Abstract
Lin28 and Lin28B, two developmentally regulated RNA-binding proteins and proto-oncogenes, selectively inhibit the maturation of let-7 family miRNAs in embryonic stem (ES) cells and certain cancers. Moreover, let-7 precursors (pre-let-7) were previously found to be terminally uridylated in a Lin28-dependent fashion. Here, we identify Zcchc11 (zinc finger, CCHC domain containing 11) as the 3′ terminal uridylyl transferase (TUTase) responsible for Lin28-mediated pre-let-7 uridylation and blockade of let-7 processing in mouse ES cells. We demonstrate that Zcchc11 activity is UTP-dependent, selective for let-7, and recruited by Lin28. Furthermore, knockdown of either Zcchc11 or Lin28, or overexpression of a catalytically inactive TUTase, relieves the selective inhibition of let-7 processing and leads to the accumulation of mature let-7 miRNAs and repression of let-7 target reporter genes. Our results establish a novel role for Zcchc11-catalyzed pre-let-7 uridylation in the control of miRNA biogenesis.
Keywords: microRNA (miRNA), Lin28, Lin-28, Zcchc11, 3′ Terminal Uridylyl Transferase, TUTase, let-7, Uridylation
MicroRNAs (miRNAs) are generated by a multistep process mediated by sequential cleavages by two RNase III endonucleases1. Primary miRNA (pri-miRNA) is cleaved by Microprocessor, generating the 60-80 nucleotide precursor miRNA (pre-miRNA). After nuclear export, Dicer cleaves the pre-miRNA to yield the ~22 nucleotide mature miRNA that associate with Argonaute proteins in the miRNA-Induced Silencing Complex (miRISC) and target mRNAs for degradation and/or translational inhibition. MicroRNAs are essential for normal development and their deregulation has been implicated in various human diseases2. For example, altered miRNA expression often correlates with clinicopathological characteristics in cancer3-5. In mice, genetic manipulations have defined a direct role for miRNAs in cancer initiation, progression, and metastasis. Hundreds of miRNAs have now been identified, many of which are expressed in a manner that is specific to the tissue and developmental stage. Under most conditions, control of miRNA expression occurs at the transcriptional level; however, emerging evidence indicates that miRNA biogenesis can also be regulated posttranscriptionally1.
Previously, we and others found that the developmentally regulated RNA binding proteins Lin28 and Lin28B block the maturation of let-7 miRNAs in mouse embryonic stem (ES) cells and in various mouse and human cancer cell lines6-9. Recent studies have provided a molecular explanation for selective inhibition of let-7 family miRNAs where Lin28 was found to bind specifically to the terminal loop region of let-7 precursors8,10. Furthermore, the loop mediates miRNA-processing inhibition in vivo and that both the Cold Shock Domain (CSD) and the CCHC-type Zinc finger domains of Lin28 are required for pre-let-7 binding and processing inhibition10.
Lin28 was originally identified in C. elegans as a heterochronic gene that controls developmental timing11. There is emerging evidence that mammalian Lin28 and Lin28B have important and widespread roles in development and disease. For example, Lin28 was identified as one of four factors that reprogram human fibroblasts into induced pluripotent stem (iPS) cells12. Also, Lin28 has an important role in primordial germ cell development and malignancies13. Genome-wide association studies have identified Lin28B as a modifier gene that affects human height, menopause timing, and onset of puberty14-19. Additionally, Lin28 and Lin28B likely function as proto-oncogenes20-23. For example, expression of Lin28B and less frequently Lin28 is activated in several cancer types and their overexpression cause cellular transformation23. This finding is consistent with the fact that many tumors are characterized by loss of the let-7 family of miRNAs that act as tumor suppressors by repressing known oncogenes, including c-Myc, Hmga2, and Ras5. Therefore, the mechanism by which Lin28 inhibits let-7 biogenesis is fundamentally important in ES cell biology, cell reprogramming, development, and cancer. Interestingly, it was recently reported that let-7 precursors (pre-let-7) are terminally uridylated in cancer cells in a Lin28-dependent fashion. Furthermore, uridylated pre-let-7 is refractory to Dicer cleavage in processing assays and is preferentially degraded by cell extracts in vitro9. To gain insight into the mechanism by which Lin28 inhibits let-7 biogenesis, we sought to identify the 3′ terminal uridylyl transferase (TUTase) responsible for the oligouridylation of pre-let-7 as well as to demonstrate the relevance of this oligonucleotide addition in controlling let-7 biogenesis. We have identified Zcchc11 as the TUTase responsible for Lin28-mediated pre-let-7 uridylation and blockade of let-7 processing in mouse ES cells.
