Abstract
The presence of 3-O-sulfated glucosamine residues in heparin or heparan sulfate plays a role in binding to antithrombin III and HSV infection. In this study, tandem mass spectrometry was used to differentiate between two heparin disaccharide isomers containing variable sulfate at C6 in a common disaccharide and C3 in a more rare one. The dissociation patterns shown by MS2 and MS3 were clearly distinguishable between the isomers, allowing their differentiation and quantitation. Using this technique, we show that an octasaccharide with 11 sulfate groups with high affinity for inflammatory chemokine CCL2 does not contain 3-O-sulfated disaccharides.
Introduction
Heparin and heparan sulfate are glycosaminoglycans (GAGs) involved in a wide array of protein binding interactions. These interactions are implicated in a diverse group of biological processes such as viral invasion [1, 2], cell growth [3, 4], blood coagulation [5], and tumor metastasis [6–10]. Over a hundred different heparin binding proteins have been identified [11]. Initially, such protein interactions were believed to be relatively non-specific. However, this perception has evolved to include selective protein binding, as research demonstrates specific GAG structural motifs can drastically alter protein binding affinity [8, 12, 13]. Despite the role of GAG structure in protein binding, exploration of specific GAG structural motifs governing protein binding remains greatly hampered due to lack of rapid and definitive means to ascertain GAG substitution patterns.
Heparin and heparan sulfate consist of a repeating disaccharide subunit containing either glucuronic acid or iduronic acid connected via a β1–4 linkage to glucosamine. These GAGs possess significant structural diversity due to sugar stereochemistry and sulfation or acetylation at numerous positions within the repeating disaccharide subunit. The glucuronic acid and iduronic acid rings may display 2-O-sulfation, and the glucosamine ring may have 6-O-sulfation or the rare 3-O-sulfation. Glucosamine may also have either sulfation or acetylation at the primary amine [8, 9]. Sulfation and acetylation alone yield 24 possible substitution patterns.
Despite the rarity of 3-O-sulfation, this modification has been implicated in a number of biologically relevant pathways. Heparin binding to the serine protease inhibitor antithrombin III (ATIII) causes inhibition of two proteases, thrombin and factor Xa, involved in blood clotting leading to an anticoagulant effect [14]. Binding depends on a particular pentasaccharide sequence responsible for binding ATIII containing a 3-O-sulfated glucosamine [15]. This 3-O-sulfate is a critical structural component of the ATIII binding sequence as removal results in a thousand fold decrease in binding affinity [13]. HSV gD also binds a 3-O-sulfated sequence of heparan sulfate enabling viral entry into host cells [2, 16].
Researchers have employed numerous different methods to elucidate GAG structure, as well as the pertinent GAG protein binding sequence. Methods include NMR, but this technique is difficult to apply due to relatively large material requirements [8]. Other methods include chromatography to identify disaccharide components [17, 18] and mass spectrometry combined with selective enzymatic digestion and desulfation [19–21]. Mass spectrometry is a particularly attractive method for structural analysis of GAGs because of minimal sample requirements and relatively fast data acquisition. Instruments capable of collision induced dissociation (CID) provide an additional advantage as they possess the potential to disassociate GAG structures enabling identification of specific sulfation sites.
A class of proteins displaying GAG binding capacity are chemokines, small secreted proteins involved in numerous biological processes such as inflammation and development [22]. Research demonstrates that GAG/chemokine interaction can be required for proper in vivo function [23, 24]. Our laboratory identified selective chemokine ligand 2 (CCL2) binding of 11 sulfated heparin octasaccharide fragments [25]. This finding provides an opportunity to evaluate the role of 3-O-sulfation in a new GAG/protein interaction using mass spectrometry.
Herein, we report on the analysis of a known chemokine binding GAG octasaccharide [25], whose disaccharide composition has been determined and probed for 3-O-sulfation. Application of successive CID events generates cross-ring dissociation events separating the primary amine, the three position, and the six position of glucosamine enabling definitive identification of the specific site of sulfation within a GAG disaccharide subunit. This method was applied to an 11 sulfated heparin octasaccharide obtained via hydrophobic trapping of CCL2/heparin complex to evaluate the role of 3-O-sulfation in the CCL2/heparin interaction.
