Abstract
Poly(ADP-ribose) is a significant nucleic acid polymer involved with diverse functions in eukaryotic cells, yet no structural information is available. A method for the synthesis of 13C, 15N- poly(ADP-ribose) (PAR) has been developed to allow characterization of the polymer using multidimensional nuclear magnetic resonance (NMR) spectroscopy. Successful integration of pentose phosphate, nicotinamide adenine dinucleotide biosynthesis, and cofactor recycling pathways with poly(ADP-ribose) polymerase-1 permitted labeling of PAR from 13C-glucose and 13C, 15N- ATP in a single pot reaction. The scheme is efficient, yielding ~ 400 nmoles of purified PAR from 5 μmoles ATP, and the behavior of the synthetic PAR is similar to data from PAR synthesized by cell extracts. The resonances for 13C, 15N-PAR were unambiguously assigned, but the polymer appears to be devoid of inherent regular structure. PAR may form an ordered macromolecular structure when interacting with proteins, and due to the extensive involvement of PAR in cell function and disease, further studies of PAR structure will be required. The labeled PAR synthesis reported here will provide an essential tool for the future study of PAR-protein complexes.
Introduction
Poly(ADP-ribose) (PAR) has been implicated in important cell processes including DNA repair 1 and the cell cycle 2, and plays a key role in cancer 3 and neurological disease 4. While the full extent of PAR activity is the subject of intense ongoing research, known roles for PAR include binding to DNA damage checkpoint proteins 5, altering the kinetics of DNA strand break rejoining 6, participating in spindle assembly and structure during cell division 7, and regulating transcription 8, 9. PAR is also a cell death signal responsible for the release of apoptosis inducing factor (AIF) from mitochondria 10. This caspase-independent PAR mediated cell death has been defined as parthanatos 11. While the specific function that PAR plays in parthanatos and other pathways remains largely unknown, PAR generally plays two roles at the molecular level, serving as a scaffold for protein binding 12 and as a post translational modification 13. Formation of PAR-protein complexes depends on the length of PAR 14 and further study may reveal the structural features specific to the various functions of PAR.
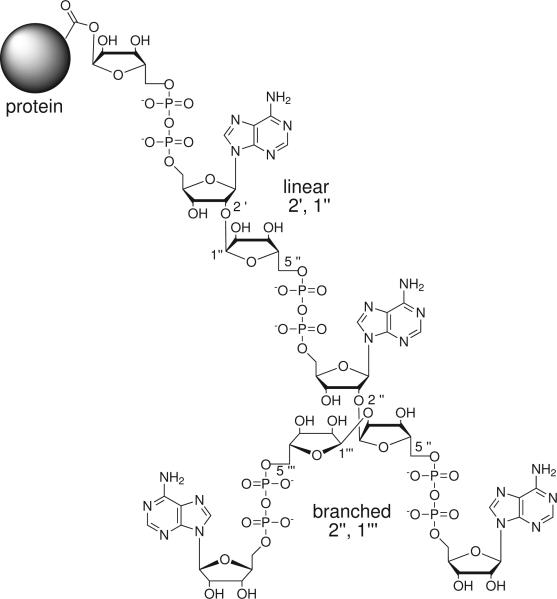
The primary structure of PAR was first elucidated using natural abundance 13C NMR in the late 1970's 15, and analysis of the polymer after phosphodiesterase digestion revealed both branched and linear linkages, as shown in Figure 116. Elaborate branched structures of PAR were observed by electron microscopy 17, and the cellular size distribution of the polymer was determined to be ~10 to 200 residues with up to six points of branching per polymer 18. Quantitative in vitro binding studies with PAR have been recently reported with different proteins including alternative splicing factor/splicing factor 2, tumor suppressor protein p53 and nucleotide excision repair protein xeroderma pigmentosum group A 14,19.
Figure 1.
Chemical structure of poly(ADP-ribose).
Poly(ADP-ribose) is synthesized by a family of eukaryotic enzymes called poly(ADP-ribose) polymerases (PARPs). Distinct subfamilies exist within the PARP family including mono-ADP-ribosylating enzymes, tankyrases which produce only linear PAR, catalytically inactive PARP's and the well studied long-chain polymerases such as PARP-1 19. Polymer synthesis by PARP-1 is initiated by formation of an ester linkage between the 1′ hydroxyl of the ADP-ribose to the side chain carboxylate of glutamate or aspartate residues on an acceptor protein. A number of different acceptor proteins have been identified, such as histones, topoisomerases, DNA ligases, DNA and RNA polymerases, high motility group proteins, tumor suppressor protein p53, and PARP-1 itself 20. The Breast Cancer 1 C-Terminal (BRCT) domain of PARP-1 can supply acceptor sites in trans for PAR synthesis by PARP-1, and PARP-1 is activated in the presence of double stranded DNA 21. Once initiated, enzymatic PAR synthesis proceeds in a linear fashion by attack of the 2′ hydroxyl of the existing ADP-ribose to the 1″ carbon of the incoming nicotinamide adenine dinucleotide (NAD+) monomer. Branches can also be introduced during PAR synthesis by attack of the 2″ hydroxyl on the 1″ carbon on the linear subunit. The chemical structure of PAR is shown in Figure 1.
This unusual nucleic acid polymer has the potential to form a regular helical structure, stabilized by base stacking and hydrogen bonding interactions, that might serve as a platform for organization of multiprotein complexes. Despite its critical role in biology, there is no three dimensional structural information available for PAR. Structural characterization of nucleic acid polymers such as RNA and DNA were revolutionized by methods developed for incorporation of stable isotopes 13C, 15N. Such labeled nucleic acids permit the use of multidimensional heteronuclear NMR experiments for resonance assignment and NMR structure determination. In order to apply these methods to analyze the three dimensional structure of PAR, we have developed an enzymatic synthesis of NAD+ that permits efficient enzymatic synthesis of 13C-15N-labeled PAR. The 1H, 13C, 15N- resonances of PAR were readily assigned using multidimensional NMR methods. However, PAR does not appear to adopt a defined regular three dimensional structure in isolation. It is possible that the polymer serves as an avidity platform for the assembly of complexes, or that it forms a regular helical structure only in the presence of the appropriate protein ligands. The availability of isotopically labeled PAR should prove useful for future studies of PAR-protein complexes.
