Abstract
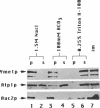
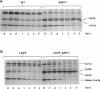
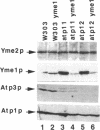
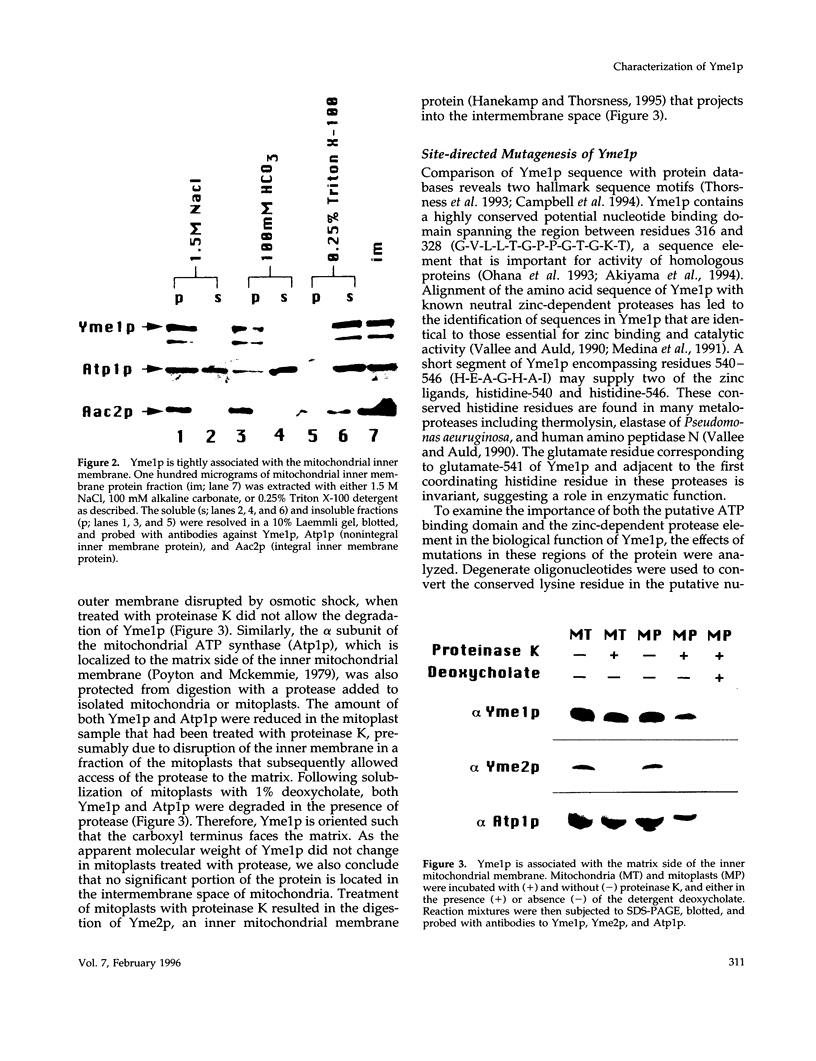
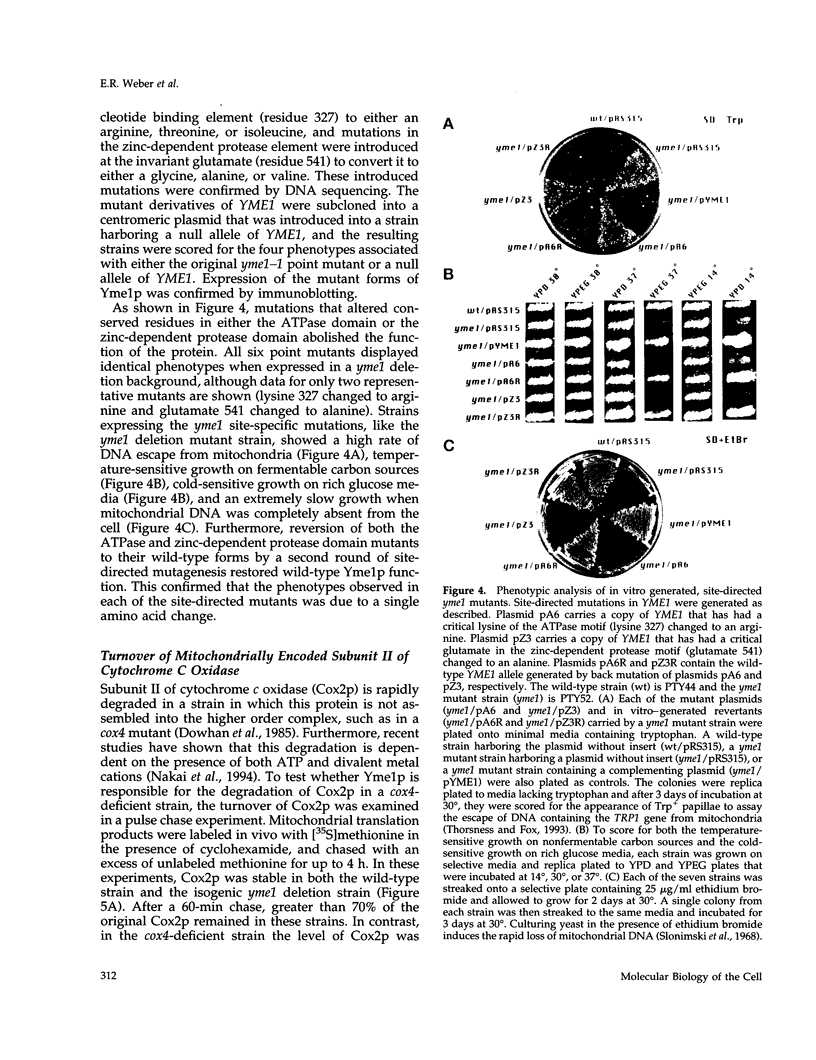
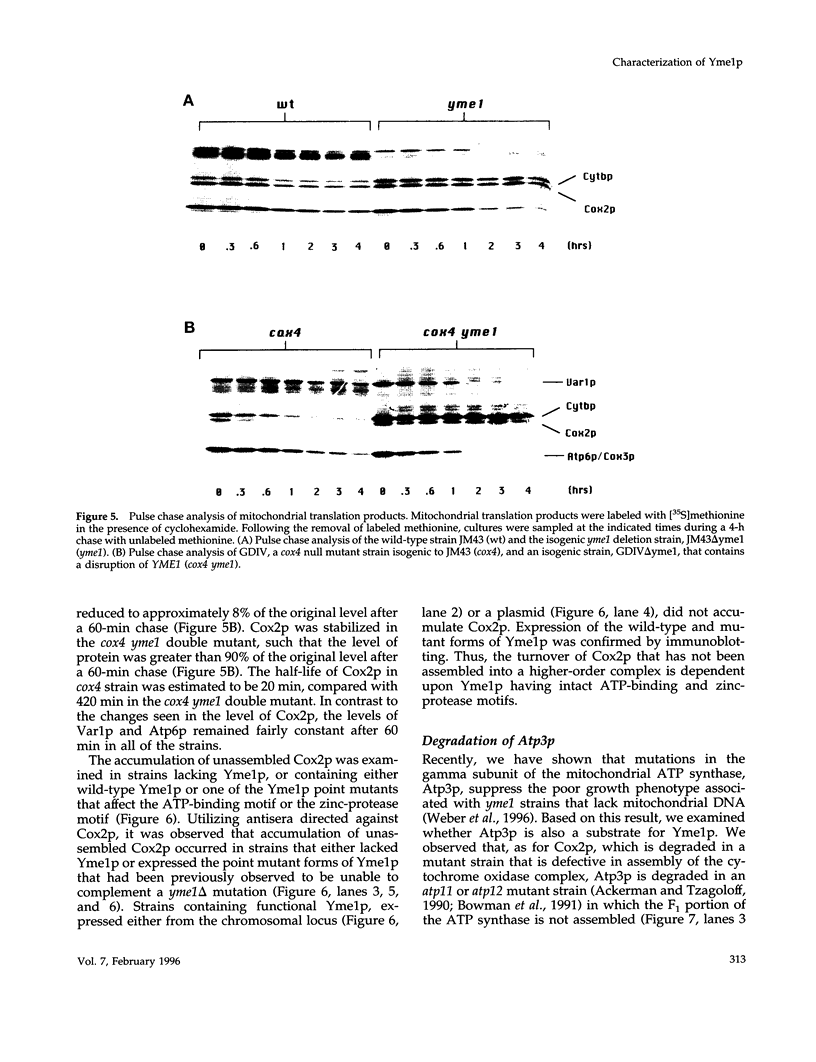
Inactivation of YME1 in yeast causes several distinct phenotypes: an increased rate of DNA escape from mitochondria, temperature-sensitive growth on nonfermentable carbon sources, extremely slow growth when mitochondrial DNA is completely absent from the cell, and altered morphology of the mitochondrial compartment. The protein encoded by YME1, Yme1p, contains two highly conserved sequence elements, one implicated in the binding and hydrolysis of ATP, and the second characteristic of active site residues found in neutral, zinc-dependent proteases. Both the putative ATPase and zinc-dependent protease elements are necessary for the function of Yme1p as genes having mutations in critical residues of either of these motifs are unable to suppress any of the phenotypes exhibited by yme1 deletion strains. Yme1p co-fractionates with proteins associated with the mitochondrial inner membrane, is tightly associated with this membrane, and is oriented with the bulk of the protein facing the matrix. Unassembled subunit II of cytochrome oxidase is stabilized in yme1 yeast strains. The data support a model in which Yme1p is an ATP and zinc-dependent protease associated with the matrix side of the inner mitochondrial membrane. Subunit II of cytochrome oxidase, when not assembled into a higher order complex, is a likely substrate of Yme1p.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackerman S. H., Tzagoloff A. Identification of two nuclear genes (ATP11, ATP12) required for assembly of the yeast F1-ATPase. Proc Natl Acad Sci U S A. 1990 Jul;87(13):4986–4990. doi: 10.1073/pnas.87.13.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama Y., Shirai Y., Ito K. Involvement of FtsH in protein assembly into and through the membrane. II. Dominant mutations affecting FtsH functions. J Biol Chem. 1994 Feb 18;269(7):5225–5229. [PubMed] [Google Scholar]