Experimental
Reagents and Instrumentation
Heparin octasaccharide library was purchased from Dextra (Reading, UK). CarboPac PA1 ion-exchange column was purchased from Dionex (Sunnyvale, CA). High-pressure liquid chromatography (HPLC) grade solvents were purchased from Fisher Scientific (Santa Clara, CA). SAX HPLC was purchased from Waters Corporation (Milford, MA). Heparin desalting was performed using 1 kDa Dispo-Biodialyzers (The Nest Group, Southborough, MA). Oasis HLB C18 column was purchased from Waters Corporation (Milford, MA). Heparinase I, II, and III were purchased from Seikagaku Corporation (East Falmouth, MA). I-S and I-P disaccharide standards were purchased from Sigma-Aldich (St. Louis, MO). Disaccharide stock solutions were prepared at a concentrations of 2 mM on the basis of their absorbance at 232 nm in 0.03 M HCl ε232 = 5500 M−1 cm−1) [26].
Mass spectra for compositional analysis were obtained by using an ESI source on an LTQ MS (ThermoFinnigan, San Jose, CA). Disaccharides were sprayed at a concentration of 20 pmol/µL in a 1:1 MeOH/H2O solution containing 10 mM NH4OH and 5 pmol/µL I-P internal standard. Samples were introduced by flow injection at 10 µL/min in 1:1 MeOH/H2O using a syringe pump (Harvard Apparatus, Holliston, MA). Spectra were obtained in negative ion mode by using a spray voltage of 3.5kV and a capillary temperature of 220°C for all experiments. For MS2 experiments, a 3 m/z isolation width was used for precursor ion selection at m/z 191.4, and the ion was activated at 20% normalized collision energy for 100 ms, and the qz value was maintained at 0.250. For MS3 experiments, a 3 Da isolation width was used for precursor ion selection at m/z 168.5, and the ion was activated at 22% normalized collision energy for 100 ms. Each mass spectrum obtained contains an average of 10 scans. Xcalibur, version 1.4 (ThermoFinnigan), was used for data acquisition and processing.
Generation of 3-O-sulfated di- and tetra-saccharides
A CHO cell line transduced with 3 copies of the human 3-OST-3A cDNA [27] were grown to confluence in 850 cm2 tissue culture roller bottles in Ham’s F-12 medium containing 50 U/ml penicillin, 50 ug/ml streptomycin, 2 mM glutamine, and 10% fetal bovine serum. For cellular GAG extraction, cells were washed once in PBS and subjected to exhaustive digestion with Pronase (Sigma/Aldrich) in PBS at 37°C for 24 hours followed by filtration through 0.22 micron cellulose acetate low protein binding membrane, anion-exchange chromatography with DEAE- sephacel (GE Healthcare, Chalfont St. Giles, UK) and ethanol precipitation as previously described [27]. The GAG chains were pelleted, washed with ice cold 70% ethanol, re-pelleted and dried. The GAG preparations were reconstituted in 100 ul HS digest buffer (40 mM ammonium acetate and 3.3 mM calcium acetate, pH 7) per roller bottle. Enzymatic depolymerization was carried out with 2 mU each of heparin lyases I, II, and III per roller bottle and digested for 16 hr at 37°C. In order to determine the retention times for specific disaccharides, a small sample of the digest was analyzed on a C18 reverse phase column (0.46 cm by 25 cm) (Grace Davison Discovery Sciences, Deerfield, IL) connected to a LCQ classic quadrupole ion trap mass spectrometer equipped with an ESI source and a quaternary HPLC pump (Thermo-Finnigan, San Jose, CA). The isocratic steps were: 100% buffer A (8 mM acetic acid, 5 mM dibutylamine) for 10 min; 17% buffer B (70% methanol, 8 mM acetic acid, 5 mM DBA) for 15 min; 32% buffer B for 15 min; 40% buffer B for 15 min; 60% buffer B for 15 min and 100% buffer B for 10 min. Eluted species were monitored in negative ion mode with the signal intensity optimized for D2S0, a representative disaccharide (GAG nomenclature described by Lawrence et al., 2008) [28]. The capillary temperature and spray voltage were kept at 140’C and 4.75 kV, respectively. Peak assignments were based on several criteria including retention time, mass, and ion adduction [27]. Preparative amounts of heparan sulfate digests were loaded onto the column and the 3-O-sulfated species were collected at the expected retention times from the divert valve attached to the mass spectrometer. Purity was checked by LC/MS using the same criteria previously mentioned and disaccharide yield approximated by comparing extracted ion current for D2S3 and D2S9 to that of a known amount of D2S6 run separately under the same conditions.