Results and Discussion
Design of the synthesis
In previous biochemical studies of PAR, the polymer was either extracted directly from tissue or synthesized in vitro using cell extracts 22. While cellular sources of PAR are convenient for many purposes, polymer extraction from cells or tissue presents significant economic disadvantage for preparation of an isotopically labeled sample. Cell culture in 13C,15N- media is extremely costly, and only a small portion of the isotope would be incorporated into PAR, necessitating large scale cultures.
Recent advances in chemical synthesis of the 2′-O-α-D-ribofuranosyladenosine monomeric unit may provide a chemical route to the ADP-ribose polymer in the future 23. However, synthetic PAR used in biochemical studies has been obtained by combining commercially available NAD+ isolated from yeast with in vitro enzymatic polymerization by PARP-1 24. Enzymatic synthesis in vitro also provides a more efficient route to convert isotopically labeled starting materials into the desired product. In addition, side reactions that consume costly precursors are minimized, and the purification of the polymer is greatly simplified.
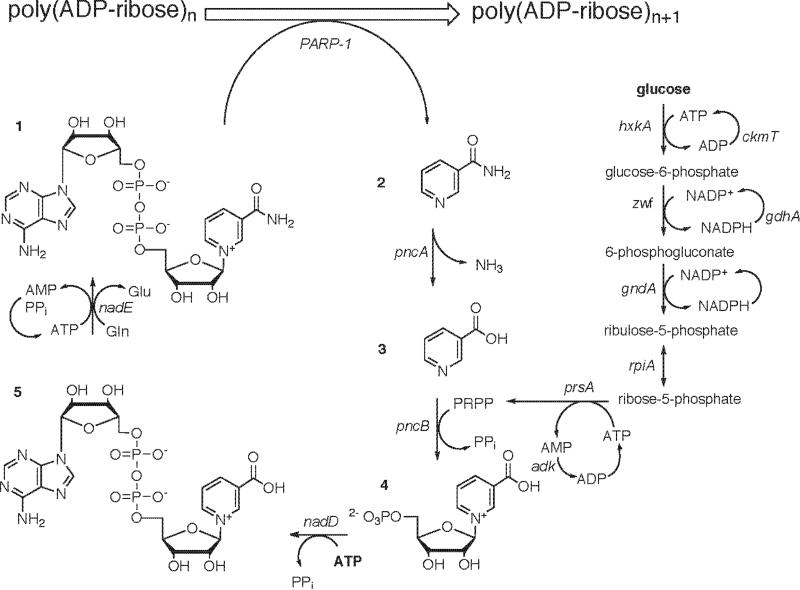
Enzymatic synthesis of PAR using PARP-1 requires stoichiometric amounts of NAD+ as a substrate, as shown in Scheme 1. Since NAD+ is not commercially available in any stable isotope labeled form, 13C, 15N- NAD+ must first be synthesized in order to produce labeled PAR using this method. Several approaches for the chemical synthesis of NAD+ have been established. Synthesis of NAD+ starting from nicotinamide triacetate and tetra-O-acetyl-β-D-ribofuranose proceeded efficiently giving a 58% isolated yield 25. Chemical synthesis of the pivotal intermediate nicotinate mononucleotide from ribose-5-phosphate and 1-(2,4-dinitrophenyl)-3-carbamoyl-pyridinium chloride followed by enzymatic conversion to NAD+ gave a 60% yield in large scale reactions 26. While these chemical reactions are efficient and robust, they are not well suited to synthesize isotopically labeled NAD+ compounds due to the unavailability of the required labeled starting materials. An enzymatic synthesis of labeled NAD+ would be ideal, and there is a good precedent for an efficient enzymatic route to NAD+. A two step in vitro enzymatic synthesis combined the readily available labeled precursors nicotinic acid, glucose and ATP to synthesize NAD+ with specific 3H, 14C, and 15N labels 27.
Scheme 1. PAR Synthesis using nicotinamide recycling and NAD+ biosynthesis.
Poly(ADP-ribose)n and NAD+ (1) are converted to poly(ADP-ribose)n+1 by the action of poly(ADP-ribose) polymerase 1 (PARP-1) with the production of nicotinamide. Nicotinamide (2) and water are converted to nicotinate by the action of nicotinamidase (pncA) with the production of an ammonium ion. Nicotinate (3) and phosphoribosyl-pyrophosphate (PRPP) are converted to nicotinate nucleotide by the action of nicotinate phosphoribosyl transferase (pncB) with the production of pyrophosphate. Nicotinate nucleotide (4) and ATP are converted to deamido-NAD+ by the action of nicotinate nucleotide adenylyl-transferase (nadD) with the production of pyrophosphate. Deamido-NAD+ (5), ATP and glutamine are converted to NAD+ by the action of NAD+ synthase (nadE) with the production of glutamate, AMP and pyrophosphate. Glucose and ATP are converted to glucose-6-phosphate by the action of hexokinase (hxkA) with the production of ADP. Glucose-6-phosphate and NADP+ are converted to 6-phosphogluconate by the action of glucose-6-phosphate dehydrogenase (zwf) with the production of NADPH. 6-Phosphogluconate and NADP+ are converted to ribulose-5-phosphate by the action of 6-phosphogluconate dehydrogenase (gndA) with the production of NADPH. Ribulose-5-phosphate is interconverted to ribose-5-phosphate by the action of phosphoriboisomerase (rpiA). Ribose-5-phosphate and ATP are converted to PRPP by the action of PRPP synthase (prsA) with the production of AMP. ATP and NADP+ recycling is achieved by creatine phosphokinase (ckmT), adenylate kinase (adk) and glutamic dehydrogenase (gdhA).
