Abstract
Sertraline and fluoxetine are selective serotonin reuptake inhibitors (SSRIs) widely-prescribed to treat depression. They exert their effects by inhibiting the presynaptic plasma membrane serotonin transporter (SERT). All SSRIs possess at specific positions halogen atoms, which are key determinants for the drugs’ specificity for SERT. For the SERT protein, however, the structural basis of its specificity for SSRIs is poorly understood. Here we report the crystal structures of LeuT, a bacterial SERT homolog, in complex with sertraline, R-fluoxetine or S-fluoxetine. The SSRI halogens all bind to exactly the same pocket within LeuT. Mutation at this halogen-binding pocket (HBP) in SERT dramatically reduces the transporter's affinity for SSRIs but not for tricyclic antidepressants. Conversely, when the only non-conserved HBP residue in both norepinephrine and dopamine transporters is mutated into that found in SERT, their affinities for all the three SSRIs increase uniformly. Thus, the specificity of SERT for SSRIs is dependent largely on interaction of the drug halogens with the protein's halogen-binding pocket.
INTRODUCTION
Selective serotonin reuptake inhibitors (SSRIs) bind directly to the serotonin transporter protein and inhibit recycling of neurotransmitter, making them effective drugs for treatment of depressive disorders 1,2. SSRIs, however, are rather promiscuous in that they bind also to the homologous norepinephrine and dopamine transporters (NET and DAT, respectively), although with much lower affinity than to their principal target SERT 3,4. The selectivity of SSRIs for SERT is intriguing. Merely one or two different functional group substitutions are sufficient to convert an SSRI into a norepinephrine reuptake inhibitor (NRI) with higher affinity to NET 5-7. It is recognized that both the position and type of substitution on an aromatic moiety of the SSRI molecule are important for the higher specificity to SERT 8,9. In particular, halogen substitutions on this ring are found to be largely responsible for SSRIs’ specificity to SERT 5,6,10. On the protein side, however, the transporter-SSRI interactions that define the specificity of SERT for these drugs have not yet been described, which hinders the development of more specific antidepressants 11.
The human SERT, NET and DAT proteins are all members of the neurotransmitter:sodium symporter (NSS) family 12-14. The same protein family also contains members from bacterial cells, and such proteins often function as amino acid transporters 15. One family member is the leucine transporter LeuT from Aquifex aeolicus. LeuT shares 20−25 % identity in primary sequence with the human neurotransmitter transporters, and the crystal structure of LeuT 16 and its transport mechanism have proven to be good model systems for the study of mammalian NSS proteins 17-20. To understand the structural basis of the serotonin transporter's specificity for SSRIs, we carried out crystallographic studies of the bacterial leucine transporter LeuT in complex with three different SSRIs. This was followed by mutagenesis and pharmacological studies of the human SERT, NET and DAT proteins at the equivalent drug-binding site.
RESULTS
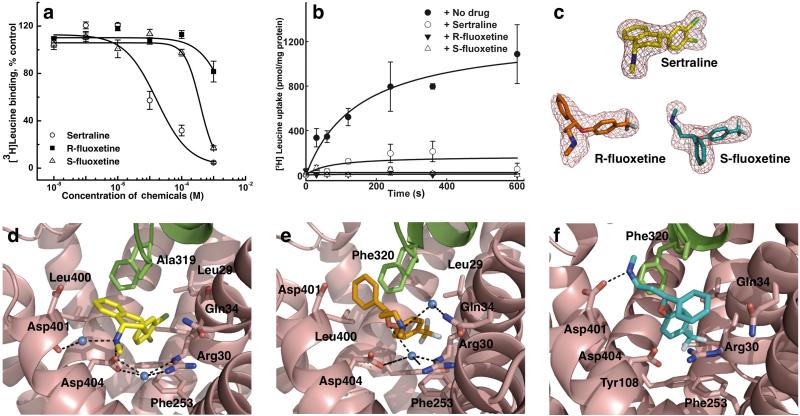
We first showed that three SSRIs — sertraline, R-fluoxetine and S-fluoxetine — all bind to LeuT (Fig. 1a), and that they also inhibit substrate transport by the protein reconstituted into proteoliposomes (Fig. 1b). We then co-crystallized LeuT with either sertraline, R-fluoxetine or S-fluoxetine (Prozac contains equal amounts of the R- and S-enantiomers, both pharmacologically active), along with substrate leucine and sodium ions, and determined these complex structures at a resolution of 2.15, 2.35 and 2.45 Å, respectively (Table 2 and Supplementary Fig. 1a). In all these crystals the overall structure of the protein-substrate complex is similar to that of the drug-free form 16. However, in all three complexes, a strong electron density peak was observed in the vestibule between the tip of the extracellular loop EL4 and the extracellular gate (Fig. 1c and Supplementary Fig. 1a), which is formed by residues Arg30, Asp404, Tyr108 and Phe253. The density was assigned to sertraline, R-fluoxetine and S-fluoxetine, respectively (Figs. 1d-f). This drug binding location is similar to the tricyclic antidepressant (TCA) binding site in LeuT 21,22, and no secondary SSRI-binding site was found in the protein.
Figure 1. Interaction of three SSRIs with LeuT and the crystal structures of their complexes.
a. Binding of SSRIs to LeuT in detergent solution was measured using a scintillation proximity assay. The IC50 values for sertraline, R-fluoxetine and S-fluoxetine to inhibit [3H]leucine binding to LeuT were determined to be 19.7 ± 9.2 μM, 2.54 ± 0.41 mM and 355 ± 46 mM, respectively. Curves show relative [3H]leucine binding, normalized to the [3H]leucine binding in the absence of inhibitors. Each point represents the mean ± S.E. (N = 3). b. LeuT transport activity measured in the absence and presence of SSRIs was measured in reconstituted proteoliposomes. Leucine transport by LeuT was completely inhibited by sertraline, R-fluoxetine and S-fluoxetine at concentration of 0.5 mM, 5 mM and 0.5 mM, respectively (N = 3). c. Fo-Fc simulated annealing omit maps of the SSRI from the three LeuT-SSRI complex structures, LeuT-sertraline at 2.15 Å resolution, LeuT-R-fluoxetine at 2.35 Å, and LeuT-S-fluoxetine at 2.45 Å resolution. The maps are contoured at 3 σ. d. Structure of the drug-binding site in the LeuT-sertraline complex at 2.15 Å resolution, viewed from within the membrane plane. e. Structure of the drug-binding site in the LeuT-R-fluoxetine complex at 2.35 Å resolution. f. Structure of the drug-binding site in the LeuT-S-fluoxetine complex at 2.45 Å resolution. The sertraline molecule is colored yellow, R-fluoxetine orange, and S-fluoxetine green. In e – f. helix TM11 in LeuT is omitted from the figures for clarity.