RESULTS
Identification of Zcchc11 as a candidate pre-let-7 TUTase
To gain insight into how Lin28 controls let-7 maturation, we analyzed by mass spectrometry affinity purified Lin28-containing protein complexes from mouse ES cells that were engineered to contain a doxycycline-inducible Flag-Lin28 transgene. We performed Flag-immunoprecipitations from control (−Dox) and induced (+Dox, 48hrs) cell extracts. We subjected the affinity eluate to SDS-polyacrylamide gel electrophoresis followed by Flag western blot (Supplementary Fig. 1a), Silver staining (Supplementary Fig. 1b) and mass spectrometric analysis (Supplementary Table 1). Significantly, our mass spectrometric analysis identified Zcchc11 as a putative novel Lin28-associated protein. Zcchc11 is predicted to contain two Poly(A)Polymerase domains, a nucleotidyltransferase (Ntr) domain and several CCHC-type Zinc finger domains (Fig. 1a), suggesting an involvement in catalyzing terminal nucleotide addition24,25. This non-canonical Poly(A)Polymerase is present in seven mouse proteins of which some have been shown to catalyze terminal uridine instead of adenosine additions. These proteins include Tut1 that is essential for U6 snRNA recycling26. Additionally, two TUTasae, Papd1 and Papd5, terminally uridylate histone H1 mRNA, leading to its turnover27. In Xenopus oocytes, the human homolog of Zcchc11 has a reported TUTase activity28. As such, research on terminal uridylation is still in its infancy and our knowledge about the functional consequences of this posttranscriptional modification is limited.
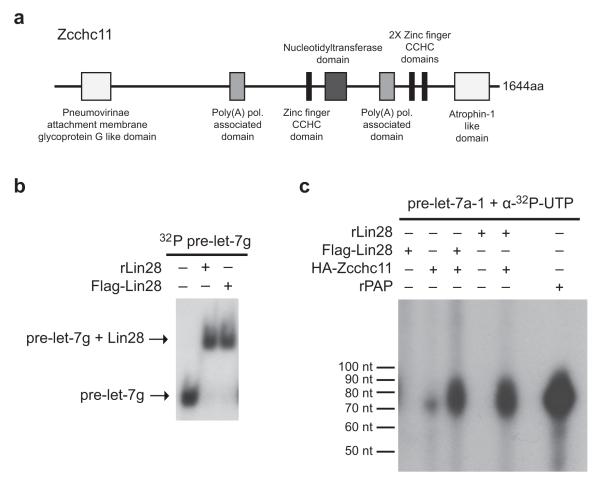
Figure 1. Identification of Zcchc11 as a candidate pre-let-7 TUTase.
(a) Diagrammatic representation of mouse Zcchc11 protein, Protein accession number (NCBI): AAI50792.1. Poly(A) Polymerase (PAP) domains are depicted in light gray, Zinc finger CCHC domains are in black, Nucleotidyltransferase (NTr) domain is in dark gray. aa, amino acids. (b) EMSA performed using radiolabeled pre-let-7g. Both recombinant Lin28 (5 μM) and Flag-Lin28 immunoprecipitated from ES cells were analyzed for ribonucleocomplex formation with pre-let-7g. (c) In vitro pre-let-7 uridylation assay. Synthetic pre-let-7a-1 was incubated with Flag-Lin28, HA-Zcchc11, r.Lin28, or a combination of Lin28 and Zcchc11 proteins. rPAP protein (8 units) was used as a positive control.