The design of a coupled enzymatic synthesis of 13C, 15N- NAD+ and 13C, 15N- PAR, from labeled glucose and ATP precursors, is illustrated in Scheme 1. Rather than synthesize and purify 13C, 15N- NAD+, it is continuously supplied as a substrate for PARP-1 during the formation of 13C, 15N- PAR. Typically, NAD+/NADH is used as a substoichiometric cofactor in enzymatic oxidation/reduction reactions as a hydrogen acceptor/donor, and the cofactor is regenerated by enzymatic coupling to a second reaction 28. In the case of PAR formation, NAD+ is stoichiometrically used as a source of ribose that is activated for nucleophilic substitution. PARP-1 adds the ADP-ribose moiety to the elongating polymer chain, effectively cleaving the nicotinamide base from NAD+. Since the nicotinamide base is not incorporated into the polymer, it can be used and recycled in unlabeled form. The NAD+ is regenerated in situ from nicotinamide and phosphoribosyl pyrophosphate (PRPP) through the action of four NAD+ biosynthesis enzymes, shown in Scheme 1. 13C, 15N- ATP and 13C-glucose provide the labeled building blocks for new ADP-ribose units. Pentose phosphate enzymes convert glucose into PRPP, while NAD+ biosynthesis enzymes add PRPP to nicotinate and utilize ATP as the precursor for the AMP moiety of NAD+. Additional ATP and NADP+ cofactor regeneration cycles are included to drive the ATP and NADP+ dependent steps forward, as shown previously 29. The design of this pathway allows the flexibility of labeling specific moieties within PAR. The separate ribose moiety can be independently labeled by including only 13C-glucose, while including only 13C, 15N-ATP will label only the AMP moiety.
Implementation of the synthesis
Based on our previous success with E. coli enzymes 30,31, the set of enzymes for the NAD+ biosynthesis pathway from E. coli was cloned into a His6-tagged expression vector and purified using nickel-nitrilo acetic acid (Ni-NTA) affinity chromatography. Commercial availability of all pertinent intermediates allowed determination of each purified enzyme's activity using a simple spectrophotometric assay. After optimizing the reaction conditions for PARP-1 activity, the addition of NAD+ biosynthesis and pentose phosphate pathway modules followed readily. The parallel synthesis of PRPP from glucose using the NADP+ cofactor was not affected by PARP-1 since NADP+ is not a substrate for the polymerization reaction. NADP+ recycling also ensured that the NAD+ remained in the oxidized form. ATP was supplied in a stoichiometric amount for PAR synthesis, and recycling enzymes were included to satisfy the energy requirements of the pentose phosphate and NAD+ biosynthesis steps. An excess of α-ketoglutarate and NH3 provided a driving force for the NADP+ recycling. Similarly, several equivalents of creatine phosphate ensured that the AMP and ADP produced were immediately charged up to ATP providing the proper substrate for the synthesis reaction to move forward.
Synthesis of 13C, 15N- PAR
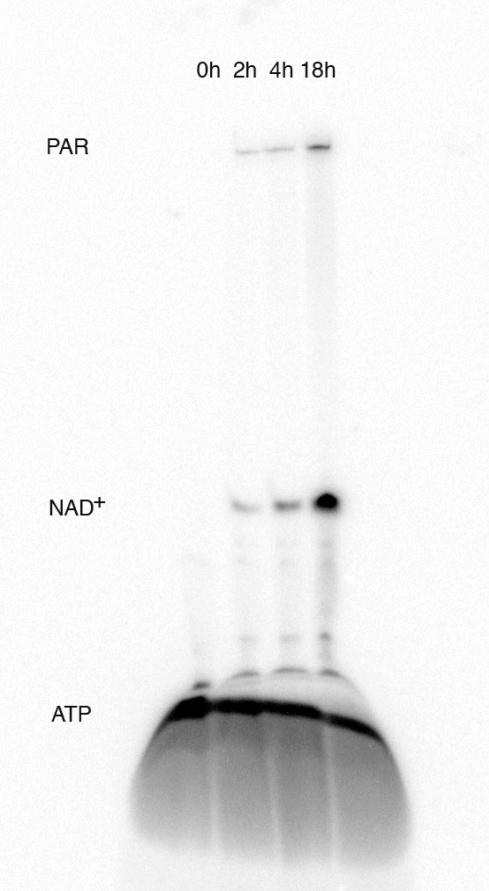
U-13C- Glucose, U-13C, 15N- ATP, unlabeled nicotinamide, phosphocreatine, α-ketoglutarate, glutamine, NH4Cl, MgCl2, catalytic NADP+, purified DNAse I treated DNA, recombinant PARP-1 BRCT domain to provide PAR acceptor sites, and BSA were combined with 14 enzymes including PARP-1 in a 2 mL buffered reaction incubated at 37°C. PAR products were labeled by including a trace amount of α-32P-ATP in the reaction, and the progress of the synthesis was monitored by denaturing polyacrylamide gel electrophoresis, and autoradiography, as shown in Figure 2. NAD+ synthesis began immediately as evidenced by the appearance of a new band at low molecular weight, and PAR formation followed shortly thereafter as evidenced by a high molecular weight species. Reactions were carried out for 18 to 72 hours, with slightly more PAR formed during the longer incubation time. PAR synthesis appears to be fairly processive since few low molecular weight intermediates are apparent during the synthesis. The length of the PAR chains is estimated to be ~100 monomers, based on partial hydrolysis of synthetic PAR using poly(ADP-ribose) glycohydrolase, as shown in Supplementary Figure 1.
Figure 2. Time course of PAR Synthesis.
Denaturing 20% polyacrylamide gel shows time point products from the 13C, 15N- PAR synthesis. PAR, NAD+ and ATP are labeled and visualized with 32P by the addition of a trace amount of α-32P-ATP to the synthesis reaction.
The polymer was purified by precipitation of the proteins with trichloroacetic acid (TCA), release of the polymer from the protein acceptor by base hydrolysis, and subsequent gel filtration chromatography, to afford ~400 nmoles of PAR from 5 μmoles of ATP, using 1 μmole of nicotinamide. PAR yield was measured using the absorbance at 260 nm and the extinction coefficient of 13.5 mM−1cm−1 for ADP-ribose 30. The overall yield for this synthesis was 8% based on ATP, and is comparable to typical 3-5% yields obtained for in vitro transcription of RNA from NTPs using T7 RNA polymerase. The overall low yield of PAR indicates that there was limited turnover and recycling of nicotinamide in the reaction. The conditions of the reaction were intentionally set to make PARP-1 activity rate limiting and this yield is not an intrinsic limitation to the NAD+ synthesis scheme.
