible RAB-D2a line confirmed that the RAB-D2a transcript was still expressed 18 days after germination on inductive medium, despite the dwarf phenotype exhibited by the seedlings (supplementary material Fig. S5).
The Rab-D2 subclass is essential in Arabidopsis but the Rab-D1 subclass is not
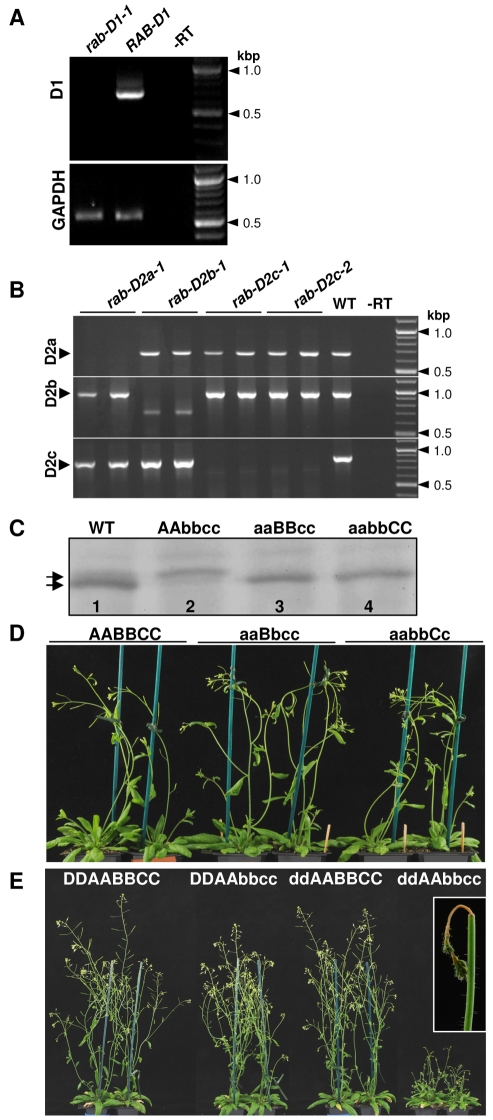
The behaviour of dominant-negative mutants had suggested that Arabidopsis Rab-D1 and Rab-D2 proteins perform distinct functions in ER-Golgi traffic, so we next used loss-of-function mutants to look at whether each subclass was essential in Arabidopsis. T-DNA insertion mutants were obtained for RAB-D1 and each of the three RAB-D2 proteins, RAB-D2a, RAB-D2b and RAB-D2c (supplementary material Fig. S6A). In each case, RT-PCR showed that transcripts were absent in the homozygous mutant lines (Fig. 5A,B). Immunoblot analysis of double-mutant combinations with anti-Rab-D2 antibodies confirmed that each insertion allele prevented protein accumulation from the three loci in this subclass (Fig. 6C).
Fig. 6.
Phenotypes of insertional mutants in the Rab-D1 and Rab-D2 subclasses. (A,B) RT-PCR analysis of Rab-D1 and Rab-D2 transcript abundance in plants carrying insertion mutants in individual genes or in wild-type plants (WT). –RT indicates PCR without reverse transcription on RNA isolated from wild-type plants. Transcripts for GAPDH served as a loading control. (C-E) Genotypes are indicated by the letters D, A, B and C denoting RAB-D1, RAB-D2a, RAB-D2b and RAB-D2c, respectively. Lower case letters (d, a, b, c) denote a mutant allele. (C) Immunoblot analysis of protein extracts from whole seedlings and T-DNA insertion mutants using a peptide antibody that recognises a conserved epitope in the Rab-D2 subclass. Extracts were prepared from wild-type plants and three double mutant genotypes that retained only a single wild-type RAB-D2 locus: lane 2, genotype AAbbcc retains only RAB-D2a; lane 3, genotype aaBBcc, retains only RAB-D2b; lane 4, genotype aabbCC retains only RAB-D2c. In the wild type (lane 1), the antibody recognises two RAB-D2-specific bands (arrows), with the upper band corresponding to RAB-D2a and the stronger lower band corresponding to RAB-D2b and RAB-D2c. (D) Phenotype of wild-type plants (AABBCC) or plants carrying a single wild-type copy of a single Rab-D2 gene, either RAB-D2b (aaBbcc) or RAB-D2c (aabbCc). (E) Phenotype of wild-type plants (DDAABBCC) or plants carrying mutations either in RAB-D1 (ddAABBCC) or in RAB-D2b and RAB-D2c (DDAAbbcc), or in all three loci (ddAAbbcc).
