Abstract
We conducted a multi-stage genome-wide association study (GWAS) of tanning response after exposure to sunlight in over 9,000 men and women of European ancestry who live in the United States. An initial analysis of 528,173 single nucleotide polymorphisms (SNPs) genotyped on 2,287 women identified LOC401937 (rs966321) on chromosome 1 as a novel locus highly associated with tanning ability, and we confirmed this association in 870 women controls from a skin-cancer case-control study with joint p-value=1.6×10−9. We further genotyped this SNP in two subsequent replication studies (one with 3,750 women and the other with 2,405 men). This association was not replicated in either of these two studies. We found that several SNPs reaching the genome-wide significance level are located in or adjacent to the loci previously known as pigmentation genes: MATP, IRF4, TYR, OCA2, and MC1R. Overall, these tanning ability-related loci are similar to those hair color-related loci reported previously in the GWAS of hair color.
Introduction
Human pigmentation shows substantial variation both within and among human populations, with high heritability (Frisancho et al., 1981; Harrison and Owen, 1964). The main contributor to human pigmentation is the melanin synthesized within melanosomes in melanocytes. There are two main types of melanin: pheomelanin (red or yellow) and eumelanin (black or brown). The tanning phenotype, combined with the hair color, skin color, and eye color, represents the visible phenotype of the human pigmentary trait. Tanning is the physiologically stimulated response to ultraviolet (UV) radiation of the solar light. UV exposure increases the production of eumelanin in an attempt to protect the skin from further damage. The sensitivity to UV radiation on the skin varies substantially both between persons at the same body site and between different body sites on the same person (Ha et al., 2003; Waterston et al., 2004). UV light is the major environmental risk factor for skin cancer in humans. Less tanning response after exposure to UV, along with lighter skin color and hair color, is a host susceptibility risk factor for skin cancer (Han et al., 2006).
It has been hypothesized that human pigmentation is tightly regulated by multiple pigmentation genes harboring a handful of genetic variants. Although more than 100 genes involved in the process of pigmentation, such as maturation, transport, and distribution of melanosomes, have been identified through animal models (Jackson, 1994), only several genes were identified to contain common genetic variants associated with human pigmentation in the normal range (Han et al., 2008; Rees, 2004; Sulem et al., 2008; Sulem et al., 2007). With new technologies that enable analysis of hundreds of thousands of single nucleotide polymorphisms (SNPs), combined with new insights into the structure of variation in the human genome (Frazer et al., 2007), it is now possible to scan the genome in an agnostic manner in search of common genetic variants associated with human pigmentation. We have previously performed a GWAS of natural hair color to identify common genetic variants associated with variation in natural diversity of human pigmentation (Han et al., 2008). In this study, we further conducted a GWAS of tanning response after exposure to UV in 2,287 U.S. women of European ancestry using data on 528,173 SNPs genotyped as part of the Cancer Genetic Markers of Susceptibility breast cancer GWAS (Hunter et al., 2007). Promising SNPs were examined in three additional studies with data on tanning response after exposure to UV: 870 U.S. women controls who were free of diagnosed skin cancer from a skin cancer case-control study within the Nurse’s Health Study (NHS); 3,750 U.S. women from a diabetes case-control study within the NHS; and 2,405 U.S. men from a diabetes case-control study within the Health Professional Follow-up Study (HPFS).
Results and Discussion
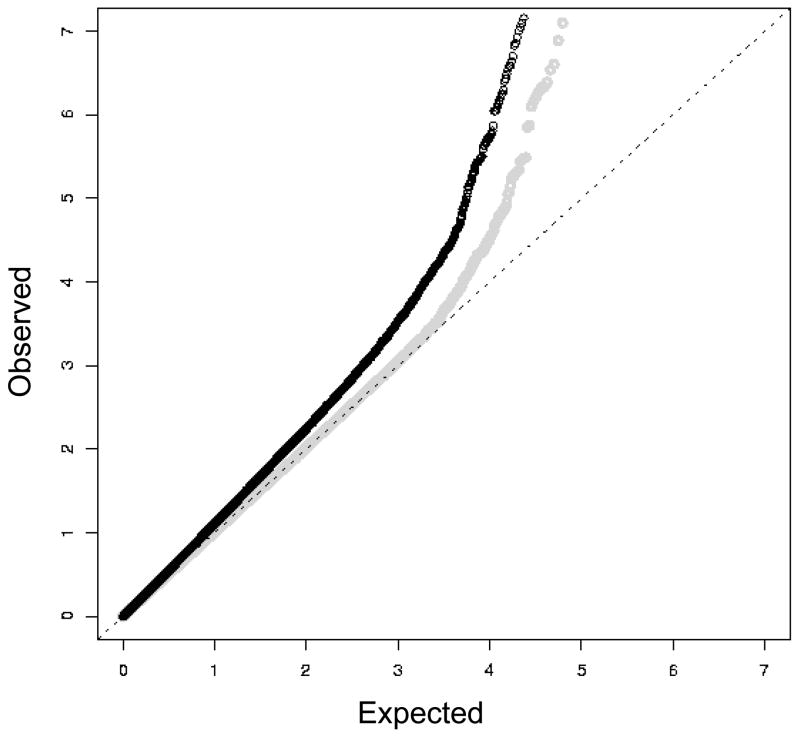
We compared the distribution of observed p-values from each of the 528,173 SNPs in the GWAS with those expected under the global null that none of the tested SNPs is associated with tanning ability to sunlight (Figure 1). The distribution of the observed p-values for the crude analyses that restricted analysis to women of self-reported European ancestry but did not further adjust for potential population stratification shows evidence for systematic bias: the genomic control inflation factor for the crude analyses (the ratio of the median observed test statistic to the theoretical median) is λGC=1.14. This systematic bias is most likely due to confounding by latent population stratification. As a major determinant of tanning ability to sunlight, skin color varies along a light-dark gradient from northern to southern Europe, so it will be associated with any SNP marker whose minor allele frequency also varies along a north-south gradient, even if that marker is not in linkage disequilibrium (LD) with a causal tanning locus (Campbell et al., 2005). Adjusting for the top four principal components of genetic variance (Price et al., 2006) eliminated most of the apparent residual confounding due to population stratification (λGC=1.02 for the adjusted analyses); further control for up to 50 principal components did not alter the λGC. All of the association results from the initial GWAS reported below are from analyses that adjusted for the top four principal components of genetic variation.