The efficiency of the NAD+ synthesis can be evaluated from the time course gel in Figure 2. It is expected that 20% NAD+ would be synthesized directly from ATP based on the mole ratio of 1:5 for nicotinamide to ATP and at 18 hours, ~26% of the radioactivity from ATP was incorporated into NAD+. This > 20% incorporation of radioactivity indicates complete conversion of the available nicotinamide into NAD+. Additional radioactivity incorporated into the PAR molecule where nicotinamide has been liberated indicates that the full recycling scheme has been utilized. The successful turnover and recycling of nicotinamide was confirmed in preliminary syntheses where only 50 nmoles of nicotinamide were included per 5 μmole scale reaction. A similar amount of PAR product was observed during syntheses using this catalytic amount of nicotinamide (data not shown). A higher concentration of nicotinamide was selected for the preparative stable isotope labeled syntheses to ensure that an excess of NAD+ was available for PAR synthesis. A sufficient quantity of PAR was obtained from the 2 mL, 5 μmole scale reaction to produce a sample for NMR analysis. Syntheses of (13C, 15N- adenosine)- PAR and (13C-ribose′)- PAR proceeded similarly, substituting unlabeled glucose and unlabeled ATP, respectively.
Analysis of the Polymer
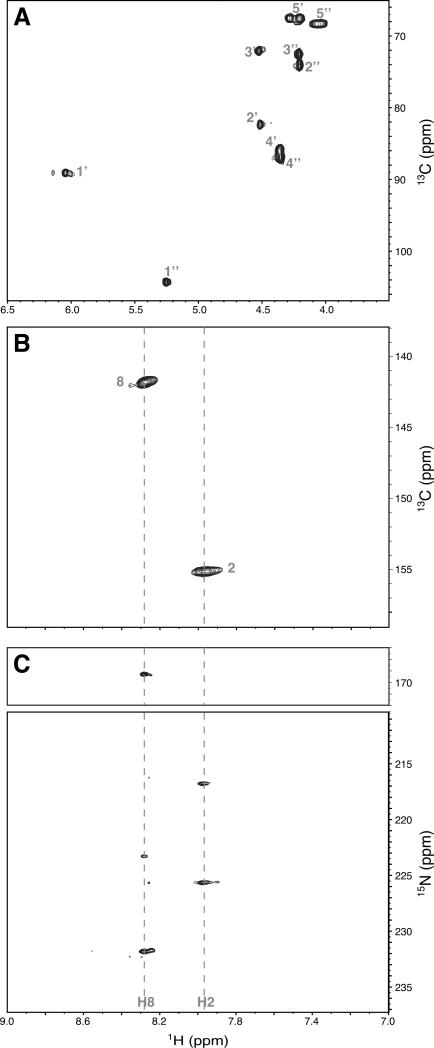
NMR spectra of the synthetic 13C, 15N-PAR exhibited the expected resonances based on the known primary structure of the linear polymer, as shown in Figure 3. A branch point (1‴) resonance expected at ~5.2 ppm 31 was not observed. Although, other groups reported ~2% branching for in vitro synthesis of PAR 18,25, and it is likely that the small number of branch points incorporated were below the threshold for detection. Analysis of the nuclease digest of the polymer did reveal evidence for a small percentage of branch points (Supplementary Figure 2). The NMR spectrum of PAR differs considerably from that of monomeric adenosine-5′-disphosphoribose (Supplementary Figure 3). The ribose regions were unambiguously assigned using heteronuclear correlation spectroscopy ((H)CCH-COSY) experiments, and the identities of the two ribose spin systems were confirmed by comparison to the spectra resulting from independent labeling of the ribose′ and adenosine conjugated ribosyl moieties (Supplementary Figure 4). Assignment of the aromatic nuclei was based on previously published spectra of the polymer subunits 34,35 and known values for the adenosine base. The chemical shifts for the poly (ADP-ribose) polymer are listed in Table 1.
Figure 3. HSQC spectra for 13C, 15N- poly(ADP-ribose).
1H-13C- correlation including assignments for each resonance is shown for the ribose (A) and aromatic (B) regions of PAR. 1H-15N- 2J coupling of adenine nitrogen atoms of PAR is shown in (C). The H8 proton is coupled to N7 and N9, while the H2 proton is coupled to N1 and N3. The dashed line indicates the chemical shift of these protons. An extra resonance at 223 ppm appears as a result of N9 signal folding in.
Table 1.
Chemical Shift Assignments for Poly(ADP-ribose)
| Adenosine | Ribose′ | ||||
|---|---|---|---|---|---|
| H2 | 7.97 | H1’ | 6.05 | H1’’ | 5.25 |
| H8 | 8.27 | H2’ | 4.52 | H2’’ | 4.21 |
| C2 | 155.1 | H3’ | 4.52 | H3’’ | 4.21 |
| C8 | 141.5 | H4’ | 4.36 | H4’’ | 4.35 |
| N1 | 225.7 | H5’ | 4.21 / 4.28 | H5’’ | 4.03 / 4.07 |
| N3 | 216.8 | C1’ | 89.1 | C1’’ | 104.3 |
| N7 | 231.8 | C2’ | 82.4 | C2’’ | 74.2 |
| N9 | 169.3 | C3’ | 72.1 | C3’’ | 72.4 |
| C4’ | 85.9 | C4’’ | 86.9 | ||
| C5’ | 67.5 | C5’’ | 68.3 | ||
To investigate the possibility of a regular structure in PAR, Nuclear Overhauser Enhancement Spectroscopy (NOESY) spectra were collected and compared to spectra collected from ADP-ribose and polyA, shown in Figure 4. Although an upfield shift of signals is observed in the spectra of PAR relative to the ADP-ribose monomer, there was no clear evidence of hydrogen bonding or base stacking in the PAR sample.