Plants homozygous for insertions in the three Rab-D2 genes were fertile and phenotypically normal under greenhouse conditions, as were the three double mutants (rab-D2a rab-D2b, rab-D2a rab-D2c and rab-D2b rab-D2c) (Fig. 6D,E). By contrast, however, we were unable to recover triple mutants that lacked all three RAB-D2 proteins. Thus, plants with the rab-D2a rab-D2b rab-D2c/+ genotype developed normally (Fig. 6D; supplementary material Fig. S6B, aabbCc) but exhibited approximately 25% ovule abortion. In 1626 PCR-genotyped progeny we recovered only rab-D2c/+ heterozygotes or wild types. Similarly, plants with the rab-D2a rab-D2b/+ rab-D2c genotype also developed normally (Fig. 6D; supplementary material Fig. S6B, aaBbcc) but exhibited approximately 25% ovule abortion and yielded no homozygous mutants amongst 1418 PCR-genotyped progeny. Thus, the Arabidopsis Rab-D2 subclass performs an essential function that cannot be provided by RAB-D1. Furthermore, this data confirms that all three members of the Rab-D2 subclass are extensively redundant because all three double mutant combinations were viable.
In contrast to the Rab-D2 subclass, however, the Rab-D1 subclass appeared to be dispensable in Arabidopsis as we were able to recover healthy fertile plants that were homozygous for either of two independent T-DNA insertions in RAB-D1 (supplementary material Fig. S6A), which is the only representative of this subclass in Arabidopsis (Pereira-Leal and Seabra, 2001; Rutherford and Moore, 2002; Vernoud et al., 2003) (Fig. 6A,E, ddAABBCC).
The Rab-D1 and Rab-D2 subclasses have overlapping function in Arabidopsis
The viability of the rab-D1 insertion alleles in Arabidopsis contrasted with the lethal phenotype associated with the expression of its dominant-negative mutant in seedlings. Thus the dominant-negative RAB-D1 [N121I] mutant must inhibit a function for which RAB-D1 is not essential, presumably by titrating a factor that performs an essential function with another protein. To investigate the possibility that these other proteins are members of the essential Rab-D2 subclass, we generated Rab-D2 mutants in a rab-D1 background. Plants homozygous for rab-D1-1 and insertions in single RAB-D2 loci were phenotypically normal (data not shown). Strikingly however, plants homozygous for insertions in RAB-D1 as well as RAB-D2b and RAB-D2c, which are the two most abundant members of the Rab-D2 subclass (Fig. 6C), exhibited a stunted bushy growth phenotype and very low fertility (Fig. 6E, ddAAbbcc). Transformation of these plants (ddAAbbcc) with a RAB-D1 cDNA under control of the CaMV 35S promoter complemented the bushy growth phenotype and infertility in 11 of 19 primary transformants (supplementary material Fig. S7A). This confirmed that both phenotypes resulted from the loss of Rab-D1 function in this background. As these phenotypes were not seen in the single rab-D1 mutant or in the rab-D2b rab-D2c double mutant (Fig. 6E, DDAAbbcc), the functions of RAB-D1 and these RAB-D2 genes must overlap. Furthermore, these data show that in the absence of RAB-D1, RAB-D2a alone is insufficient for normal development. The bushy phenotype of these plants arose in part because the upper region of the primary inflorescence stem became necrotic a few millimetres below the apex (see Fig. 6E, inset). Notably, the same phenotype was observed in inflorescences of Arabidopsis plants expressing the dominant-negative RAB-D2a [N121I] mutant (supplementary material Fig. S7B).
Discussion
Higher plants are unusual in that their Rab1/Ypt1-related proteins can be divided into two clades, Rab-D1 and Rab-D2, that might represent subclasses with distinct functions in membrane trafficking (Pereira-Leal and Seabra, 2001; Rutherford and Moore, 2002; Vernoud et al., 2003). Here, we show that the single Arabidopsis Rab-D1 protein, RAB-D1, targets YFP to the same Golgi-associated punctate compartments as RAB-D2a and that, like RAB-D2a, it acts in membrane traffic between the ER and Golgi, consistent with the activities of other Rab1/Ypt1-related proteins. However, based on the behaviour of dominant-negative RAB-D1 and RAB-D2a mutants in reciprocal rescue experiments, we propose that Rab-D1 and Rab-D2 proteins act in ER-Golgi traffic via distinct biochemical interactions. Thus, the inhibition of membrane traffic by dominant-negative RAB-D1 in tobacco leaf epidermis or Arabidopsis seedlings could be substantially restored by coexpressing wild-type RAB-D1 but not wild-type RAB-D2a. Similarly, inhibition of ER-Golgi traffic by dominant-negative RAB-D2a could be restored by coexpression of wild-type RAB-D2a but not of RAB-D1.
Because the dominant-negative mutants are likely to act by competitive titration of essential cofactors, we propose that RAB-D1 and RAB-D2 preferentially interact with and titrate different factors. When overexpressed, the cognate wild-type protein can compete with the dominant-negative mutant and restore the missing function, whereas the incorrect partner either cannot compete with the dominant-negative mutant or cannot fulfil the same function if it does interact.
