Abstract
Leukotrienes are arachidonic acid derivatives long known for their inflammatory properties and their involvement with a number of human diseases, most particularly asthma. Recently, leukotriene-based inflammation has also been shown to play an important role in atherosclerosis: ALOX5AP and LTA4H, both genes in the leukotriene biosynthesis pathway, have individually been shown to be associated with various cardiovascular disease (CVD) phenotypes. To assess the role of the leukotriene pathway in CVD pathogenesis, we performed genetic association studies of ALOX5AP and LTA4H in a family based study of early onset coronary artery disease (EOCAD) (GENECARD, 1,101 families) and in a non-familial dataset of EOCAD (CATHGEN, 656 cases and 405 controls). We found weak to moderate association between single nucleotide polymorphisms (SNPs) in ALOX5AP and LTA4H with EOCAD. The previously reported four-SNP haplotype (HapA) in ALOX5AP showed association with EOCAD in CATHGEN (P = 0.02), while controlling for age, race and CVD risk factors. HapK, the previously reported ten-SNP haplotype in LTA4H was associated with EOCAD in CATHGEN (P = 0.04). Another previously reported four-SNP haplotype in ALOX5AP (HapB) was not significant in our sample (P = 0.39). The overall lack of (or weak) association of single SNPs as compared with the haplotype results demonstrates the need for analyzing multiple SNPs within each gene in such studies. Interestingly, we detected an association of SNPs in ALOX5 (P < 0.05), the target of ALOX5AP, with CVD. Using a pathway-based approach, we also detected statistical evidence for interactions among ALOX5, ALOX5AP and LTA4H using RNA expression data from a collection of freshly harvested human aortas with varying degrees of atherosclerosis. The GENECARD families did not demonstrate evidence for linkage or association with ALOX5, ALOX5AP or LTA4H. Our results support a modest role for the leukotriene pathway in atherosclerosis pathogenesis, reveal important genomic interactions within the pathway, and suggest the importance of using pathway-based modeling for evaluating the genomics of atherosclerosis susceptibility.
Introduction
Cardiovascular disease (CVD) is a major burden on health care in the United States and remains the leading cause of morbidity and mortality in Western society (Zhao and Funk 2004). Coronary artery disease (CAD), the most common manifestation of CVD, is characterized by atherosclerotic lesions in the epicardial coronary arteries. Risk factors for atherosclerosis, including smoking, dyslipidemia, hypertension, diabetes and obesity, have been identified to be important in many large scale epidemiological and intervention studies (Spanbroek et al. 2003). However, despite consistent evidence of a strong heritable nature to CAD risk, the underlying genetic architecture remains largely elusive. Understanding the etiology of complex disease traits such as atherosclerosis involves modeling multiple factors or variables that include genetic variation, intermediate risk factors, family history, biomarkers, environmental conditions and the potential interaction of all of these.
It is known that the processes of atherosclerotic plaque formation and rupture are driven by inflammation. Plaque rupture correlates with increased inflammation within the plaque, implicating the genes involved in inflammatory processes as excellent candidates for study (Cipollone et al. 2005). The 5-lipoxygenase (5-LO) cascade leads to formation of leukotrienes, which exhibit strong pro-inflammatory activities in cardiovascular tissue (Spanbroek et al. 2003). Several genetic linkage and associations studies as well as gene expression studies have shown an association of the ALOX5/ALOX5AP pathway to CAD (Dwyer et al. 2004; Helgadottir et al. 2004, 2005a, b; Lohmussaar et al. 2005; Shah et al. 2008).
Helgadottir et al. (2004) performed a genome-wide scan in search of myocardial infarction (MI) susceptibility genes using 1,068 microsatellite markers in 296 multiplex Icelandic families including 713 individuals. While no regions resulted in genome-wide significance, the most promising observation (lod score 2.86) was on chromosome 13q12–13. Within this region, a four-single nucleotide polymorphism (SNP) haplotype (HapA) spanning the ALOX5AP gene conferred a nearly two times greater risk of MI and stroke in a separate case–control cohort. The ALOX5AP gene is mapped to a locus on chromosome 13q12.3 and encodes a protein that, with 5-LO (ALOX5), is required for leukotriene synthesis. The same group also published an ALOX5AP haplotype (HapB) that conferred risk in a United Kingdom case–control cohort, although HapA was not significant in this additional sample (Helgadottir et al. 2004). In addition, a ten-SNP haplotype (HapK) spanning the LTA4H gene encoding leukotriene A4 hydrolase (LTA4H) on chromosome 12q23.1, a protein in the same biological pathway as ALOX5AP, was shown to confer a modest risk of MI in the Icelandic cohort (Helgadottir et al. 2005b).
In a separate study, Seo et al. (2004) analyzed human aorta samples with varying degrees of atherosclerosis to identify gene expression patterns that predict well-defined aortic atherosclerosis. The two diseased groups (minimally and severely) had significantly different pathological severity of atherosclerosis, as determined by raised lesions and Sudan IV staining. Among the genes predictive of severity of atherosclerosis was ALOX5, but not ALOX5AP or LTA4H.
Given these previous results, the goal of our study was to determine the association between CVD phenotypes, expression and previously reported ALOX5AP and LTA4H haplotypes. We attempted to validate the genetic association findings in both a family based dataset of early onset coronary artery disease (EOCAD) (GENECARD) as well as a non-familial CAD dataset of EOCAD (CATHGEN), including both MI and more general classification of CAD as outcomes. Further, to understand the role of the leukotriene pathway in atherosclerosis pathophysiology, we used expression and clinical data from human donor aortas to evaluate correlations among gene expression patterns in the leukotriene biosynthetic cascade and severity of histologically determined atherosclerosis. Our study takes a comprehensive approach to assess genetic association, genotype–phenotype correlation and gene interactions in three previously implicated candidate genes in the leukotriene pathway.
Methods
Three sample sets were available for the genetic association analysis: CATHGEN, GENECARD and AORTA. Descriptive statistics for the CATHGEN and GENECARD samples (Table 1) were generated using the summary. formula function in the Hmisc library (R Statistical Computing).
Table 1.