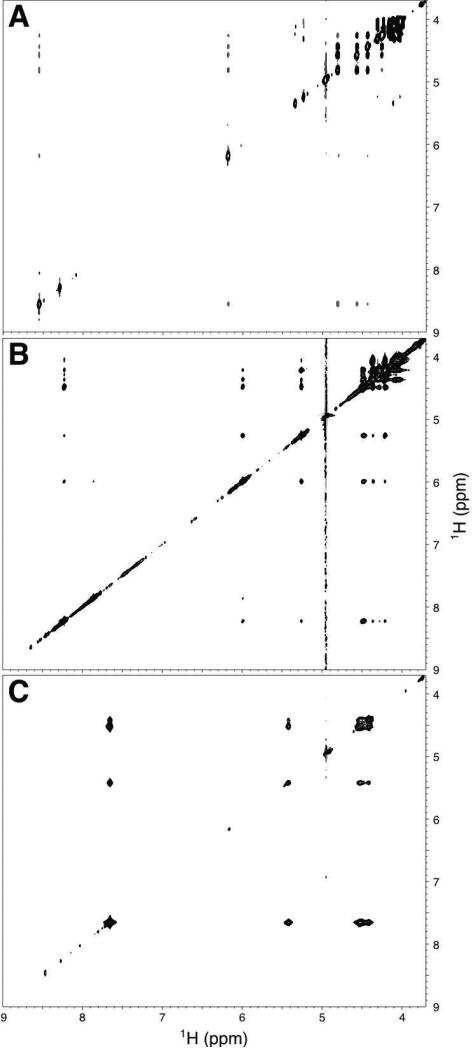
Figure 4. Through space correlation of protons in Adenosine compounds.
(A) Homonuclear ROESY of ADP-ribose monomer. The homonuclear NOESY spectra of poly(ADP-ribose) (B) and poly(Adenosine) (C) are also shown.
Nevertheless, the number of NOE correlations between H8 (8.27 1H ppm) and the ribose protons (4.21 – 4.52 1H ppm) suggests that the adenine base is in the anti conformation and accessible for making other interactions. Structural information about the polymer is limited due to the degeneracy of proton resonances, and the only inter-ribose NOE correlations are associated with protons near the 2′ - 1″ linkage (4.52, 5.25 1H ppm). The only unambiguous medium range correlation is between the H8 of adenine (8.27 1H ppm) and the 5″ protons (4.03/4.07 1H ppm).
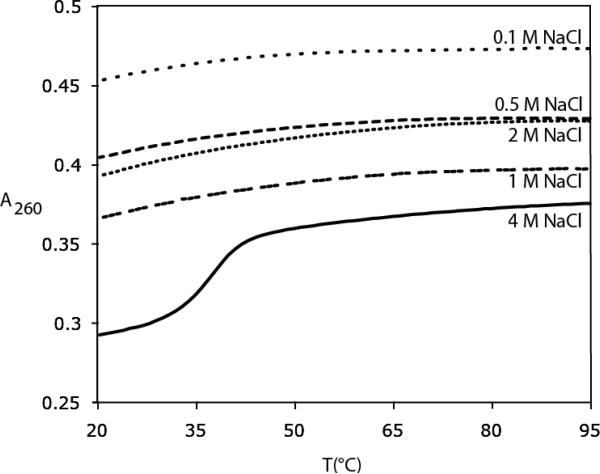
Although PAR appears to exist in an extended conformation in low salt buffers, there is previous circular dichroism evidence that the polymer may adopt some structure under different buffer conditions, including: 0.1 mM spermine, 0.5 mM CaCl2, 0.5 mM MgCl2, NaCl > 3 M and pH > 5 32. Thermal denaturation curves were recorded over a wide range of pH, Mg++ and NaCl concentrations, monitoring the absorbance at 260 nm to look for evidence of a cooperative hyperchromic helix-coil transition. A complete list of experiments and conditions is included in Supplementary Table 1. The melting curves for PAR over a range of NaCl concentration are shown in Figure 5. No melting transitions are observed at low salt, but a transition with melting temperature of 37.4 °C was observed in 4 M NaCl. The full profile of melting behavior from 0.1 M NaCl to 4 M NaCl is consistent with behavior seen previously with PAR synthesized by liver nuclei 36,37.
Figure 5. Thermal Denaturation of PAR at various concentrations of NaCl.
The transition in absorbance at 4 M NaCl suggests that poly(ADP-ribose) is more structured under these conditions.
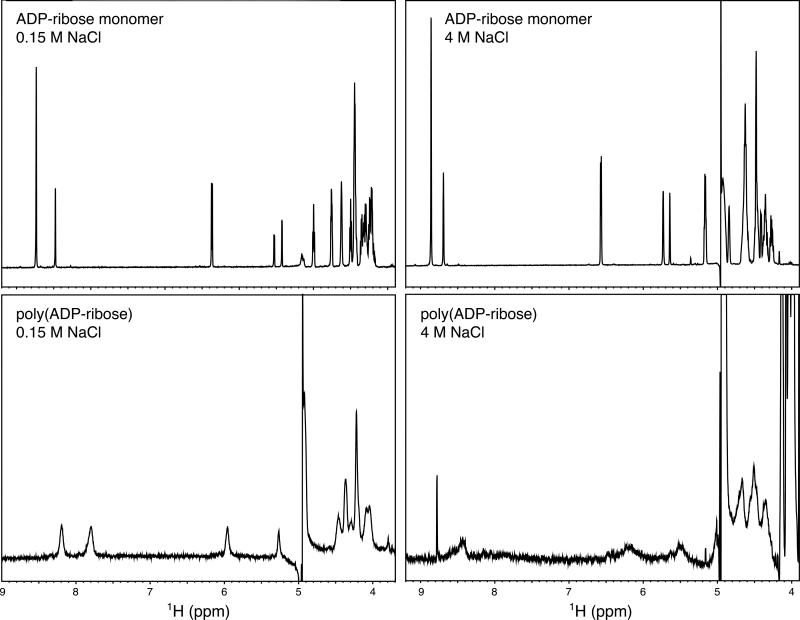
Proton spectra of the ADP-ribose monomer and PAR at 0.15 M NaCl and 4 M NaCl are compared in Figure 6. A distinct change is observed in the spectra of PAR at high salt, including broadening of the peaks and decrease in the signal intensity. These data suggest the possibility of a structured conformation stabilized at high salt concentrations. Comparison of the spectra of ADP-ribose monomer at low and high salt confirms that this change is not a simple salt effect. The NMR data and melting data taken together do not indicate any evidence for a regular helical structure in isolated preparations of PAR under near-physiological conditions.
Figure 6. Comparison of ADP-ribose monomer and polymer at low and high salt.
In both cases, the polymer linewidths are broader than those of the monomer, as expected. The increased broadening of signals at 4 M NaCl suggests that PAR may adopt a macromolecular structure.
