Abstract
During intracellular membrane trafficking and remodeling, protein complexes known as the ESCRTs interact with membranes and are required for budding processes directed away from the cytosol, including the budding of intralumenal vesicles to form multivesicular bodies, for the budding of some enveloped viruses, and for daughter cell scission in cytokinesis. Here we found that the ESCRT-III proteins CHMP2A and CHMP3 could assemble in vitro into helical tubular structures that expose their membrane interaction sites on the outside of the tubule while the AAA-type ATPase VPS4 could bind on the inside of the tubule and disassemble the tubes upon ATP hydrolysis. CHMP2A and CHMP3 co-polymerized in solution and their membrane targeting was cooperatively enhanced on planar lipid bilayers. Such helical CHMP structures could thus assemble within the neck of an inwardly-budding vesicle, catalyzing late steps in budding under the control of VPS4.
ESCRT (Endosomal Sorting Complexes Required for Transport) complexes 0, I, II, III and accessory proteins regulate cell surface receptor sorting into intralumenal endosomal vesicles, generating multivesicular bodies (MVB) (1-3). ESCRTs are also recruited during budding of some enveloped viruses (4) and cytokinesis (5, 6), processes that are topologically similar to vesicle budding into endosomes.
Yeast expresses six ESCRT-III-like proteins (7) whereas mammalian cells express ten, known as charged multivesicular body protein (CHMP) 1 to 6 (8). C-terminally truncated CHMP3 has a four helical bundle core and two regions that are important for CHMP polymerization and membrane targeting (9). CHMPs exist in an auto-inhibited state in the cytosol (10, 11); removal of autoinhibition induces membrane targeting (9, 12, 13) and ESCRT-III assembly into a putative protein lattice (1, 2). Over-expression of SNF7/CHMP4 in mammalian cells produces filaments that induce outward buds in the presence of catalytically inactive VPS4 (14), but little is known about heteromeric polymerization by ESCRT-III proteins.
The recruitment of the AAA-type ATPase VPS4 is essential for the termination of the budding process and catalyzes disassembly of the complex (2, 15, 16). The central role of ESCRT-III and VPS4 in all known ESCRT catalyzed budding events is further underlined by the inhibitory effects of dominant negative mutants of ESCRT-III (8, 9, 17, 18) and VPS4 (2, 5, 18) on MVB formation, HIV-1 budding and cytokinesis.
Yeast Vps2p (CHMP2) and Vps24p (CHMP3) form a subcomplex (7) consistent with the hetero-dimerization potential suggested by the CHMP3 crystal structure (9). Such subcomplexes may thus represent the building blocks for polymerization. We thus set out to study the interactions and polymerization mode of CHMP2 and CHMP3 in vitro. Interactions between full length, auto-inhibited CHMP3 and CHMP2A could not be detected, and so both proteins were produced as C-terminal truncations (fig S1A; CHMP2AΔC and CHMP3ΔC) in their proposed activated forms (9, 10, 13). While CHMP3ΔC was monodisperse in solution (9), CHMP2AΔC fused to the maltose binding protein (MBP) formed monomers and concentration-dependent aggregates (fig S1B). Co-incubation of monomeric CHMP2AΔC and CHMP3ΔC led to the formation of oligomers, migrating to the bottom fraction of a sucrose gradient (Fig. 1A). CHMP2AΔC and CHMP3ΔC assembled into long tubular structures (Fig. 1B) with the majority of the particles revealing a diameter of ~40 nm and a small fraction showing deviations up to 70 nm in negative staining electron microscopy (EM).
Figure 1.
CHMP2AΔC-CHMP3ΔC polymer formation. (A) Sucrose density gradient analysis of CHMP2AΔC-CHMP3ΔC complex formation.
(B) Negative staining EM of tubular structures formed by CHMP2AΔC and CHMP3ΔC.
(C) Negative staining EM of CHMP2AΔC and (D) of CHMP2AΔC after removal of MBP. (Scale bars, 100 nm; insets 50 nm).
Although neither CHMP3, CHMP3ΔC nor CHMP2A formed higher order oligomers on their own, CHMP2AΔC sedimented in a sucrose gradient (fig S1C) and formed ring-like structures (Fig. 1C). Removal of MBP produced particles with inner and outer diameters of ~12 nm and ~ 30 nm, respectively. Increased aggregation upon MBP cleavage and the exclusive face-up orientations (Fig. 1D) hindered determination of the ring thickness.
