Abstract
We report here on the characterization of a three-generation Chinese family with aminoglycoside-induced and nonsyndromic hearing impairment. Ten of 17 matrilineal relatives exhibited bilateral and sensorineural hearing impairment. Of these, nine matrilineal relatives, who had a history of exposure to aminoglycosides, exhibited variable severity and audiometric configuration of hearing loss. The dose and age at the time of drug administration seemed to be correlated with the severity of the hearing loss experienced by affected individuals. Sequence analysis of the complete mitochondrial genome in the pedigree showed the presence of homoplasmic A1555G mutation and 37 variants belonging to haplogroup D4a. Of those variants, the G7444A mutation is of special interest as the mutation at this position results in a read-through of the stop condon AGA of the COI message, thereby adding three amino acids (Lys-Gln-Lys) to the C-terminal of the polypeptide. Alternatively, the G7444A mutation is adjacent to the site of 3’ end endonucleolytic processing of L-strand RNA precursor, spanning tRNASer(UCN) and ND6 mRNA. Thus, the G7444A mutation, similar to the deafness-associated A7445G mutation, may lead to a defect in the processing of the L-strand RNA precursor, thus influencing the phenotypic expression of the A1555G mutation. These data also imply that nuclear background plays a role in the aminoglycoside ototoxicity associated with the A1555G mutation in this Chinese pedigree.
Keywords: aminoglycoside ototoxicity, nonsyndromic hearing loss, mitochondrial DNA mutation, 12S rRNA, tRNASer(UCN), penetrance, modulation, haplogroup, Chinese, processing, precursor, COI
Introduction
Mitochondrial 12S rRNA and tRNASer(UCN) genes have been shown to be the hot spots for mutations associated with hearing loss [Fischel-Ghodsian, 1999; Guan, 2004]. Of these, four non-syndromic deafness-associated mutations: A7445G [Reid et al., 1994; Fischel-Ghodsian et al., 1995], 7472insC [Verhoeven et al., 1999; Jacobs et al., 2005], T7510C [Hutchin et al., 2000; del Castillo et al., 2002] and T7511C [Sue et al., 1999; Li et al., 2005a], have been identified in the tRNASer(UCN) gene. The C1494T mutation in the highly conserved decoding site of the 12S rRNA has been associated with both aminoglycoside-induced and nonsyndromic hearing loss in a large Chinese family [Zhao et al., 2004a; Zhao et al., 2005]. Mutations at position 961 of the 12S rRNA have been implicated to be associated with both aminoglycoside-induced and nonsyndromic hearing loss [Casano et al., 1999; Li et al., 2004a]. Furthermore, the T1095C mutation in the same rRNA has also been shown to be associated with hearing impairment in several genetically unrelated families [Thyagarajan et al., 2002; Zhao et al., 2004b; Li et al., 2005c]. In contrast to those deafness-associated mitochondrial DNA (mtDNA) mutations reported only in a small number of families from different ethnic origins, the homoplasmic A1555G mutation in the highly conserved decoding site of the 12S rRNA has been associated with both aminoglycoside-induced and nonsyndromic hearing loss in many families of different ethnic origins [Prezant et al., 1993; Hutchin et al., 1993; Matthijis et al., 1996; Pandya et al., 1997; Estivill et al., 1998; del Castillo et al., 2003; Li et al., 2004a, 2004b; 2005b;Young et al., 2005]. Matrilineal relatives within families or in other families carrying the A1555G mutation exhibited incomplete penetrance and variable expressivity including severity and age-of-onset in hearing loss [Prezant et al., 1993; Estivill et al., 1998; Li et al., 2004b; Young et al., 2005]. Incomplete penetrance and variable expressivity of hearing loss as well as a mild biochemical defect associated with this mutation indicated that the A1555G mutation itself is not sufficient to produce the clinical phenotype [Prezant et al., 1993; Estivill et al., 1998; Li et al., 2004b; Li et al., 2005c; Young et al., 2005; Guan et al., 1996; 2001]. Therefore, other modifier factors including aminoglycosides, nuclear modifier genes and mitochondrial haplotypes modulate the expressivity and penetrance of deafness associated with the A1555G mutation [Guan et al., 1996; 2000; 2001].
With the aim of investigating a role of mitochondrial halogroups in the phenotypic manifestation of the A1555G mutation, a systematic and extended mutational screening of the 12S rRNA has been initiated at the Institute of Otolaryngology of the Chinese PLA General Hospital [Zhao et al., 2004a; 2004b;Young et al., 2005]. In the previous investigation, we showed extremely low penetrance of hearing loss in four Chinese families carrying the A1555G mutation [Young et al., 2005]. In the present study, clinical and genetic evaluation showed a high penetrance of hearing loss with variable severity, audiometric configuration and age-at-onset in a three-generation Chinese family with aminoglycoside-induced and nonsyndromic hearing loss. Molecular analysis demonstrated the coexistence of the homoplasmic 12S rRNA A1555G mutation and G7444A mutation in the COI/the precursor of tRNASer(UCN) genes in this family. To elucidate the role of mitochondrial haplotype in the phenotypic expression of the A1555G mutation, we performed PCR-amplification of fragments spanning the entire mitochondrial genome and subsequent DNA sequence analysis in the matrilineal relatives of this family.