Preparation of 11-Sulfated Heparin Octasaccharide
Heparin octasaccharide containing 11 sulfates was isolated from the heparin octasaccharide library by using strong anion-exchange HPLC and a method similar to that described previously [29]. Approximately 400 µg of an octasaccharide library was injected onto a CarboPac PA1 column and SAX HPLC was performed at a flow rate of 1 mL/min using Solvent A (30 mM ammonium acetate, pH 3.5) and Solvent B (3 M NaCl, 30 mM ammonium acetate pH 3.5). A typical gradient consisted of: (1) 1–11 min, 0% B, (2) 11–110 min, 0–100% B, and (3) 111–120 min, 0% B. Chromatograms were recorded by monitoring the UV absorbance at 232 nm. Fractions corresponding to the 11-sulfated octasaccharide were collected, lyophilized and extensively desalted by dialysis against water using 1 kDa Dispo-Biodialyzers.
Preparation of CCL2
The CCL2 protein was expressed, purified, and processed as previously described [30]. Briefly, CCL2 was expressed using a codon-optimized construct in TAP302 cells. Soluble CCL2 was purified by ion-exchange chromatography followed by reverse-phase high-performance liquid chromatography (RP-HPLC). The N-terminal methionine was removed by aminopeptidase, leaving authentic human CCL2 that was then subject to re-purification by RP-HPLC.
CCL2 Affinity Enrichment of 11-Sulfated Octasaccharide
Octasaccharide was enriched by incubating 40 µM CCL2 with 200 µM 11-sulfate octasaccharide in 100 µL of 100 mM NH4OAc solution (pH 6.8). The solution was applied to an Oasis solid-phase extraction (SPE) cartridge (Waters, Milford, MA) that had been previously conditioned using 1 mL of methanol and then 1 mL of H2O. The solid-phase SPE cartridge was flushed three times, each using 1 mL 200 mM NH4OAc solution (pH 6.8) and then eluted by 1 mL 760 mM NH4OAc (pH 6.8). The three washing fractions and the final elution faction were collected and desalted by dialyzing against water using a dialyzer with a molecular weight cutoff (MWCO) of 1 kDa.
Compositional Analysis of Affinity Purified Heparin Octasaccharide
Complete enzymatic digestion of heparin octasaccharide to constituent disaccharides was carried out with 5 µg heparin in 20 µL of 20 mM ammonium acetate buffer, pH 7.5, 2 mM calcium acetate, and 0.001 unit each of heparinase I, heparinase II, and heparinase III, which was incubated at 37°C for 16 h [20]. The reaction was quenched yielding a final solution containing 20 pmol/µL heparin disaccharide, 1:1 MeOH/H2O, 5 pmol/µL internal standard I-P, and 10 mM ammonium hydroxide. Samples were then analyzed using a combination of MS2 at m/z 191.4 and MS3 at 168.5 on an LTQ mass spectrometer.
Results and Discussion
Mass spectra were collected for both the D2S6 disaccharide and the D2S3 disaccharide. As expected for positional isomers, the MS1 data for each of the disaccharides were virtually indistinguishable; in both cases the triply charged ion at m/z 191.4, representing a molecular weight of 574.43 Da, was the principle ion detected (Supplementary Figure 1). These ions were isolated and studied utilizing tandem mass spectrometry via CID.