Clinical characteristics of the CATHGEN EOCAD cases–controls and GENECARD probands; percentages and (mean ± SD)
| Descriptive statistics by EOCAD |
CATHGEN EOCAD subjects (n = 656) |
CATHGEN controls (n = 405) |
Test statistic significance EOCAD versus controls |
GENECARD (US probands) (n = 759) |
|---|---|---|---|---|
| Age of onset | (46 ± 6) | NA | (48 ± 10) | |
| Age at sampling | (52 ± 9) | (69 ± 7) | P < 0.001a | (50 ± 7) |
| Self reported race | P < 0.002a | |||
| Black | 22% (143) | 19% (77) | 8% (58) | |
| Native American | 5% (35) | 3% (14) | 3% (21) | |
| Other | 2% (13) | 6% (25) | 4% (31) | |
| White | 71% (465) | 71% (289) | 86% (649) | |
| Male sex | 79% (519) | 43% (174) | P < 0.0001b | 69% (522) |
| Positive family history of CAD | 55% (358) | 27% (108) | P < 0.001b | 100% (759) |
| Body mass index (kg/m2) | (31 ± 6) | (29 ± 7) | P = 0.001a | (30 ± 7) |
| Positive smoking history (ever-smoked) | 69% (454) | 40% (160) | P < 0.001b | 27% (202) |
| Positive history of diabetes | 33% (219) | 21% (86) | P < 0.001b | 25% (188) |
| Positive history of hypertension | 70% (457) | 67% (272) | P = 0.40b | 63% (477) |
| History of myocardial infarction | 52% (341) | 0% (0) | NA | 63% (481) |
| History of coronary artery bypass graft | 40% (262) | 0% (0) | NA | 49% (349) |
| Systolic blood pressure (mmHg) | (141 ± 24) | (150 ± 23) | P < 0.001a | (142 ± 25) |
| Diastolic blood pressure (mmHg) | (78 ± 14) | (77 ± 14) | P = 0.30a | (78 ± 14) |
| Total cholesterol (mg/dL) | (193 ± 62) | (192 ± 49) | P = 0.30a | (200 ± 58) |
| LDL (mg/dL) | (109 ± 43) | (107 ± 36) | P = 0.90a | (122 ± 59) |
| HDL (mg/dL) | (40 ± 12) | (52 ± 18) | P < 0.001a | (44 ± 32) |
| Triglycerides (mg/dL) | (223 ± 261) | (157 ± 126) | P < 0.001a | (212 ± 253) |
The test statistic significance provided is for the CATHGEN EOCAD versus unaffected controls
x ± s represents X̄ ± 1 SD
Numbers after percents are frequencies
Wilcoxon test
Pearson test
CATHGEN sample
DNA samples used in this study were collected through the Cardiac Catheterization Laboratories at Duke University Hospital (Durham, NC, USA) under a protocol approved by the Duke Institutional Review Board. Beginning in January 2001, all individuals presenting to the catheterization laboratory for cardiac catheterization were invited to participate in the CATHGEN study and signed informed consent to give a blood sample and allow abstraction of medical record information. Collected samples were later joined to the diagnostic and outcome information stored in the Duke Information System for Cardiovascular Care database maintained at the Duke Clinical Research Institute (Durham, NC, USA). DNA samples and clinical data from over 2,000 individuals were available for this study. From these, 1,061 subjects were selected for the CATHGEN study on the basis of extent of CAD measured by the coronary artery disease index (CADi) and age-at-catheterization. CADi is a numerical summary of coronary angiographic data that incorporates the extent and anatomical distribution of coronary disease (Smith et al. 1991). For the CATHGEN sample, we defined EOCAD as having an age-at-catheterization of 55 years of age or less and significant CAD (CADi ≥ 32) (n = 656). The unaffected control group was defined as older than 60 with insignificant CAD (CADi ≤ 23) and no individual epicardial coronary artery with clinically significant (i.e. >50% stenosis) (n = 405). In addition, the unaffected controls had no history of documented cerebro-vascular or peripheral vascular disease, cardiac transplant, MI or interventional cardiac catheterization procedure. We selected a subgroup of individuals who experienced MI from the overall CATHGEN sample. Any subject regardless of age who has ever had documentation of an MI, either prior to the index catheterization, or subsequent to the index catheterization was classified as a case (n = 483) and compared to the unaffected control group as defined above (n = 405). This includes thrombolytic therapy for an MI in the past or follow-up.
The GENECARD family study
The primary goal of the GENECARD study was to provide a genome-wide linkage scan to identify genetic factors for CAD by linkage analysis. The study was coordinated through Duke University and the study design has been previously described (Hauser et al. 2003). To be eligible for the linkage study, families were required to include at least two siblings, each of whom met the diagnostic criteria for EOCAD (Hauser et al. 2003). In addition, individuals were recruited if they had been diagnosed before the age of 51 years in men and 56 years in women. A total of 420 families were included in the linkage analysis. For the purpose of association analysis we also included families with only one member meeting the diagnostic criteria but with living parents or a living older unaffected sibling. Blood samples were obtained and DNA extraction was performed by a standard protocol at the Duke Center for Human Genetics (CHG). Medical history and risk-factor information was obtained by interviewing patients and by abstracting information from medical records (Hauser et al. 2003). ACS is a serious manifestation of CAD which includes the subgroup of MI and is diagnosed by the presence of at least two of the three signs/symptoms: (1) Chest pain typical of CAD; (2) Changes on electrocardiogram; and (3) A positive serum biomarkers for MI. In addition, an ACS family must have two or more members that qualify individually for ACS (n = 428). Results from the initial genome scan have been reported (Hauser et al. 2004).
AORTA sample
Aorta samples from heart donors with varying degrees of atherosclerosis were harvested in University of Wisconsin solution on ice to minimize postmortem changes (Seo et al. 2004). RNA processing methods are referenced in Seo et al. (2004). Early atherosclerotic plaques were assessed with image processing by quantifying the area of Sudan IV staining and advanced disease was quantified as area of raised lesion using PDAY methodologies (Cornhill et al. 1985). The ratio of affected area over total surface of the studied section was used as the disease burden outcome.
Genotyping
We selected a total of 31 SNPs in three genes: ALOX5, ALOX5AP and LTA4H. For the ALOX5 gene, a minimal set of haplotype tagging SNPs (htSNPs) using minor allele frequency >0.05 and r2 > 0.7, were selected using the SNPselector program (Xu et al. 2005) to cover the predicted linkage disequilibrium (LD) structure in both Caucasian and African American populations. SNPselector incorporates available information from HapMap (http://www.hapmap.org), The SNP Consortium (http://snp.cshl.org), Japanese SNP database (http://snp.ims.u-tokyo.ac.jp/index.html), and the Affymetrix 120K SNP 20 (http://www.affymetrix.com/index.affx) to generate the most likely LD bins and determines the optimal tag SNP for each bin. The ALOX5AP and LTA4H SNPs were selected on the basis of the candidate haplotypes: HapA, HapB and HapK.
Single nucleotide polymorphism probe and primer construction were performed and purchased from Applied Biosystems (AB) for the TaqMan® colorimetric microtiter-plate based assay. The AB 7900 HT sequence detection system (SDS) was used for high-throughput genotyping for all three samples. The scoring of the genotypes is performed using SDS 7.1 software provided by AB. A total of 15 quality control samples, composed of six reference genotype controls in duplicate, two Center d’Etude du Polymorphisme Humain pedigree individuals, and one no-template sample were included in each quadrant of the 384-well plate. Quality control consisted of evaluation of duplicate genotypes for mismatches, genotyping efficiencies and Hardy–Weinberg equilibrium (HWE). All SNPs examined were successfully genotyped for 95% or more of the individuals in the study. Error rate estimates for SNPs meeting QC benchmarks were <0.2%.
Statistical analysis
Allele and gene frequencies
All markers had a minor allele frequency greater than 0.05. Haploview was used to assess LD between SNPs (Barrett et al. 2005). A two marker EM method as implemented in Haploview was used to estimate the maximum-likelihood values of D′ and r2. Association between single SNPs or haplotypes with EOCAD was assessed through logistic regression models, using an additive allele model. In addition to the term for the genotype, the basic model included adjustment for race and sex and the full model included adjustment for known CAD risk factors including history of diabetes, history of smoking, body mass index, hypertension and history of dyslipidemia.
Haplotype analysis
The Haplo.stats package through R Statistical Computing was used to identify haplotypes and to provide a measure (haplo.score) of association to disease (Schaid et al. 2002). Haplo.stats expands on the likelihood approach to account for phase ambiguity in case–control studies by using a generalized linear model (GLM) to test for haplotype association which allows for adjustment of non-genetic covariables (Schaid et al. 2002). This method derives a score statistic to test the null hypothesis of no association of the trait with the genotype, H0: β = 0. In addition to a global statistic, haplo.stats computes score statistics for the components of the genetic vectors, such as individual haplotypes (Schaid et al. 2002). Because we wanted to assess the role of the previously identified haplotypes, we tested these individual haplotypes for association. Models were controlled for age, sex, race, and CAD risk factors as described above. We also performed race stratified analyses to control for potential confounding by race as well as to evaluate the previously reported race-specific results (Helgadottir et al. 2005b). Both Gaussian and binomial traits were considered depending on the phenotype.