Subjects and Methods
Subjects and audiological examinations
As the part of genetic screening program for hearing loss, a three-generation Chinese family, as shown in Figure 1, was ascertained through the Otology Clinic at Chinese PLA General Hospital. A comprehensive history and physical examination were performed to identify any syndromic findings, the history of the use of aminoglycosides, genetic factors related to the hearing impairment in members of this pedigree. An age-appropriate audiological examination was performed and this examination included pure-tone audiometry (PTA) and/or auditory brainstem response (ABR), immittance testing and distortion product otoacoustic emissions (DPOAE). The PTA was calculated from the sum of the audiometric thresholds at 500, 1000 and 2000, 4000 and 8000 Hz. The severity of hearing impairment was classified into five grades: normal <26dB; mild =26–40 dB; moderate=41–70 dB; severe=71–90 dB; and profound >90 dB. Informed consent was obtained from participants prior to their participation in the study, in accordance with the Cincinnati Children’s Hospital Medical Center Institutional Review Board and Ethnics Committee of Chinese PLA General Hospital.
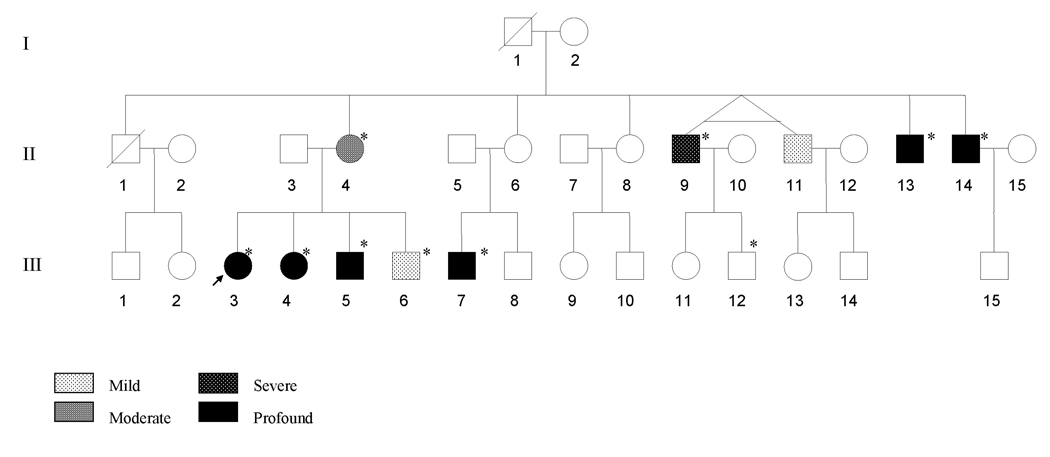
Figure 1. A three-generation Chinese pedigree with aminoglycoside-induced and nonsyndromic hearing impairment.
Hearing impaired individuals are indicated by filled symbols. Arrow denotes proband. Asterisks denote individuals who had a history of exposure to aminoglycosides.
Mutational screening of the mitochondrial genome
Genomic DNA was isolated from whole blood of participants using the Puregene DNA Isolation Kits (Gentra Systems). First, affected and control subject’s DNA fragments spanning the 12S rRNA gene or tRNASer(UCN) gene were amplified by PCR using oligodeoxynucleotides corresponding to the mtDNA at positions 618–635 and 1988–2007 and 7148–7167 and 8076–8095 [Rieder et al., 1998], respectively. Each fragment was purified and subsequently analyzed by direct sequencing in an ABI 3700 automated DNA sequencer using the Big Dye Terminator Cycle sequencing reaction kit. mtDNA sequence alignments were carried out using seqweb program GAP (GCG).
The entire mitochondrial genome of one affected matrilineal relative III-3 was PCR amplified in 24 overlapping fragments by use of sets of the light-strand and the heavy strand oligonucleotide primers, as described elsewhere [Rieder et al., 1998]. Each fragment was purified and subsequently submitted for sequence analysis as described above. The resultant sequence data were compared with the updated consensus Cambridge sequence (GenBank accession number: NC_001807) [Anderson et al., 1981].
Quantification of the mtDNA mutations
For the detection of the A1555G mutation, the amplified segments were digested with a restriction enzyme BsmAI [Li et al., 2004a; 2004b; Li et al., 2005b]. For the analysis of G7444A mutation, DNA fragments were amplified by PCR using oligodeoxynucleotides corresponding to the mtDNA at positions 7148–7167 and 8076–8095, and the resultant PCR fragments were digested with a restriction enzyme XbaI as this G7444A mutation abolishes a site for XbaI [Pandya et al., 1999]. Equal amounts of various digested samples were then analyzed by electrophoresis through 1.5% agarose gel. The proportions of digested and undigested PCR product were determined by using the IMAGE-QUANT program after ethidium bromide staining to determine if the A1555G or G7444A mutation is in the homoplasmy in these subjects.