Table 2.
Data collection and refinement statistics for LeuT in complex with various SSRIs
| LeuT-sertraline | LeuT-R-fluoxetine | LeuT-S-fluoxetine | |
|---|---|---|---|
| Data collection | |||
| Space group | C2 | C2 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 88.34, 86.85, 81.31 | 87.81, 87.39, 81.15 | 86.35, 86.28, 80.62 |
| α, β, γ (°) | 90, 95.65, 90 | 90, 95.67, 90 | 90, 95.16, 90 |
| Resolution (Å) | 2.15 (2.22−2.15) * | 2.35 (2.43−2.35) * | 2.45 (2.55−2.45) * |
| Rsym | 0.079 (0.300) | 0.064 (0.212) | 0.063 (0.186) |
| I / σI | 20.7 (2.6) | 15.6 (2.4) | 24.0 (3.8) |
| Completeness (%) | 91.2 (63.2) | 91.9 (65.5) | 90.6 (53.8) |
| Redundancy | 3.4 (2.4) | 3.0 (1.6) | 3.5 (2.6) |
| Refinement | |||
| Resolution (Å) | 50.00−2.15 | 40.00−2.35 | 50.00−2.45 |
| No. reflections | 29,371 | 22,191 | 18,572 |
| Rwork / Rfree | 0.203/0.227 | 0.201/0.232 | 0.204/0.226 |
| No. atoms | |||
| Protein | 4,044 | 3,963 | 3,991 |
| Ligand/ion | 29/2 | 31/2 | 31/2 |
| Water | 100 | 70 | 23 |
| B-factors | |||
| Protein | 33.7 | 38.3 | 47.1 |
| Leucine | 19.8 | 20.3 | 32.3 |
| Sodium | 24.7 | 24.9 | 35.6 |
| Drug | 31.6 | 60.1 | 56.1 |
| Water | 38.2 | 42.7 | 43.6 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.008 | 0.007 |
| Bond angles (°) | 0.931 | 1.008 | 0.949 |
A single crystal was used for each structure.
Values in parentheses are for highest-resolution shell.
Sertraline-binding site in LeuT
Although the position of binding to the protein is similar to that observed for TCAs, how the SSRIs bind to the protein is markedly different. In the LeuT-sertraline structure, the drug molecule binds to the extracellular vestibule in the protein in such a way that the two chlorine atoms on the phenyl ring insert into a pocket that is formed by Leu25, Gly26, Leu29, Arg30, Tyr108, Ile111 and Phe253 (Fig. 1d), and make additional van der Waals contact with Leu29, Tyr108 and Phe253 (Fig. 2a and Supplementary Fig. 2a). The amino acid sequence in this halogen-binding pocket (HBP) is highly conserved between LeuT and SERT. Importantly, four of the halogen-binding residues, Leu25, Gly26, Tyr108 and Phe253, also interact at the opposite side of the polypeptide chain with the leucine (Supplementary Fig. 1b) or another bound-substrate 23, demonstrating that the drug-binding site and the substrate-binding site, although not sharing the same physical space, do share several common amino acid residues. At the other end of the sertraline molecule, the dichlorophenyl ring is approximately perpendicular to the tetrahydronaphthalene ring, which is tilted slightly from the membrane plane. In addition to being in contact with Leu400, Asp401 and Thr409 of transmembrane α-helix 10 (TM10), the tetrahydronaphthalene rings interact with residues Ala319 of the EL4 hairpin loop and Arg30 and Gln34 of TM1, whereas the amine tail points toward the cytoplasm. Notably, the amine nitrogen forms a salt bridge (named salt bridge 1) with the carboxyl group of Asp404 (Fig. 2b) and, simultaneously, interacts with the backbone oxygen of Asp401 via a bound water molecule. Another water molecule, located in the same position between Arg30 and Asp404 as one observed in the drug-free structure of LeuT 16, mediates the two residues’ interactions with the SSRI. The bound sertraline molecule itself has its dichlorophenyl ring rotated about the C4-C13 bond by 180° (Supplementary Fig. 3a) compared to the free drug 24. Otherwise, the sertraline molecule is rather rigid, indicating that the drug maintains its low-energy configuration upon binding to its protein target.
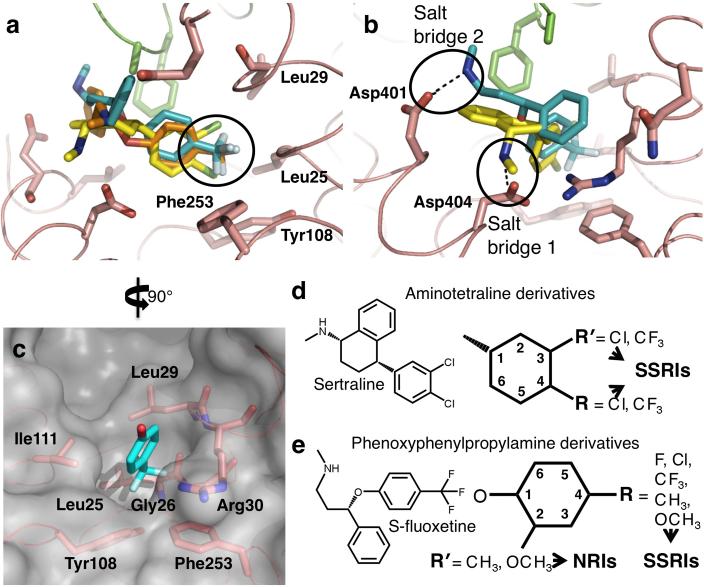
Figure 2. Comparison of common features of SSRI binding to LeuT and key determinants for specificity for SSRIs.
a and b. Superposition of the three LeuT-SSRI structures at the drug binding site. c. Surface presentation of the halogen-binding pocket in the LeuT-S-fluoxetine structure 10. d. Illustration showing halogen substitutions at one or both of the 3rd and 4th positions in the phenyl ring of aminotetraline with Cl- or CF3- yield an SSRI. e. Illustration showing phenoxyphenylpropylamine-based antidepressants. Substitutions at the 4- substitution in the phenoxy ring with F-, Cl-, CF3-, CH3- or OCH3- produce SSRIs, including R- and S-fluoxetines, whereas substitutions at the 2nd position with CH3- or OCH3- yield NRIs like tomoxetine and nisoxetine 6. In all the panels, the SSRIs are colored as in Fig.1.