Conclusions
We have designed and implemented an efficient enzymatic synthesis of 13C, 15N-poly(ADP-ribose). Our synergistic enzymatic approach combining NAD+ biosynthesis, PARP-1, pentose phosphate enzymes, and cofactor regeneration allowed us to recycle nicotinamide while labeling PAR from commercially available isotopically labeled precursors: 13C- glucose and 13C, 15N-ATP. Successful integration of eukaryotic and prokaryotic biochemical pathways in one pot proved to be a robust and efficient method to synthesize PAR in vitro. Notably, our synthetic PAR behaved similarly to PAR formed by liver nuclei.
The synthesis of isotopically labeled PAR allowed us to unambiguously assign the NMR spectra for PAR, and to characterize the limited structural features inherent in the polymer under physiological conditions. It is possible that PAR will form specific molecular interactions with associated proteins, and also that helical structures may form upon protein binding. Due to the important and numerous roles of PAR in the cell, additional insights into PAR function will emerge over the coming years. The enzymatic synthesis of isotopically labeled PAR reported here should provide useful tools for the future structural studies of PAR and its associated biomolecules.
Experimental
Materials
Glucose (U-13C, 99%) and ATP (U-13C, 98%, U-15N, 98%) were purchased from Cambridge Isotope Laboratories. All other chemicals were purchased from Sigma unless otherwise noted. Hexokinase, glucose-6-phosphate dehydrogenase, and glutamic dehydrogenase from baker's yeast, phosphoriboisomerase from spinach, adenylate kinase from chicken muscle, and creatine phosphokinase from rabbit muscle were purchased from Sigma. PRPP synthase and inorganic pyrophosphatase were prepared as previously described 29. PARP-1, activated DNA and PARP-1 BRCT domain were the generous gift of Valérie Schreiber of the Université de Strasbourg. Human PARP-1 was prepared as previously described 33 and purified DNAse I treated DNA was purchased from Alexis Biochemicals, Lausen Switzerland. The DNA fragment encoding human PARP-1 BRCT domain (residues 385-524) was amplified by PCR and subcloned into the NdeI/BamHI sites of pET15b vector (Invitrogen). Expression was induced in E. coli BL21 cells by 0.3 mM IPTG for 4 hours at 31 °C. His-tagged recombinant protein was purified on Ni-Sepharose beads (GE Healthcare) and stored at 1μg/μl in 50mM Tris-HCl pH8, 200 mM NaCl, 60 mM imidazole, 0.5 mM phenylmethysulfonyl fluoride (PMSF), 10% Glycerol.
NAD+ Biosynthesis Enzymes Cloning and Purification
The genes for pncA, pncB, nadD and nadE were cloned from E. coli K12 MRE600 genome, based on the reported sequences in Genbank using standard procedures. Each gene was amplified from genomic DNA using PCR with gene-specific primers containing compatible restriction sites of BamHI and EcoRI for cloning into pET22-HT (derived from pET22b, Novagen), which adds an N-terminal hexahistidine tag. All constructs were verified by DNA sequencing. Plasmids were transformed to BL21(DE3) strain for protein production. Enzymes were purified by the standard method previously described for his-tagged E. coli enzymes 29. Yields and specific activities are reported in Table 2.
Table 2.
NAD+ Biosynthesis Enzymes
| Enzyme | EC | Gene | Yield1 | Specific Activity2 |
|---|---|---|---|---|
| Nicotinamidase | 3.5.1.19 | pncA | 7 | 0.5 |
| Nicotinate phosphoribosyl transferase | 2.4.2.11 | pncB | 2 | 30 |
| Nicotinate nucleotide adenylyl transferase | 2.7.7.18 | nadD | 13 | 1 |
| NAD synthase | 6.3.1.5 | nadE | 2 | 0.5 |
Yield is given in mg of protein per L of culture.
Specific activity is given by U/mg.
Enzymatic Assays
Enzymatic activity is reported in terms of units (U), where 1 U is the amount of enzyme required to convert 1 μmol min −1 of substrate into product, and the specific activity is reported as U mg −1. Enzymatic activities were determined by coupling the particular reaction to ATP consumption, which is monitored by the change in A340 due to the action of pyruvate kinase and lactate dehydrogenase on ADP and NADH 34.Activities for each enzyme except PARP and inorganic phosphatase were determined using this method. To assay inorganic pyrophosphatase, the phosphate detection method of Michelson was used 35. PARP-1 activity was determined as described previously 36.
13C, 15N- PAR Synthesis
Substrates: 1.9 mg (10 μmoles) U-13C-glucose, 2.75 mg (5 μmoles) U-13C, 15N-ATP, 1.2 mg (1 μmole) nicotinamide, 2 mg (7.3 μmoles) glutamine, 9 mg (40 μmoles) α-ketoglutarate, 20 mg (78 μmoles) phosphocreatine and 0.1 mg BRCT were combined in a solution of 20 ng/μL activated DNA, 2 mM DTT, 10 mM MgCl2, 40 mM NH4Cl, 0.2 mg/mL BSA and 50 mM Tris-HCl pH 8.0. Enzymes were added to the solution: 0.044 U hexokinase (hxkA), 0.048 U glucose-6-phosphate dehydrogenase (zwf), 0.053 U 6-phosphogluconate dehydrogenase (gndA), 0.059 U ribose phosphate isomerase (rpiA), 0.064 U PRPP synthase (prsA), 0.224 U glutamic dehydrogenase (gdhA), 0.011 U nicotinamidase (pncA), 0.012 U nicotinate phosphoribosyl transferase (pncB), 0.013 U nicotinate nucleotide adenylyl transferase (nadD), 0.015 U NAD+ synthase (nadE), 0.5 U creatine phosphokinase (ckmT), 0.1 U adenylate kinase (adk), 0.1 U inorganic pyrophosphatase (ppa), 0.01 U PARP-1, giving a final volume of 2 mL.
PAR and NAD+ formation were monitored by inclusion 5 μCi of α-32P-ATP (Perkin-Elmer) in 100 μL of reaction mixture run in parallel to the main reaction. Aliquots of 5 μL were withdrawn at various timepoints and quenched with an equal volume of formamide/ EDTA loading dye and stored at −20°C until use. Half of each sample was analyzed on a denaturing 20% polyacrylamide gel, run at 20 W for 1.5 h. The dried gel was exposed to a phosphor screen for 4 hours and then scanned on a STORM 820 phosphoimager (Molecular Dynamics).