Figure 1.
Quantile-quantile plot of the -log10 p-values from an analysis of the initial GWAS that did not adjust for principal components of genetic variation (black dots) and an analysis that did adjust for the four largest principal components (graydots). P-values smaller than 10−8 are not plotted.
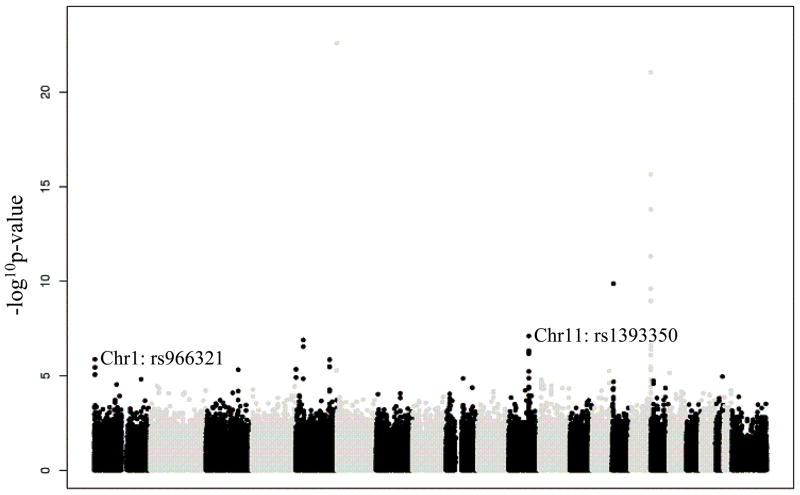
The GWAS identified several genomic locations as potentially associated with tanning response after exposure to UV (Figure 2). Of 528,173 SNPs tested, the 60 SNPs with the most extreme p-values associated with tanning response after exposure to UV are listed in Table 1.
Figure 2.
−log10 p-values from the test of association with tanning response after exposure to UV in the initial GWAS, by position along chromosome. Only p-values smaller than 0.05 are plotted.
Table 1.
Sixty SNPs with the smallest p-values of the 528,173 tested for association of tanning ability in the initial GWAS of in 2,287 women of European ancestry.
| SNP | Chromosome | Location | Gene Neighborhood | WT/VT | MAF | beta | s.e. | p value | Presented in the GWAS of hair color* | Selected for replication |
|---|---|---|---|---|---|---|---|---|---|---|
| rs12203592 | 6 | 341321 | IRF4 | C/T | 0.17 | 0.34 | 0.03 | 2.5E-23 | Yes | |
| rs258322 | 16 | 88283404 | MC1R | C/T | 0.09 | 0.41 | 0.04 | 8.8E-22 | Yes | |
| rs8049897 | 16 | 88551703 | MC1R | G/A | 0.15 | 0.30 | 0.04 | 5.8E-17 | Yes | |
| rs4785763 | 16 | 88594437 | MC1R | C/A | 0.33 | 0.21 | 0.03 | 9.9E-15 | Yes | |
| rs4238833 | 16 | 88578190 | MC1R | T/G | 0.37 | 0.19 | 0.03 | 3.7E-12 | Yes | |
| rs12913832 | 15 | 26039213 | HERC2/OCA2 | G/A | 0.25 | −0.19 | 0.03 | 1.1E-10 | Yes | |
| rs4408545 | 16 | 88571529 | MC1R | T/C | 0.50 | 0.16 | 0.03 | 2.0E-10 | Yes | |
| rs164741 | 16 | 88219799 | MC1R | C/T | 0.30 | 0.17 | 0.03 | 9.4E-10 | Yes | |
| rs1393350 | 11 | 88650694 | TYR | G/A | 0.26 | 0.16 | 0.03 | 8.0E-08 | No | Yes |
| rs28777 | 5 | 33994716 | MATP | A/C | 0.03 | −0.41 | 0.08 | 1.3E-07 | Yes | |
| rs11648785 | 16 | 88612062 | MC1R | C/T | 0.32 | −0.14 | 0.03 | 2.5E-07 | No | Yes |
| rs35391 | 5 | 33991430 | MATP | C/T | 0.03 | −0.42 | 0.08 | 2.9E-07 | No | Yes |
| rs464349 | 16 | 88183752 | MC1R | C/T | 0.47 | −0.13 | 0.03 | 4.1E-07 | No | Yes |
| rs10831496 | 11 | 88197639 | GRM5 | A/G | 0.31 | −0.14 | 0.03 | 4.7E-07 | No | Yes |
| rs10765198 | 11 | 88609422 | TYR | T/C | 0.28 | 0.15 | 0.03 | 4.7E-07 | LD with rs1393350 | |
| rs7119749 | 11 | 88154670 | GRM5 | G/A | 0.31 | −0.14 | 0.03 | 5.3E-07 | LD with rs10831496 | |
| rs10765770 | 11 | 88153284 | GRM5 | A/G | 0.31 | −0.14 | 0.03 | 6.1E-07 | LD with rs10831496 | |
| rs2169660 | 11 | 88179832 | GRM5 | G/A | 0.31 | −0.14 | 0.03 | 6.9E-07 | LD with rs10831496 | |
| rs7188458 | 16 | 88253985 | MC1R | G/A | 0.43 | 0.13 | 0.03 | 8.0E-07 | Yes | |
| rs966321 | 1 | 4225577 | LOC401937 | A/C | 0.48 | −0.12 | 0.03 | 1.4E-06 | No | Yes |
| rs32579 | 5 | 149191041 | PPARGC1B | G/A | 0.30 | −0.14 | 0.03 | 1.4E-06 | No | Yes |
| rs109075 | 5 | 149175116 | PPARGC1B | T/C | 0.30 | −0.13 | 0.03 | 3.3E-06 | LD with rs32579 | |
| rs154659 | 16 | 88194838 | MC1R | T/C | 0.26 | 0.14 | 0.03 | 3.3E-06 | No | Yes |
| rs109077 | 5 | 149176875 | PPARGC1B | T/G | 0.30 | −0.13 | 0.03 | 3.5E-06 | LD with rs32579 | |