- Bowman S., Ackerman S. H., Griffiths D. E., Tzagoloff A. Characterization of ATP12, a yeast nuclear gene required for the assembly of the mitochondrial F1-ATPase. J Biol Chem. 1991 Apr 25;266(12):7517–7523. [PubMed] [Google Scholar]
- Burgess S. M., Delannoy M., Jensen R. E. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994 Sep;126(6):1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C. L., Tanaka N., White K. H., Thorsness P. E. Mitochondrial morphological and functional defects in yeast caused by yme1 are suppressed by mutation of a 26S protease subunit homologue. Mol Biol Cell. 1994 Aug;5(8):899–905. doi: 10.1091/mbc.5.8.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G., Böhni P. C., Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982 Nov 10;257(21):13028–13033. [PubMed] [Google Scholar]
- Dowhan W., Bibus C. R., Schatz G. The cytoplasmically-made subunit IV is necessary for assembly of cytochrome c oxidase in yeast. EMBO J. 1985 Jan;4(1):179–184. doi: 10.1002/j.1460-2075.1985.tb02334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakle K. A., Bernstein M., Emr S. D. Characterization of a component of the yeast secretion machinery: identification of the SEC18 gene product. Mol Cell Biol. 1988 Oct;8(10):4098–4109. doi: 10.1128/mcb.8.10.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Oppliger W., Schatz G. Both ATP and an energized inner membrane are required to import a purified precursor protein into mitochondria. EMBO J. 1987 Apr;6(4):1073–1077. doi: 10.1002/j.1460-2075.1987.tb04860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R., Wiebel F. F., Flessau A., Rytka J., Beyer A., Fröhlich K. U., Kunau W. H. PAS1, a yeast gene required for peroxisome biogenesis, encodes a member of a novel family of putative ATPases. Cell. 1991 Feb 8;64(3):499–510. doi: 10.1016/0092-8674(91)90234-p. [DOI] [PubMed] [Google Scholar]
- Guelin E., Rep M., Grivell L. A. Sequence of the AFG3 gene encoding a new member of the FtsH/Yme1/Tma subfamily of the AAA-protein family. Yeast. 1994 Oct;10(10):1389–1394. doi: 10.1002/yea.320101016. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawson J. E., Douglas M. G. Separate genes encode functionally equivalent ADP/ATP carrier proteins in Saccharomyces cerevisiae. Isolation and analysis of AAC2. J Biol Chem. 1988 Oct 15;263(29):14812–14818. [PubMed] [Google Scholar]