Results
Clinical presentation
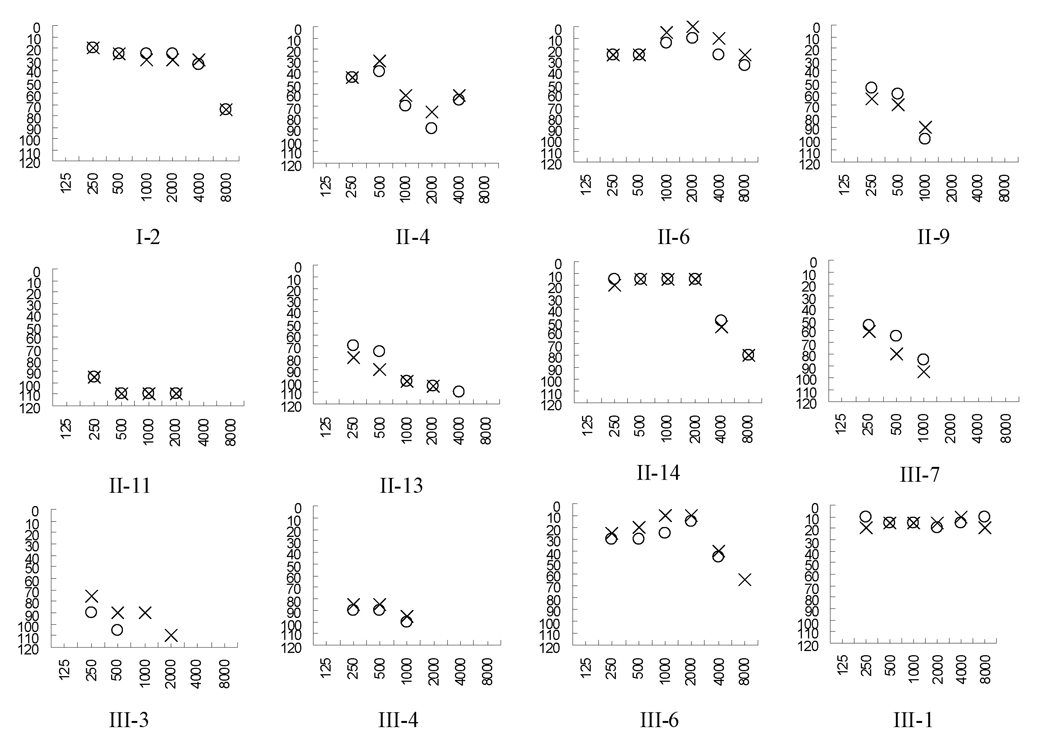
The proband (III-3) was a young woman at the age of 26 years, from Jiangxi Province, Southern China. She had been administered gentamicin (3–5 mg/kg/dose every 8 hours) for 7 days for fever at the age of one year. She developed bilateral hearing impairment within one month after drug administration. As illustrated in Figure 2, audiological evaluation showed that she exhibited profound bilateral hearing impairment (108 dB at right ear, 97 dB at left ear; with a flat-shaped pattern). Also she had no other significant medical history. A comprehensive history and physical examination as well as audiological examination were performed to identify any syndromic findings, the history of the use of aminoglycosides, genetic factors related to the hearing impairment in 29 members of the three-generation family. As shown in Table I and Figure 2, none of the offspring of either deaf father has hearing impairment; 10 of 17 matrilineal relatives, who were offspring of subject I-2, suffered from bilateral and sensorineural hearing impairment as the sole clinical symptom; other members of this family exhibited normal hearing. Thus, these data suggested a maternal inheritance of hearing impairment in this family.
Figure 2. Air conduction audiogram of some members in the Chinese family.
Symbols: X-left, O-right ear.
Table I.
Summary of clinical data for maternal members in the Chinese pedigree examined for mutations
| Subject | Gender | Age at test (yrs) | Age at onset (yrs) | Use of aminoglycoside | PTA(dB) Right Ear | PTA (dB) Left Ear | Audiometric configuration | Level of hearing impairment |
|---|---|---|---|---|---|---|---|---|
| I-2 | F | 72 | - | No | 25 | 26 | - | Normal |
| II-4 | F | 49 | 27 | Yes | 67 | 55 | Sloping | Moderate |
| II-6 | F | 47 | - | No | 17 | 10 | - | Normal |
| II-8 | F | 44 | - | No | 15 | 17 | - | Normal |
| II-9 | M | 41 | 8 | Yes | 90 | 90 | Sloping | Severe |
| II-11 | M | 41 | - | No | 28 | 28 | Sloping | Mild |
| II-13 | M | 38 | 2 | Yes | 110 | 110 | Flat | Profound |
| II-14 | M | 31 | 3 | Yes | 93 | 98 | Sloping | Profound |
| III-3 | F | 26 | 1 | Yes | 108 | 97 | Flat | Profound |
| III-4 | F | 21 | 1 | Yes | 100 | 100 | Flat | Profound |
| III-5 | M | 19 | 1 | Yes | 105 | 100 | Flat | Profound |
| III-6 | M | 17 | 3 | Yes | 27 | 28 | Sloping | Mild |
| III-7 | M | 24 | 2 | Yes | 87 | 95 | Sloping | Profound |
| III-8 | M | 19 | - | No | 17 | 20 | - | Normal |
| III-9 | F | 21 | - | No | 15 | 15 | - | Normal |
| III-10 | M | 18 | - | No | 13 | 18 | - | Normal |
| III-11 | F | 18 | - | No | 12 | 5 | - | Normal |
| III-12 | M | 16 | - | Yes | 17 | 15 | - | Normal |
| III-13 | F | 19 | - | No | 0 | 5 | - | Normal |
| III-14 | M | 15 | - | No | 10 | 7 | - | Normal |
| III-15 | M | 8 | - | No | 5 | 10 | - | Normal |
Interestingly, nine matrilineal relatives of this family, who had a history of exposure to gentamicin and/or streptomycin, had a variable severity of hearing impairment in this maternal kindred, ranging from profound hearing impairment (II-13, II-14, III-3, III-4, III-5, III-7), to severe hearing impairment (II-9), to moderate hearing impairment (II-4), to mild hearing impairment (III-6). These subjects also exhibited variable patterns of audiometric configuration, including the flat-pattern (subjects II-13, III-3, III-4 and III-5) and the sloping pattern (subjects II-4, II-9, II-14, III-6, and III-7). All affected individuals showed the loss of the high frequencies and their hearing impairment was symmetric. The age at the time of administration varied among those individuals. Six individuals (II-13, II-14, III-3, III-4, III-5, III-7), who received a regular dose of aminoglycosides (3 5 mg/kg/dose every 8 h for gentamicin or 15 25 mg/kg/dose every 12 h for streptomycin) for over three days at an age below 3 years, developed profound hearing loss. In particular, subject III-5, who was administered only gentamicin for a week at the age of 1 year, developed profound hearing loss within one month. However, the subject III-6, who was given only one dose of gentamicin at the age of 3, exhibited a mild hearing impairment. In addition, subject II-6 suffered from moderate hearing loss after administration of a regular dose of streptomycin for three days at the age of 27 years, whereas subject II-9, who received a regular dose of streptomycin over three days at the age of 8 years, developed severe hearing impairment. Thus, the dose and the age at the time of drug administration seemed to correlate with the severity of the hearing loss experienced by affected individuals. Notably, subject II-11, who had a history of the exposure to noise but not to aminoglycoside, exhibited a mild hearing impairment. Furthermore, subject III-12, who was not a matrilineal relative, exhibited normal hearing despite receiving a regular dose of kanamycin for 3 days at the age of 6 years. Comprehensive family medical histories of these subjects showed no other clinical abnormalities, including diabetes, muscular diseases, visual dysfunction, and neurological disorders.