R-fluoxetine-binding site in LeuT
R-fluoxetine binds to the same extracellular vestibule in LeuT as sertraline (Fig. 1e). The halogen atoms (in this case three fluorines of the methylphenoxy ring) insert into the same HBP as the two sertraline chlorines, making contact with Leu25, Gly26, Leu29, Arg30 and Tyr108 (Fig. 2a and Supplementary Fig. 2b). At the opposite end of the drug, the phenyl ring is surrounded by Ala319 and Phe320 from the tip of the EL4 hairpin and Leu400 and Asp401 from TM10 on the extracellular side. Similar to sertraline, the amine tail in R-fluoxetine points toward the cytoplasm and interacts via a bound water molecule with Gln34. As observed in the LeuT-sertraline structure, a water molecule, located between Arg30 and Asp404, mediates interaction of these two residues with the methylphenoxy ring of the drug (Fig. 1e). In this LeuT-bound form, the same ring rotates about the O5-C6 bond by 46° compared to that of the free drug 25 (Supplementary Fig. 3b).
S-fluoxetine-binding site in LeuT
The fluorine atoms on the phenoxy ring of S-fluoxetine also insert into the same HBP in LeuT (Fig. 1f), making van der Waals contact with Leu25, Gly26, Leu29, Arg30, Tyr108 and Phe253 (Figs. 2a and c). As observed in the LeuT-sertraline- and LeuT-R-fluoxetine complexes (Supplementary Fig. 1b), Leu25, Gly26, Tyr108 and Phe253 are shared between the drug-binding site and the substrate-binding site. In contrast to R-fluoxetine, however, due to its opposite chirality, the rest of the S-fluoxetine molecule is reversed in the binding pocket, with the phenyl ring interacting with the gate, the amine tail pointing to the extracellular space and the nitrogen atom of the amine group forming a salt bridge (named salt bridge 2) with Asp401 (Figs. 1f and 2b). Moreover, the Arg30 side chain flips towards Asp404, forming a salt bridge with it. As a result, both the S-fluoxetine rings, the positively charged guanidinium group of Arg30 and the phenyl ring of Phe253 form a stable cation-π cluster similar to that observed in the LeuT-TCA complexes 21,22. Like R-fluoxetine, the methylphenoxy ring of S-fluoxetine rotates about the O5-C6 bond upon binding to LeuT, but by a smaller angle (19°) (Supplementary Fig. 3c).
Common features of SSRI binding to LeuT
All three SSRIs bind to LeuT at the same position and in a very similar manner. The electronic dipole moment in each drug, which arises due to the positively-charged amine group and the electronegative halogen atoms, points in approximately the same direction, from TM10 to TM1. Not only are the dichlorophenyl group of sertraline and the trifluoromethylphenoxy groups of both fluoxetine enantiomers roughly superimposable, but all three SSRIs have their halogen atoms inserted into the same HBP in LeuT and interact with the same residues (Fig. 2a). Conversely, the difference in their binding lies with the manner in which the other parts of the molecules interact with the protein - the amine group points either toward the extracellular space and interacts with the N-terminal end of TM10 (for S-fluoxetine), or toward the cytoplasm where it interacts with the extracellular gate of the transporter (for sertraline and R-fluoxetine) (Fig. 2b). Although both R-fluoxetine and S-fluoxetine show certain degrees of flexibility, the structure of the protein itself in all the three LeuT-SSRI complexes is also rather rigid; besides the rotation of Arg30 in the LeuT-S-fluoxetine complex, only the side chains of Gln34, Ala319, Phe320, Leu400 and Asp401 shift slightly upon drug binding (Supplementary Fig. 4).
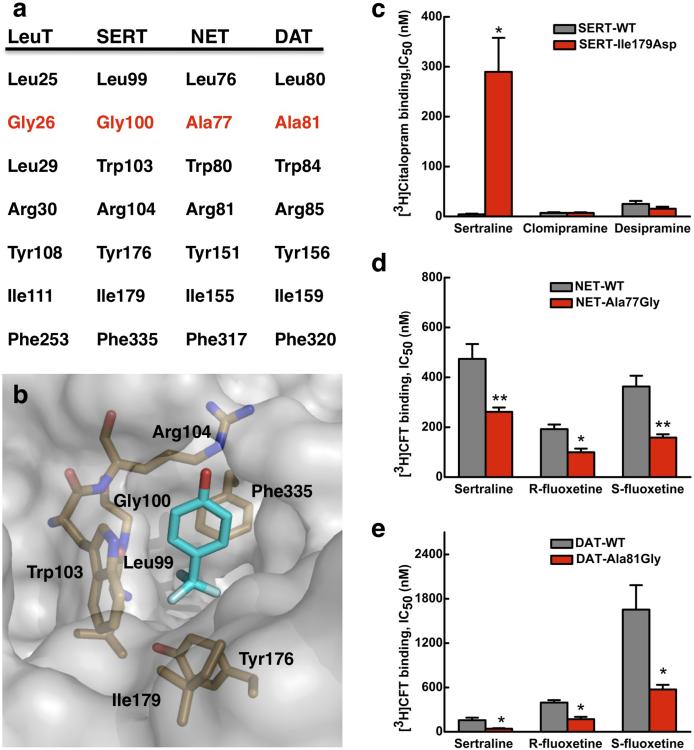
It is of note that all the shared key features of SSRI molecules are important for binding to LeuT (Fig. 1). The observation that halogen-substituted phenyl rings and particularly the halogen atoms themselves — the most important determinants for SSRI specificity — bind at the same position in LeuT (Fig. 2a) immediately suggests that in the human SERT these SSRIs may also bind both at the same position and in a similar manner. Whereas the other parts of the drug molecules will likely bind to SERT in different ways given the diversity in their molecular structures, the binding site for their halogen atoms is invariant. Indeed, amino acids around the presumed extracellular vestibule in the human serotonin transporter SERT are largely homologous with those in LeuT, particularly within the halogen-binding pocket (Figs. 2a, 2c, 3a and 3b) where six of the seven residues (Leu99, Gly100, Arg104, Tyr176, Ile179 and Phe335 in SERT) are conserved; only Trp103 (Leu29 in LeuT) is not.