| rs2411738 | 1 | 4236741 | LOC401937 | A/G | 0.50 | −0.12 | 0.03 | 3.6E-06 | LD with rs966321 | |
| rs11133935 | 5 | 2244437 | C/T | 0.38 | −0.12 | 0.03 | 4.6E-06 | No | Yes | |
| rs12493507 | 3 | 140744316 | RBP1 | C/T | 0.11 | 0.19 | 0.04 | 4.8E-06 | No | Yes |
| rs352935 | 16 | 88176081 | MC1R | A/G | 0.49 | 0.12 | 0.03 | 5.0E-06 | No | Yes |
| rs12210050 | 6 | 420489 | EXOC2 | C/T | 0.18 | 0.16 | 0.03 | 5.4E-06 | No | Yes |
| rs17094273 | 14 | 96173560 | G/A | 0.10 | 0.20 | 0.04 | 5.5E-06 | No | Yes | |
| rs11018528 | 11 | 88570025 | TYR | A/G | 0.28 | 0.13 | 0.03 | 5.9E-06 | LD with rs1393350 | |
| rs9960018 | 18 | 3783080 | DLGAP1 | C/T | 0.14 | −0.17 | 0.04 | 7.2E-06 | No | Yes |
| rs1908490 | 1 | 4222578 | LOC401937 | A/G | 0.50 | −0.11 | 0.03 | 8.5E-06 | LD with rs966321 | |
| rs6677984 | 1 | 4240660 | LOC401937 | G/A | 0.49 | −0.11 | 0.03 | 8.8E-06 | LD with rs966321 | |
| rs7279297 | 21 | 42100984 | PRDM15 | A/G | 0.28 | −0.13 | 0.03 | 1.1E-05 | No | Yes |
| rs2897241 | 5 | 2255185 | G/A | 0.38 | −0.12 | 0.03 | 1.3E-05 | LD with rs11133935 | ||
| rs1847134 | 11 | 88644901 | TYR | A/C | 0.30 | 0.12 | 0.03 | 1.4E-05 | LD with rs1393350 | |
| rs1409937 | 9 | 75220958 | A/G | 0.47 | 0.11 | 0.03 | 1.4E-05 | No | Yes | |
| rs40132 | 5 | 33986460 | MATP | T/C | 0.42 | −0.47 | 0.11 | 1.5E-05 | No | Yes |
| rs12750212 | 1 | 206070491 | G/A | 0.10 | −0.19 | 0.04 | 1.5E-05 | No | Yes | |
| rs1805761 | 12 | 8990800 | M6PR | A/G | 0.45 | 0.11 | 0.03 | 1.6E-05 | No | Yes |
| rs2241039 | 16 | 88615938 | MC1R | C/T | 0.38 | −0.12 | 0.03 | 1.6E-05 | Yes | |
| rs1805733 | 12 | 8980539 | M6PR | T/G | 0.45 | 0.11 | 0.03 | 1.7E-05 | LD with rs1805761 | |
| rs10852800 | 17 | 11825241 | ZNF18 | C/T | 0.19 | 0.14 | 0.03 | 1.8E-05 | No | Yes |
| rs7204478 | 16 | 88322986 | MC1R | C/T | 0.44 | 0.11 | 0.03 | 1.9E-05 | Yes | |
| rs1805721 | 12 | 9038836 | KLRG1 | C/T | 0.45 | 0.11 | 0.03 | 2.0E-05 | LD with rs1805761 | |
| rs7495174 | 15 | 26017833 | OCA2 | A/G | 0.08 | −0.21 | 0.05 | 2.1E-05 | Yes | |
| rs735408 | 14 | 96167406 | G/A | 0.09 | 0.19 | 0.05 | 2.4E-05 | LD with rs17094273 | ||
| rs12449769 | 17 | 11809087 | DNAH9 | A/G | 0.19 | 0.14 | 0.03 | 2.5E-05 | LD with rs10852800 | |
| rs1990236 | 17 | 11806187 | DNAH9 | G/A | 0.19 | 0.14 | 0.03 | 2.7E-05 | LD with rs10852800 | |
| rs12821842 | 12 | 9000251 | M6PR | G/A | 0.45 | 0.11 | 0.03 | 2.7E-05 | LD with rs1805761 | |
| rs1805723 | 12 | 9033564 | KLRG1 | A/G | 0.45 | 0.11 | 0.03 | 2.9E-05 | LD with rs1805761 | |
| rs1028889 | 1 | 98518442 | C/T | 0.26 | 0.12 | 0.03 | 2.9E-05 | No | Yes | |
| rs11054623 | 12 | 7626586 | G/T | 0.09 | −0.19 | 0.04 | 3.2E-05 | No | Yes | |
| rs7974991 | 12 | 19082600 | LOC90193 | G/A | 0.06 | 0.23 | 0.06 | 3.3E-05 | No | Yes |
| rs7969151 | 12 | 52445544 | LOC440100 | G/A | 0.21 | 0.13 | 0.03 | 3.3E-05 | No | Yes |
| rs11170681 | 12 | 52451280 | LOC440100 | T/C | 0.21 | 0.13 | 0.03 | 3.3E-05 | LD with rs7969151 | |
| rs1345151 | 2 | 30286837 | YPEL5 | G/A | 0.20 | 0.14 | 0.03 | 3.5E-05 | No | Yes |
| rs11931790 | 4 | 187715837 | LOC285441 | C/T | 0.24 | −0.13 | 0.03 | 3.7E-05 | No | Yes |
| rs10830236 | 11 | 88540464 | TYR | C/T | 0.32 | 0.11 | 0.03 | 3.9E-05 | No | Yes |
Detailed results for the SNPs indicated “yes” are presented in Table S1.