Mitochondrial genome analysis
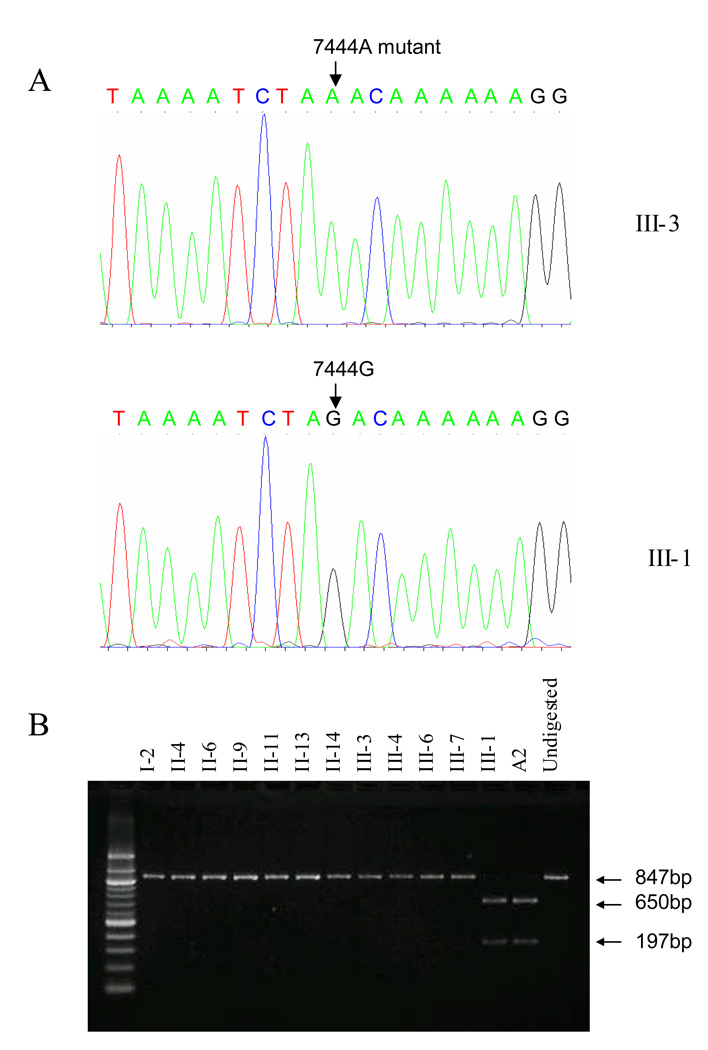
The maternal transmission of hearing impairment in this family suggested the mitochondrial involvement and led us to analyze mtDNA of matrilineal relatives. First, DNA fragments spanning 12S rRNA and tRNASer(UCN) genes were PCR amplified from derived from four matrilineal relatives (proband III-3, his mother II-4, affected female II-6, unaffected male II-9) and two unrelated Chinese controls. Each fragment was purified and subsequently analyzed by DNA sequencing. We failed to detect the presence of the T7510C, T7511C, 7472insC and A7445G mutations in the tRNASer(UCN) gene and mutations at positions 961, 1095 and 1494 in the 12S rRNA gene. However, the A1555G mutation in the 12S rRNA was found in these subjects. The restriction enzyme digestion and subsequent electrophoresis analysis indicated that the A1555G mutation was indeed present in homoplasmy in those subjects and other matrilineal relatives of this family but not other members including III-12 of this family (Figure 3). Furthermore, the G7444A mutation in the COI and the precursor of tRNASer(UCN) genes, as shown in Figure 4A, was identified to co-segregate with the A1555G mutation in those subjects. In fact, this mutation was previously identified to coexist with the A1555G mutation in six Mongolian subjects with aminoglycoside-induced deafness [Pandya et al., 1999]. To further determine the presence and amount of the G7444A mutation in these matrilineal relatives, PCR amplification was performed as detailed in Materials and Methods section, and the resultant PCR segments were then digested by a restriction enzyme XbaI, and separated them by electrophoresis. As can be seen in Figure 4B, the G7444A mutation appears to be homoplasmy in these matrilineal relatives of this Chinese family but to be absent in PCR products derived from other members in this family.
Figure 3. Qualification of the A1555G mutation of matrilineal relatives in this Chinese family.