These observations support the phylogenetic prediction that the plant Rab-D1 and Rab-D2 clades have acquired distinct functions in membrane traffic (Pereira-Leal and Seabra, 2001; Rutherford and Moore, 2002). Further support is provided by insertional mutants that demonstrated: (i) that all three members of the Arabidopsis Rab-D2 subclass are extensively redundant; and (ii) that the Rab-D2 subclass performs an essential function that cannot be provided by RAB-D1. Attempts to investigate whether the inability of RAB-D1 to substitute for Rab-D2 function arises from differences in the coding sequences or in expression patterns were unsuccessful because RAB-D2 cDNAs did not complement the triple RAB-D2 mutant when expressed from their own promoter (data not shown). In contrast to Rab-D2, the Rab-D1 subclass is apparently dispensable for normal development. Nevertheless these two putative subclasses exhibit some overlap of function because Arabidopsis plants lacking the two most highly expressed members of the Rab-D2 subclass (RAB-D2b and RAB-D2c) were phenotypically wild type but were stunted and highly infertile when RAB-D1 was also disrupted. Consistent with this, the expression of dominant-negative RAB-D2a [N121I] phenocopied the rab-D1 rab-D2b rab-D2c plants with respect to the stunting of growth and the necrosis of the apical region of the stem. This is the region of the stem that exhibits the highest rate of elongation (Fukaki et al., 1996), which might not be sustainable when Rab-D1 and Rab-D2 function is compromised by the dominant-negative mutant.
The overlap in function between Rab-D1 and Rab-D2 subclasses might account for the contradictory observations regarding the lethality of RAB-D1 dominant-negative and loss-of-function mutants: whereas rab-D1 plants were apparently normal and fertile, expression of dominant-negative RAB-D1 [N121I] resulted in severe growth defects and eventually in cell death in both Arabidopsis and tobacco. Thus, the dominant-negative mutant must inhibit activities in which Rab-D1 is not essential. Nevertheless, the membrane trafficking and cell viability phenotypes caused by dominant-negative RAB-D1 can be rescued specifically by coexpression with wild-type RAB-D1, suggesting that this protein can fulfil the essential functions that are inhibited by the mutant.
A simple resolution to this apparent paradox is suggested by the overlap in function between Rab-D1 and Rab-D2 proteins discussed above. Thus, even though proteins of the Rab-D2 subclass might be unable to compete with the dominant-negative RAB-D1 [N121I] mutant they might nevertheless be able to interact productively with the usual partners of RAB-D1 when RAB-D1 is simply missing (rab-D1 T-DNA alleles). This interpretation is consistent with biochemical data that show that small GTPase mutants of the NI class (equivalent to N116I of Ras) bind tightly to essential regulators such as nucleotide exchange factors (Jones et al., 1995; Olkkonen and Stenmark, 1997; Walch-Solimena et al., 1997; Goh et al., 2007) to the extent that their dominant-inhibitory effect is not always alleviated by coexpression of the wild-type protein (Tisdale et al., 1992; Pind et al., 1994; Jones et al., 1995; Yoo et al., 1999). The interpretation is also consistent with the previous proposal that Rab-D1 and Rab-D2 proteins interact preferentially with different interactors and that each interaction contributes an essential function. In support of this, when the two most highly expressed Rab-D2 loci, RAB-D2b and RAB-D2c, were disrupted along with RAB-D1, the single remaining Rab-D2 gene (RAB-D2a) is apparently insufficient to fulfil Rab-D1 function (these `ddAAbbcc' plants were stunted and infertile) even though it can evidently fulfil all essential Rab-D2-specific functions (rab-D2b rab-D2c plants, `DDAAbbcc' were healthy and fertile but triple mutants in the Rab-D2 subclass were not viable).
We cannot rule out the alternative explanations that dominant-negative RAB-D1 [N121I] titrates an essential factor that does not normally interact with wild-type RAB-D1, or that the titrated factor has additional functions that are required for cell viability independently of RAB-D1. Indeed the fact that wild-type RAB-D1 does not completely restore trafficking when coexpressed with RAB-D1 [N121I] in tobacco leaf epidermis could be interpreted as evidence of this. It is also possible that, in this context, restoration is partial simply because of the stochasticity of transient coexpression of wild-type and mutant forms of the Rab protein in individual epidermal cells with this system (Samalova et al., 2006).