Specific labeled PAR Syntheses
Synthesis of 13C, 15N- adenosine PAR followed the same protocol as above except unlabeled glucose was substituted for the 13C-glucose. Similarly, 13C-ribose′ PAR followed the same synthesis protocol, using 13C-glucose and substituting unlabeled ATP for the U-13C, 15N-ATP.
PAR Purification
The 2 mL reactions were stopped and the proteins and covalently linked PAR were precipitated by addition of 650 μL of 6.1 N TCA, with gentle rocking at 4°C for 1 h. The precipitate was pelleted by centrifugation at 4°C for 30 m at 30000 g, and the supernatant containing ATP, NAD+, and non-protein buffer components was removed. The pellet was resuspended in 0.6 mL of 1 M KOH with 0.05 M EDTA and incubated at 60°C for 1 h to cleave the polymer from the protein. Micro P30 spin columns (Bio-Rad) were equilibrated with water using 5 column volumes, following the manufacturer instructions. The released polymer solution was loaded to the columns (75 μL solution/ column) and after centrifugation, the flowthrough containing PAR was concentrated by vacuum, and the dried polymer was resuspended in 0.5 mL NMR buffer: 150 mM NaCl, 10 mM NaPi pH 6.5. The polymer was precipitated by addition of 4.5 volumes of ethanol. After incubation overnight at −20°C, the ethanol pellet was collected by centrifugation, then dried and resuspended in 500 μL water/D2O for dialysis into the appropriate buffer.
Thermal Denaturation Experiments
PAR was prepared as described above for use in thermal melts. After the last step, the polymer was desalted using a 3000 MWCO microcon (Millipore), lyophilized and resuspended in the appropriate buffer. (See Supplementary Table 1 for the list of conditions.) UV absorbance at 260 nm was observed and recorded in 0.5°C increments from 20°C to 95°C using a Cary-1Bio temperature controlled spectrophotometer (Varian). Each run started at 20°C ramping to 95°C, returned back to 20°C and increased again to 95°C at a rate of 1° per minute. Averaged data from the second increase in temperature is shown.
NMR Experiments
Homonuclear NOESY spectra (Tmix=150ms) were collected on poly(ADP-ribose) and poly(A) (Sigma), and compared to the homonuclear Rotating-frame Overhauser Effect Spectroscopy (ROESY) (Tmix=300ms) spectrum collected for ADP-ribose monomer (adenosine 5-diphosphoribose, Sigma). One dimensional proton spectra were also collected for poly(ADP-ribose) and ADP-ribose monomer at 0.15 M and 4 M NaCl. For all homonuclear experiments, water suppression was achieved with presaturation pulses. Two bond coupled 1H, 15N-HSQC correlation experiments were collected using an INEPT transfer delay time (2Δ) of 20 μs 37. Water suppression was achieved through the use of flip-back pulses and the Watergate scheme38, 39.1H, 13C-HSQC spectra were collected using echo/antiecho gradient selection. Assignment of the sugar resonances was enabled by the collection of a 2D double resonance (H)CCH-COSY spectra 40. Spectra were collected at 10°C on a 500 MHz Bruker Avance spectrometer equipped with a 5mm TXI-HCN triple resonance probe with a z-axis gradient, referenced to 2,2-dimethyl-2-silapentane-5-sulfonate 41, processed with NMRPipe and viewed in NMRDraw 42.
Supplementary Material
Acknowledgements
The authors thank Valérie Schreiber of the Université de Strasbourg for the generous gift of PARP-1 and its activators. This work was supported by a grant from the National Institute of Health, GM-53320 to J.R.W.
Footnotes
Supporting Information Available:
Supplementary figures of spectra for ADP-ribose and specifically labeled PAR are included, limited digests of the polymer, and a supplementary table listing conditions for structural characterization experiments. This material is free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451(7174):81–5. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 2.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432(7017):645–9. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 3.Haince JF, Rouleau M, Hendzel MJ, Masson JY, Poirier GG. Targeting poly(ADP-ribosyl)ation: a promising approach in cancer therapy. Trends Mol Med. 2005;11(10):456–63. doi: 10.1016/j.molmed.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: A key event in parthanatos. Exp Neurol. 2009 doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275(52):40974–80. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 6.Ryabokon NI, Goncharova RI, Duburs G, Hancock R, Rzeszowska-Wolny J. Changes in poly(ADP-ribose) level modulate the kinetics of DNA strand break rejoining. Mutat Res. 2008;637(12):173–81. doi: 10.1016/j.mrfmmm.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nature Cell Biology. 2005;7(11):1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 8.Oei SL, Griesenbeck J, Ziegler M, Schweiger M. A novel function of poly(ADP-ribosyl)ation: silencing of RNA polymerase II-dependent transcription. Biochemistry. 1998;37(6):1465–9. doi: 10.1021/bi9727390. [DOI] [PubMed] [Google Scholar]
- 9.Olabisi OA, Soto-Nieves N, Nieves E, Yang TT, Yang X, Yu RY, Suk HY, Macian F, Chow CW. Regulation of transcription factor NFAT by ADP-ribosylation. Mol Cell Biol. 2008;28(9):2860–71. doi: 10.1128/MCB.01746-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci U S A. 2006;103(48):18314–9. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harraz MM, Dawson TM, Dawson VL. Advances in neuronal cell death 2007. Stroke. 2008;39(2):286–8. doi: 10.1161/STROKEAHA.107.511857. [DOI] [PubMed] [Google Scholar]