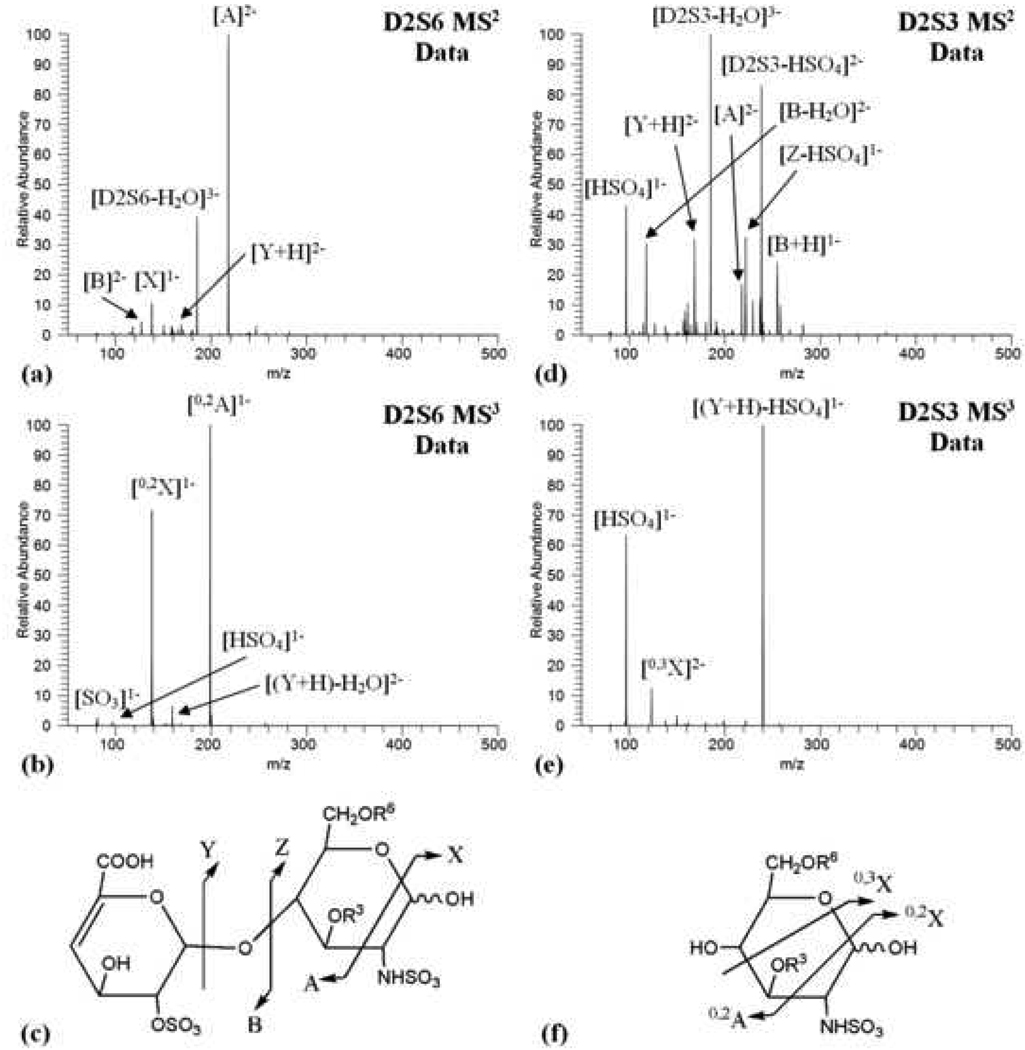
The MS2 data for the disaccharide isomers displayed different dissociation patterns, and are clearly distinguishable (Figure 1). The D2S6 CID spectra (Figure 1a) was dominated by cross-ring cleavage yielding ion A at m/z 218.0, and by a water loss ion at m/z 185.3 (Figure 1c) (dissociation nomenclature described by Domon et al., 1988) [31]. Dissociation also resulted in the formation of X, Y, and B ions. Ion X, at m/z 138.0, is singly charged and results from a cross ring cleavage of the glucosamine residue, while ions Y and B, at m/z 168.5 and m/z 127.0 respectively, are the glycosidic bond cleavage ions. The D2S3 CID spectra (Figure 1d) displayed greater diversity. The two major species resulted again from loss of water at m/z 185.3 and a HSO4 loss ion appearing at m/z 238.5. Additional abundant ions include a singly charged HSO4 at m/z 97 and ion B with a water loss at m/z 118.0. Trace amounts of ion A at m/z 218.0 were also detected as well as other dissociation species annotated in Figure 1c.
Figure 1.
(a) D2S6 MS2 data (b) D2S6 MS3 data (c) cross ring and glycosidic cleavage pattern observed in MS2 dissociation (d) D2S3 MS2 data (e) D2S3 MS3 data (f) cross ring cleavage pattern observed in MS3 dissociation
While the MS2 data of the D2S6 and D2S3 disaccharides display clear differences in dissociation, thus preliminary differentiation is possible, none of the observed fragments correspond to a cross ring cleavage separating the 6 and 3 positions of the glucosamine ring enabling definitive identification of the 3-O-sulfation site. It is most intriguing that one simple modification of 3-O versus 6-O-sulfation results in such drastically different spectra. This effect was also quite pronounced in earlier investigation of disaccharides carrying C-3 modifications [32, 33]. Since both D2S3 and D2S6 produced the Y ion at m/z 168.5 in the MS2 data, corresponding to the glucosamine ring containing the structural variation, this ion was isolated and subjected to further dissociation in an MS3 experiment. Similar to the MS2 data, the MS3 data displayed differing dissociation patterns. The MS3 data of the D2S6 disaccharide (Figure 1b) revealed ion 0,2A at m/z 199.0 and ion 0,2X at m/z 138.0 with high abundance. The parent ion with water loss was also observed at m/z 159.5. Only trace HSO4 ions at m/z 97.0 and SO3 ions at m/z 80.0 were detected. The MS3 of the D2S3 disaccharide (Figure 1e) contained two major ions; the precursor ion with HSO4 loss at m/z 240.0 and the HSO4 ion at m/z 97.0. The less abundant 0,3X ion appears at m/z 123.5. The 0,3X ion corresponds to an ion containing an N-sulfate and a 3-O-sulfate, resulting from cross-ring cleavage separating the 0,3X ion from the 6 position of the glucosamine ring.