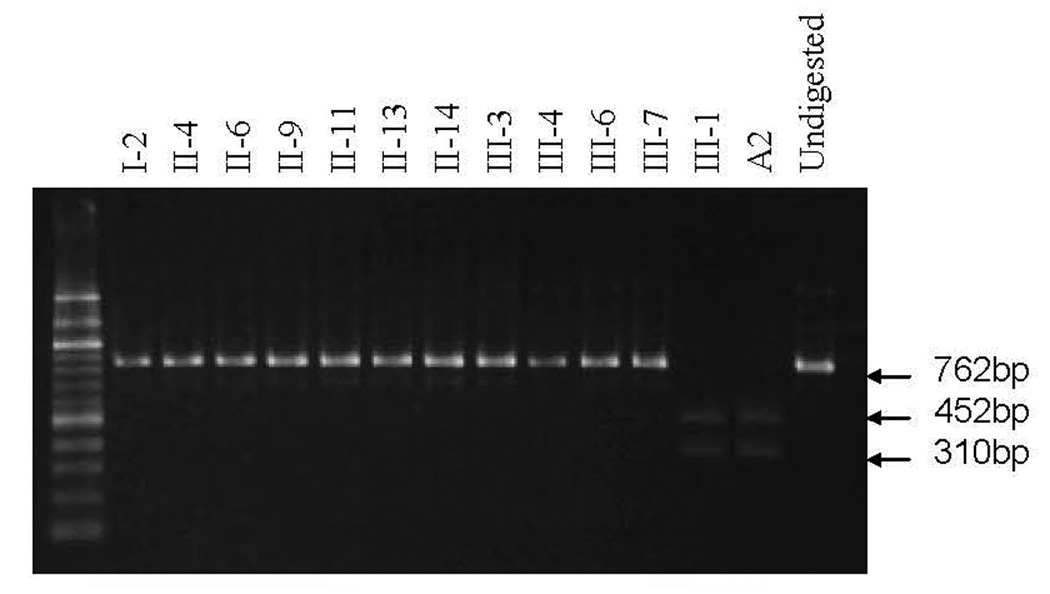
PCR products around A1555G region of mtDNA were digested with BsmAI and analyzed by electrophoresis in a 1.5% agarose gel stained with ethidium bromide. A2 is a Chinese control subject [Zhao et al., 2004a].
Figure 4. Identification and qualification of G7444A mutation.
(A) Partial sequence chromatograms of COI/tRNASer(UCN) genes from affected individual III-3 and a married-in-control III-1. An arrow indicates the location of the base changes at position 7444; (B) Quantification of G7444A mutation of matrilineal relatives and control subjects derived from the Chinese family. PCR products around G7444A region of mitochondrial genome were digested with XbaI and analyzed by electrophoresis in a 1.5% agarose gel stained with ethidium bromide.
To determine the role of mitochondrial haplotypes in the phenotypic manifestation of the A1555G mutation, the DNA fragments spanning the entire mtDNA of an affected patient III-3 were PCR amplified. Each fragment was purified and subsequently analyzed by direct sequence. The comparison of the resultant sequences with the Cambridge consensus sequence [Anderson et al., 1981] identified a total of 38 nucleotide changes as shown in Table II. All of those nucleotide changes were verified in 3 additional matrilineal relatives of this family (II-4, II-6 and II-9) by sequence analysis and appeared to be homoplasmy. Sequence analysis confirmed the presence of the A1555G and G7444A mutations in matrilineal relatives of this family. Of other nucleotide changes in this mitochondrial genome, there are eight variants in the D-loop, 14 silent mutations in the protein encoding genes, 2 known variants in 12S rRNA gene and 4 known variants in the 16S rRNA gene [Brandon et al., 2005]. Furthermore, as shown in Table II, eight amino acid substitutions caused by corresponding mtDNA variants occurred in different polypeptides in this matrilineal relative. These variants in rRNAs and polypeptides were further evaluated by phylogenetic analysis of these variants and sequences from other organisms including mouse [Bibb et al., 1981], bovine [Gadaleta et al., 1989] and Xenopus laevis [Roe et al., 1985]. However, none of variants in the polypeptides were highly evolutionarily conserved and implicated to have significantly functional consequence.
Table II.
mtDNA mutations in the Chinese family with hearing loss
| Gene | Position | Replacement | Conservationa (H/B/M/X) | Previously reportedb |
|---|---|---|---|---|
| D-loop | 73 | A to G | yes | |
| 152 | T to C | yes | ||
| 217 | T to C | yes | ||
| 263 | A to G | yes | ||
| 16129 | G to A | yes | ||
| 16223 | C to T | yes | ||
| 16362 | T to C | yes | ||
| 16519 | T to C | yes | ||
| 12S rRNA | 750 | A to G | A/A/A/- | yes |
| 1438 | A to G | A/A/A/G | yes | |
| 1555 | A to G | A/A/A/A | yes | |
| 16S rRNA | 1811 | A to G | A/A/T/A | yes |
| 2706 | A to G | A/G/A/A | yes | |
| 3010 | G to A | G/G/A/A | yes | |
| 3206 | C to T | C/A/T/A | yes | |
| ND1 | 3324 | C to T | yes | |
| ND2 | 4769 | A to G | yes | |
| 4883 | C to T | yes | ||
| 5178 | C to A (Leu-Met) | L/T/T/T | yes | |
| COI | 7028 | C to T | yes | |
| 7444 | G to A (Ter-Lys) | yes | ||
| A8 | 8414 | C to T (Leu-Phe) | L/F/M/W | yes |
| 8473 | T to C | yes | ||
| A6 | 8701 | A to G (Thr-Ala) | T/S/L/Q | yes |
| 8860 | A to G (Thr-Ala) | T/A/A/T | yes | |
| COIII | 9540 | T to C | yes | |
| ND3 | 10398 | A to G (Thr-Ala) | T/T/T/A | yes |
| 10400 | C to T | yes | ||
| ND4 | 10873 | T to C | yes | |
| 11719 | G to A | yes | ||
| ND5 | 12705 | C to T | yes | |
| ND6 | 14668 | C to T | yes | |
| CYTB | 14766 | C to T (Thr-Ile) | T/S/T/S | yes |
| 14783 | T to C | yes | ||
| 14979 | T to C (Ile-Thr) | I/I/L/L | yes | |
| 15043 | G to A | yes | ||
| 15301 | G to A | yes | ||
| 15326 | A to G (Thr-Ala) | T/M/I/I | yes |
Conservation of amino acid for polypetides or nucleotide for RNAs, in human (H), bovine (B), mouse (M) and Xenopus laevis (X).