To examine whether Zcchc11 can directly catalyze the terminal oligonucleotide addition to pre-let-7 and to assess the potential functional interaction between Lin28 and Zcchc11, we developed an in vitro uridylation assay. First, pre-let-7 RNA binding was monitored by electromobility shift assays (EMSAs) to confirm that both the affinity-purified Flag-Lin28 from ES cells and recombinant His6-Lin28, were functional in pre-let-7 binding and to ensure that sufficient amounts of Lin28 from these two sources were used in subsequent uridylation assays (Fig. 1b). For in vitro uridylation assays, we incubated synthetic pre-let-7 RNA with the affinity-purified proteins in the presence of [α-32P]-UTP. E. coli Poly(A) Polymerase served as a positive control in these reactions, since this enzyme can also catalyze 3′ terminal uridine addition in the absence of ATP. Though we observed the weak appearance of some uridylated pre-let-7a when HA-Zcchc11 alone was added to the reaction, this activity was greatly enhanced by addition of either Flag-Lin28 purified from ES cells or recombinant Lin28, thus indicating that Lin28 directly recruits the Zcchc11 TUTase activity to pre-let-7a (Fig. 1c). Neither Flag-Lin28 nor recombinant Lin28 alone possessed any detectable uridylation activity, confirming that the appearance of the uridylated product was dependent on the presence of Zcchc11 in the reaction. We also resolved products of the in vitro uridylation assays on high resolution sequencing gels. This analysis indicated that purified HA-Zcchc11 when incubated with recombinant Lin28 primarily catalyzed the addition of 4-8 uridines to the synthetic pre-let-7a-1 RNA (Supplementary Fig. 2).
Characterization of Zcchc11 TUTase activity
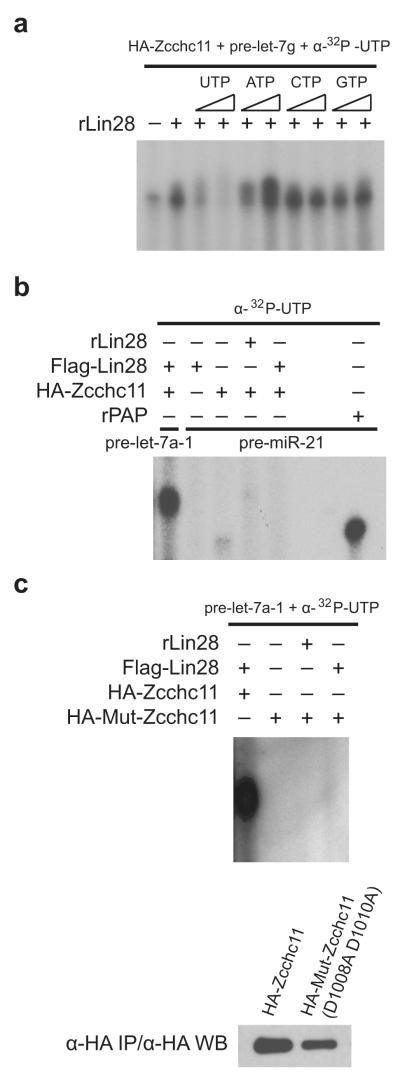
To elucidate the ribonucleotide selectivity of Zcchc11, we performed uridylation assays in the presence of excess non-radiolabeled nucleotides. These competition assays performed with synthetic pre-let-7g RNA revealed that unlabeled UTP is able to compete effectively in these reactions and leads to a diminution of detectable TUTase activity whereas the presence of ATP, GTP, and CTP did not. These results demonstrate that the Zcchc11 preferentially utilizes UTP for its catalytic activity (Fig. 2a). Next, we generated a mutant version of the Zcchc11 enzyme in which two aspartate residues (D1008, D1010) from the catalytic triad in the Ntr domain were mutated to alanines. Unlike wild-type Zcchc11, the mutant protein showed no uridylation activity towards pre-let-7a, showing that the pre-let-7 uridylation requires a catalytically active Zcchc11 enzyme in these reactions (Fig. 2b). To explore the substrate specificity of Lin28-mediated terminal uridylation catalyzed by Zcchc11, reactions were performed using a synthetic pre-miR-21 as a substrate. As with pre-let-7a and pre-let-7g, we observed a minimal background uridylase activity in reactions containing Zcchc11 alone; however, unlike reactions performed with pre-let-7, this activity was not enhanced by the addition of Lin28. In control reactions, pre-miR-21 was oligouridylated by recombinant Poly(A)Polymerase (Fig. 2c). These experiments illustrate that Lin28-mediated TUTase activity is selective for pre-let-7 family members.
Figure 2. Zcchc11 TUTase activity is Lin28-dependent.
(a) Uridylation assays performed with pre-let-7g in the presence of unlabelled UTP, ATP, CTP or GTP. (b) TUTase assay performed with pre-let-7a with a catalytically inactive Zcchc11 protein. A mutant Zcchc11 protein with two mutated aspartate residues (D1008A, D1010A in the NTr) was used in the uridylation assay. c, Uridylation assay with synthetic pre-miR-21 as a substrate in the uridylation assay as in Fig 1(c).