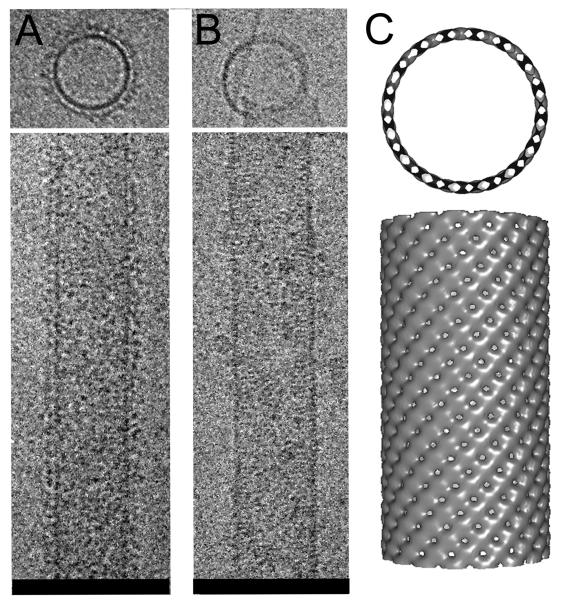
Cryo-EM of the CHMP2AΔC-CHMP3ΔC polymer showed tubes with a fuzzy surface (Fig. 2A). Removal of MBP from CHMP2AΔC rendered the surface smooth, revealing striations perpendicular to the longitudinal axis of the tube (Fig. 2B). Because image analyses of MBP-cleaved tubes was hindered by their aggregation, the iterative helical real space reconstruction algorithm (19) was applied to reconstruct volumes from the tubes containing MBP attached to CHMP2AΔC (20). The 32 Å pitch of the helical assembly was determined by the Fourier transform of the images (20). The calculated helical structure contains 16.57 repeating units per turn, with inner and outer diameters of 43 nm and 52 nm, respectively (Fig. 2C). The CHMP2A-CHMP3 dimer model could fit into the repeating unit of the EM map (20) (fig. S2), exposing the membrane targeting surface to the outside and the VPS4B interaction site towards the inside of the tube (fig. S2D and E).
Figure 2.
Cryo EM of CHMP2ΔC-CHMP3ΔC tubes (A); (B) tubes after removal of MBP from CHMP2ΔC.
(C) The EM reconstruction model showing the 45 Å width of the helical structure produced by the CHMP lattice (top view). (Scale bars 100 nm).
The organization of the CHMP2AΔC-CHMP3ΔC polymer did not indicate how other CHMP proteins could participate in the same polymer. In fact, monomeric CHMP4BΔC did not integrate into CHMP2AΔC-CHMP3ΔC tubes (fig. S3), despite similarities in polymerization (14). Although CHMP4 may participate in CHMP2A-CHMP3 tube formation as a CHMP4-CHMP6 subcomplex (7), different ESCRT-III complexes may be formed in vivo that interact (7) and act sequentially.
Because the VPS4B-CHMP interaction requires an intact C-terminus (21, 22) full-length proteins were employed for tube formation. While CHMP2AΔC and CHMP3 formed similar tubes to those formed with CHMP3ΔC (Figs. 1B and 3A), combinations of CHMP2A and CHMP3 or CHMP2A and CHMP3ΔC did not polymerize. In sucrose gradient centrifugation, isolated forms of CHMP2AΔC, CHMP3 and VPS4B floated at similar positions (Figs. 3B, C, D), consistent with monomeric or dimeric VPS4B in the absence of ATP (23). Mixing all three proteins recruited them to the pellet fraction, corroborating the CHMP3-VPS4B interaction (Fig. 3E). Negative staining EM of the CHMP2AΔC-CHMP3 tubes assembled in the presence of VPS4B revealed additional electron dense material along the longitudinal axis of the tubes (Fig. 3G). Radial density profiles of cross sections obtained by cryo-EM (fig. S4B) showed a broad central peak, confirming the VPS4B presence inside the tubes, followed by a sharp peak corresponding to the CHMP protein lattice and a smaller peak corresponding to MBP attached to CHMP2AΔC (Fig. 3H and figs.S4D). In contrast, in cross sections of tubes without VPS4B (fig.S4A), the central peak of the density profile did not rise above the background signal (Fig. 3I and fig. S4C). Thus, VPS4B used CHMP2AΔC-CHMP3 tubes as a scaffold for assembly, which apparently produced disordered VPS4B oligomers in the absence of ATP. Addition of ATP and Mg2+ to the CHMP2AΔC-CHMP3-VPS4B tubular structures induced disassembly of the tubes (Fig. 3F and J). Incubation of fluorescein-labeled tubes (fig. S5) with buffer, VPS4B or VPS4B plus adenylyl-imidodiphoshate (AMP-PNP) Mg2+ showed no change in emission intensity as a function of time (Fig. 3K). In contrast, dequenching indicating disassembly was measured as an increase in emission intensity when the tubes were incubated with VPS4B plus ATP Mg2+ (Fig. 3K). Thus, VPS4B induced tube disassembly in vitro.