Cellular functions of Rab-D1 and Rab-D2 subclasses
Rab1 in mammalian cells is associated principally with cis-Golgi membranes and ER-Golgi intermediates, consistent with its function in the early stages of biosynthetic traffic (Saraste et al., 1995). In plant cells, however, we found that Rab-D1 and Rab-D2 proteins targeted YFP to the TGN as well as the Golgi stack, suggesting that they might function on late as well as early Golgi compartments. There is evidence that Ypt1 function is required in yeast for efficient recycling of membrane proteins from the post-Golgi endosome via the Golgi (Lafourcade et al., 2004) and it has been suggested that the Arabidopsis TGN is homologous to the yeast post-Golgi endosome (Chow et al., 2008). The TRAPP complex that acts as an exchange factor for Ypt1 functions across the whole Golgi (Sacher et al., 2008) and in rat liver, Rab1a has been associated with transcytotic vesicles as well as ER-Golgi intermediates (Jin et al., 1996). The strong accumulation of plant Rab1-homologues at trans-rather than cis-Golgi membranes might perhaps reflect the fact that most of the material secreted by plants cells originates within the Golgi rather than the ER (Staehelin and Moore, 1995). As a result, there might be more active recycling within the Golgi and between the Golgi and plasma membrane than between the Golgi and ER.
Whereas the genetic evidence suggests distinct roles for the Rab-D1 and Rab-D2 subclasses in ER-Golgi traffic, the cellular and biochemical basis of this difference is unclear. We do not have any evidence that Rab-D1 and Rab-D2 proteins have spatially distinct functions because proteins of each subclass targeted fluorescent proteins to the same Golgi-associated structures. The two subclasses could potentially act either in parallel biosynthetic pathways between ER and Golgi, in opposing pathways in ER-Golgi cycling, or at different points on a single pathway. In yeast, Ypt1p is required for the selective packaging of GPI-anchored cargo at the ER (Morsomme and Reizman, 2002). The observation that dominant-negative mutants of both RAB-D1 and RAB-D2a acted similarly on the trafficking and distribution of the GPI-anchored fusion protein HIPL1-GFP, the heterologous artificial secretory protein secGFP, and the COPII-vesicle component YFP-SEC24 argues that both proteins are required for the default pathway. This is supported by the observation that both types of mutant also inhibited trafficking of luminal and membrane markers of the vacuole as well as a marker of a newly described vacuolar degradation pathway (Foresti et al., 2008).
Mammalian Rab1 and yeast Ypt1p have been implicated in distinct trafficking steps at the Golgi: targeting of ER-derived COPII vesicles to the cis-face and a subsequent intra-Golgi transport step in which the ArfGEF GBF1 is implicated (Plutner et al., 1991; Jedd et al., 1995; Alvarez et al., 2003; Cai et al., 2008). Ypt1 is also required for retrograde transport between Golgi and ER in yeast and for endocytic recycling from the post-Golgi-endosome to the Golgi complex (Lafourcade et al., 2004; Kamena et al., 2008). One possibility is that in plants, Rab-D1 and Rab-D2 proteins have become specialised to act preferentially in one or other of these processes. Confocal microscopy did not reveal consistent differences between the effects of RAB-D1 and RAB-D2a N121I mutants on any of the markers we analysed, but such differences might not be resolvable by these methods or might be confounded by secondary effects. The identification of RAB-D1 and RAB-D2a interactors could allow this hypothesis to be tested experimentally.
Materials and Methods
Plant material
Arabidopsis plants expressing YFP fusions to RAB-D1 and RAB-D2a, lines WAVE 25y and 29y, respectively, were generated as part of the WAVE collection (http://www.unil.ch/dbmv/page52928_en.html#2). The T-DNA insertion alleles described here correspond to the following stocks: rab-D1-1, SALK_033433; rab-D1-2 SALK_054734; rab-D2a-1, SALK_099495; rab-D2b-1, TMRI 151_C11; rab-D2c-1, SALK_034873; rab-D2c-2, SALK_054626. SAIL (Syngenta Arabidopsis Insertion Library) lines were obtained from TMRI (Torrey Mesa Research Institute, San Diego, CA). SALK Institute Genomic Analysis Laboratory (SIGnAL) lines were obtained from NASC or ABRC (Arabidopsis Biological Resource Center). Other plant materials and bacterial strains are described in the relevant sections below.