- McEwen J. E., Ko C., Kloeckner-Gruissem B., Poyton R. O. Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae. Characterization of mutants in 34 complementation groups. J Biol Chem. 1986 Sep 5;261(25):11872–11879. [PubMed] [Google Scholar]
- Medina J. F., Wetterholm A., Rådmark O., Shapiro R., Haeggström J. Z., Vallee B. L., Samuelsson B. Leukotriene A4 hydrolase: determination of the three zinc-binding ligands by site-directed mutagenesis and zinc analysis. Proc Natl Acad Sci U S A. 1991 Sep 1;88(17):7620–7624. doi: 10.1073/pnas.88.17.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai T., Mera Y., Yasuhara T., Ohashi A. Divalent metal ion-dependent mitochondrial degradation of unassembled subunits 2 and 3 of cytochrome c oxidase. J Biochem. 1994 Oct;116(4):752–758. doi: 10.1093/oxfordjournals.jbchem.a124592. [DOI] [PubMed] [Google Scholar]
- Nakai T., Yasuhara T., Fujiki Y., Ohashi A. Multiple genes, including a member of the AAA family, are essential for degradation of unassembled subunit 2 of cytochrome c oxidase in yeast mitochondria. Mol Cell Biol. 1995 Aug;15(8):4441–4452. doi: 10.1128/mcb.15.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., Fox T. D., Walter P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science. 1993 Dec 24;262(5142):1997–2004. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- Ohana B., Moore P. A., Ruben S. M., Southgate C. D., Green M. R., Rosen C. A. The type 1 human immunodeficiency virus Tat binding protein is a transcriptional activator belonging to an additional family of evolutionarily conserved genes. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):138–142. doi: 10.1073/pnas.90.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic A., Tauer R., Feldmann H., Neupert W., Langer T. Yta10p is required for the ATP-dependent degradation of polypeptides in the inner membrane of mitochondria. FEBS Lett. 1994 Oct 17;353(2):201–206. doi: 10.1016/0014-5793(94)01046-3. [DOI] [PubMed] [Google Scholar]