Figure 3. Halogen-binding pocket in SERT and its critical importance in the protein's specificity for antidepressant.
a. Sequence alignment at the HBP between NSS proteins. The only difference in primary sequence between SERT and NET or DAT is at Gly100 in SERT, which is an alanine in the latter two proteins. b. Docking of R-fluoxetine to a SERT homology model 18. Only the trifluoromethylphenoxy group of the drug is shown for clarity. c – e. Changes in affinity (IC50) for antidepressants of human SERT, NET and DAT when a residue at their HBP is mutated, relative to the appropriate wild-type transporter, as measured in binding assays with HEK293 cells. c. Binding of [3H]citalopram to the SERT-Ile179Asp mutant in the presence of sertraline, chlomipramine and desipramine. d. Binding of [3H]CFT to the NET-Ala77Gly mutant and to the DAT-Ala81Gly mutant (e) both in the presence of sertraline, R-fluoxetine and S-fluoxetine. Values are the Mean ± S.E.M. of three to five experiments. The significance of measurements is indicated by * (P < 0.05) and ** (P < 0.005) compared with wild-type (Student's t-test).
SSRI binding to SERT mutants
We tested our hypothesis of conservation of both the position and the mode of SSRI binding between SERT and LeuT by performing mutagenesis around the tentative SSRI-binding site in the human protein, followed by two types of independent measurements using uptake (Table 1) and binding inhibition assays (Fig. 3) in HEK293 cells. Based on an homology model of SERT 18, we chose three areas for investigation that are nonlinear in three-dimensional space. We first focused on the HBP. Ile179 in SERT (Ile111 in LeuT) was examined for its charge and size: when mutated to alanine, phenylalanine, or aspartate, there was a dramatic reduction in the protein's affinity to sertraline as measured by transport inhibition, with increases in IC50 ranging from 1080 fold for Ile179Phe to 310 fold for Ile179Ala (Table 1). Such observed changes in SERT affinity are as large as previously reported for other mutants 26-28. For two of our mutants (Ile179Ala and Ile179Asp) the affinities for R- and S-fluoxetines also decreased, although less dramatically (by 7 − 20 fold) (Table 1). Mutations in two residues near the HBP also reduced the affinity of SERT to sertraline. Mutation of Tyr175 to a negatively-charged glutamate caused an 8-fold decrease in affinity, and Ile108 when mutated to a leucine (as found in the DAT sequence) yielded a 180-fold lower affinity. We chose one of these mutants, Ile179Asp, and further tested how it affected SERT's affinity for sertraline using a binding assay. This mutation at the HBP was found to dramatically reduce the protein's affinity to sertraline (Fig. 3c, Supplementary Figs. 5a-c and Table 1), but had no effect on the affinities to the TCAs clomipramine (with a halogen substitution at a different position, Supplementary Fig. 6) or desipramine (without halogen substitution). These results strongly support our hypothesis that it is indeed the halogen atoms in SSRIs that bind to the HBP site. The larger changes in affinity to sertraline than the fluoxetines observed in our work are probably due to the fact that the chlorine substitutions in sertraline are much more rigid with respect to the phenyl ring than the trimethylfluorine substitution in fluoxetines (Supplementary Fig. 3) 25, and therefore are much less adaptive to changes in the HBP. The above hypothesis also agrees with previous mutagenesis experiments on SERT where the Gly100Ala and Thr178Ala mutatations at the HBP reduced affinity of the protein to citalopram 29. The mutation of Gly100 to the larger alanine, the residue found in the NET and DAT sequences, probably interferes with interactions between the citalopram fluorine and the HBP. This explains why an SSRI does not bind as tightly to NET or DAT.
Table 1.
Inhibition of [3H]serotonin uptake into HEK-293 cells expressing SERT mutants by SSRIs
| Mutation | Sertraline (%)a | R-fluoxetine (%)a | S-fluoxetine (%)a |
|---|---|---|---|
| Wild-type | 99.7 ± 15.7 | 100 ± 13 | 100 ± 15 |
| Ile108Leu | 17,946 ± 5,944 ** | 89.5 ± 14.34 | 63.0 ± 13.1 |
| Tyr175Glu | 880 ± 333 ** | 175 ± 21 | 154 ± 25 |
| Ile179Ala | 31,057 ± 17,095 ** | 660 ± 372 ** | 1,130 ± 424 ** |
| Ile179Phe | 108,105 ± 28,821 ** | 72.1 ± 36.7 | 152 ± 28 |
| Ile179Asp | 50,319 ± 12,464 ** | 818 ± 263 ** | 2,042 ± 793 ** |
| ΔAla401 | 20,944 ± 3,537 ** | 75.0 ± 17.5 | 154 ± 72 |
| Val489Phe | 272 ± 29.5 * | 87.0 ± 11.3 | 103 ± 20 |
| Val489Leu | 1,654 ± 348 ** | 114 ± 21 | 38.2 ± 12.9 |
| Lys490Thr | 9,796 ± 2,449 ** | 112 ± 22 | 45.3 ± 10.4 |
| Glu493Asp | 250 ± 53.6 * | 188 ± 29 | 59.0 ± 25.2 |
| Glu493Gln | 1,537 ± 371 ** | 334 ± 110 ** | 73.1 ± 34.6 |
Note:
Data are IC50 values expressed as % of wild-type, shown as mean ± SEM of at least three experiments. The significance of measurements are indicated by * (P ≤ 0.05) and ** (P ≤ 0.005) compared with the wild-type (one-way analysis of variance, followed by the Holm-Sidak multiple comparisons test).