Raw gene expression data transformation and normalization
The AORTA sample microarray signal intensity values were normalized using the justRMA function in Bioconductor. We analyzed cis and trans effects of variants in the four Affymetrix tags representing the three genes of interest. The second phenotype for the AORTA study was the location of the section (distal and proximal) within the thoracic aorta as a surrogate for disease susceptibility (Seo et al. 2004). Because some subjects had multiple aorta samples, each individual and sample was treated separately. The expression level of each tag was modeled using multiple linear regression including age, sex, race and additive genotype. CAD risk factors were not available for the AORTA sample. To account for the multiple aorta samples per subject we fit a mixed model, which adjusts for the correlation between aorta samples from the same individual. In a mixed model, linear combinations of the fixed and random effects can be formed from linear combinations of the conditional means (Littell et al. 2006). The random effect for an aorta with a distal and proximal section collected is subject-specific. We did not see a significant change in results when controlling for repeated measures.
Gene set enrichment analysis
Using prior knowledge of the leukotriene biosynthesis pathway provided by Funk and derived from KEGG, we generated a custom gene set (Funk 2005; Ogata et al. 1999). We created a leukotriene biosynthesis pathway gene set incorporating ALOX5, ALO5AP and LTA4H and adding the following genes: (1) Leukotriene-C4 synthase (LTC4S); (2) Gamma-glutamyltransferase-like activity 1 (GGTLA1); and (3) Leukotriene-B4 20-monooxygenase (CYP4F2). Supplemental Figure 4 illustrates the correlation structure for all the genes in the pathway derived from the aorta specimens. As input, gene set enrichment analysis (GSEA) uses a sorted correlation metric between expression and phenotype. An enrichment score (ES) is calculated that reflects the degree to which a gene set is overrepresented at the extremes of the entire list (Subramanian et al. 2005). In addition to our custom set, we also analyzed the C2 gene sets curated from a large number of online pathway data bases (Subramanian et al. 2005). We generated correlation coefficients for all tags using raised lesion mapping and Sudan IV staining as the phenotypes. Tags without gene assignments (including 33153_AT) were removed. Statistical significance was assessed using empirical P values estimated by randomly sampling gene sets of the same size and correlation coefficient sign.
Family based analyses
Non-parametric relative pairs linkage analysis as implemented in Multipoint Engine for Rapid Likelihood INference (MERLIN) was used to assess two-point linkage of each SNP in theGENECARDstudy (Abecasis et al. 2002). To assess family based association with MI and EOCAD in GENECARD, association in the presence of linkage (APL), the pedigree disequilibrium test (PDT) and geno-PDT were used (Martin et al. 2001). PDT is a family based association test for extended pedigrees. It will perform allele-specific analysis and genotype-specific analysis for single markers. APL provides a novel test for APL that also correctly infers missing parental genotypes by estimating identity-by-descent (IBD) parameters (Chung et al. 2006). It provides options for single locus and multiple locus haplotype analysis. Simulations show APL has more power than PDT and FBAT/haplotype version of the family based association test (HBAT) in nuclear family data. However, unlike PDT, APL does not consider extended pedigrees, using only one nuclear family from each pedigree. Given the varied family structures in GENECARD, we used both PDT and APL to maximize power for detecting association in these families. For haplotype analysis, we used HBAT. This program uses data from nuclear families, sibships, or a combination of the two, to provide a general-purpose family based testing strategy for allelic association between phenotypes and haplotypes (Laird et al. 2000).
Power calculations
The application nQuery Advisor 4.0 was used to estimate the power with our sample size, and the effect size generated from the case–control proportions for HapA, HapB and HapK published by Helgadottir et al. (2005a, b). We used the continuity corrected chi-square test with α= 0.05 two-sided significance level to detect the difference between proportions given our sample size for the MI and CAD groups. Power estimates for the GENECARD study design have been reported (Hauser et al. 2003).
Results
Table 1 depicts the clinical characteristics for the GENECARD probands and CATHGEN case and control subjects. The clinical characteristics of the affected individuals from GENECARD and from CATHGEN include increased prevalence of common CAD risk factors including poor lipid profiles, diabetes, hypertension, overweight, and male gender compared to the controls. These differences suggest the need for adjustment for these common clinical risk factors in understanding genetic risk.
We first addressed the previously published association results. With prior knowledge of haplotype references for both the ALOX5AP and LTA4H genes, we analyzed SNPs in the case–control (CATHGEN) and family based samples (GENECARD) which included all SNPs in HapA, HapB and HapK (Helgadottir et al. 2004, 2005b). We also analyzed SNPs from ALOX5 to assess further association in genes from the leukotriene biosynthesis pathway. Supplemental Figure 1A illustrates the location of the HapA and HapB SNPs within the ALOX5AP gene. Supplemental Figure 1B illustrates the location of HapK SNPs within the LTA4H gene. All SNPs in the haplotypes are non-coding SNPs located in introns, or 5′ and 3′ regions. Supplemental Figure 1C illustrates the ALOX5 htSNPs identified using the SNPselector program (see “Methods”). All SNPs are intronic except for rs2228065 which is a missense mutation.
Linkage disequilibrium plots stratified by race and EOCAD affection status in the CATHGEN sample for ALOX5AP, LTA4H and ALOX5 show minimal LD between SNPs with the strongest correlation being between rs4769874 and rs9315050 members of HapA and HapB, respectively (Supplemental Figure 2A). As often noted, LD is reduced in African Americans compared to European Americans. For the CATHGEN sample, there was significant LD between SNPs in LTA4H. For the Caucasians, the novel downstream SNP SG12S16 was moderately correlated to rs17677715, rs2247570, and rs2660890 with r2 values of 0.67, 0.70 and 0.63, respectively, but there was only weak correlation in African Americans. All markers met HWE expectations when stratified by race.
Single marker and haplotype association with measures of EOCAD
Our initial goal was to test the previously reported association of ALOX5AP, LTA4H and ALOX5 SNPs with atherosclerosis. Each dataset has unique measures of atherosclerosis; however, we attempted to match the MI phenotype from the previously published associations in the datasets as well as to broaden the phenotype to include more general EOCAD. The phenotypes for the GENECARD and CATHGEN datasets were EOCAD and MI, while the phenotypes for the aorta dataset were proportion raised lesion and Sudan IV staining as measures of atherosclerosis burden.
Single SNP analyses
Table 2A lists the odds ratio (OR) estimates and significance levels for each marker comprising Haplotypes A and B within ALOX5AP. Marker rs17216473 demonstrated evidence of association with EOCAD (P = 0.01) and rs17222842, demonstrated evidence of association with MI (P = 0.02); these SNPs are members of HapA and HapB, respectively. There was suggestive association (P = 0.06) with rs10507391 and the Sudan IV scoring phenotype. None of the SNPs in ALOX5AP demonstrated significant family based association in GENECARD (data not shown).
Table 2.