Molecular cloning
Molecular cloning was performed according to standard protocols as described previously (Batoko et al., 2000; Craft et al., 2005). The RAB-D1 cDNA sequence was amplified using primers P1 and P2 to introduce BamHI and SalI sites upstream and a BamHI site downstream of the coding sequence. The S22N, Q67L and N121I mutations were introduced by overlapping PCR (Batoko et al., 2000) using primers P1 and P2 together with primers P3 and P4, P5 and P6, or P7 and P8, respectively. These mutagenic primer pairs also contained silent mutations that introduced an AccI site into the S22N mutant, a PvuII site into the Q71L mutant and a HinfI site into the N121I mutant and disrupted a HindIII site in the N121I mutant. All RAB-D1 sequences were introduced as SalI and BamHI fragments into the expression vector pVKH18-En6-GUS (Batoko et al., 2000) for Agrobacterium-mediated transient expression in tobacco leaf epidermis and for complementation of T-DNA insertion mutants in Arabidopsis. The same fragments were inserted into the SalI and BamHI sites of pV-TOP (Craft et al., 2005) for dexamethasone-inducible expression via the pOp/LhGR system in the Arabidopsis driver line 4C-S5/7 (Craft et al., 2005) and of pVKHEn6-HAvenus (Zheng et al., 2005) to generate N-terminal fusions to the Venus variant of YFP. The RAB-D2a and RAB-D2a [N121I] cDNA sequences were inserted into pV-TOP and pVKHEn6-HAvenus as XhoI-BamHI fragments from pBluescript SK+ derivatives described previously (Batoko et al., 2000) and introduced into the Arabidopsis driver line 4C-S5/7 (Craft et al., 2005). The GFP fusion to RAB-D2a was a kind gift of Ulla Neumann and Chris Hawes (Oxford Brookes University, Oxford, UK).
Growth and RT-PCR analysis of dexamethasone-inducible Rab-D expression in Arabidopsis
Seedlings were germinated and grown on medium supplemented with 20 μM dexamethasone or with an equivalent volume of DMSO as described (Craft et al., 2005). RT-PCR was performed on individual F1 seedlings of crosses between lines expressing RAB-D1 or RAB-D1 [N121I] from pV-TOP in the Arabidopsis driver line 4C-S5/7 (Craft et al., 2005) and on seedlings of the parental generation. Total AtRabD1 transcript was amplified from total RNA as described previously (Craft et al., 2005) using the intron-spanning primers P9 and P10 and digested with HindIII to reveal the 350 bp fragment specific to transcripts of the RAB-D1 [N121I] transgene. Endogenous AtRabD1 transcript was amplified using the intron-spanning primers P9 and P11.
RT-PCR of knock-out lines
RNA extracted from 100 mg of leaves of 12-day-old seedlings homozygous for T-DNA insertion mutants using the Qiagen RNeasy Kit (Qiagen, Crawley, UK) was analysed by RT-PCR. RT reactions were performed as follows: a mixture of 0.5 μl 10x diluted deoxyribonuclease I (DNaseI, Invitrogen), 0.9 μl 10x DNase I buffer, 0.5 μl RNase inhibitor (RNasin, Promega), 2.1 μl RNase-free water and 5 μl of sample RNA (2.5 μg diluted to 5 μl) was left for 15 minutes at room temperature. EDTA (1 μl of 25 mM) was added and reactions were incubated at 65°C for 10 minutes. Primer P12 (1 μl of 100 mM) and 1 μl of a mix of dNTPs at 10 mM was added and the reaction was left for a further 5 minutes at 65°C. On ice, 2 μl of 0.1M DTT, 4 μl 5× First Strand buffer (Invitrogen) and 1 μl RNasin was added and the reaction was incubated at 42°C for 2 minutes. SuperScript II RNase H-RT (1 μl) from Invitrogen was added and the tubes were incubated at 42°C for 1 hour and then at 70°C for 15 minutes. Final cDNA was diluted five- to tenfold and 1 μl was used for initial PCR reactions. PCR was performed using the following primers: P9 + P11 primers to amplify the AtRabD1 sequence; P13 + P14 to amplify the RAB-D2a sequence; P15 + P16 to amplify the RAB-D2b sequence; P17 + P18 to amplify the RAB-D2c sequence; and P19 + P20 primers to amplify the GAPDH sequence. Amplification was performed at 94°C for 5 minutes, followed by 25 cycles of 94°C for 30 seconds, 58°C for 30 seconds and 72°C for 130 seconds, and finally at 72°C for 10 minutes. Products were analysed by agarose gel electrophoresis in the presence of ethidium bromide and imaged under UV illumination.
Genotyping T-DNA insertion mutants
The following primers were used to identify wild-type and insertion alleles: T-DNA rab-D1-1, P2 + P21; WT rab-D1-1, P2 + P7; T-DNA rab-D1-2, P9 + P21; WT rab-D1-2, P2 + P9; T-DNA rab-D2a, P21 + P22; WT rab-D2a, P22 + P23; T-DNA rab-D2b, P24 + P25; WT rab-D2b, P25 + P26; T-DNA rab-D2c-1 and rab-D2c-2, P21 + P27; WT rab-D2c-1 and rab-D2c-2, P27 + P28. PCR was performed with Taq polymerase using 50 mM Tris-HCl pH 9.1, 14 mM (NH4)2SO4 and 2.5 mM MgCl2 buffer with the following cycle: 94°C for 5 minutes; then 30-35 cycles of 94°C for 30 seconds, 55°C for 30 seconds and 72°C for 130 seconds; and finally 72°C for 10 minutes. Plants carrying a single remaining wild-type Rab-D2 allele were maintained as heterozygotes and were all genotyped by PCR in each generation and following Agrobacterium-mediated transformation by floral dip.