We next investigated SERT-SSRI interaction at the salt bridge 1 region because the aspartate residue for salt bridge 2 is not conserved in SERT. In LeuT the gate residue Asp404 forms an ion pair with the amine nitrogen in sertraline (Figs. 1d & 2b) and it also interacts with the amine nitrogen atom in R-fluoxetine via a bound water molecule (Fig. 1e). The amine nitrogen atom in S-fluoxetine, in contrast, points toward the extracellular space (Fig. 1f). When the equivalent gate residue in the salt bridge 1 region in SERT, Glu493, was mutated to another acidic amino acid aspartate, its affinity to the three SSRIs in uptake assays changed little; when mutated to an uncharged glutamine, however, the protein's affinity to sertraline and R-fluoxetine was reduced by 15- and 3-fold, respectively. As expected, the affinity to S-fluoxetine which has an amine group that points towards to the extracellular space did not change. These results are consistent with the idea that sertraline and R-fluoxetine, but not S-fluoxetine, interact strongly with the gate residue Glu493 as was observed in the LeuT-SSRI complexes.
The third region examined was the N-terminal end of TM10 and the tip of the EL4 hairpin, both on the extracellular side of the bound drugs. When Lys490 in SERT was mutated to a threonine (as found in the NET and DAT sequences), the protein's affinity to sertraline decreased by close to 100-fold (Table 1). Mutations at the adjacent residue, V489F and V489L (Phe is found in DAT and Leu in NET), reduced the affinities of SERT to sertraline by 16- and 3-fold, respectively. Similarly, when Ala401 in SERT was deleted to make the EL4 hairpin tip more like that of the NET and DAT sequences (Supplementary Fig. 7), SERT bound sertraline with a 210-fold lower affinity. Concomitantly, little or no change in affinity for R- or S-fluoxetine was observed in these mutants. These results indicate the rigid 3-ring sertraline molecule interacts directly with the N-terminal end of TM10 and the EL4 hairpin tip, whereas the 2-ring R- and S-fluoxetines probably bind deeper toward the gate residues in SERT. The above experimental results on three non-linear areas in the extracellular vestibule of SERT strongly suggest that SSRIs bind to the same site in the human serotonin transporter as found in LeuT.
SSRI binding to NET and DAT mutants
Human SERT binds SSRIs with much greater affinity than either NET or DAT does, underlining the specificity of SERT for these antidepressants. The only difference in primary sequence at the halogen-binding pocket between SERT and NET or DAT is at Gly100 in the former (Figs. 3a and 3b), which is an alanine in the other two proteins. If the HBP plays a key role in determining SERT's specificity for SSRIs, our model predicts explicitly that mutation of the alanine to glycine at that position in both NET and DAT would substantially increase their affinities for SSRIs. As that glycine residue is essential and its mutation abolished the proteins’ transport activity 29, to verify the above prediction we measured the SSRI affinities using binding assays. The validity of such a binding assay has been shown previously 4,30 and reiterated here by the agreement between the results of transport inhibition assays and binding assays for the I179D SERT mutant (Table 1, Fig. 3c).
Remarkably, when the key alanine in the HBP was mutated to a glycine as found in the SERT sequence (Figs. 3a and 3b), the human NET-Ala77Gly construct showed markedly increased affinities to all three SSRIs tested in binding assays. The IC50s for sertraline, R-fluoxetine and S-fluoxetine decreased to 55%, 51% and 43%, respectively, of that of the wild-type NET (Fig. 3d, Supplementary Figs. 5d-f and Table 1). Similarly, for the human DAT protein, the Ala81Gly mutation reduced its IC50s for sertraline, R-and S-fluoxetine, to 26%, 43% and 35% of the respective wild-type values (Fig. 3e, Supplementary Figs. 5g-i and Table 1). Such gain-of-function mutagenesis results are in total agreement with the prediction made by our model. The fact that when both NET and DAT, the two very neurotransmitter transporters that SSRIs are developed to select against, showed such uniformly increased affinities for all three SSRIs tested when their HBP was mutated to that of the SERT sequence, strongly suggests that the halogen-binding pocket in SERT is directly involved SSRI binding and functions as one key determinant for this protein's antidepressant specificity.
DISCUSSION
SSRI binding and TCA binding to LeuT
Previously it has been observed that tricyclic antidepressants bind to the extracellular vestibule and it is this binding that inhibits transport 21,22. Now we have shown that three different SSRI molecules can bind to the same position. However, the interactions of SSRIs with LeuT are very different from those of TCAs. Although the TCA amine group is located at the same position as that in S-fluoxetine and also interacts with Asp401 (Supplementary Fig. 6a), none of the three rings of any TCA possess substitutions that penetrate into the HBP; the chlorine atom on clomipramine, a TCA that has a much higher affinity to SERT than to NET 3, points in a different direction rather than inserting itself into the HBP (Supplementary Fig. 6b). Similar to TCAs bound to LeuT 21,22, the sertraline molecule, which possesses three rings as opposed to the two found in the fluoxetines, displays higher structural rigidity (Supplementary Fig. 3). The agreement of position of the SSRI-binding site with the TCA-binding site indicates a common inhibition mechanism of transport 21,22. In contrast to the binding of a second substrate at this vestibule, which triggers the cytoplasmic release of the substrate at the primary site 31,32, TCAs and SSRIs bind to distinct regions of the vestibule and prevent substrate release by stabilizing the so-called occluded conformation (Supplementary Figs. 1b and 6). This manner of inhibition differs from that by tryptophan, which binds at the primary leucine site 23.