Single SNP odds ratio estimates and P values using logistic regression for the CATHGEN subjects with EOCAD (n = 656) versus unaffected controls (n = 405), all affected with myocardial infarction (MI) (n = 483) versus unaffected controls (n = 405), and the AORTA case–control samples (raised lesion mapping and Sudan IV staining)
| A. |
ALOX5AP Haplotype A, B |
EOCAD (n = 1061) |
MI (n = 888) |
Raised lesion (n = 201) |
Sudan IV (n = 150) |
307_AT (ALOX5) |
33153_AT (ALOX5) |
37099_at (ALOX5AP) |
38081_at (LTA4H) |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | P value | Odds ratio | P value | Odds ratio | P value | Odds ratio | P value | Odds ratio | P value | P value | P value | P value | |
| RS17222814* | 0.08 | 0.72 | 0.21 | 0.75 | 0.71 | 0.8 | 0.48 | 0.72 | 0.49 | 0.52 | 0.41 | 0.60 | |
| RS17216473* | 0.01 | 1.47 | 0.13 | 1.31 | 0.95 | 1.03 | 0.35 | 0.7 | 0.12 | 0.98 | 0.45 | 0.86 | |
| RS10507391*, † | 0.26 | 1.12 | 0.6 | 0.94 | 0.25 | 1.58 | 0.06 | 1.78 | 0.01 | 0.75 | 0.54 | 0.20 | |
| RS4769874* | 0.3 | 1.29 | 0.51 | 1.22 | 0.56 | 0.63 | 0.68 | 0.78 | 0.01 | 0.39 | 0.09 | 0.60 | |
| RS9551963† | 0.8 | 0.97 | 0.63 | 1.06 | 0.4 | 1.36 | 0.81 | 0.94 | 0.26 | 0.13 | 0.03 | 0.66 | |
| RS9315050† | 0.59 | 1.09 | 0.85 | 1.04 | 0.56 | 0.71 | 0.24 | 0.55 | 0.06 | 0.64 | 0.25 | 0.37 | |
| RS17222842† | 0.10 | 1.41 | 0.02 | 1.77 | 0.2 | 2.49 | 0.52 | 1.41 | 0.32 | 0.93 | 0.15 | 0.27 | |
| B. |
LTA4H Haplotype K |
EOCAD (n = 1061) |
MI (n = 888) |
Raised lesion (n = 201) |
Sudan IV (n = 150) |
307_AT (ALOX5) |
33153_AT (ALOX5) |
37099_at (ALOX5AP) |
38081_at (LTA4H) |
||||
| SNP | P value | Odds ratio | P value | Odds ratio | P value | Odds ratio | P value | Odds ratio | P value | P value | P value | P value | |
| 12P0557 | 0.87 | 1.02 | 0.52 | 1.10 | 0.74 | 1.14 | 0.19 | 1.52 | 0.73 | 0.61 | 0.94 | 0.22 | |
| RS2660880 | 0.96 | 1.01 | 0.55 | 0.86 | 0.36 | 1.91 | 0.39 | 1.55 | 0.25 | 0.67 | 0.29 | 0.49 | |
| RS6538697 | 0.40 | 0.86 | 0.10 | 0.7 | 0.27 | 0.50 | 0.85 | 1.10 | 0.36 | 0.89 | 0.05 | 0.05 | |
| RS1978331 | 0.08 | 1.19 | 0.65 | 0.95 | 0.60 | 1.19 | 0.08 | 1.61 | 0.17 | 0.34 | 0.90 | 0.40 | |
| RS17677715 | 0.65 | 1.06 | 0.23 | 1.21 | 0.48 | 0.73 | 0.97 | 0.99 | 0.34 | 0.94 | 0.29 | 0.12 | |
| RS2247570 | 0.14 | 1.18 | 0.33 | 0.88 | 0.88 | 1.06 | 0.51 | 1.20 | 0.36 | 0.06 | 0.82 | 0.11 | |
| RS2660898 | 0.82 | 0.98 | 0.81 | 0.97 | 0.58 | 0.82 | 0.35 | 1.32 | 0.43 | 0.41 | 0.24 | 0.85 | |
| RS2540482 | 0.95 | 0.99 | 1.00 | 1.00 | 0.23 | 0.63 | 0.89 | 1.04 | 0.98 | 0.79 | 0.40 | 0.48 | |
| RS2660845 | 0.83 | 0.98 | 0.64 | 0.94 | 0.34 | 0.72 | 0.88 | 0.96 | 0.45 | 0.39 | 0.63 | 0.79 | |
| RS2540475 | 0.98 | 1.00 | 0.63 | 0.93 | 0.54 | 1.3 | 0.64 | 1.18 | 0.77 | 0.62 | 0.27 | 0.42 | |
| C. | ALOX5 | EOCAD (n = 856) |
MI (n = 698) |
Raised lesion (n = 75) |
Sudan IV (n = 50) |
307_AT (ALOX5) |
33153_AT (ALOX5) |
37099_at (ALOX5AP) |
38081_at (LTA4H) |
||||
| SNP | P value | Odds ratio | P value | Odds ratio | P value | Odds ratio | P value | Odds ratio | P value | P value | P value | P value | |
| RS1864414 | 0.32 | 1.19 | 0.21 | 1.26 | 0.96 | 1.04 | 0.15 | 0.47 | 0.60 | 0.66 | 0.56 | 0.01 | |
| RS3824613 | 0.36 | 1.18 | 0.3 | 1.21 | 0.94 | 1.06 | 0.19 | 0.50 | 0.70 | 0.80 | 0.50 | 0.03 | |
| RS2029253 | 0.74 | 1.04 | 0.97 | 1.00 | 0.51 | 1.63 | 0.05 | 3.17 | 0.02 | 0.11 | 0.96 | 0.31 | |
| RS1369214 | 0.53 | 0.92 | 0.43 | 0.9 | 0.23 | 2.64 | 0.01 | 8.05 | 0.25 | 0.25 | 0.91 | 0.64 | |
| RS2115819 | 0.58 | 0.93 | 0.47 | 0.91 | 0.25 | 2.58 | 0.01 | 10.40 | 0.04 | 0.31 | 0.80 | 0.99 | |
| RS10900215 | 0.05 | 1.39 | 0.17 | 1.27 | 0.53 | 0.63 | 0.23 | 0.52 | 0.92 | 0.24 | 0.08 | 0.64 | |
| RS892691 | 0.42 | 0.89 | 0.96 | 0.99 | 0.03 | 4.39 | 0.93 | 0.96 | 0.24 | 0.59 | 0.72 | 0.26 | |
| RS3780906 | 0.08 | 0.78 | 0.21 | 0.83 | 0.07 | 3.1 | 0.95 | 0.98 | 0.44 | 0.90 | 0.93 | 0.26 | |
| RS3740107 | 0.04 | 0.74 | 0.30 | 0.85 | 0.08 | 3.5 | 0.89 | 1.06 | 0.33 | 0.73 | 0.88 | 0.45 | |
| RS1487562 | 0.03 | 1.46 | 0.07 | 1.39 | 0.93 | 0.92 | 0.12 | 0.34 | 0.44 | 0.20 | 0.54 | 0.85 | |
| RS2242332 | 0.33 | 0.87 | 0.56 | 0.92 | 0.12 | 2.81 | 0.81 | 1.13 | 0.78 | 0.78 | 0.89 | 0.06 | |
| RS2242334 | 0.26 | 0.85 | 0.44 | 0.89 | 0.33 | 1.89 | 0.96 | 0.98 | 0.75 | 0.34 | 0.20 | 0.03 | |
The relationship between expression level of each tag and SNP was modeled using multiple linear regression
SNPs represented are in candidate haplotypes HapA(*) and HapB(†)
P values ≤0.05 are shown in bold
Table 2B shows results for LTA4H; we detected no evidence for single SNP associations with EOCAD, MI or either AORTA phenotypes. However, one SNP in the LTA4H gene provided significant results in the GENECARD family based association analysis; rs6538697 was significant for both acute coronary syndrome (ACS-MI) (P = 0.006) and CAD (P = 0.0098). Table 2C shows single SNP association results for ALOX5. Three SNPs in ALOX5 were significant in the EOCAD sample including rs10900215 (P = 0.05), rs3740107 (P = 0.04) and rs1487562 (P = 0.03). For the AORTA samples, ALOX5 SNPs showed significance rs892691 was significant for the raised lesion phenotype, while three SNPs were significant in the Sudan IV staining phenotype: rs2029253 (P = 0.05), rs1369214 (P = 0.01) and rs2115819 (P = 0.01). SNPs rs3780902 and rs2228065 had minor allele frequencies <0.05 and we were unable to estimate an accurate OR for aorta phenotypes. Again, the family based association analysis showed no significant ALOX5 SNPs in GENECARD.