The p-values are based on primary association test adjusted for top four principal components of genetic variance.
The regression parameter beta refers to the mean change in tanning ability scoring per copy of the SNP minor allele.
Among the 60 SNPs, we selected 40 SNPs for further study in an independent sample (Table S1). The remaining 20 SNPs were in strong LD (r2>0.8) with one of these 40 SNPs (Table 1). Of the 40 SNPs selected, 13 SNPs located in the MATP, IRF4, HERC2/OCA2, and MC1R genes were previously evaluated in the GWAS of natural hair color by our group (Table 1 and Table S1) (Han et al., 2008). Notably, the most significant loci (rs12203592 in the IRF4 gene and rs12913832 in the HERC2/OCA2 gene) for hair color were the same as those for tanning ability. It has been shown that the HERC2/OCA2 locus is associated with human pigmentary variation and the SNP rs12913832 in this region has been identified as a determinant for human blue-brown eye color and hair color (Eiberg et al., 2008; Han et al., 2008; Shekar et al., 2008; Sturm et al., 2008). We finally selected 27 SNPs for further study in an independent sample (Table 2). The sample consisted of 870 controls of European ancestry from a nested case-control study of skin cancer within the NHS.
Table 2.
27 selected SNPs in the controls of the skin cancer study within the NHS and pooled data.
| in skin cancer controls |
in pooled data |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chromosome | Location | Gene Neighborhood | WT/VT | beta | s.e. | p value | beta | s.e. | p value |
| rs966321 | 1 | 4225577 | LOC401937 | A/C | −0.14 | 0.05 | 3.8E-03 | −0.14 | 0.02 | 1.6E-09 |
| rs1028889 | 1 | 98518442 | C/T | 0.03 | 0.05 | 0.57 | 0.10 | 0.03 | 1.4E-04 | |
| rs12750212 | 1 | 206070491 | G/A | −0.02 | 0.08 | 0.78 | −0.16 | 0.04 | 3.0E-05 | |
| rs1345151 | 2 | 30286837 | YPEL5 | G/A | 0.05 | 0.06 | 0.33 | 0.12 | 0.03 | 2.0E-05 |
| rs12493507 | 3 | 140744316 | RBP1 | C/T | 0.05 | 0.07 | 0.45 | 0.16 | 0.04 | 1.0E-05 |
| rs11931790 | 4 | 187715837 | LOC285441 | C/T | −0.04 | 0.05 | 0.48 | −0.10 | 0.03 | 1.2E-04 |
| rs11133935 | 5 | 2244437 | C/T | −0.03 | 0.04 | 0.45 | −0.09 | 0.02 | 1.3E-04 | |
| rs40132 | 5 | 33986460 | MATP | T/C | −0.44 | 0.18 | 0.01 | −0.53 | 0.09 | 1.5E-08 |
| rs35391 | 5 | 33991430 | MATP | C/T | −0.24 | 0.13 | 0.06 | −0.44 | 0.07 | 3.2E-10 |
| rs32579 | 5 | 149191041 | PPARGC1B | G/A | −0.03 | 0.05 | 0.49 | −0.11 | 0.02 | 3.6E-06 |
| rs12210050 | 6 | 420489 | EXOC2 | C/T | 0.28 | 0.06 | 5.7E-07 | 0.22 | 0.03 | 5.5E-14 |
| rs1409937 | 9 | 75220958 | A/G | 0.06 | 0.04 | 0.16 | 0.10 | 0.02 | 2.0E-05 | |
| rs10831496 | 11 | 88197639 | GRM5 | A/G | −0.08 | 0.05 | 0.09 | −0.14 | 0.02 | 4.7E-09 |
| rs10830236 | 11 | 88540464 | TYR | C/T | 0.06 | 0.05 | 0.22 | 0.12 | 0.02 | 6.2E-07 |
| rs1393350 | 11 | 88650694 | TYR | G/A | 0.16 | 0.05 | 7.3E-04 | 0.19 | 0.03 | 2.4E-13 |
| rs11054623 | 12 | 7626586 | G/T | 0.04 | 0.08 | 0.61 | −0.13 | 0.04 | 1.1E-03 | |
| rs1805761 | 12 | 8990800 | M6PR | A/G | 0.04 | 0.04 | 0.41 | 0.10 | 0.02 | 1.0E-05 |
| rs7974991 | 12 | 19082600 | LOC90193 | G/A | −0.08 | 0.09 | 0.40 | 0.13 | 0.05 | 7.2E-03 |
| rs7969151 | 12 | 52445544 | LOC440100 | G/A | 0.10 | 0.05 | 0.06 | 0.13 | 0.03 | 1.5E-06 |
| rs17094273 | 14 | 96173560 | G/A | 0.19 | 0.07 | 0.01 | 0.20 | 0.04 | 8.6E-08 | |
| rs352935 | 16 | 88176081 | MC1R | A/G | 0.11 | 0.04 | 0.01 | 0.12 | 0.02 | 7.4E-08 |
| rs464349 | 16 | 88183752 | MC1R | C/T | −0.12 | 0.04 | 4.9E-03 | −0.14 | 0.02 | 3.1E-09 |
| rs154659 | 16 | 88194838 | MC1R | T/C | 0.08 | 0.05 | 0.13 | 0.14 | 0.03 | 7.4E-08 |
| rs11648785 | 16 | 88612062 | MC1R | C/T | −0.10 | 0.05 | 0.04 | −0.14 | 0.02 | 2.7E-09 |
| rs10852800 | 17 | 11825241 | ZNF18 | C/T | 0.04 | 0.06 | 0.47 | 0.12 | 0.03 | 7.0E-05 |
| rs9960018 | 18 | 3783080 | DLGAP1 | C/T | −0.03 | 0.06 | 0.64 | −0.15 | 0.03 | 1.0E-05 |
| rs7279297 | 21 | 42100984 | PRDM15 | A/G | −0.03 | 0.05 | 0.49 | −0.12 | 0.03 | 2.7E-06 |