SSRI Halogens and the halogen-binding pocket
The common features of the binding of all three SSRIs to LeuT, particularly the superposition of their halogen atoms in the overlapping halogen-substituted phenyl ring, are clearly reminiscent of previously well-characterized determinants of antidepressant specificity — key features that make an SSRI specific for SERT versus those that make an NRI specific for NET. All SSRIs have a halogen substitution at the 3rd or 4th position of the phenyl or phenoxy ring (Fig. 2), and it is this characteristic that appears largely responsible for the drugs’ specificity to SERT over NET 5,6,8,10. In sertraline-related compounds, other types of halogen substitutions with Cl- or CF3- at one or both positions in aminotetraline also yield an SSRI (Fig. 2d) 10. Similarly, for phenoxyphenylpropylamine derivative-based antidepressants, besides the 4-CF3- substitution found in R- and S-fluoxetines, other kinds of substitutions at this position (F-, Cl-, CH3- or OCH3-) also produce SSRIs (Fig. 2e) 5,6. In contrast, substitutions with CH3- or OCH3- at the 2nd position in the phenoxy ring yield NRIs like tomoxetine and nisoxetine (Fig. 2e). As an important drug component, fluorine substituents in various drugs have been shown to be critical for the drugs’ affinity and selectivity by undergoing multipolar interactions with protein backbone in a hydrophobic environment as well as with the positively charged guanidinium side chain of arginine 33. Not restricted to the aforementioned aminotetraline and phenoxyphenylpropylamine derivatives, substitutions with an electronegative halogen at the 4th position in the phenyl or phenoxy ring are found in every other SSRI currently on the market — CF3- in fluvoxamine, and F- in paroxetine and citalopram 5,7,34. Strikingly, halogen substitutions at the same ring positions are also found in serotonin-norepinephrine reuptake inhibitors (sibutramine and nefazodone) 35,36 as well as the recently discovered serotonin-norepinephrine-dopamine triple reuptake inhibitors (brasofensine, tesofensine, indatraline and DOV21,947) 37-40 but, in stark contrast, not in NRIs 8,34.
SERT's specificity for SSRIs
Given that it is the halogen atoms of SSRIs that confer specificity of these drugs to SERT 5,6,10, that structurally dissimilar SSRIs bind to the same extracellular vestibule in LeuT with their halogen atoms overlapping, that the halogen-binding pocket is highly conserved between LeuT and SERT, that mutations of residues in the SERT HBP result in drastic changes in affinity to SSRIs and, particularly, that uniformly significant gain-of-function data was obtained for both NET and DAT with three SSRIs, it is likely that in SERT SSRIs bind at the same extracellular vestibule as in LeuT and that the specificity of the human transporter to this class of antidepressants is defined in large part by interaction of the drug halogens with the protein's halogen-binding pocket. Given the size of the SSRI molecules, additional structural determinants for SERT's specificity for the drugs must also exist. Nonetheless, the better understanding of the structural basis of SERT's drug specificity provided by the current work should aid in designing more specific antidepressants.
METHODS
LeuT preparation and activity analysis
LeuT was overexpressed and purified as described previously 21. Cells were disrupted using an EmulsiFlex-C3 (Avestin) at 20,000 psi. Following membrane solubilization in 1% (w/v) n-dodecyl-β-D-maltoside, LeuT was purified using Ni2+-affinity chromatography for binding and transport activity studies. A scintillation proximity assay (SPA) 21,41 was carried out for binding studies of [3H]leucine to LeuT. To determine the mechanism of drug inhibition, a “varying hot” method was applied (0.3 nM to 1 μM [3H]leucine) in the presence of 10 μM sertraline, 1 mM R-fluoxetine or 0.1 mM S-fluoxetine, respectively. To measure IC50 values of SSRI inhibition to leucine binding, the concentrations of sertraline, R-fluoxetine, S-fluoxetine were varied in the range of 10 nM to 1 mM. For transport activity measurements, LeuT was reconstituted into liposomes 21,42,43 of Escherichia coli polar lipid extract (10 mg per ml) using 1.5 % (w/v) n-octyl-β-D-glucopyranoside. The resulting proteoliposomes were incubated with 0.5 mM sertraline, 5 mM R-fluoxetine or 0.5 mM S-fluoxetine for 1 hour on ice, followed by [3H] leucine transport assays. All transport experiments were performed in triplicate at room temperature.
Crystallization and structure determination
For co-crystallization experiments with SSRIs, LeuT was further purified using size exclusion chromatography in the presence of 1.2 % (w/v) n-octyl-β-D-glucopyranoside. Peak fractions were concentrated to 6 mg/mL and combined with either 2 mM sertraline, or 40 mM R-fluoxetine, or 10 mM S-fluoxetine. After incubation at 4°C for 30 mins, crystallization drops were set up by hanging-drop vapor diffusion at 20°C as described previously 16,21. X-ray diffraction data were collected from frozen crystals, and the data sets were processed with HKL2000 44. Crystals of LeuT in complex with sertraline, R-fluoxetine and S-fluoxetine complex diffracted to 2.15 Å, 2.35 Å and 2.45 Å resolution, respectively (Table S1). These three types of SSRI-LeuT crystals were indexed in the space group C2 with unit cell dimensions that varied slightly: a = 86.4 − 88.3 Å, b = 86.3 − 87.4 Å, c = 80.6 − 81.3 Å, β = 95.2−95.7°. For all three data sets, Rsym ranged from 6.3% to 7.9% with a mean redundancy of 3.0 to 3.5. The structures were refined initially against the LeuT model (PDB ID 2A65) with the ligands omitted using CCP4 45 with Rfree sets containing 5.1% of the reflections. L-leucine, 2 Na+, SSRI, and H2O molecules were added into sigma-weighted 2Fo-Fc and Fo-Fc maps for model building in Coot 46. Simulated-annealing Fo-Fc omit maps were also calculated using CNS 47 to assist manual rebuilding. The model refinement was conducted until Rwork (∼20%) and Rfree (∼23%) converged. The root-mean-square deviations of bond lengths and bond angles of the final models have reasonable values. In a Ramachandran plot, about 96% of the residues are in preferred regions as shown in Table S1. Structure figures were generated with Pymol 48.
Docking of drug molecules into the SERT model
After superposition of the two protein models, R- fluoxetine was manually fitted into the SERT homology model 18 based upon the position of R-fluoxetine in the LeuT co-crystal structure, and the resultant structure was energy minimized.
[3H]serotonin uptake assays of SERT in human embryonic cells
SERT mutants were constructed by using the QuikChange Mutagenesis Kit (Stratagene). The sequences of all mutants were confirmed by DNA sequencing. The full-length SERT cDNAs were cloned into the pIRESneo3 vector (Clontech). Transient transfection of HEK-293 cells with LipofectAMINE 2000 (Invitrogen) was carried out as described previously 21. Uptake of [3H]serotonin into SERT expressing cells was measured as in our previous work 21 with high sodium, low potassium, glucose and tropolone containing buffer (for details see 49). Nonspecific uptake was defined by 100 μM citalopram. The significance of observed differences between mutants and wild-type transporter was analyzed by applying one-way analysis of variance to the entire data set for each drug, with data log-transformed when needed for normality. This was followed by the Holm-Sidak multiple comparisons test, which compared each separate mutant only with the wild-type. The two-sided significance levels are indicated at p ≤ 0.05 and p ≤ 0.005.