Haplotype analyses
The haplotype-specific results for MI and EOCAD phenotypes and the haplotype frequencies for cases and controls are shown in Table 3. Table 1 of Electronic Supplementary Material contains the power estimates for our samples given the case–control haplotype frequencies and effect sizes reported by Helgadottir et al.
Table 3.
Haplotype A, B and K association P values (individual and global) and case–control frequencies for the MI, EOCAD and AORTA phenotypes; results for MI and EOCAD are presented overall and stratified by race; expression tags for ALOX5, ALOX5AP and LTA4H were analyzed as a continuous variable
| Haplotype | Phenotype | Race | Haplotype frequency | P value | Global P value | |
|---|---|---|---|---|---|---|
| Control | Case | |||||
| ALOX5AP HapA | MI | All | 0.12 | 0.14 | 0.11 | 0.75 |
| AA | 0.11 | 0.09 | 0.33 | 0.48 | ||
| CA | 0.12 | 0.15 | 0.07 | 0.48 | ||
| EOCAD | All | 0.12 | 0.15 | 0.02 | 0.45 | |
| AA | 0.11 | 0.08 | 0.67 | 0.78 | ||
| CA | 0.12 | 0.17 | 0.01 | 0.25 | ||
| Raised lesion | All | 0.18 | 0.20 | 0.06 | 0.40 | |
| Sudan IV | 0.21 | 0.17 | 0.11 | 0.08 | ||
| 307_AT (ALOX5) | NAa | 0.06 | 0.97 | |||
| 33153_AT (ALOX5) | 0.31 | 0.55 | ||||
| 37099_at (ALOX5AP) | 0.03 | 0.23 | ||||
| 38081_at (LTA4H) | 0.16 | 0.77 | ||||
| ALOX5AP HapB | MI | All | 0.08 | 0.09 | 0.64 | 1.00 |
| AA | 0.14 | 0.14 | 0.87 | 0.80 | ||
| CA | 0.06 | 0.08 | 0.61 | 0.0001 | ||
| EOCAD | All | 0.08 | 0.10 | 0.39 | 0.72 | |
| AA | 0.14 | 0.16 | 0.37 | 0.89 | ||
| CA | 0.06 | 0.08 | 0.44 | 0.65 | ||
| Raised lesion | All | 0.08 | 0.07 | 0.76 | 0.00001 | |
| Sudan IV | 0.09 | 0.58 | 0.04 | |||
| 307_AT (ALOX5) | NAa | 0.38 | 0.11 | |||
| 33153_AT (ALOX5) | 0.41 | 0.08 | ||||
| 37099_at (ALOX5AP) | 0.24 | 0.21 | ||||
| 38081_at (LTA4H) | 0.18 | 0.37 | ||||
| LTA4H HapK | MI | All | 0.12 | 0.14 | 0.57 | 0.07 |
| AA | 0.08 | 0.07 | 0.63 | 0.16 | ||
| CA | 0.14 | 0.15 | 0.79 | 0.83 | ||
| EOCAD | All | 0.12 | 0.13 | 0.04 | 0.33 | |
| AA | 0.07 | 0.05 | 0.27 | 0.62 | ||
| CA | 0.14 | 0.16 | 0.03 | 0.54 | ||
| Raised lesion | All | 0.19 | 0.10 | 0.32 | 0.06 | |
| Sudan IV | 0.18 | 0.17 | 0.71 | 0.02 | ||
| 307_AT (ALOX5) | NAa | 0.41 | 0.18 | |||
| 33153_AT (ALOX5) | 0.16 | 0.07 | ||||
| 37099_at (ALOX5AP) | 0.94 | 0.13 | ||||
| 38081_at (LTA4H) | 0.27 | 0.0009 | ||||
Two statistical tests were performed, one comparing the frequency of the particular haplotype of interest in cases and controls and the second performing the global test of haplotype association testing the null hypothesis that there were no haplotype differences between cases and controls. The alternative hypothesis was that at least one of the haplotypes (not necessarily the haplotype of interest) was different
Expression values analyzed as a continuous variable
P values ≤0.05 are shown in bold
HapA in ALOX5AP
HapA comprises the SNP markers rs17222814 (G), rs10507391 (T), rs4769874 (G) and rs9551963 (A). Given the HapA case–control frequencies and effect size (RR = 1.79; cases 0.158, controls 0.095) with MI as the clinical endpoint found in the Helgadottir et al. (2004) Icelandic cohort, we estimated that we had 73% power with our CATHGEN sample of 819 subjects (414 MI subjects and 405 older controls). Assuming the same effect size for CAD, we had 82% power for the EOCAD sample (n = 1061). HapA showed a trend for association with MI in Caucasians (P = 0.07) with case– control frequencies of 0.147 and 0.119. However, we did not observe a significant association with MI in our African American sample (P = 0.33) with haplotype frequencies of 0.092 and 0.108 in cases and controls, respectively. We observed a significant HapA association in the EOCAD Caucasian sample (P = 0.01; case 0.166, control 0.118). HapA was not significant in the EOCAD African Americans (P = 0.67; cases 0.084, controls 0.107). The overall test (global) of all haplotype association with the HapA markers was not significant for either ethnic group.
HapB in ALOX5AP
HapB comprises the SNP markers rs17216473 (A), rs10507391 (A), rs9315050 (A) and rs17222842 (G). With our CATHGEN sample of 819 subjects, we had 50% power to detect the HapB results (RR = 1.95; cases 0.075, controls 0.040) reported by Helgadottir et al. (2004). We did not detect an association of HapB with neither EOCAD nor MI in the overall sample or when stratified by race. There were no significant single haplotype results for differences between HapB cases and controls. However, we observed a significant global P value for MI in Caucasians (P = 0.0001) suggesting overall differences in haplotype frequencies with that combination of SNPs. In almost every test for association, the haplotypes with the A allele in place of the G allele for marker rs17222842 were significant. This difference in the associated haplotypes supports the significant global P value and suggests the potential for additional associated haplotypes in this sample or may indicate further divergence of the haplotypes from an ancestral haplotype containing the disease mutation detected in the Icelandic population.
HapK in LTA4H
HapK comprises the SNP markers SG12S16 (deCODE) (C), rs2660880 (G), rs6538697 (T), rs1978331 (A), rs17677715 (T), rs2247570 (T), rs2660898 (T), rs2540482 (C), rs2660845 (G) and rs2540475 (G). The CATHGEN sample of 656 Caucasians subjects with MI provided 27% power to detect reported the HapK case– control frequencies (RR = 1.37; cases 0.186, controls 0.143) (1). Given our sample of African American subjects with MI, we had 49% power to detect the HapK case–control frequencies (RR = 6.50; cases 0.103, controls 0.017) found in the Philadelphia African American sample (Helgadottir et al. 2005b). We did not observe HapK as a significant risk haplotype in the sample of Caucasians with MI (P = 0.79; cases 0.154, controls 0.136) nor in the African Americans with MI (P = 0.63; cases 0.069, controls 0.075). As in the case for HapA, when we expanded the phenotype to EOCAD, the Caucasians showed the largest difference in haplotypes frequencies between cases and controls (P = 0.03; cases 0.158, controls 0.137).