The 0,3X ion permits definitive identification of the D2S3 sulfation pattern in the MS3 data, and validates CID as a rapid, effective tool to identify 3-O-sulfation in GAG disaccharide structures with minimal sample material. Interestingly, the D2S3 disaccharide standard also displayed a high abundance of HSO4 ions in contrast to the D2S6 disaccharide standard suggesting the 3-O-sulfate is more labile than sulfates on other positions when the species undergo CID. Previously, our laboratory developed a quantitative method to determine GAG disaccharide composition [20] including all non-3-O-sulfated species. Identification of several diagnostic ions, such as m/z 238.5 in MS2 and m/z 240 in MS3, in D2S3 samples enables incorporation of this 3-O-sulfated species into existing mass spectrometry methodology for GAG quantitative compositional analysis.
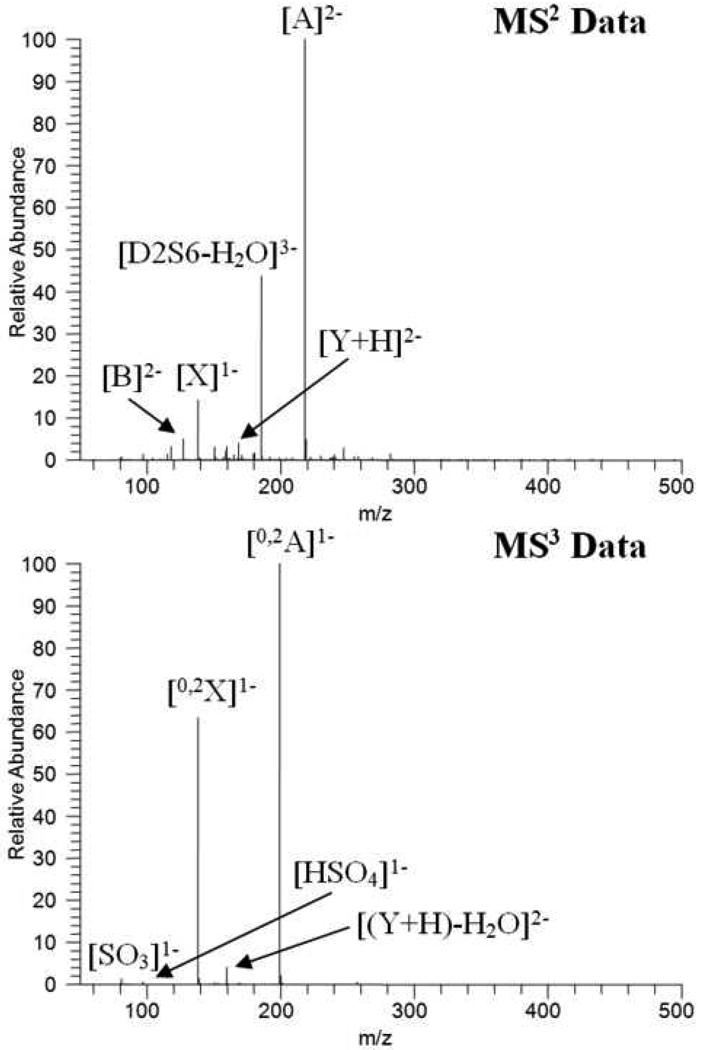
This CID based method was used to explore the possible involvement of 3-O-sulfation in the heparin/CCL2 interaction. A fraction of 11-sulfated heparin octasaccharide, previously determined to bind to CCL2 [25], was obtained via strong anion exchange chromatography and incubated with CCL2 protein. The protein was bound to a hydrophobic trapping column, and an 11-sulfated heparin fraction enriched for CCL2 affinity was eluted from the column. This enriched eluent, and a control 11-sulfated heparin octasaccharide fraction were enzymatically digested down to disaccharide constituents [29]. The sulfated disaccharide ion obtained at m/z 191.4 in the MS1 data was subjected to two successive rounds of CID to probe for the presence of 3-O-sulfation. The MS2 and MS3 data obtained from both reaction mixtures were virtually identical to the MS2 and MS3 data obtained from the D2S6 disaccharide standard (Figure 2) asserting that the 3-sulfated disaccharide subunits possessed a D2S6 structure. The absence of numerous diagnostic ions observed in the D2S3 standard, such as m/z 238.5 in the MS2 data, m/z 240.0 in the MS3 data, and the 0,3X ion in the MS3 data, illustrates that the D2S3 structure was not present. The D0S9 disaccharide is not a possibility since all three sulfates reside in the reducing ring, and the mass of the prominent glycosidic bond cleavage ion clearly precludes this possibility. The D2H9 isomer is also eliminated since the mass of the A ion does not include the extra sulfate. It is clear that neither of these 2 isomers is present.