Eight of these 27 SNPs showed evidence of significant associations with tanning ability among the 870 controls (p<0.05) (Table 2). Those SNPs were located on chromosome 1 (rs966321), chromosome 5 (rs40132), chromosome 6 (rs12210050), chromosome 11 (rs1393350), chromosome 14 (rs17094273), and chromosome 16 (rs352935, rs464349, and rs11648785), respectively. These eight SNPs showed very strong evidence of associations with tanning ability in a pooled analysis of the initial GWAS and the replication sample (p<9.5×10−8=0.05/528,173) (Table 2). Of these eight SNPs, six of them (rs40132, rs12210050, rs17094273, rs352935, rs464349, and rs11648785) were located in or adjacent to the four hair color-related loci reported previously in the GWAS of hair color conducted by our group (Han et al., 2008): MATP, EXOC2, SLC24A4, and MC1R. One SNP (rs1393350) in the TYR gene was previously found to be associated with skin color and tanning ability from a candidate gene approach by our group. The remaining one novel SNP rs966321 located on chromosome 1 (LOC401937) was strongly associated with tanning ability in the initial GWAS and the follow-up study (pooled p-value for trend = 1.6×10−9). We genotyped rs966321 in an additional 6,155 subjects of predominantly European ancestry from the United States, including 3,750 women from the NHS and 2,405 men from the HPFS. This significant association was not reproduced in subsequent replication studies. The p-values were 0.59 (regression parameter beta (β), −0.01) and 0.16 (β, 0.05), respectively. Using the same three subsequent replication studies, we successfully replicated the associations between hair color and tanning ability and some SNPs identified from our previous GWAS for hair color (Han et al., 2008). It appears that the SNP rs966321 is not a robust variant influencing tanning ability, which further underlines that the replication in independent studies is the key to confirm robust associations in genetic association studies.
We identified the SNP rs40132 in the MATP gene from the GWAS, and the association with tanning ability was confirmed in the follow-up study (pooled p-value=1.5×10−8). Three SNPs in the MATP gene have been associated with human pigmentation: rs16891982 (Phe374Leu), rs26722 (Glu272Lys), and rs13289 C/G (−1721 in the promoter region) (Graf et al., 2005; Graf et al., 2007). We previously evaluated these three SNPs for associations with pigmentary phenotypes in the controls of skin cancer study. None of the three SNPs were in strong LD with rs40132 (r2<0.08), which is an intronic SNP. A multivariate analysis mutually adjusting for rs40132, rs16891982, rs26722, and rs13289 simultaneously showed that only rs16891982 remained significant in the model (p-value=0.02) and other SNPs became non-significant (p-value>0.05). The MATP, a membrane-associated transporter protein, has been considered as a sodium-hydrogen exchanger of melanosomes, regulating tyrosinase activity in human melanocyte (Smith et al., 2004). These data suggested that rs16891982 is most likely to be the causal variant or in strong LD with the causal variant in the MATP gene.
The SNP rs12210050 in the EXOC2 (SEC5L1) gene was strongly associated with tanning ability in the initial GWAS and was confirmed in the follow-up study (pooled p-value =5.5×10−14). On the same chromosome 6, 79.2 kb telomeric from the EXOC2 rs12210050, a SNP (rs12203592) in the intron 4 of the IRF4 gene, has been strongly associated with pigmentary phenotypes, such as hair color, tanning ability, and skin color in the GWAS of hair color (Han et al., 2008). These two SNPs are in weak LD (r2=0.26). We mutually adjusted these two significant SNPs and found that only rs12203592 remained significant (p-value=8.3×10−8 in the follow-up study). Sulem et al. identified two SNPs (rs4959270 and rs1540771) between the EXOC2 and IRF4 genes in relation to hair color and skin sensitivity to sun with much weaker associations than those of the SNP rs12203592 in the IRF4 gene (Sulem et al., 2007).