- 12.Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36(22):6959–76. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hakme A, Wong HK, Dantzer F, Schreiber V. The expanding field of poly(ADP-ribosyl)ation reactions. ‘Protein Modifications: Beyond the Usual Suspects’ Review Series. EMBO Rep. 2008;9(11):1094–100. doi: 10.1038/embor.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35(21):e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miwa M, Saito H, Sakura H, Saikawa N, Watanabe F, Matsushima T, Sugimura T. A 13C NMR study of poly(adenosine diphosphate ribose) and its monomers: evidence of alpha-(1” leads to 2') ribofuranosy1 ribofuranoside risidue. Nucleic Acids Res. 1977;4(11):3997–4005. doi: 10.1093/nar/4.11.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miwa M, Saikawa N, Yamaizumi Z, Nishimura S, Sugimura T. Structure of poly(adenosine diphosphate ribose): identification of 2'-[1”-ribosyl-2”-(or 3”-)(1'”-ribosyl)]adenosine-5',5”,5'”-tris(phosphate) as a branch linkage. Proc Natl Acad Sci U S A. 1979;76(2):595–9. doi: 10.1073/pnas.76.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Murcia G, Jongstra-Bilen J, Ittel ME, Mandel P, Delain E. Poly(ADP-ribose) polymerase auto-modification and interaction with DNA: electron microscopic visualization. Embo J. 1983;2(4):543–8. doi: 10.1002/j.1460-2075.1983.tb01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez-Gonzalez R, Jacobson MK. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987;26(11):3218–24. doi: 10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- 19.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32(1):57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 20.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–68. [PMC free article] [PubMed] [Google Scholar]
- 21.Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35(22):7665–75. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juarez-Salinas H, Levi V, Jacobson EL, Jacobson MK. Poly(ADP-ribose) has a branched structure in vivo. J Biol Chem. 1982;257(2):607–9. [PubMed] [Google Scholar]
- 23.Mikhailov SN, Kulikova IV, Nauwelaerts K, Herdewijn P. Synthesis of 2'-O-alpha-D-ribofuranosyladenosine, monomeric unit of poly(ADP-ribose). Tetrahedron. 2008;64(12):2871–2876. [Google Scholar]
- 24.Kiehlbauch CC, Aboul-Ela N, Jacobson EL, Ringer DP, Jacobson MK. High resolution fractionation and characterization of ADP-ribose polymers. Anal Biochem. 1993;208(1):26–34. doi: 10.1006/abio.1993.1004. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Churchil H, Choi WB, Lynch JE, Roberts FE, Volante RP, Reider PJ. A chemical synthesis of nicotinamide adenine dinucleotide (NAD(+)). Chemical Communications. 1999;(8):729–730. [Google Scholar]
- 26.Walt DR, Findeis M. a., Riosmercadillo VM, Auge J, Whitesides GM. An Efficient Chemical and Enzymatic-Synthesis of Nicotinamide Adenine-Dinucleotide (Nad+). Journal of the American Chemical Society. 1984;106(1):234–239. [Google Scholar]
- 27.Rising K. a., Schramm VL. Enzymatic-Synthesis of Nad(+) with the Specific Incorporation of Atomic Labels. Journal of the American Chemical Society. 1994;116(15):6531–6536. [Google Scholar]
- 28.Chenault HK, Whitesides GM. Regeneration of nicotinamide cofactors for use in organic synthesis. Appl Biochem Biotechnol. 1987;14(2):147–97. doi: 10.1007/BF02798431. [DOI] [PubMed] [Google Scholar]
- 29.Schultheisz HL, Szymczyna BR, Scott LG, Williamson JR. Pathway engineered enzymatic de novo purine nucleotide synthesis. ACS Chem Biol. 2008;3(8):499–511. doi: 10.1021/cb800066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shah GM, Poirier D, Duchaine C, Brochu G, Desnoyers S, Lagueux J, Verreault A, Hoflack JC, Kirkland JB, Poirier GG. Methods for biochemical study of poly(ADP-ribose) metabolism in vitro and in vivo. Anal Biochem. 1995;227(1):1–13. doi: 10.1006/abio.1995.1245. [DOI] [PubMed] [Google Scholar]
- 31.Miwa M, Ishihara M, Takishima S, Takasuka N, Maeda M, Yamaizumi Z, Sugimura T, Yokoyama S, Miyazawa T. The branching and linear portions of poly(adenosine diphosphate ribose) have the same alpha(1 leads to 2) ribose-ribose linkage. J Biol Chem. 1981;256(6):2916–21. [PubMed] [Google Scholar]
- 32.Minaga T, Kun E. Probable helical conformation of poly(ADP-ribose). The effect of cations on spectral properties. J Biol Chem. 1983;258(9):5726–30. [PubMed] [Google Scholar]
- 33.Giner H, Simonin F, Demurcia G, Menissierdemurcia J. Overproduction and Large-Scale Purification of the Human Poly(Adp-Ribose) Polymerase Using a Baculovirus Expression System. Gene. 1992;114(2):279–283. doi: 10.1016/0378-1119(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 34.Scott LG, Tolbert TJ, Williamson JR. Preparation of specifically 2H- and 13C-labeled ribonucleotides. Methods Enzymol. 2000;317:18–38. doi: 10.1016/s0076-6879(00)17004-1. [DOI] [PubMed] [Google Scholar]
- 35.Michelson OB. Photometric Determination of Phosphorous as Molybdovanadophosphoric Acid. Analytical Chemistry. 1957;29:60–62. [Google Scholar]
- 36.Ame JC, Hakme A, Quenet D, Fouquerel E, Dantzer F, Schreiber V. Detection of the Nuclear Poly(ADP-ribose)-Metabolizing Enzymes and Activities in Response to DNA Damage. Methods Mol Biol. 2009;464:267–83. doi: 10.1007/978-1-60327-461-6_15. [DOI] [PubMed] [Google Scholar]
- 37.Sklenar V, Peterson RD, Rejante MR, Feigon J. Correlation of nucleotide base and sugar protons in a 15N-labeled HIV-1 RNA oligonucleotide by 1H-15N HSQC experiments. J Biomol NMR. 1994;4(1):117–22. doi: 10.1007/BF00178339. [DOI] [PubMed] [Google Scholar]
- 38.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2(6):661–5. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 39.Grzesiek S, BAX A. The Importance of Not Saturating H2O in Protein Nmr - Application to Sensitivity Enhancement and Noe Measurements. Journal of the American Chemical Society. 1993;115(26):12593–12594. [Google Scholar]
- 40.Gehring K, Ekiel I. H(C)CH-COSY and (H)CCH-COSY experiments for C-13-labeled proteins in H2O solution. Journal of Magnetic Resonance. 1998;135(1):185–193. doi: 10.1006/jmre.1998.1543. [DOI] [PubMed] [Google Scholar]
- 41.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6(2):135–40. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 42.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.