Figure 2.
Spectra of CCL2 affinity enriched 11-sulfated heparin fraction showing same product ions as observed for the D2S6 isomer (a) MS2 data (b) MS3 data
Several GAG/protein interactions depend on 3-O-sulfation [2, 13, 15, 16]. In contrast, our exploration of the heparin/CCL2 interaction, using mass spectrometry combined with CID, did not reveal measurable levels of 3-O-sulfate containing species, suggesting that 3-O-sulfation may not be involved in chemotactic GAG/protein interactions. Subsequently, while 3-O-sulfation may be crucial for certain interactions, its function could be limited to specific GAG/protein complexes instead of playing a more universal role in chemotaxis or protein binding.
Conclusion
We have utilized a mass spectrometry method using CID to create cross-ring cleavage events separating the numerous potential sulfation positions within the disaccharide subunit structure. This method allows for robust, rapid detection and identification of individual 3-O-sulfate containing heparin structures, such as D2S3, within a heterogeneous disaccharide mixture via identification of specific diagnostic ions. Exploration of GAG/protein interaction has been greatly hampered due to lack of effective means to investigate GAG structure, and this tool presents a means to improve our understanding of the relationship between GAG structure and protein binding affinity for individual GAG/protein interactions.
This method was used to probe the possible role of 3-O-sulfation in the CCL2/GAG interaction, from a heparin library. An 11-sulfated heparin octasaccharide enriched for CCL2 binding affinity did not reveal the presence 3-O-sulfation at a detectable level suggesting 3-O-sulfation is not a prominent factor governing CCL2/GAG binding.
Supplementary Material
MS 1 data
Table 1.
Ions observed in the MS2 and MS3 data
| MS2 Spectra of D2S6 Standard | MS3 Spectra of D2S6 Standard | ||||
|---|---|---|---|---|---|
| m/z | Ion | Charge | m/z | Ion | Charge |
| 127.0 | B | −2 | 80.0 | SO3 | −1 |
| 138.0 | X | −1 | 97.0 | HSO4 | −1 |
| 168.5 | Y+H | −2 | 138.0 | 0,2X | −1 |
| 185.3 | Parent-H2O | −3 | 159.5 | (Y+H)-H2O | −2 |
| 218.0 | A | −2 | 199.0 | 0,2A | −1 |
| MS2 Spectra of D2S3 Standard | MS3 Spectra of D2S3 Standard | ||||
| m/z | Ion | Charge | m/z | Ion | Charge |
| 97.0 | HSO4 | −1 | 97.0 | HSO4 | −1 |
| 118.0 | B-H2O | −2 | 123.5 | 0,3X | −2 |
| 168.5 | Y+H | −2 | 240.0 | (Y+H)-HSO4 | −1 |
| 185.3 | Parent-H2O | −3 | |||
| 218.0 | A | −2 | |||
| 222.0 | Z-HSO4-H | −1 | |||
| 238.5 | Parent-HSO4 | −2 | |||
| 255.0 | B+H | −1 | |||
Acknowledgments
The authors thank the National Institute of Health (GM33063 and GM063581) for their financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM. Dengue Virus Infectivity Depends on Envelope Protein Binding to Target Cell Heparan Sulfate. Nature Medicine. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 2.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. A Novel Role for 3-O-Sulfated Heparan Sulfate in Herpes Simplex Virus 1 Entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 3.Castellot JJ, Hoover RL, Harper PA, Karnovsky MJ. Heparin and Glomerular Epithelial Cell-Secreted Heparin-Like Species Inhibit Mesangial-Cell Proliferation. The American Journal of Pathology. 1985;120:427–435. [PMC free article] [PubMed] [Google Scholar]