Tobacco leaf infiltrations and confocal microscopy
The quantitative secGFP accumulation assay in tobacco leaf epidermis and detection of fluorescent proteins by confocal microscopy was as described by Zheng and colleagues (Zheng et al., 2005). Ratiometric imaging constructs and simultaneous imaging of GFP with mRFP1 or YFP were as described by Samalova and colleagues (Samalova et al., 2006).
Immunoblot analysis
Protein isolation and blotting was performed as described by Samalova and colleagues (Samalova et al., 2006). Proteins were extracted from tobacco leaf samples 48 hours after Agrobacterium infiltration. OD 0.03 and 0.01 was used for the RAB-D and ST-GFP Agrobacterium cultures, respectively. Affinity-purified polyclonal antibody for RAB-D2 was raised in rabbits against the peptide antigen acetyl-NARPPTVQIRGQPVAQKNGC-amide (Cambridge Research Biochemicals, Cambridge, UK). Full-length GFP antibody was obtained from Molecular Probes (Leiden, The Netherlands). Each was used at 1/1000 dilution to probe the membrane overnight at 4°C. Alkaline-phosphatase-conjugated secondary antibodies (anti-rabbit IgG; Sigma) were used at 1/10,000 dilution for 1-hour incubation at room temperature.
List of oligonucleotides
Oligonucleotides were synthesised by MWG-Biotech (Germany) or Sigma-Genosys (Cambridge, UK). See supplementary material Table S1.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/20/3749/DC1
This work was supported by BBSRC research grant BBS/B/03904 to I.M. and by a BBSRC CASE studentship to H.P. The Wave lines were generated with support from an HFSP fellowship and the Swiss National Science Foundation to N.G. and from the HHMI, USDA and NIH to J.C. We thank Ulla Neumann and Chris Hawes for Agrobacterium strains expressing GFP:RAB-D2a, YFP-SEC24 and ERD2-GFP; Allessandro Vitale (CNR_IGV, Portici, Italy) and Michele Bellucci (Istituto di Genetica Vegetale-Sezione di Perugia, Perugia, Italy) for zein-GFP; and Paul Dupree (University of Cambridge, Cambridge, UK) for HIPL1-GFP. John Baker for photography, and Caroline O'Brien for technical assistance. We thank the Torrey Mesa Research Institute and SALK Institute Genomic Analysis Laboratory for the T-DNA insertion lines from the SAIL and SIGnAL collections, some of which were obtained from the Nottingham Arabidopsis Stock Centre, UK, or the Arabidopsis Biological Resource Centre, Columbus, OH. Deposited in PMC for release after 12 months.
References
- Alvarez, C., Garcia-Mata, R., Brandon, E. and Sztul, E. (2003). COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol. Biol. Cell 14, 2116-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko, H., Zheng, H. Q., Hawes, C. and Moore, I. (2000). A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12, 2201-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard, M., Satoh, A., Shorter, J. and Warren, G. (2005). A cryptic Rab1-binding site in the p115 tethering protein. J. Biol. Chem. 280, 25840-25848. [DOI] [PubMed] [Google Scholar]
- Behnia, R. and Munro, S. (2005). Organelle identity and the signposts for membrane traffic. Nature 438, 597-604. [DOI] [PubMed] [Google Scholar]
- Biermann, B., Randall, S. K. and Crowell, D. N. (1996). Identification and isoprenylation of plant GTP-binding proteins. Plant Mol. Biol. 31, 1021-1028. [DOI] [PubMed] [Google Scholar]
- Bock, J. B., Matern, H. T., Peden, A. A. and Scheller, R. H. (2001). A genomic perspective on membrane compartment organization. Nature 409, 839-841. [DOI] [PubMed] [Google Scholar]
- Borner, G. H. H., Sherrier, D. J., Weimar, T., Michaelson, L. V., Hawkins, N. D., MacAskill, A., Napier, J. A., Beale, M. H., Lilley, K. S. and Dupree, P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. evidence for plasma membrane lipid rafts. Plant Physiol. 137, 104-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y., Chin, H. F., Lazarova, D., Menon, S., Fu, C., Cai, H., Sclafani, A., Rodgers, D. W., De La Cruz, E. M., Ferro-Novick, S. et al. (2008). The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell 133, 1202-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, C. M., Neto, H., Foucart, C. and Moore, I. (2008). Rab-A2 and -A3 GTPases define a trans-Golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20, 101-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool, R. H., Schmidt, G., Lenzen, C. U., Prinz, H., Vogt, D. and Wittinghofer, A. (1999). The Ras mutant D119N is both dominant negative and activated. Mol. Cell. Biol. 19, 6297-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft, J., Samalova, M., Baroux, C., Townley, H., Martinez, A., Jepson, I., Tsiantis, M. and Moore, I. (2005). New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 41, 899-918. [DOI] [PubMed] [Google Scholar]