A SNP (rs1393350) in the TYR gene showed a significant association with tanning ability in the initial GWAS and was confirmed in the follow-up study (pooled p-value=2.4×10−13). On the same chromosome 11, we identified two SNPs (TYR rs10830236 and GRM5 rs10831496) associated with tanning ability in the initial GWAS, but not in the follow-up study. Neither of these two SNPs, rs10831496 and rs10830236, was in the LD with rs1393350 (r2=0.06 and 0.64, respectively). Only rs1393350 remained significant after adjusting for these three SNPs mutually in the follow-up study of skin cancer controls (p-value=2.6×10−3). Sulem et al. recently reported a pigmentation GWAS in the Icelandic population and showed a strong association between the variant rs1393350 in the TYR gene and eye color, freckles, and skin sensitivity to sun (Sulem et al., 2007). The rs1393350 is in strong LD with a non-synonymous SNP rs1126809 (Arg402Gln) in the TYR gene (D′=1 and r2=0.86), a common polymorphism of tyrosinase. Tyrosinase is a critical enzyme during melanosomal maturation and its high activity leads to the formation of eumelanosome (Jimbow et al., 2000; Spritz, 1994). It has been reported that the TYR Arg402Gln was correlated with reduced pigmentation of the retina and iris resulting from low tyrosinase activity (Fukai et al., 1995).
We identified that the three SNPs (rs352935, rs464349, and rs11648785) on chromosome 16 that showed significant associations with tanning ability in the initial GWAS and were confirmed in the follow-up study. Pooled p-values for rs352935, rs464349, and rs11648785 were 7.4×10−8, 3.1×10−9, and 2.7×10−9, respectively. These SNPs are adjacent to the MC1R (melanocortin 1 receptor), a well-established pigmentation gene encoding a 317-amino acid 7-pass-transmembrane G protein-coupled receptor. As the rate-limiting step in the activation of the cAMP pathway in terms of melanin production, MC1R has been strongly associated with pigmentary phenotypes, especially with red-hair color phenotype. We had previously genotyped seven common MC1R variants among the NHS skin cancer controls (Han et al., 2006). The analysis mutually adjusting for these 10 SNPs in the controls of the skin cancer study showed that the significant associations with tanning response after exposure to UV that we observed for three SNPs (rs352935, rs464349, and rs11648785) were eliminated by inclusion of the three MC1R red-hair color alleles (Arg151Cys, Arg160Trp, and Asp294His). This result suggests that the signals that we identified on chromosome 16 were explained by the functional variants in the MC1R gene, although the LD between the MC1R variants and surrounding highly significant SNPs was relatively low. Similar results were noted in the GWAS of hair color (Han et al., 2008). We observed that the highly significant associations between the SNPs on chromosome 16 and hair color phenotype were eliminated after adjusting for functional MC1R variants.
There is some evidence that determinants of human pigmentation may act along different phenotypic axes. For example, alleles at the MC1R locus primarily determine presence or absence of red hair (Rees, 2004). Hence, we additionally evaluated the associations of 27 selected SNPs with tanning response after excluding individuals with red hair color. The association patterns were similar to those shown in the analyses including red haired individuals (Table S2).
One limitation of this study was the self-reported tanning information. Self-report has been shown to be an appropriate and widely-used method of assessing risk factors for skin cancer. Test-retest reliability of collecting phenotypic factors from questionnaires is moderate to substantial, including skin color, tanning/burning tendency, and sunburn history(Branstrom et al., 2002; Glanz et al., 2003; Westerdahl et al., 1996).
In our study of individuals of European ancestry, we focused on the most statistically significant associations from our GWAS, identifying and confirming the loci previously known as pigmentation genes, such as MATP, IRF4, TYR, OCA2, and MC1R. The strongest loci for tanning ability were the same as those for hair color. Because a subset of true associations would be weakly associated with outcome in any given GWAS, large-scale replication is necessary for confirmation, and some true associations may be missed if they are not carried forward into replication studies (Chanock et al., 2007).
Materials and Methods
Nurses’ Health Study (NHS)
The NHS was established in 1976, when 121,700 female U.S. registered nurses between the ages of 30 and 55, residing in 11 larger U.S. states, completed and returned the initial self-administered questionnaire on their medical histories and baseline health-related exposures, forming the basis for the NHS cohort. Biennial questionnaires with collection of exposure information on risk factors and (every 4 years since 1980) nutritional assessments have been collected prospectively. Along with exposures every 2 years, outcome data with appropriate follow-up of reported disease events, including melanoma and non-melanoma skin cancers, are collected. Overall, follow-up has been very high; after more than 20 years approximately 90% of participants continue to complete questionnaires. From May 1989 through September 1990, we collected blood samples from 32,826 participants in the NHS cohort. The information on tanning response after exposure to UV in childhood and adolescence was collected in the 1982 prospective questionnaire. The question was “as a child or adolescent, after repeated sun exposures, e.g., a two-week vacation outdoors, what kind of tan would you get?”, and the multiple choices were “practically none, light tan, average tan, and deep tan”.