- 4.Perrimon N, Bernfield M. Specificities of Heparan Sulphate Proteoglycans in Developmental Processes. Nature. 2000;404:725–728. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- 5.Marcum JA, McKenney JB, Galli SJ, Jackman RW, Rosenberg RD. Anticoagulantly Active Heparin-Like Molecules from Mast Cell-Deficient Mice. The American Journal of Physiology. 1986;250:H879–H888. doi: 10.1152/ajpheart.1986.250.5.H879. [DOI] [PubMed] [Google Scholar]
- 6.Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I, Spector L, Pecker I. Mammalian Heparanase: Gene Cloning, Expression and Function in Tumor Progression and Metastasis. Nature Medicine. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 7.Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Cloning of Mammalian Heparanase, an Important Enzyme in Tumor Invasion and Metastasis. Nature Medicine. 1999;5:803–809. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 8.Rabenstein DL. Heparin and Heparan Sulfate: Structure and Function. Natural Product Reports. 2002;19:312–331. doi: 10.1039/b100916h. [DOI] [PubMed] [Google Scholar]
- 9.Bishop JR, Schuksz M, Esko JD. Heparan Sulphate Proteoglycans Fine-Tune Mammalian Physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 10.Esko JD, Selleck SB. Order out of Chaos: Assembly of Ligand Binding Sites in Heparan Sulfate. Annual Review of Biochemistry. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 11.Conrad HE. Heparin-Binding Proteins. San Diego, CA: Academic press; 1998. pp. 1–5. 183–202. [Google Scholar]
- 12.Lindahl U, Kusche-Gullberg M, Kjellén L. Regulated Diversity of Heparan Sulfate. Journal of Biological Chemistry. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 13.Atha DH, Lormeau JC, Petitou M, Rosenberg RD, Choay J. Contribution of Monosaccharide Residues in Heparin Binding to Antithrombin Iii. Biochemistry. 1985;24:6723–6729. doi: 10.1021/bi00344a063. [DOI] [PubMed] [Google Scholar]
- 14.de Kort M, Buijsman RC, van Boeckel CA. Synthetic Heparin Derivatives as New Anticoagulant Drugs. Drug Discovery Today. 2005;10:769–779. doi: 10.1016/S1359-6446(05)03457-4. [DOI] [PubMed] [Google Scholar]
- 15.Petitou M, Casu B, Lindahl U. 1976–-1983, a Critical Period in the History of Heparin: The Discovery of the Antithrombin Binding Site. Biochimie. 2003;85:83–89. doi: 10.1016/s0300-9084(03)00078-6. [DOI] [PubMed] [Google Scholar]
- 16.Tiwari V, Clement C, Xu D, Valyi-Nagy T, Yue BY, Liu J, Shukla D. Role for 3-O-Sulfated Heparan Sulfate as the Receptor for Herpes Simplex Virus Type 1 Entry into Primary Human Corneal Fibroblasts. Journal of Virology. 2006;80:8970–8980. doi: 10.1128/JVI.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Liu J. Characterization of the Structure of Antithrombin-Binding Heparan Sulfate Generated by Heparan Sulfate 3-O-Sulfotransferase 5. Biochimica et Biophysica Acta. 2005;1725:190–200. doi: 10.1016/j.bbagen.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Shriver Z, Blaiklock P, Yoshida K, Sasisekharan R, Rosenberg RD. Heparan Sulfate D-Glucosaminyl 3-O-Sulfotransferase-3a Sulfates N-Unsubstituted Glucosamine Residues. Journal of Biological Chemistry. 1999;274:38155–38162. doi: 10.1074/jbc.274.53.38155. [DOI] [PubMed] [Google Scholar]
- 19.Venkataraman G, Shriver Z, Raman R, Sasisekharan R. Sequencing Complex Polysaccharides. Science. 1999;286:537–542. doi: 10.1126/science.286.5439.537. [DOI] [PubMed] [Google Scholar]
- 20.Saad OM, Leary JA. Compositional Analysis and Quantification of Heparin and Heparan Sulfate by Electrospray Ionization Ion Trap Mass Spectrometry. Analytical Chemistry. 2003;75:2985–2995. doi: 10.1021/ac0340455. [DOI] [PubMed] [Google Scholar]