GENECARD family based association analyses
We used HBAT to test for family based association, but we found no significant (P < 0.05) global or specific candidate- haplotype results for ACS or EOCAD (Laird et al. 2000). Given that there was no evidence for linkage in the GENECARD genome-wide linkage analysis, perhaps it is not surprising that there was no evidence for the haplotypes with EOCAD and ACS in this dataset (Hauser et al. 2004). The linkage lod scores in the regions around the genes were highest for ALOX5AP but were still very low (max multipoint lod score = 0.01 and 0.12 in ACS families for ALOX5AP; max multipoint lod score = 0.25 and 0.27 in ACS-MI families for LTA4H; and max multipoint lod score = 0.09 and 0.13 in ACS-MI families for ALOX5. The power to detect linkage in 400 affected sibpairs (ASPs) is over 80%for recurrence ratios of 1.4 or greater and is over 80% for a variety of genetic models with 250 families of 2 ASPs and one affected sib at an α = 0.001 (Chung et al. 2006). The estimated HapA frequencies are 0.170 for EOCAD and ACS. For HapB, the estimated frequencies are 0.070 for EOCAD and ACS. The estimated HapK frequencies are 0.140 for EOCAD and 0.170 for ACS, frequencies consistent with those observed in the CATHGEN case–control sample.
In summary, the genetic associations provide some support of the published results for ALOX5AP and LTA4H with additional support for ALOX5. None of these results would be significant with most multiple test corrections and thus individually the studies of these genes provide weak validation in the MI and general EOCAD phenotypes; however, the multiplicity of even these weak results within the leukotriene biosynthesis pathway do support the evidence of association with this pathway as a whole. Our next step was to evaluate correlations and interactions among the three genes.
Expression results for the AORTA sample
In the AORTA dataset previously used in the genetic association studies, 122 aorta tissue sections (proximal-1A and distal-4B) from 78 subjects were assayed for gene expression using the Affymetrix HG-U95Av2 chip. Both LTA4H and ALOX5AP have one representative tag and ALOX5 has two tags (307_at and 33153_at). Tag 307_at represents the same strand (+) and same region as the ALOX5 gene while 33153_at probes a short region of the complement (−) at the 3′ end. Figure 1 illustrates the RMA normalized expression values for LTA4H, ALOX5 and ALOX5AP. The diagonal presents a histogram of the individual RNA expression values. Pairwise XY scatter plots are illustrated below the diagonal and the calculated correlation coefficients are found above the diagonal. Tags ALOX5_307_AT and ALOX_ 33153_AT were negatively correlated (r = −0.19, P = <0.04) despite their physical proximity suggesting that these tags are identifying different transcripts. ALOX5AP and LTA4H expression levels were significantly correlated to ALOX5_307_AT (r = 0.54, P = <0.0001 and r = 0.29, P = 0.0015, respectively) and to each other (r = 0.42, P = <0.0001). As expected, ALOX5AP and LTA4H expression levels were negatively correlated with the ALOX5_33153_AT tag (r = −0.29, P = 0.001 and r = −0.35, P <0.0001, respectively). We then separately considered the AORTA phenotypes and the expression values as a predictor. In support of the findings of Seo et al., we found that expression values for ALOX5 had a strong effect on raised lesion mapping (point estimate = 7.33, P = 0.0005) and Sudan IV staining (point estimate = 4.65, P = 0.007). No other tags demonstrated significant association with the AORTA phenotypes.
Fig. 1.
Pairwise scatterplots (below the diagonal), histograms (across the diagonal) and pairwise correlation coefficients (above the diagonal) using RMA normalized gene expression data for ALOX5, ALOX5AP and LTA4H; all correlation coefficients were statistically significant (P ≤ 0.05)
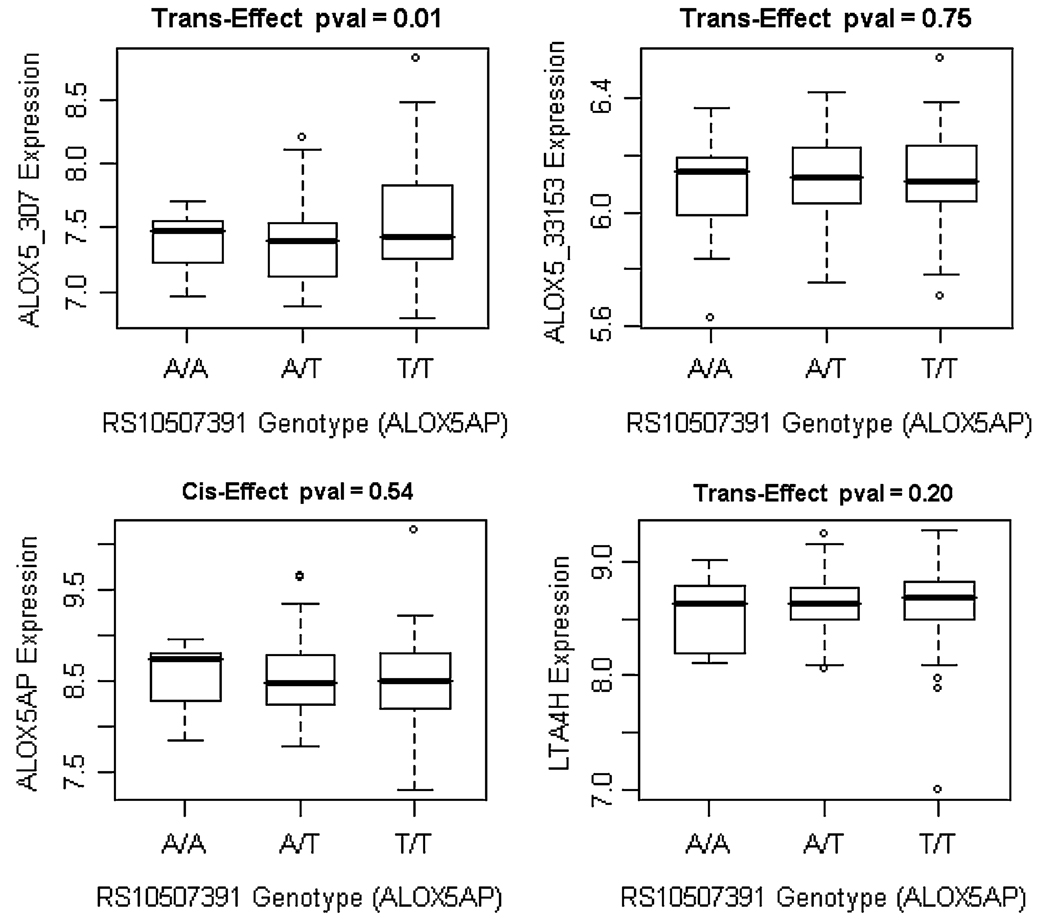
We examined genotype and specific expression for ALOX5AP, LTA4H and ALOX5 using the previously studied SNPs found in HapA, HapB and HapK as well as SNPs found in the ALOX5 gene. In addition to looking at cis-effects (intra-gene/SNP interaction) on gene expression, we also considered trans-effects (inter-gene/SNP interaction), as a model of the leukotriene biosynthesis pathway using a biological systems approach. Table 2 lists the P values for the SNP effects on the four tags representing ALOX5, ALOX5AP and LTA4H. To visualize possible interactions and to highlight cis versus trans effects we have illustrated the genotype and haplotype-specific expression association results from Table 2 in Supplemental Figure 3. All three genes have at least one SNP with a cis-and trans-effect as shown in Supplemental Figure 3 and Table 2. Interestingly, all effects for a given gene had a single gene target in terms of expression value. For example, SNPs in ALOX5AP only have trans effects on the expression values for ALOX5, but not for LTA4H. SNP rs10507391 in ALOX5AP demonstrated one of the most significant associations with mRNA expression levels (Fig. 2). We chose to illustrate this SNP because of its trans-effect significance (P = 0.01) on ALOX_307 expression level. We also considered a dominant model and the association was also significant (P = 0.009); although the goodness of fit determined by the log likelihood was not different for the two genetic models. This SNP is the only marker shared by both HapA and HapB; however, different alleles are included in the HapA and HapB risk haplotypes. There was no effect on the ALOX_33153 espression which seems to serve as a natural control, nor was there an effect on the ALOX5AP (cis) or LTA4H (trans) expression. Given an additive haplotype effect model, HapA was associated with expression for ALOX5AP (P = 0.03) and potentially with ALOX5 (P = 0.06). Neither HapK nor HapB were associated with ALOX5, ALOX5AP or LTA4H expression values.