Initial GWAS
We initially performed genotyping in a nested case-control study of postmenopausal invasive breast cancer within the NHS cohort (Tworoger et al., 2007) using the Illumina HumanHap550 array, as part of the National Cancer Institute’s Cancer Genetic Markers of Susceptibility (CGEMS) Project (Hunter et al., 2007). We performed our initial genome-wide analysis on 528,173 SNPs in 2,287 women (Hunter et al., 2007). All cases and controls were self-described as being of European ancestry. Four samples were excluded because of evidence of intercontinental admixture. Controlling for breast cancer case-control status made no material difference to the GWAS results.
Detailed methods related to the initial GWAS were published previously (Hunter et al., 2007), including genotyping and quality control, initial assessment of sample completion rates, assessment of SNP call rates, concordance rate, deviation from Hardy Weinberg proportions in control DNA, and final sample selection and exclusion for association analysis.
The controls in the skin cancer nested case-control study within the NHS
The promising SNPs from the initial GWAS were further genotyped among 870 controls in the skin cancer nested case-control study within the NHS. The distribution of risk factors for skin cancer in the subcohort of those who donated blood samples was very similar to that in the overall cohort (Han et al., 2006). A common control series was randomly selected from participants who gave a blood sample and were free of diagnosed skin cancer up to and including the questionnaire cycle in which the corresponding case was diagnosed.
Health Professional Follow-up Study (HPFS)
In 1986, 51,529 men from all 50 U.S. states in health professions (dentists, pharmacists, optometrists, osteopath physicians, podiatrists, and veterinarians) aged 40–75 answered a detailed mailed questionnaire, forming the basis of the study. Between 1993 and 1994, 18,159 study participantsprovided blood samples by overnight courier. The information on tanning response after exposure to UV was asked in the 1992 questionnaire.
The diabetes nested case-control studies within the NHS and HPFS
Two additional studies were used to genotype novel pigmentation loci: 3,750 samples from the nested case-control study of diabetes in the NHS and 2,405 samples from the nested case-control study of diabetes in the HPFS. All samples that we used were cases and controls from these two studies. Cases were incident cases of diabetes after blood collection, and controls were matched on age and history of cardiovascular disease. Controlling for case-control status made no material difference to the results.
There was no sample overlap among the initial GWAS, the skin cancer case-control study, and the two diabetes case-control studies. The study protocol was approved by the Institutional Review Board of Brigham and Women’s Hospital and Harvard School of Public Health. Informed consent was obtained from all patients.
Statistical analysis
For the primary analysis of tanning response after exposure to UV we regressed an ordinal coding for tanning ability (1=deep tan; 2=average tan; 3=light tan; and 4=no tan) on an ordinal coding for genotype (0, 1, or 2 copies of SNP minor allele) separately for each SNP that passed quality control filters (Hunter et al., 2007). Crude analyses that did not adjust for any other variables showed evidence of systematic bias (see Results and Discussion section). However, this bias was greatly reduced by adjusting for the four largest principal components of genetic variation. These principal components were calculated for all subjects on the basis of ca. 10,000 unlinked markers using the EIGENSTRAT software (Hunter et al., 2007; Price et al., 2006). The top four eigenvectors were chosen on the basis of significant (p<0.05) Tracy-Wisdom tests (Patterson et al., 2006). Adjusting for up to the top 50 principal components did not further reduce the genomic control inflation factor λGC. We chose markers for genotyping in subsequent validation studies based on the p-values for association from the primary analysis. The regression parameter beta refers to the mean change in tanning ability scoring per copy of the SNP minor allele. Pooled analyses of multiple studies were conducted by merging data sets and including separate baseline parameters for each study.
Genotyping in follow-up studies
The 27 promising SNPs from the initial GWAS were genotyped in the skin cancer controls using TaqMan/BioTrove assays at the Dana Farber/Harvard Cancer Center Polymorphism Detection Core. A novel locus rs966321 on chromosome 1 was further genotyped in diabetes samples in the NHS and HPFS studies using the Taqman assay. Laboratory personnel were blinded to the case-control status, and 10% blinded quality control samples were inserted to validate genotyping procedures; concordance for the blinded samples was 100%. Primers, probes, and conditions for genotyping assays are available upon request.
Supplementary Material
Acknowledgments
We thank the investigators involved in the CGEMS project, including Kevin B. Jacobs at Bioinformed Consulting Services, Gaithersburg, MD; Sholom Wacholder, Nick Orr, Kai Yu, Nilanjan Chatterjee, Joseph F. Fraumeni, Jr., Richard B. Hayes, Margaret Tucker, and Daniela S. Gerhard at the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Department of Health and Human Services, Bethesda, MD; and Meredith Yeager, the late Robert Welch, Amy Hutchinson, Andrew Crenshaw, and Zhaoming Wang at the Core Genotyping Facility, Advanced Technology Program, SAIC Frederick, Inc, NCI-Frederick, Frederick, MD. We thank Pati Soule and Dr. Hardeep Ranu of the Dana Farber/Harvard Cancer Center High-Throughput Polymorphism Detection Core for sample handling and genotyping of the NHS samples. We are also indebted to the participants in all of these studies. This work was funded in part by NCI grants CA128080 and CA122838.