- 21.Saad OM, Ebel H, Uchimura K, Rosen SD, Bertozzi CR, Leary JA. Compositional Profiling of Heparin/Heparan Sulfate Using Mass Spectrometry: Assay for Specificity of a Novel Extracellular Human Endosulfatase. Glycobiology. 2005;15:818–826. doi: 10.1093/glycob/cwi064. [DOI] [PubMed] [Google Scholar]
- 22.Rossi D, Zlotnik A. The Biology of Chemokines and Their Receptors. Annal Review of Immunology. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 23.Proudfoot AE, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TN, Kosco-Vilbois MH. Glycosaminoglycan Binding and Oligomerization Are Essential for the in Vivo Activity of Certain Chemokines. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson Z, Kosco-Vilbois MH, Herren S, Cirillo R, Muzio V, Zaratin P, Carbonatto M, Mack M, Smailbegovic A, Rose M, Lever R, Page C, Wells TN, Proudfoot AE. Interference with Heparin Binding and Oligomerization Creates a Novel Anti-Inflammatory Strategy Targeting the Chemokine System. Journal of Immunology. 2004;137:5776–5785. doi: 10.4049/jimmunol.173.9.5776. [DOI] [PubMed] [Google Scholar]
- 25.Yu Y, Sweeney MD, Saad OM, Crown SE, Hsu AR, Handel TM, Leary JA. Chemokine-Glycosaminoglycan Binding: Specificity for Ccr2 Ligand Binding to Highly Sulfated Oligosaccharides Using Fticr Mass Spectrometry. Journal of Biological Chemistry. 2005;280:32200–32208. doi: 10.1074/jbc.M505738200. [DOI] [PubMed] [Google Scholar]
- 26.Desai UR, Wang HM, Linhardt RJ. Specificity Studies on the Heparin Lyases from Flavobacterium Heparinum. Biochemistry. 1993;32:8140–8145. doi: 10.1021/bi00083a012. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence R, Kuberan B, Lech M, Beeler DL, Rosenberg RD. Mapping Critical Biological Motifs and Biosynthetic Pathways of Heparan Sulfate. Glycobiology. 2004;14:467–479. doi: 10.1093/glycob/cwh057. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence R, Lu H, Rosenberg RD, Esko JD, Zhang L. Disaccharide Structure Code for the Easy Representation of Constituent Oligosaccharides from Glycosaminoglycans. Nature Methods. 2008;5:291–292. doi: 10.1038/nmeth0408-291. [DOI] [PubMed] [Google Scholar]
- 29.Sweeney MD, Yu Y, Leary JA. Effects of Sulfate Position on Heparin Octasaccharide Binding to Ccl2 Examined by Tandem Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2006;17:1114–1119. doi: 10.1016/j.jasms.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 30.Paavola CD, Hemmerich S, Grunberger D, Polsky I, Bloom A, Freedman R, Mulkins M, Bhakta S, McCarley D, Wiesent L, Wong B, Jarnagin K, Handel TM. Monomeric Monocyte Chemoattractant Protein-1 (Mcp-1) Binds and Activates the Mcp-1 Receptor Ccr2b. Journal of Biological Chemistry. 1998;273:33157–33165. doi: 10.1074/jbc.273.50.33157. [DOI] [PubMed] [Google Scholar]
- 31.Domon B, Costello CE. A Systematic Nomenclature for Carbohydrate Fragmentations in Fab-Ms/Ms Spectra of Glycoconjugates. Glucoconjugate Journal. 1988;5:397–409. [Google Scholar]
- 32.Staempfli A, Zhou Z, Leary JA. Gas-Phase Dissociation Mechanisms of Dilithiated Disaccharides: Tandem Mass Spectrometry and Semiempirical Calculations. Journal of Organic Chemistry. 1992;57:3590–3594. [Google Scholar]
- 33.Leary JA, Zhou Z, Ogden S, Williams TD. Gas Phase Investigation of Lithiated Peptide Adducts: Tandem Mass Spectrometry and Semiempirical Studies. Journal of the American Society for Mass Spectrometry. 1990;1:473. doi: 10.1016/1044-0305(90)85030-P. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MS 1 data