Fig. 2.
Boxplots of RS10507391 genotype-specific expression levels of RMA normalized transcripts in 122 aorta-sections. The x-axis annotates the genotype for RS10507391 and the y-axis annotates the expression levels for ALOX5 (trans-effect), ALOX5AP (cis-effect) and LTA4H (trans-effect)
For a pathway-based approach of association using the expression data, we used GSEA (Subramanian et al. 2005). Using the prior-based biological knowledge of the leukotriene biosynthesis pathway, we generated a custom gene set to test concordance. The two phenotypes analyzed include raised lesion mapping and Sudan IV staining. The genes were ranked using the correlation between each individual tag and the phenotype variable as a score. For the raised lesion mapping, our custom gene set for leukotriene biosynthesis had a positive ES of 0.6897 and an estimated significance level of P = 0.004. The Sudan IV phenotype produced a positive ES of 0.6349 and an estimated significance level of P = 0.009. We view this strong enrichment for the leukotriene pathway as a whole as supportive of the modest genetic association results shown above for individual SNPs. These results support the hypothesis that these genes act in concert to increase risk for CAD, both at the level of affected tissue as well as at the level of clinical disease.
Discussion
We replicated several of the results observed by Helgadottir et al. in our sample: both HapA (ALOX5AP) and HapK (LTA4H) were significantly associated with CAD in the CATHGEN case–control subjects. Previously published results for association with MI, CAD and stroke combined with our results suggest that these SNPs and haplotypes are associated with vascular disease (Dwyer et al. 2004; Helgadottir et al. 2004, 2005a, b; Lohmussaar et al. 2005; Shah et al. 2008). Overall, the case–control comparison of EOCAD in Caucasians demonstrated the strongest association for all three haplotypes with significant results for HapA and HapK. We were unable to detect an association with HapB. While we were able to determine a general association with CAD, we did not see significant specific association results for the MI phenotypes in our case–control groups.
According to the LTA4H results produced by Helgadottir et al., HapK is more strongly associated with MI in Philadelphia African Americans (RR = 6.50, P = 0.0001) when compared to Caucasians (RR = 1.37, P = 0.010) although both groups demonstrate significant association (Helgadottir et al. 2005b). Similar results were found in the Cleveland and Atlanta samples, although with somewhat lower ORs (Helgadottir et al. 2005b). HapK was not significant in our African American sample, but was associated in the Caucasians. Overall we observed similar frequencies found in the Helgadottir et al. study, but the case and control frequencies were not significantly different in our study, likely due to the low power to detect this difference. Another reason for differences in statistical results between Helgadottir et al. and our results for association with MI may be due to subtle differences in case and control definitions.
In addition to the evidence from prior studies about candidate genes and a metabolic pathway, we had prior knowledge of haplotypes and SNPs to test for association with our CVD sample. In the context of our ongoing candidate gene analyses, the P values from the logistic regression tests for these SNPs and the haplotype global associations would not have suggested pursuing these as candidates nor would they have been detected in a genomewide association approach. The comparison of the single SNP and haplotype results shows the need for analysis of multiple SNPs per gene, even in a replication setting, and in the utility of additional genomic information such as the aorta expression study, along with knowledge of biological pathways.
Having multiple levels of genetic evidence gives us confidence in the observations that support the leukotriene–CVD association findings of Helgadottir et al. and other groups (Dwyer et al. 2004; Helgadottir et al. 2004, 2005a, b; Lohmussaar et al. 2005; Shah et al. 2008). Given that genes with the strongest effects will be observed in more than one context, taking advantage of independent sources of genetic information is useful. As demonstrated in Supplemental Figure 3, the one-on-one trans relationship between the SNP genotypes and the expression values allows for reconstructing gene networks. Our results suggest that HapA and the relationship to expression levels for ALOX5AP (cis) and ALOX5 (trans) may be an important feature that links the genetic and genomic results. Our GSEA results for our custom leukotriene biosynthesis gene set suggest another approach for assessing pathway association.
Our results support previous association studies; however, the relationship between genetic variation and CVD outcomes is not driven by a single gene or SNP. By exploring the pathway in terms of risk for atherosclerosis, CAD and MI along with genotypic effects on expression, rather than the looking at each component in isolation, we observe significant complexity in the interactions that may alter risk profiles related to genetic variation. For example, it may be necessary to measure changes in allele-specific expression of ALOX5 when evaluating leukotriene inhibitors for potential primary or secondary prevention of MI. This pathway approach should be considered for evaluation of other studies of genetic variation in CAD.
Supplementary Material
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-008-0619-0) contains supplementary material, which is available to authorized users.
Contributor Information
David R. Crosslin, Center for Human Genetics, Duke University Medical Center, 595 LaSalle Street, Durham, NC 27710, USA david.crosslin@duke.edu
Svati H. Shah, Center for Human Genetics, Duke University Medical Center, 595 LaSalle Street, Durham, NC 27710, USA Division of Cardiovascular Medicine, Duke University Medical Center, Durham, NC 27710, USA.
Sarah C. Nelson, Center for Human Genetics, Duke University Medical Center, 595 LaSalle Street, Durham, NC 27710, USA
Carol S. Haynes, Center for Human Genetics, Duke University Medical Center, 595 LaSalle Street, Durham, NC 27710, USA
Jessica J. Connelly, Center for Human Genetics, Duke University Medical Center, 595 LaSalle Street, Durham, NC 27710, USA
Shera Gadson, Center for Human Genetics, Duke University Medical Center, 595 LaSalle Street, Durham, NC 27710, USA.
Pascal J. Goldschmidt-Clermont, Miller School of Medicine, University of Miami, Miami, FL 33136, USA
Jeffery M. Vance, Institute of Human Genomics, University of Miami, Miami, FL 33136, USA
Jason Rose, Center for Human Genetics, Duke University Medical Center, 595 LaSalle Street, Durham, NC 27710, USA.
Chris B. Granger, Division of Cardiovascular Medicine, Duke University Medical Center, Durham, NC 27710, USA
David Seo, Miller School of Medicine, University of Miami, Miami, FL 33136, USA.