Abbreviations
- OR
odds ratio
- CI
confidence interval
- UV
ultraviolet
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- Branstrom R, Kristjansson S, Ullen H, Brandberg Y. Stability of questionnaire items measuring behaviours, attitudes and stages of change related to sun exposure. Melanoma Res. 2002;12:513–519. doi: 10.1097/00008390-200209000-00014. [DOI] [PubMed] [Google Scholar]
- Campbell CD, Ogburn EL, Lunetta KL, Lyon HN, Freedman ML, Groop LC et al. Demonstrating stratification in a European American population . Nat Genet. 2005;37:868–872. doi: 10.1038/ng1607. Epub 2005 Jul 2024. [DOI] [PubMed] [Google Scholar]
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Eiberg H, Troelsen J, Nielsen M, Mikkelsen A, Mengel-From J, Kjaer KW, et al. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum Genet. 2008;123:177–187. doi: 10.1007/s00439-007-0460-x. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisancho AR, Wainwright R, Way A. Heritability and components of phenotypic expression in skin reflectance of Mestizos from the Peruvian lowlands. Am J Phys Anthropol. 1981;55:203–208. doi: 10.1002/ajpa.1330550207. [DOI] [PubMed] [Google Scholar]
- Fukai K, Holmes SA, Lucchese NJ, Siu VM, Weleber RG, Schnur RE, et al. Autosomal recessive ocular albinism associated with a functionally significant tyrosinase gene polymorphism. Nat Genet. 1995;9:92–95. doi: 10.1038/ng0195-92. [DOI] [PubMed] [Google Scholar]
- Glanz K, Schoenfeld E, Weinstock MA, Layi G, Kidd J, Shigaki DM. Development and reliability of a brief skin cancer risk assessment tool. Cancer Detect Prev. 2003;27:311–315. doi: 10.1016/s0361-090x(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Graf J, Hodgson R, van Daal A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutat. 2005;25:278–284. doi: 10.1002/humu.20143. [DOI] [PubMed] [Google Scholar]
- Graf J, Voisey J, Hughes I, van Daal A. Promoter polymorphisms in the MATP (SLC45A2) gene are associated with normal human skin color variation. Hum Mutat. 2007;28:710–717. doi: 10.1002/humu.20504. [DOI] [PubMed] [Google Scholar]
- Ha T, Javedan H, Waterston K, Naysmith L, Rees JL. The relationship between constitutive pigmentation and sensitivity to ultraviolet radiation induced erythema is dose-dependent. Pigment Cell Res. 2003;16:477–479. doi: 10.1034/j.1600-0749.2003.00076.x. [DOI] [PubMed] [Google Scholar]
- Han J, Colditz GA, Hunter DJ. Risk factors for skin cancers: a nested case-control study within the Nurses’ Health Study. Int J Epidemiol. 2006;35:1514–1521. doi: 10.1093/ije/dyl197. [DOI] [PubMed] [Google Scholar]
- Han J, Kraft P, Nan H, Guo Q, Chen C, Qureshi A, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison GA, Owen JJ. Studies on the Inheritance of Human Skin Colour. Ann Hum Genet. 1964;28:27–37. doi: 10.1111/j.1469-1809.1964.tb00457.x. [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. Epub 2007 May 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson IJ. Molecular and developmental genetics of mouse coat color. Annu Rev Genet. 1994;28:189–217. doi: 10.1146/annurev.ge.28.120194.001201. [DOI] [PubMed] [Google Scholar]
- Jimbow K, Hua C, Gomez PF, Hirosaki K, Shinoda K, Salopek TG, et al. Intracellular vesicular trafficking of tyrosinase gene family protein in eu- and pheomelanosome biogenesis. Pigment Cell Res. 2000;13(Suppl 8):110–117. doi: 10.1034/j.1600-0749.13.s8.20.x. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. Epub 2006 Jul 2023. [DOI] [PubMed] [Google Scholar]
- Rees JL. The genetics of sun sensitivity in humans. Am J Hum Genet. 2004;75:739–751. doi: 10.1086/425285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekar SN, Duffy DL, Frudakis T, Sturm RA, Zhao ZZ, Montgomery GW, et al. Linkage and association analysis of spectrophotometrically quantified hair color in Australian adolescents: the effect of OCA2 and HERC2. J Invest Dermatol. 2008;128:2807–2814. doi: 10.1038/jid.2008.147. [DOI] [PubMed] [Google Scholar]
- Smith DR, Spaulding DT, Glenn HM, Fuller BB. The relationship between Na(+)/H(+) exchanger expression and tyrosinase activity in human melanocytes. Exp Cell Res. 2004;298:521–534. doi: 10.1016/j.yexcr.2004.04.033. [DOI] [PubMed] [Google Scholar]
- Spritz RA. Molecular genetics of oculocutaneous albinism. Hum Mol Genet. 1994;3 doi: 10.1093/hmg/3.suppl_1.1469. Spec No:1469–1475. [DOI] [PubMed] [Google Scholar]
- Sturm RA, Duffy DL, Zhao ZZ, Leite FP, Stark MS, Hayward NK, et al. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am J Hum Genet. 2008;82:424–431. doi: 10.1016/j.ajhg.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Jakobsdottir M, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008;40:835–837. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- Tworoger SS, Eliassen AH, Sluss P, Hankinson SE. A prospective study of plasma prolactin concentrations and risk of premenopausal and postmenopausal breast cancer. J Clin Oncol. 2007;25:1482–1488. doi: 10.1200/JCO.2006.07.6356. Epub 2007 Mar 1419. [DOI] [PubMed] [Google Scholar]
- Waterston K, Naysmith L, Rees JL. Physiological variation in the erythemal response to ultraviolet radiation and photoadaptation. J Invest Dermatol. 2004;123:958–964. doi: 10.1111/j.0022-202X.2004.23411.x. [DOI] [PubMed] [Google Scholar]
- Westerdahl J, Anderson H, Olsson H, Ingvar C. Reproducibility of a self-administered questionnaire for assessment of melanoma risk. Int J Epidemiol. 1996;25:245–251. doi: 10.1093/ije/25.2.245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.