Figure 3.
CHMP2ΔC-CHMP3 tube disassembly by VPS4B.
(A) Negative staining EM of tubes formed by CHMP2AΔC and CHMP3.
(B) Sucrose gradient analysis of CHMP2AΔC; (C) CHMP3; (D) VPS4B; (E) CHMP2AΔC-CHMP3-VPS4B complex formation (F) CHMP2AΔC-CHMP3-VPS4B complexes after incubation with ATP Mg2+.
(G) Negative staining EM of CHMP2AΔC-CHMP3 tubes revealing VPS4B on the inside.
(H) Radial density profile of a CHMP2AΔC-CHMP3-VPS4B and (I) of CHMP2AΔC-CHMP3 tubes calculated across the cross section of the tube.
(J) Negative staining EM after adding ATP Mg2+ to CHMP2AΔC-CHMP3-VPS4B tubes.
(K) Disassembly of fluorescein-labeled CHMP2AΔC-CHMP3 tubes measured by change in emission intensity upon addition of (i) HBS (magenta), (ii) 10 μM VPS4B (blue), (iii) 10 μM VPS4B plus 100μM AMP-PNP Mg2+ (green), (iv) 5 μM VPS4B plus 50μM ATP Mg2+ (black) and, (v) 10 μM VPS4B plus 100μM ATP Mg2+ (red). Inset shows the fluorescein-labeled CHMP2AΔC and CHMP3 visualized on an SDS-PAGE. (Scale bars 100 nm).
CHMPs are selectively targeted to cellular membranes (7, 13, 24), which requires an extended basic surface in case of CHMP3 (9). Using Reflectometric interference spectroscopy (RIfS) (20) CHMP2AΔC, CHMP3ΔC and their complex showed no significant mass deposition (< 0.10 ng/mm2) on silica supported bilayers composed of 1-Stearoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine (SOPC) lipids (Fig.4A). However, CHMP3ΔC bound to 1,2-Dioleoyl-sn-Glycero-3-Phosphoserine (DOPS):SOPC bilayers (fig. S6) with an equilibrium binding amplitude (Γeq) of 1.08 ng/mm2 and dissociated with a kd of ~ 0.3 s−1 while CHMP2AΔC (4 μM) showed Γeq of 1.36 ng/mm2 and a kd of 0.081 s−1 (Fig.4B). When the two proteins were mixed, a Γeq of 5.95 ng/mm2 was reached. An apparent kd of > 0.0008 s−1 was determined by fitting a mono-exponential function to the dissociation phase, which is by 3 and 2 orders of magnitude lower as compared to the single proteins (fig. S7). Both SOPC and DOPS:SOPC bilayers showed no interaction with the control protein MBP (Figs. 4A and B). Dissociation of bound CHMP2AΔC and CHMP3ΔC in HEPES buffered saline (HBS) containing 1 M NaCl revealed a dissociation kd of ≥ 1 s−1, which was much faster than dissociation in HBS alone. In contrast, the CHMP2AΔC-CHMP3ΔC polymer did not dissociate with a higher rate (kd of ≤ 0.00046 s−1) in the presence of 1 M NaCl (fig. S7), indicating resistance to change in ionic strength. Once assembled on membranes CHMP2AΔC-CHMP3ΔC did not exchange with soluble or membrane-bound CHMP3ΔC (fig. S8). Thus, CHMP2A-3 complexes assembled on membranes in vitro in the absence of CHMP4-6 subcomplexes, even though yeast Snf7-Vps20 (CHMP4-6) complexes may recruit Vps2-Vps24 (CHMP2A-3) complexes to membranes in vivo (7).
Figure 4.
CHMP protein membrane interaction.
(A) Binding curves measured by RIfS on a SOPC bilayer; (i) MBP (black), (ii) CHMP3ΔC (magenta), (iii) CHMP2AΔC (green); (iv) CHMP2AΔC-CHMP3ΔC (blue).
(B) Binding curves measured by RIfS on a DOPS:SOPC bilayer; (i) MBP (black); (ii) CHMP3ΔC (magenta); (iii) CHMP2AΔC (green); (iv) CHMP2AΔC-CHMP3ΔC (blue). Protein injections were followed by injection of 1M NaCl in HBS.
(C) CHMP2AΔC-CHMP3ΔC tubes with and (D) without MBP attached to CHMP2AΔC.
(E) CHMP2AΔC-CHMP3ΔC tubes assembled in the presence of SOPC LUVs.