Inhibition of Zcchc11 leads to elevated mature let-7 levels in embryonic cells
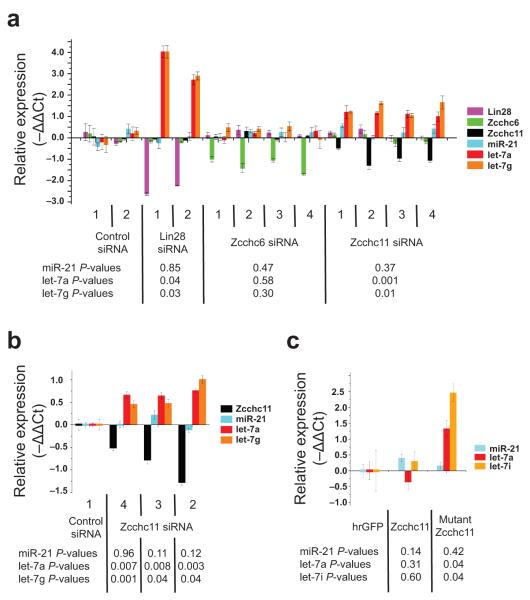
Next, we performed knockdown experiments in mouse embryonic cells and monitored the effect of Zcchc11 depletion on the levels of mature miRNAs by real-time PCR. We transfected P19 embryonal carcinoma (EC) cells with at least two siRNAs for Lin28, Zcchc11, or Zcchc6 (a closely related family member). Upon either Lin28 or Zcchc11 knockdown, we found that the levels of mature let-7a and let-7g increased while the control miRNA, miR-21, was unaffected (Fig. 3a). Zcchc6 knockdown had no effect on levels of the miRNAs analyzed (Fig. 3a). Importantly, we verified that knockdown of Zcchc11 does not affect Lin28 levels, thereby excluding the possibility that the observed effect on let-7 levels is due to Zcchc11 regulation of Lin28 expression (Fig. 3a). In multiple independent experiments (Fig. 3, 4, Supplementary Fig. 3, and data not shown) we observed a greater increase in let-7 levels upon Lin28 depletion compared to that of Zcchc11 knockdowns. This observation could be explained by the more efficient knockdown that was achieved for Lin28 than for Zcchc11. However, this observation is also consistent with the notion that Lin28 can block let-7 processing at multiple steps of the let-7 processing pathway, some of which may be independent of Zcchc11 function. For example, Lin28 has been reported to inhibit the Microprocessor leading to a block at the pri-let-7 to pre-let-7 processing step6-9. We extended our studies using a panel of siRNAs to target Lin28, Lin28B, and five of the seven known or predicted TUTases (Zcchc11, Zcchc6, Pols, Papd4, and Papd5)24,25. We excluded Tut1, the U6 snRNA-specific TUTase, from our analysis due to the reported cell lethality caused by its depletion by RNAi26. Also, mtPap was not investigated due to its mitochondrial localization29. This analysis confirmed that only reduction in Lin28 or Zcchc11 reproducibly and substantially elevated let-7a levels (Supplementary Fig. 3). This result highlights that Zcchc11 is the key TUTase responsible for regulating levels of let-7a in P19 cells. We explored the relevance of this regulatory pathway in mouse ES cells using siRNAs to deplete Zcchc11. We found that Zcchc11 depletion led to an increase in mature let-7a and let-7g levels, but had no effect on levels of miR-21 (Fig. 3b). Moreover, overexpression of the catalytically inactive mutant Zcchc11 where two of three aspartates of the catalytic triad are mutated to alanine also resulted in increased let-7 levels (Fig. 3c). These results showed that Zcchc11 mediates the selective inhibition of let-7 maturation in ES cells.
Figure 3. Inhibition of Zcchc11 selectively elevates mature let-7 levels in embryonic cells.
(a) Real-time PCR analysis of mRNA and miRNA levels in response to the indicated siRNA transfection of mouse P19 cells. Data is plotted as −ΔΔCt. Relative fold change equals 2(−ΔΔCt). Positive and negative −ΔΔCt values reflect upregulation and loss, respectively. (b) Knockdown of Zcchc11 in mouse ES cells. Depicted errors bars are ±s.d. (c) Mature microRNA levels upon HA-Zcchc11 and HA-mutant Zcchc11 overexpression in P19 cells. Presented p-values are based on comparisons between control siRNA versus siRNA set for a given gene knockdown. Depicted errors bars are ±s.d.