Simon G. Gregory, Center for Human Genetics, Duke University Medical Center, 595 LaSalle Street, Durham, NC 27710, USA
William E. Kraus, Division of Cardiovascular Medicine, Duke University Medical Center, Durham, NC 27710, USA
Elizabeth R. Hauser, Center for Human Genetics, Duke University Medical Center, 595 LaSalle Street, Durham, NC 27710, USA, elizabeth.hauser@duke.edu
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Chung RH, Hauser ER, Martin ER. The APL test: extension to general nuclear families and haplotypes and examination of its robustness. Hum Hered. 2006;61:189–199. doi: 10.1159/000094774. [DOI] [PubMed] [Google Scholar]
- Cipollone F, Mezzetti A, Fazia ML, Cuccurullo C, Iezzi A, Ucchino S, Spigonardo F, Bucci M, Cuccurullo F, Prescott SM, Stafforini DM. Association between 5-lipoxygenase expression and plaque instability in humans. Arterioscler Thromb Vasc Biol. 2005;25:1665–1670. doi: 10.1161/01.ATV.0000172632.96987.2d. [DOI] [PubMed] [Google Scholar]
- Cornhill JF, Barrett WA, Herderick EE, Mahley RW, Fry DL. Topographic study of sudanophilic lesions in cholesterol-fed minipigs by image analysis. Arteriosclerosis. 1985;5:415–426. doi: 10.1161/01.atv.5.5.415. [DOI] [PubMed] [Google Scholar]
- Dwyer JH, Allayee H, Dwyer KM, Fan J, Wu H, Mar R, Lusis AJ, Mehrabian M. Arachidonate 5-lipoxygenase promoter genotype, dietary arachidonic acid, and atherosclerosis. N Engl J Med. 2004;350:29–37. doi: 10.1056/NEJMoa025079. [DOI] [PubMed] [Google Scholar]
- Funk CD. Leukotriene modifiers as potential therapeutics for cardiovascular disease. Nat Rev Drug Discov. 2005;4:664–672. doi: 10.1038/nrd1796. [DOI] [PubMed] [Google Scholar]
- Hauser ER, Crossman DC, Granger CB, Haines JL, Jones CJ, Mooser V, McAdam B, Winkelmann BR, Wiseman AH, Muhlestein JB, Bartel AG, Dennis CA, Dowdy E, Estabrooks S, Eggleston K, Francis S, Roche K, Clevenger PW, Huang L, Pedersen B, Shah S, Schmidt S, Haynes C, West S, Asper D, Booze M, Sharma S, Sundseth S, Middleton L, Roses AD, Hauser MA, Vance JM, Pericak-Vance MA, Kraus WE. A genomewide scan for early-onset coronary artery disease in 438 families: the GENE-CARD study. Am J Hum Genet. 2004;75:436–447. doi: 10.1086/423900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser ER, Mooser V, Crossman DC, Haines JL, Jones CH, Winkelmann BR, Schmidt S, Scott WK, Roses AD, Pericak-Vance MA, Granger CB, Kraus WE. Design of the Genetics of Early Onset Cardiovascular Disease (GENECARD) study. Am Heart J. 2003;145:602–613. doi: 10.1067/mhj.2003.13. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Gretarsdottir S, St Clair D, Manolescu A, Cheung J, Thorleifsson G, Pasdar A, Grant SF, Whalley LJ, Hakonarson H, Thorsteinsdottir U, Kong A, Gulcher J, Stefansson K, MacLeod MJ. Association between the gene encoding 5-lipoxygenase-activating protein and stroke replicated in a Scottish population. Am J Hum Genet. 2005a;76:505–509. doi: 10.1086/428066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgadottir A, Manolescu A, Helgason A, Thorleifsson G, Thorsteinsdottir U, Gudbjartsson DF, Gretarsdottir S, Magnusson KP, Gudmundsson G, Hicks A, Jonsson T, Grant SF, Sainz J, O’brien SJ, Sveinbjornsdottir S, Valdimarsson EM, Matthiasson SE, Levey AI, Abramson JL, Reilly MP, Vaccarino V, Wolfe ML, Gudnason V, Quyyumi AA, Topol EJ, Rader DJ, Thorgeirsson G, Gulcher JR, Hakonarson H, Kong A, Stefansson K. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat Genet. 2005b;38(1):68–74. doi: 10.1038/ng1692. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Manolescu A, Thorleifsson G, Gretarsdottir S, Jonsdottir H, Thorsteinsdottir U, Samani NJ, Gudmundsson G, Grant SFA, Thorgeirsson G, Sveinbjornsdottir S, Valdimarsson EM, Matthiasson SE, Johannsson H, Gudmundsdottir O, Gurney ME, Sainz J, Thorhallsdottir M, Andresdottir M, Frigge ML, Topol EJ, Kong A, Gudnason V, Hakonarson H, Gulcher JR, Stefansson K. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet. 2004;36:233–239. doi: 10.1038/ng1311. [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19 Suppl 1:S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Littell RC, Milliken GA, Stroup WW, Wolfinger RD, Schabenberger O. SAS for Mixed Models. 2nd edn. Cary, NC: SAS Institute Inc.; 2006. [Google Scholar]
- Lohmussaar E, Gschwendtner A, Mueller JC, Org T, Wichmann E, Hamann G, Meitinger T, Dichgans M. ALOX5AP gene and the PDE4D gene in a central European population of stroke patients. Stroke. 2005;36:731–736. doi: 10.1161/01.STR.0000157587.59821.87. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Kaplan NL. Correcting for a potential bias in the pedigree disequilibrium test. Am J Hum Genet. 2001;68:1065–1067. doi: 10.1086/319525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo D, Wang T, Dressman H, Herderick EE, Iversen ES, Dong C, Vata K, Milano CA, Rigat F, Pittman J, Nevins JR, West M, Goldschmidt-Clermont PJ. Gene expression phenotypes of atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24:1922–1927. doi: 10.1161/01.ATV.0000141358.65242.1f. [DOI] [PubMed] [Google Scholar]
- Shah SH, Hauser ER, Crosslin D, Wang L, Haynes C, Connelly J, Nelson S, Johnson J, Gadson S, Nelson CL, Seo D, Gregory S, Kraus WE, Granger CB, Goldschmidt-Clermont P, Newby LK. ALOX5AP variants are associated with in-stent restenosis after percutaneous coronary intervention. Atherosclerosis. 2008;201:148–154. doi: 10.1016/j.atherosclerosis.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LR, Harrell FE, Jr, Rankin JS, Califf RM, Pryor DB, Muhlbaier LH, Lee KL, Mark DB, Jones RH, Oldham HN. Determinants of early versus late cardiac death in patients undergoing coronary artery bypass graft surgery. Circulation. 1991;84:III245–III253. [PubMed] [Google Scholar]
- Spanbroek R, Grabner R, Lotzer K, Hildner M, Urbach A, Ruhling K, Moos MP, Kaiser B, Cohnert TU, Wahlers T, Zieske A, Plenz G, Robenek H, Salbach P, Kuhn H, Radmark O, Samuelsson B, Habenicht AJ. Expanding expression of the 5-lipoxygenase pathway within the arterial wall during human atherogenesis. Proc Natl Acad Sci USA. 2003;100:1238–1243. doi: 10.1073/pnas.242716099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Gregory SG, Hauser ER, Stenger JE, Pericak-Vance MA, Vance JM, Zuchner S, Hauser MA. SNPselector: a web tool for selecting SNPs for genetic association studies. Bioinformatics. 2005;21:4181–4186. doi: 10.1093/bioinformatics/bti682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Funk CD. Lipoxygenase pathways in atherogenesis. Trends Cardiovasc Med. 2004;14:191–195. doi: 10.1016/j.tcm.2004.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.