(F) CHMP2AΔC-CHMP3ΔC tubes assembled in the presence of 0.5 mg/ml (left panel) and 1.5mg/ml (right panel) SOPC: DOPS LUVs are shorter; (G) reveal single helical coils and (H) are often cone shaped. (Scale bars 100 nm).
To assess the influence of CHMP2AΔC-CHMP3ΔC tubes on membrane shapes, large unilamellar vesicles (LUVs) composed of DOPS:SOPC were used. LUV incubation with either CHMP2AΔC or CHMP3ΔC had no effect on their floatation in sucrose gradients (figs. S9A and B), while preformed CHMP2AΔC-CHMP3ΔC tubes restricted LUV floatation to the middle of the gradient (figs. S9C and D). Negative staining EM confirmed CHMP2AΔC-CHMP3ΔC tube membrane interaction via their outer surface (Fig.4C and D). However, no systematic remodeling of the LUV membranes was observed. Potential membrane remodeling by the CHMP co-polymer or vice versa was further explored by assembling the polymer in the presence of LUVs. While CHMP2AΔC-CHMP3ΔC assembly in the presence of SOPC LUVs had no effect on tube morphology (Fig.4E), the presence of DOPS:SOPC LUVs produced shorter tubes (Fig.4F), displaying loose helical coils (Fig. 4G) and cone shaped tubes that appeared “closed” at the narrower end (Fig.4H). This thus suggests a mechanism where lipid interaction affects CHMP protein polymerization.
Because modified VPS4 and CHMP3 exert dominant negative effects on HIV-1 budding (8, 9, 17, 25) and cytokinesis (6, 18) CHMP2A-CHMP3-VPS4 complexes may catalyze a common step such as membrane fission. The CHMP2A-CHMP3 polymer presents a membrane binding topology that is inverse to that of dynamin membrane complexes, (26) which catalyze endocytotic vesicle abscission (27). ESCRT-III coupled to VPS4 may exert a similar role in budding processes directed away from the cytosol. We thus propose that a helical CHMP2A-CHMP3 polymer assembles on the inside of a membrane bud which may induce membrane deformation leading to constriction and eventually abscission when coupled to VPS4 activity, the only energy-providing candidate in the pathway (2, 15)(fig. S10).
Supplementary Material
Acknowledgments
We thank X. Siebert (IBS) for advice on UROX. This work was supported by Deutsche Forschungsgemeinschaft (SPP 1175) (W. W.), by the ANRS (W. W.), University Joseph Fourier (W. W.), the ANR (G. S.), the CNRS (G. S.), the NIH (grant AI29873, H.G.) and post-doctoral fellowships from EMBO (S. L.) and the International Human Frontier Science Program Organization (S. L.). The EM map has been deposited at the European Bioinformatics Institute, accession code EMD-1536.
References and Notes
- 1.Williams RL, Urbe S. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 2.Saksena S, Sun J, Chu T, Emr SD. Trends Biochem Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Hurley J. Curr Opin Cell Biol. 2008;20(1):4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniasz PD. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 5.Carlton JG, Martin-Serrano J. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 6.Morita E, et al. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 8.von Schwedler UK, et al. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 9.Muziol T, et al. Dev Cell. 2006;10:821–30. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Zamborlini A, et al. Proc. Natl. Acad. Sci. USA. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lata S, et al. J Mol Biol. 2008;378:818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. J Biol Chem. 2005;280:12799–809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 13.Shim S, Kimpler LA, Hanson PI. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 14.Hanson PI, Roth R, Lin Y, Heuser JE. J Cell Biol. 2008;180(2):389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babst M, Wendland B, Estepa EJ, Emr SD. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishop N, Woodman P. Mol. Biol. Cell. 2000;11:227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strack B, Calistri A, Popova E, Gőttlinger H. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 18.Dukes JD, Richardson JD, Simmons R, Whitley P. Biochem J. 2008;411(2):233–9. doi: 10.1042/BJ20071296. [DOI] [PubMed] [Google Scholar]
- 19.Egelman EH. Ultramicroscopy. 2000;85:225–34. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 20.Materials and methods are available as supporting materials on Science Online.
- 21.Stuchell-Brereton MD, et al. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 22.Obita T, et al. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 23.Scott A, et al. EMBO J. 2005 [Google Scholar]
- 24.Whitley P, et al. J. Biol. Chem. 2003;278:38786–38795. doi: 10.1074/jbc.M306864200. [DOI] [PubMed] [Google Scholar]
- 25.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Proc Natl Acad Sci U S A. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sweitzer S, Hinshaw J. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 27.Takei K, McPherson PS, Schmid SL, De Camilli P. Nature. 1995;374:186–90. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.