Discussion
In the present study, we have performed the clinical, genetic and molecular characterization of a three-generation Chinese pedigree with aminoglycoside-induced and nonsyndromic hearing impairment. Hearing impairment as a sole clinical phenotype was only present in the maternal lineage of those pedigrees, suggesting that the mtDNA mutation is the molecular basis for this disorder. Molecular analysis led to the identification of the A1555G mutation present to be homoplasmy in matrilineal relatives of this Chinese family. Indeed, this Chinese family exhibited 59% penetrance (10/17) of hearing loss (9 of 10 affected matrilineal relatives had aminoglycoside-induced hearing loss), in contrast with the fact that four Chinese pedigrees carrying the A1555G mutation exhibited only an average 8% penetrance of hearing loss [Young et al., 2005]. Strikingly, nine matrilineal relatives with aminoglycoside ototoxicity in this Chinese family exhibited variable severity and audiometric configuration of hearing impairment, although these subjects share some common features: bilateral and sensorineural hearing impairment. This discrepancy likely reflects the dosage and the age at the time of aminoglycoside administration, and different nuclear backgrounds in those individuals. In particular, there was a considerable variability in sensitivity to aminoglycosides in lymphoblastoid cell lines derived from two asymptomatic members and four symptomatic members of the Chinese family carrying the C1494T mutation [Zhao et al., 2004a]. However, the very significant/nearly identical increase in the ratio of doubling times in a medium in the presence/absence of high concentration of paromomycin was observed in cybrids derived from three symptomatic and two asymptomatic individuals carrying the C1494T mutation under a constant nuclear background [Zhao et al., 2005]. Therefore, nuclear background seems to play a role in the aminoglycoside ototoxicity associated with the A1555G mutation in this family.
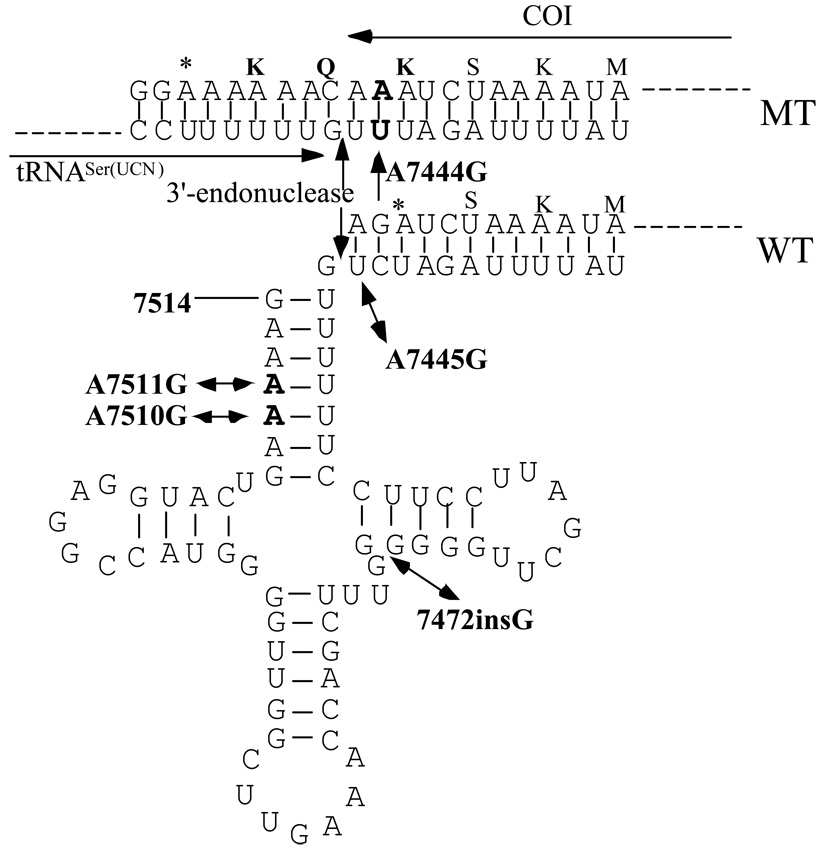
Furthermore, the background sequences (haplotype) of mtDNA have been shown to influence the penetrance of hearing loss associated with primary mtDNA mutations. In particular, mtDNA mutations at positions 4216 and 13708 labeled as second LHON mutations were implicated to increase the penetrance of the deafness-associated A7445G mutation [Guan et al., 1998], while the ND1 T3308C and tRNAAla T5655C mutations likely contribute to the higher penetrance of deafness in an African pedigree than Japanese and French families carrying the T7511C mutation [Li et al., 2004c]. Apart from the A1555G and G7444A mutations, 36 variants in this mitochondrial genome, belonging to the Eastern Asian haplogroup D4a [Yao et al., 2002], showed no evolutionary conservation. The G7444A mutation in the COI gene/the precursor of tRNASer(UCN) gene was implicated to influence the penetrance and expressivity of the A1555G mutation [Pandya et al., 1999] and the primary LHON-associated mtDNA mutations [Brown et al., 1995]. As shown in Figure 5, on the H strand of mtDNA, this mutation results in a read-through of the stop condon AGA of the COI message, thereby adding three amino acids (Lys-Gln-Lys) to the C-terminal of the polypeptide. Thus, the mutated polypeptide may retain a partial function. Alternatively, the G7444A mutation is adjacent to the site of 3’ end endonucleolytic processing of the L-strand RNA precursor, spanning tRNASer(UCN) and ND6 mRNA [Guan et al., 1998; Levinger et al., 2004]. Our previous data showed that the A7445G mutation in the precursor of tRNASer(UCN) led to a failure in the processing of the L-strand RNA precursor, thereby causing a marked decrease of the steady-state levels of tRNASer(UCN) and ND6 mRNA [Guan et al., 1998; Li et al., 2005b]. Thus, the G7444A mutation, similar to the A7445G mutation, may also cause a defect in the processing of the L-strand RNA precursor, thus causing mitochondrial dysfunctions. Although aminoglycoside is the predominant modifier factor for hearing impairment, the G7444A mutation may also play a role in the phenotypic expression of the A1555G mutation in this Chinese family. Therefore, a functional analysis is necessary to determine if the biochemical defect caused by the G7444A mutation influences the phenotypic manifestation of the A1555G mutation.