- Dettmer, J., Hong-Hermesdorf, A., Stierhof, Y. D. and Schumacher, K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18, 715-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte, P., Pissarra, J. and Moore, I. (2008). Processing and trafficking of a single isoform of the aspartic proteinase cardosin A on the vacuolar pathway. Planta 227, 1255-1268. [DOI] [PubMed] [Google Scholar]
- Foresti, O., De Marchis, F., de Virgilio, M., Klein, E., Arcioni, S., Belluci, M. and Vitale, A. (2008). Protein domains involved in assembly in the endoplasmic reticulum promote vacuolar delivery when sused to secretorey GFP, indicating a quality control pathway for degradation in the plant vacuole. Mol. Plant 1, 1067-1076. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., Fujisawa, H. and Tasaka, M. (1996). Gravitropic response of inflorescence stems in Arabidopsis thaliana. Plant Physiol. 110, 933-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner, N., Dénervaud-Tendon, V., Hyman, D. L., Mayer, U., Stierhof, Y. D. and Chory, J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59, 169-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, T., Uchida, W., Arakawa, S., Ito, E., Dainobu, T., Ebine, K., Takeuchi, M., Sato, K., Ueda, T. and Nakano, A. (2007). VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell 19, 3504-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe, M., Xu, J., Mobius, W., Ueda, T., Nakano, A., Geuze, H. J., Rook, M. B. and Scheres, B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13, 1378-1387. [DOI] [PubMed] [Google Scholar]
- Grosshans, B. L., Ortiz, D. and Novick, P. (2006). Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA 103, 11821-11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd, G., Richardson, C., Litt, R. and Segev, N. (1995). The Ypt1 GTPase is essential for the first 2 steps of the yeast secretory pathway. J. Cell Biol. 131, 583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, M., Saucan, L., Farquhar, M. G. and Palade, G. E. (1996). Rab1a and multiple other Rab proteins are associated with the transcytotic pathway in rat liver. J. Biol. Chem. 271, 30105-30113. [DOI] [PubMed] [Google Scholar]
- Jones, S., Litt, R. J., Richardson, C. J. and Segev, N. (1995). Requirement of nucleotide exchange factor for Ypt1 GTPase mediated protein-transport. J. Cell Biol. 130, 1051-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamena, F., Diefenbacher, M., Kilchert, C., Schwarz, H. and Spang, A. (2008). Ypt1p is essential for retrograde Golgi-ER transport and for Golgi maintenance in S. cerevisiae. J. Cell Sci. 121, 1293-1302. [DOI] [PubMed] [Google Scholar]
- Kotzer, A. M., Brandizzi, F., Neumann, U., Paris, N., Moore, I. and Hawes, C. (2004). AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J. Cell Sci. 117, 6377-6389. [DOI] [PubMed] [Google Scholar]
- Lafourcade, C., Galan, J. M., Gloor, Y., Haguenauer-Tsapis, R. and Peter, M. (2004). The GTPase-activating enzyme Gyp1p is required for recycling of internalised membrane material by inactivation of the Rab/Ytp GTPase Ypt1p. Mol. Cell. Biol. 24, 3815-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, B., Batoko, H., Duby, G. and Boutry, M. (2004). Targeting of a Nicotiana plumbaginifolia H+-ATPase to the plasma membrane is not by default and requires cytosolic structural determinants. Plant Cell 16, 1772-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markgraf, D. F., Peplowska, K. and Ungerman, C. (2007). Rab cascades and tethering factors in the endomembrane system. FEBS Lett. 581, 2125-2130. [DOI] [PubMed] [Google Scholar]
- Moore, I., Schell, J. and Palme, K. (1995). Subclass-specific sequence motifs identified in Rab GTPases. Trends Biochem Sci. 20, 10-12. [DOI] [PubMed] [Google Scholar]
- Morsomme, P. and Riezman, H. (2002). The Rab GTPase Ypt1p and tethering factors couple protein sorting at the ER to vesicle targeting to the Golgi apparatus. Dev. Cell 2, 307-317. [DOI] [PubMed] [Google Scholar]
- Olkkonen, V. M. and Stenmark, H. (1997). Role of rab GTPases in membrane traffic. In International Review of Cytology: A Survey of Cell Biology, vol. 176 (ed. K. W. Jeon), pp. 1-85. New York: Academic Press. [DOI] [PubMed] [Google Scholar]