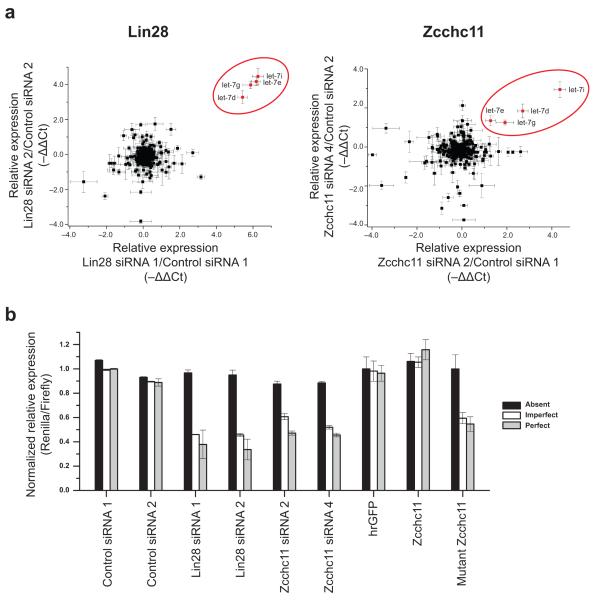
Figure 4. Zcchc11 inhibition exclusively affects let-7 biogenesis and leads to repression of let-7 target gene expression.
(a) The TaqMan Rodent MicroRNA A Array v2.0 was used to examine global miRNA levels. Independent siRNA sets were used as indicated in the axis legends and data plotted as −ΔΔCt. Left and right panels depict the results for Lin28 and Zcchc11, respectively. For each panel, the upper right quadrant contains those miRNAs that are reproducibly upregulated by both siRNA knockdown sets. Depicted errors bars are ±s.e.m. (b) Luciferase reporter assays to evaluate the in vivo relevance of elevated mature let-7 levels. Renilla luciferase lacked (termed “Absent”), contained an imperfect, or had a perfect let-7 binding site in its 3′UTR. Depicted errors bars are ±s.d.
Zcchc11 depletion exclusively affects let-7 biogenesis
To investigate more broadly the impact of Zcchc11 on miRNA regulation, global profiling of mature miRNA expression was used to analyze RNA isolated from both Lin28 and Zcchc11 knockdowns (Fig. 4a and Supplementary Table 2). We found that multiple let-7 family members (let-7b, let-7d, let-7g, and let-7i) were highly and reproducibly elevated by knockdown of both Lin28 and Zcchc11. This global analysis demonstrates unequivocally that Zcchc11 is dependent in large measure for Lin28-mediated repression.
Zcchc11 inhibition leads to repression of let-7 target gene expression
To address further the in vivo significance of this regulatory pathway, we used reporter assays to monitor the repression of let-7 responsive target genes. In these experiments, we co-transfected P19 embryonal carcinoma cells with Renilla luciferase constructs containing either fully complementary, partially complementary, or mutant let-7 binding sites in the 3′UTR, together with a Firefly luciferase control plasmid. siRNA-mediated depletion of either Lin28 or Zcchc11 in these cells led to the specific repression of reporter constructs containing let-7 target sites. No effect was observed on reporter expression that contained the mutant let-7 binding site. Hence, the specific accumulation of mature let-7 miRNA we observed by either Lin28 or Zcchc11 depletion (Fig. 3 and 4a) leads to the effective repression of reporter constructs containing let-7 target sites. We found that the extent of reporter gene repression was comparable between Lin28-depleted and Zcchc11-depleted cells. Furthermore, overexpression of the inactive mutant Zcchc11 protein also resulted in repression of the let-7 responsive reporter constructs, whereas overexpression of wild-type Zcchc11 did not (Fig. 4b). These data demonstrate that the presence of catalytically active Zcchc11 is essential for the posttranscriptional control of let-7 maturation and may provide embryonic cells with a mechanism that enables expression of let-7 target genes.