Figure 5. Location of deafness-associated mutations in the tRNASer(UCN) and adjacent COI.
Arrows indicate the position of the T7510C, T7511C and 7472insC mutations in the tRNASer(UCN) and the A7445G and G7444A mutation in the precursor of this tRNA and adjacent sequence of COI from wild-type (WT) and mutant (MT). The processing site for the 3-end of tRNASer(UCN) precursor, determined by and 3’-endonuclease [Guan et al., 1998], was indicated by arrow.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grants DC04958 and DC05230 from the National Institute on Deafness and Other Communication Disorders and a grant from National Basic Research Priorities Program of China 2004CCA02200 to M.X.G., and a Research Foundation grant 03YZJJ003 from Chinese PLA General Hospital to. H.Y. We are grateful to Jinzhong Sun, Bin Nie, Ming Li, Defu Xie, Fenglin Liu, Nongxiang Zhang for their assistance in blood sample collection, Chuck Loftice, Li Yang and Terri Wallace for skilled technical and clerical assistance.
REFERENCES
- Anderson S, Bankier AT, Barrell BG, deBruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Rose BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young I. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Brandon MC, Lott MT, Nguyen KC, Spolim S, Navathe SB, Baldi P, Wallace DC. MITOMAP: a human mitochondrial genome database--2004 update. Nucleic Acids Res. 2005;33:D611–D613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Torroni A, Reckord CL, Wallace DC. Phylogenetic analysis of Leber’s hereditary optic neuropathy mitochondrial DNA’s indicates multiple independent occurrences of the common mutations. Hum Mut. 1995;6:311–325. doi: 10.1002/humu.1380060405. [DOI] [PubMed] [Google Scholar]
- Casano RA, Johnson DF, Bykhovskaya Y, Torricelli F, Bigozzi M, Fischel-Ghodsian N. Inherited susceptibility to aminoglycoside ototoxicity: genetic heterogeneity and clinical implications. Am J Otolaryngol. 1999;20:151–156. doi: 10.1016/s0196-0709(99)90062-5. [DOI] [PubMed] [Google Scholar]
- del Castillo FJ, Villamar M, Moreno-Pelayo MA, Almela JJ, Morera C, Adiego I, Moreno F, del Castillo I. Maternally inherited nonsyndromic hearing impairment in a Spanish family with the 7510T>C mutation in the mitochondrial tRNASer(UCN) gene. J Med Genet. 2002;39:e82. doi: 10.1136/jmg.39.12.e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo FJ, Rodriguez-Ballesteros M, Martin Y, Arellano B, Gallo-Teran J, Morales-Angulo C, Ramirez-Camacho R, Cruz Tapia M, Solanellas J, Martinez-Conde A, Villamar M, Moreno-Pelayo MA, Moreno F, del Castillo I. Heteroplasmy for the 1555A>G mutation in the mitochondrial 12S rRNA gene in six Spanish families with non-syndromic hearing loss. J Med Genet. 2003;40:632–636. doi: 10.1136/jmg.40.8.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estivill X, Govea N, Barcelo E, Badenas C, Romero E, Moral L, Scozzari R, D'Urbano L, Zeviani M, Torroni A. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment with aminoglycosides. Am J Hum Genet. 1998;62:27–35. doi: 10.1086/301676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischel-Ghodsian N. Mitochondrial deafness mutations reviewed. Hum Mut. 1999;13:261–270. doi: 10.1002/(SICI)1098-1004(1999)13:4<261::AID-HUMU1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisa E, Saccone C. The complete nucleotide sequence of the Rattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol. 1989;28:497–516. doi: 10.1007/BF02602930. [DOI] [PubMed] [Google Scholar]
- Guan MX. Molecular pathogenetic mechanism of maternally inherited deafness. Ann NY Acad Sci. 2004;1011:259–271. doi: 10.1007/978-3-662-41088-2_25. [DOI] [PubMed] [Google Scholar]
- Guan MX, Fischel-Ghodsian N, Attardi G. Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum Mol Genet. 1996;5:963–971. doi: 10.1093/hmg/5.7.963. [DOI] [PubMed] [Google Scholar]
- Guan MX, Fischel-Ghodsian N, Attardi G. A biochemical basis for the inherited susceptibility to aminoglycoside ototoxicity. Hum Mol Genet. 2000;9:1787–1793. doi: 10.1093/hmg/9.12.1787. [DOI] [PubMed] [Google Scholar]
- Guan MX, Fischel-Ghodsian N, Attardi G. Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Hum Mol Genet. 2001;10:573–580. doi: 10.1093/hmg/10.6.573. [DOI] [PubMed] [Google Scholar]