- Paul M. F., Ackerman S., Yue J., Arselin G., Velours J., Tzagolof A., Ackermann S [corrected to Ackerman S. ]. Cloning of the yeast ATP3 gene coding for the gamma-subunit of F1 and characterization of atp3 mutants. J Biol Chem. 1994 Oct 21;269(42):26158–26164. [PubMed] [Google Scholar]
- Pearce D. A., Sherman F. Degradation of cytochrome oxidase subunits in mutants of yeast lacking cytochrome c and suppression of the degradation by mutation of yme1. J Biol Chem. 1995 Sep 8;270(36):20879–20882. doi: 10.1074/jbc.270.36.20879. [DOI] [PubMed] [Google Scholar]
- Poyton R. O., McKemmie E. Post-translational processing and transport of the polyprotein precursor to subunits IV to VII of yeast cytochrome c oxidase. J Biol Chem. 1979 Jul 25;254(14):6772–6780. [PubMed] [Google Scholar]
- Rechsteiner M., Hoffman L., Dubiel W. The multicatalytic and 26 S proteases. J Biol Chem. 1993 Mar 25;268(9):6065–6068. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall R., Mannhaupt G., Stucka R., Tauer R., Ehnle S., Schwarzlose C., Vetter I., Feldmann H. Identification of a set of yeast genes coding for a novel family of putative ATPases with high similarity to constituents of the 26S protease complex. Yeast. 1994 Sep;10(9):1141–1155. doi: 10.1002/yea.320100903. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonimski P. P., Perrodin G., Croft J. H. Ethidium bromide induced mutation of yeast mitochondria: complete transformation of cells into respiratory deficient non-chromosomal "petites". Biochem Biophys Res Commun. 1968 Feb 15;30(3):232–239. doi: 10.1016/0006-291x(68)90440-3. [DOI] [PubMed] [Google Scholar]
- Takeda M., Chen W. J., Saltzgaber J., Douglas M. G. Nuclear genes encoding the yeast mitochondrial ATPase complex. Analysis of ATP1 coding the F1-ATPase alpha-subunit and its assembly. J Biol Chem. 1986 Nov 15;261(32):15126–15133. [PubMed] [Google Scholar]
- Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992 Oct;119(2):301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness P. E., Fox T. D. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics. 1993 May;134(1):21–28. doi: 10.1093/genetics/134.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness P. E., White K. H., Fox T. D. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Sep;13(9):5418–5426. doi: 10.1128/mcb.13.9.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T., Gamer J., Bukau B., Kanemori M., Mori H., Rutman A. J., Oppenheim A. B., Yura T., Yamanaka K., Niki H. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor sigma 32. EMBO J. 1995 Jun 1;14(11):2551–2560. doi: 10.1002/j.1460-2075.1995.tb07253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T., Yamanaka K., Murata K., Suzaki T., Bouloc P., Kato A., Niki H., Hiraga S., Ogura T. Topology and subcellular localization of FtsH protein in Escherichia coli. J Bacteriol. 1993 Mar;175(5):1352–1357. doi: 10.1128/jb.175.5.1352-1357.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T., Yuki T., Morimura S., Mori H., Yamanaka K., Niki H., Hiraga S., Ogura T. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J Bacteriol. 1993 Mar;175(5):1344–1351. doi: 10.1128/jb.175.5.1344-1351.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Yue J., Jang J., Paul M. F. A new member of a family of ATPases is essential for assembly of mitochondrial respiratory chain and ATP synthetase complexes in Saccharomyces cerevisiae. J Biol Chem. 1994 Oct 21;269(42):26144–26151. [PubMed] [Google Scholar]
- Vallee B. L., Auld D. S. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990 Jun 19;29(24):5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- Weber E. R., Rooks R. S., Shafer K. S., Chase J. W., Thorsness P. E. Mutations in the mitochondrial ATP synthase gamma subunit suppress a slow-growth phenotype of yme1 yeast lacking mitochondrial DNA. Genetics. 1995 Jun;140(2):435–442. doi: 10.1093/genetics/140.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. W., Wilcox C. A., Flynn G. C., Chen E., Kuang W. J., Henzel W. J., Block M. R., Ullrich A., Rothman J. E. A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature. 1989 Jun 1;339(6223):355–359. doi: 10.1038/339355a0. [DOI] [PubMed] [Google Scholar]
- Yaffe M. P., Jensen R. E., Guido E. C. The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J Biol Chem. 1989 Dec 15;264(35):21091–21096. [PubMed] [Google Scholar]