Binding assays of SERT, NET, and DAT in human cells
. Mutations of Gly100 to other residues were found to abrogate transport activities of SERT. Therefore, a binding inhibition assay was developed to study the effects of mutations at that site for SERT as well as at the equivalent sites in NET and DAT. The NET and DAT mutants were constructed as described above for SERT. Radioligand binding to cells transiently expressing the transporters was measured with the same buffer and procedures as described above for the transport assays. The radioligands were [3H]citalopram for SERT and [3H]2β-carbomethoxy-3β-(4-fluorophenyl)tropane (CFT) for NET and DAT. Binding assays were conducted at room temperature for 1 hr (SERT and NET) or 15 min (DAT). Nonspecific binding was defined by 100 μM cocaine for SERT, 1 μM desipramine for NET, and 1 μM CFT for DAT. Statistical analysis was with the Student's t-test (two-tailed).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of the X12B, X25 and X29 beamlines at National Synchrotron Light Source in Brookhaven National Laboratory for assistance in x-ray diffraction data collection. We are grateful to L.R. Forrest and B. Honig of Columbia University for providing the coordinates of the human SERT homology model, and to B. Czyzewski, M.R. Li, I. Parrington and J. Wu for assistance and helpful discussions. N.K.K. thanks American Heart Association and the NIH for postdoctoral fellowships. This work was financially supported by the NIH Roadmap (GM075936 to D.N.W.), and the NIH (MH083840 to D.N.W. and M.E.A.R., DA019676 and DA013261 to M.E.A.R.).
Footnotes
Accession codes. Atomic coordinates and structure factors have been deposited with the Protein Data Bank under access codes 3GWU (LeuT-sertraline), 3GWV (LeuT-R-fluoxetine) and 3GWW (LeuT-R-fluoxetine).
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
References
- 1.Chang AS, et al. Cloning and expression of the mouse serotonin transporter. Mol Brain Res. 1996;43:185–92. doi: 10.1016/s0169-328x(96)00172-6. [DOI] [PubMed] [Google Scholar]
- 2.Baldessarini RJ. Drug therapy of depression and anxiety disorders. In: Brunton LL, Lazoskip JS, Parker KL, editors. Goodman and Gilman's the Pharmacological Basis of Therapeutics. McGraw-Hill; Columbus, OH: 2005. pp. 429–459. [Google Scholar]
- 3.Tatsumi M, Groshan K, Blakely RD, Richelson E. Pharmacological profile of antidepressants and related compounds at human monoamine transporters. Eur J Pharmacol. 1997;340:249–58. doi: 10.1016/s0014-2999(97)01393-9. [DOI] [PubMed] [Google Scholar]
- 4.Eshleman AJ, et al. Characteristics of drug interactions with recombinant biogenic amine transporters expressed in the same cell type. J. Pharmacol. Exp. Ther. 1999;289:877–85. [PubMed] [Google Scholar]
- 5.Wong DT, Bymaster FP. Development of antidepressant drugs. Fluoxetine (Prozac) and other selective serotonin uptake inhibitors. Adv Exp Med Biol. 1995;363:77–95. [PubMed] [Google Scholar]
- 6.Wong DT, Bymaster FP, Engleman EA. Prozac (fluoxetine, Lilly 110140), the first selective serotonin uptake inhibitor and an antidepressant drug: twenty years since its first publication. Life Sci. 1995;57:411–41. doi: 10.1016/0024-3205(95)00209-o. [DOI] [PubMed] [Google Scholar]
- 7.Eildal JN, et al. From the selective serotonin transporter inhibitor citalopram to the selective norepinephrine transporter inhibitor talopram: synthesis and structure-activity relationship studies. J Med Chem. 2008;51:3045–8. doi: 10.1021/jm701602g. [DOI] [PubMed] [Google Scholar]
- 8.Pinder RM, Wieringa JH. Third-generation antidepressants. Med Res Rev. 1993;13:259–325. doi: 10.1002/med.2610130304. [DOI] [PubMed] [Google Scholar]
- 9.Roman DL, Walline CC, Rodriguez GJ, Barker EL. Interactions of antidepressants with the serotonin transporter: a contemporary molecular analysis. Eur J Pharmacol. 2003;479:53–63. doi: 10.1016/j.ejphar.2003.08.056. [DOI] [PubMed] [Google Scholar]
- 10.Welch WM, Kraska AR, Sarges R, Koe BK. Nontricyclic antidepressant agents derived from cis- and trans-1-amino-4-aryltetralins. J Med Chem. 1984;27:1508–15. doi: 10.1021/jm00377a021. [DOI] [PubMed] [Google Scholar]
- 11.Iversen L. Neurotransmitter transporters and their impact on the development of psychopharmacology. Br. J. Pharmacol. 2006;147:S82–8. doi: 10.1038/sj.bjp.0706428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudnick G. Mechanisms of biogenic amine neurotransmitter transporters. In: Reith MEA, editor. Neurotransmitter Transporters: Structure, Function, and Regulation. Humana Press; Totowa, NJ: 2002. pp. 25–52. [Google Scholar]
- 13.Torres GE, Gainetdinov RR, Caron MG. Plasma membrane monoamine transporters: structure, regulation and function. Nat. Rev. Neurosci. 2003;4:13–25. doi: 10.1038/nrn1008. [DOI] [PubMed] [Google Scholar]
- 14.Kanner BI, Zomot E. Sodium-coupled neurotransmitter transporters. Chem Rev. 2008;108:1654–68. doi: 10.1021/cr078246a. [DOI] [PubMed] [Google Scholar]
- 15.Androutsellis-Theotokis A, et al. Characterization of a functional bacterial homologue of sodium-dependent neurotransmitter transporters. J Biol Chem. 2003;278:12703–9. doi: 10.1074/jbc.M206563200. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 2005;437:215–23. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 17.Henry LK, Defelice LJ, Blakely RD. Getting the message across: a recent transporter structure shows the way. Neuron. 2006;49:791–6. doi: 10.1016/j.neuron.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Forrest LR, Tavoulari A, Zhang YW, Rudnick G, Honig B. Identification of a chloride ion binding site in Na+/Cl−-dependent transporters. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12761–6. doi: 10.1073/pnas.0705600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zomot E, et al. Mechanism of chloride interaction with neurotransmitter:sodium symporters. Nature. 2007;449:726–30. doi: 10.1038/nature06133. [DOI] [PubMed] [Google Scholar]