- Park, M., Kim, S. J., Vitale, A. and Hwang, I. (2004). Identification of the protein storage vacuole and protein targeting to the vacuole in leaf cells of three plant species. Plant Physiol. 134, 625-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal, J. B. and Seabra, M. C. (2000). The mammalian Rab family of small GTPases: Definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J. Mol. Biol. 301, 1077-1087. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal, J. B. and Seabra, M. C. (2001). Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 313, 889-901. [DOI] [PubMed] [Google Scholar]
- Pind, S. N., Nuoffer, C., McCaffery, J. M., Plutner, H., Davidson, H. W., Farquhar, M. G. and Balch, W. E. (1994). Rab1 and Ca2+ are required for the fusion of carrier vesicles mediating endoplasmic-reticulum to Golgi transport. J. Cell Biol. 125, 239-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutner, H., Cox, A. D., Pind, S., Khosravi-Far, R., Bourne, J. R., Schwaninger, R., Der, C. J. and Balch, W. E. (1991). Rab1B regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J. Cell Biol. 115, 31-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, C. J., Jones, S., Litt, R. J. and Segev, N. (1998). GTP hydrolysis is not important for Ypt1 GTPase function in vesicular transport. Mol. Cell. Biol. 18, 827-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, S., Geldner, N., Schrader, J., Wolters, H., Stierhof, Y. D., Rios, G., Koncz, C., Robinson, D. G. and Jürgens, G. (2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448, 488-492. [DOI] [PubMed] [Google Scholar]
- Rutherford, S. and Moore, I. (2002). The Arabidopsis Rab GTPase family: another enigma variation. Curr. Opin. Plant Biol. 5, 518-528. [DOI] [PubMed] [Google Scholar]
- Sacher, M., Kim, Y. G., Lavie, A., Oh, B. H. and Segev, N. (2008). The TRAPP complex: insights into its architecture and function. Traffic 9, 2023-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore, C. M., Evins, J., Batoko, H., Brandizzi, F., Moore, I. and Hawes, C. (2002). Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29, 661-678. [DOI] [PubMed] [Google Scholar]
- Samalova, M., Fricker, M. and Moore, I. (2006). Ratiometric fluorescence-imaging assays of plant membrane traffic using polyproteins. Traffic 7, 1701-1723. [DOI] [PubMed] [Google Scholar]
- Saraste, J., Lahtinen, U. and Goud, B. (1995). Localization of the small GTP-binding protein rab1p to early compartments of the secretory pathway. J. Cell Sci. 108, 1541-1552. [DOI] [PubMed] [Google Scholar]
- Staehelin, L. A. and Moore, I. (1995). The plant Golgi apparatus-Structure, functional organization and trafficking mechanisms. Annu. Rev. Plant Physiol. Mol. Biol. 46, 261-288. [Google Scholar]
- Stefano, G., Renna, L., Chatre, L., Hanton, S. L., Moreau, P., Hawes, C. and Brandizzi, F. (2006). In tobacco leaf epidermal cells, the integrity of protein export from the endoplasmic reticulum and of ER export sites depends on active COPI machinery. Plant J. 46, 95-110. [DOI] [PubMed] [Google Scholar]
- Teh, O. K. and Moore, I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448, 493-496. [DOI] [PubMed] [Google Scholar]
- Tisdale, E. J., Bourne, J. R., Khosravifar, R., Der, C. J. and Balch, W. E. (1992). GTPbinding mutants of Rab1 and Rab2 are potent inhibitors of vesicular transport from the endoplasmic-reticulum to the Golgi-complex. J. Cell Biol. 119, 749-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, T., Uemura, T., Sato, M. H. and Nakano, A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 4, 783-789. [DOI] [PubMed] [Google Scholar]
- Vernoud, V., Horton, A. C., Yang, Z. and Nielsen, E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131, 1191-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena, C., Collins, R. N. and Novick, P. J. (1997). Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol. 137, 1495-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollard, A. A. D. and Moore, I. (2008). The functions of Rab GTPases in plant membrane traffic. Curr. Opin. Plant Biol. 11, 610-619. [DOI] [PubMed] [Google Scholar]
- Yoo, J. S., Grabowski, R., Xing, L., Trepte, H. H., Schmitt, H. D. and Gallwitz, D. (1999). Functional implications of genetic interactions between genes encoding small GTPases involved in vesicular transport in yeast. Mol. Gen. Genet. 261, 80-91. [DOI] [PubMed] [Google Scholar]
- Zheng, H., Kunst, L., Hawes, C. and Moore, I. (2004). A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J. 37, 398-414. [DOI] [PubMed] [Google Scholar]
- Zheng, H. Q., Camacho, L., Wee, E., Henri, B. A., Legen, J., Leaver, C. J., Malho, R., Hussey, P. J. and Moore, I. (2005). A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell 17, 2020-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.