DISCUSSION
Here, we describe the first example of a specific terminal uridylyl transferase (TUTase) that controls mature miRNA levels. We demonstrate that Zcchc11 is the enzyme responsible for Lin28-mediated pre-let-7 uridylation and blockade of let-7 processing in ES cells. We have shown that the pre-let-7 TUTase activity of Zcchc11 is Lin28 dependent and that Zcchc11 exclusively regulates the expression of the let-7 miRNA family. Work from Miska and colleagues has identified a homologous pathway in C. elegans indicating that this novel miRNA gene regulatory mechanism is evolutionarily conserved (REF). Based on the substoichiometric and salt sensitive nature of the Lin28-Zcchc11 interaction, we propose a model in which Lin28 binding to pre-let-7 recruits Zcchc11 to add the U-tail. Future biochemical dissection of these proteins will likely yield additional mechanistic insight. There is emerging evidence supporting extensive posttranscriptional control of miRNA expression in various developmental and disease contexts. However, in most cases the mechanisms responsible have so far remained elusive. It will therefore be of interest to explore whether this type of regulatory mechanism involving the recruitment of a specific TUTase to miRNA precursors is relevant to other miRNAs and to identify the enzymes responsible.
Previously, the only example of terminal uridylation shown to control the levels of an endogenous mammalian transcript was that of Papd1 and Papd5 which leads to histone mRNA degradation25,27. Similarly in S. pombe, it has recently been demonstrated that Cid1-mediated uridylation leads to mRNA decapping and accelerated decay30. We envision a similar downstream pathway leading to targeted decay of the uridylated pre-let-7. Consistent with an earlier report9, we have been unable to detect robustly uridylated pre-let-7 in vivo on freshly isolated RNA. This observation likely reflects the high instability of the uridylated precursor miRNA. Although pre-let-7 with 3′-terminal U-tails can inhibit Dicer processing activity in vitro, we believe that based on the relative scarcity of this species in vivo, that the normal pathway likely involves the recruitment of specific nucleases leading to the rapid decay of the uridylated pre-let-7 RNAs. Understanding the exact downstream mechanism(s) and identification of the nucleases involved remain important avenues for future investigation.
Our results provide novel insight into the role of pre-let-7 terminal uridylation as part of the Lin28-mediated blockade of let-7 miRNA processing in ES cells and may serve as a paradigm for other miRNA families. Given the recent description of both Lin28 and Lin28B as proto-oncogenes20-23 and the corresponding role of the let-7 miRNAs as tumor suppressors, our discovery of the TUTase Zcchc11 suggests a putative novel therapeutic target in cancers in which Lin28 or Lin28B restrict the expression of the tumor suppressor let-7 miRNAs, and potentially, other diseases driven by let-7 miRNA dysregulation.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to S. Viswanathan and G. Daley (Children’s Hospital Boston) for providing Flag-Lin28 expressing ES cells, Heather Rusk and Robert LaPierre for technical assistance, and the Proteomics Center at Children’s Hospital Boston for expertise in the microcapillary HPLC/mass spectrometry. We thank Eric Miska (University of Cambridge) for helpful discussions. R.I.G is supported by lab start-up funds from The Children’s Hospital Boston and grants from NIGMS: (1R01GM086386-01A1), The Harvard Stem Cell Institute, The March of Dimes Basil O’Conner award, and the Emerald Foundation. R.I.G is a Pew Research Scholar.
Footnotes
COMPETING INTERESTS STATEMENT The authors declare no competing financial interests.
REFERENCES
- 1.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 2.Blakaj A, Lin H. Piecing together the mosaic of early mammalian development through microRNAs. J Biol Chem. 2008;283:9505–8. doi: 10.1074/jbc.R800002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 4.Hagan JP, Croce CM. MicroRNAs in carcinogenesis. Cytogenet Genome Res. 2007;118:252–9. doi: 10.1159/000108308. [DOI] [PubMed] [Google Scholar]
- 5.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–9. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–93. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 8.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 10.Piskounova E, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–4. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 11.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–16. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 12.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 13.West JA, et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009 doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lettre G, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–91. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulem P, et al. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet. 2009 doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- 16.Ong KK, et al. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet. 2009 doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stolk L, et al. Loci at chromosomes 13, 19 and 20 influence age at natural menopause. Nat Genet. 2009 doi: 10.1038/ng.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He C, et al. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009 doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry JR, et al. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet. 2009 doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dangi-Garimella S, et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–58. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Chang TC, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci U S A. 2009;106:3384–9. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viswanathan SR, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–8. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–49. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilusz CJ, Wilusz J. New ways to meet your (3′) end oligouridylation as a step on the path to destruction. Genes Dev. 2008;22:1–7. doi: 10.1101/gad.1634508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trippe R, et al. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–7. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32:6001–14. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.