- 20.Ravna AW, Sylte I, Dahl SG. Structure and localisation of drug binding sites on neurotransmitter transporters. J Mol Model. 2009 doi: 10.1007/s00894-009-0478-1. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Z, et al. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science. 2007;317:1390–3. doi: 10.1126/science.1147614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh SK, Yamashita A, Gouaux E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 2007;448:952–6. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- 23.Singh SK, Piscitelli CL, Yamashita A, Gouaux E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 2008;322:1655–60. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caruso F, Besmer A, Rossi M. The absolute configuration of sertraline (Zoloft) hydrochloride. Acta Cryst. 1999;C55:1712–1714. [Google Scholar]
- 25.Robertson DW, Jones ND, Swartzendruber JK, Yang KS, Wong DT. Molecular structure of fluoxetine hydrochloride, a highly selective serotoninuptake inhibitor. J Med Chem. 1988;31:185–9. doi: 10.1021/jm00396a030. [DOI] [PubMed] [Google Scholar]
- 26.Barker EL, Blakely RD. Identification of a single amino acid, phenylalanine 586, that is responsible for high affinity interactions of tricyclic antidepressants with the human serotonin transporter. Mol Pharmacol. 1996;50:957–65. [PubMed] [Google Scholar]
- 27.Henry LK, et al. Tyr-95 and Ile-172 in transmembrane segments 1 and 3 of human serotonin transporters interact to establish high affinity recognition of antidepressants. J Biol Chem. 2006;281:2012–23. doi: 10.1074/jbc.M505055200. [DOI] [PubMed] [Google Scholar]
- 28.Walline CC, Nichols DE, Carroll FI, Barker EL. Comparative molecular field analysis using selectivity fields reveals residues in the third transmembrane helix of the serotonin transporter associated with substrate and antagonist recognition. J Pharmacol Exp Ther. 2008;325:791–800. doi: 10.1124/jpet.108.136200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plenge P, Wiborg O. High- and low-affinity binding of S-citalopram to the human serotonin transporter mutated at 20 putatively important amino acid positions. Neurosci Lett. 2005;383:203–8. doi: 10.1016/j.neulet.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 30.Owens MJ, Knight DL, Nemeroff CB. Second-generation SSRIs: human monoamine transporter binding profile of escitalopram and R-fluoxetine. Biol Psychiatry. 2001;50:345–50. doi: 10.1016/s0006-3223(01)01145-3. [DOI] [PubMed] [Google Scholar]
- 31.Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA. The mechanism of a neurotransmitter:sodium symporter--inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30:667–77. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quick M, et al. Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0811322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller K, Faeh C, Diederich F. Fluorine in pharmaceuticals: looking beyond intuition. Science. 2007;317:1881–6. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- 34.Iversen L. Antidepressants. In: Abraham DJ, editor. Burger's Medical Chemistry and Drug Discovery. Vol. 6. Wiley; 2003. pp. 483–524. [Google Scholar]
- 35.Heal DJ, et al. Sibutramine: a novel anti-obesity drug. A review of the pharmacological evidence to differentiate it from d-amphetamine and dfenfluramine. Int J Obes Relat Metab Disord. 1998;22(Suppl 1):S18–28. [PubMed] [Google Scholar]
- 36.Sanchez C, Hyttel J. Comparison of the effects of antidepressants and their metabolites on reuptake of biogenic amines and on receptor binding. Cell Mol Neurobiol. 1999;19:467–89. doi: 10.1023/a:1006986824213. [DOI] [PubMed] [Google Scholar]
- 37.Pearce RK, et al. The monoamine reuptake blocker brasofensine reverses akinesia without dyskinesia in MPTP-treated and levodopa-primed common marmosets. Mov Disord. 2002;17:877–86. doi: 10.1002/mds.10238. [DOI] [PubMed] [Google Scholar]
- 38.Lehr T, et al. Population pharmacokinetic modelling of NS2330 (tesofensine) and its major metabolite in patients with Alzheimer's disease. Br J Clin Pharmacol. 2007;64:36–48. doi: 10.1111/j.1365-2125.2007.02855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothman RB, et al. Neurochemical neutralization of methamphetamine with high-affinity nonselective inhibitors of biogenic amine transporters: a pharmacological strategy for treating stimulant abuse. Synapse. 2000;35:222–7. doi: 10.1002/(SICI)1098-2396(20000301)35:3<222::AID-SYN7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 40.Skolnick P, Popik P, Janowsky A, Beer B, Lippa AS. Antidepressant-like actions of DOV 21,947: a “triple” reuptake inhibitor. Eur J Pharmacol. 2003;461:99–104. doi: 10.1016/s0014-2999(03)01310-4. [DOI] [PubMed] [Google Scholar]
- 41.Quick M, Javitch J. Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fann MC, Busch A, Maloney PC. Functional characterization of cysteine residues in GlpT, the glycerol 3-phosphate transporter of Escherichia coli. J. Bacteriol. 2003;185:3863–70. doi: 10.1128/JB.185.13.3863-3870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Law CJ, et al. Salt-bridge dynamics control substrate-induced conformational change in the membrane transporter GlpT. J Mol Biol. 2008;378:828–39. doi: 10.1016/j.jmb.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Miror W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzym. 1997;276(Part A):307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Collaborative-Computational-Project-Number-4 The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.DeLano WL. The PyMOL User's Manual. DeLano Scientific; San Carlos, CA: 2002. p. 1. [Google Scholar]
- 49.Chen N, Rickey J, Berfield JL, Reith ME. Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation and cocaine binding. J. Biol. Chem. 2004;279:5508–19. doi: 10.1074/jbc.M306294200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.