- Guan MX, Enriquez JA, Fischel-Ghodsian N, Puranam R, Lin CP, Marion MA, Attardi G. The Deafness-associated mtDNA 7445 mutation, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase ND6 subunit gene expression. Mol Cell Biol. 1998;l18:5868–5879. doi: 10.1128/mcb.18.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchin TP, Haworth I, Higashi K, Fischel-Ghodsian N, Stoneking M, Saha N, Arnos C, Cortopassi G. A molecular basis for human hypersensitivity to aminoglycoside antibiotics. Nucleic Acids Res. 1993;21:4174–4179. doi: 10.1093/nar/21.18.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchin T, Parker MJ, Young ID, Davis AC, Pulleyn JL, Deeble J, Lench NJ, Markham AF, Muller RF. A novel mutation in the mitochondrial tRNASer(UCN) gene in a family with non-syndromic sensorineural hearing impairment. J Med Genet. 2000;37:692–694. doi: 10.1136/jmg.37.9.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs HT, Hutchin TP, Kappi T, Gillies G, Minkkinen K, Walker J, Thompson K, Rovio AT, Carella M, Melchionda S, Zelante L, Gasparini P, Pyykko I, Shah ZH, Zeviani M, Mueller RF. Mitochondrial DNA mutations in patients with postlingual, nonsyndromic hearing impairment. Eur J Hum Genet. 2005;13:26–33. doi: 10.1038/sj.ejhg.5201250. [DOI] [PubMed] [Google Scholar]
- Levinger L, Morl M, Florentz C. Mitochondrial tRNA 3' end metabolism and human disease. Nucleic Acids Res. 2004;32:5430–5441. doi: 10.1093/nar/gkh884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Greinwald JH, Yang L, Choo DI, Wenstrup RJ, Guan MX. Molecular analysis of mitochondrial 12S rRNA and tRNASer(UCN) genes in paediatric subjects with nonsyndromic hearing loss. J Med Genet. 2004a;41:615–620. doi: 10.1136/jmg.2004.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Xing G, Yan M, Cao X, Liu XZ, Bu X, Guan MX. Cosegregation of C-insertion at position 961 with A1555G mutation of mitochondrial 12S rRNA gene in a large Chinese family with maternally inherited hearing loss. Am J Med Genet. 2004b;124A:113–117. doi: 10.1002/ajmg.a.20305. [DOI] [PubMed] [Google Scholar]
- Li R, Ishikawa K, Deng JH, Yang L, Tamagawa Y, Bai Y, Ichimura K, Guan MX. Maternally inherited nonsyndromic hearing loss is associated with the mitochondrial tRNASer(UCN) 7511C mutation in a Japanese family. Biochem Biophys Res Commun. 2005a;328:32–37. doi: 10.1016/j.bbrc.2004.12.140. [DOI] [PubMed] [Google Scholar]
- Li X, Fischel-Ghodsian N, Schwart F, Yan Q, Friedman RA, Guan MX. Biochemical characterization of the mitochondrial tRNASer(UCN) T7511C mutation associated with nonsyndromic deafness. Nucleic Acids Res. 2004c;32:867–877. doi: 10.1093/nar/gkh226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang LS, Fischel-Ghodsian N, Guan MX. Biochemical characterization of the deafness-associated mitochondrial tRNASer(UCN) A7445G mutation in osteosarcoma cell cybrids. Biochem Biophys Res Commun. 2005b;328:491–498. doi: 10.1016/j.bbrc.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Li Z, Li R, Chen J, Liao Z, Zhu Y, Qian Y, Xiong S, Heman-Ackah S, Wu J, Choo DI, Guan MX. Mutational analysis of the mitochondrial 12S rRNA gene in Chinese pediatric subjects with aminoglycoside induced and non-syndromic hearing loss. Hum Genet. 2005c;117:9–15. doi: 10.1007/s00439-005-1276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijs G, Claes S, Longo-Bbenza B, Cassiman J-J. Non-syndromic deafness associated with a mutation and a polymorphism in the mitochondrial 12S ribosomal RNA gene in a large Zairean pedigree. Eur J Hum Genet. 1996;4:46–51. doi: 10.1159/000472169. [DOI] [PubMed] [Google Scholar]
- Pandya A, Xia X, Radnaabazar J, Batsuuri J, Dangaansuren B, Fischel-Ghodsian N, Nance WE. Mutation in the mitochondrial 12S ribosomal-RNA gene in 2 families from Mongolia with matrilineal aminoglycoside ototoxicity. J Med Genet. 1987;34:169–172. doi: 10.1136/jmg.34.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya A, Xia XJ, Erdenetungalag R, Amendola M, Landa B, Radnaabazar J, Dangaasuren B, Van Tuyle G, Nance WE. Heterogenous point mutations in the mitochondrial tRNASer(UCN) precursor coexisting with the A1555G mutation in deaf students from Mongolia. Am J Hum Genet. 1999;65:1803–1806. doi: 10.1086/302658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, Shohat M, Fischel-Ghodsian N. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- Reid FM, Vernham GA, Jacobs HT. A novel mitochondrial point mutation in a maternal predigree with sensorineural deafness. Hum. Mut. 1994;3:243–247. doi: 10.1002/humu.1380030311. [DOI] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Tobe VO, Nickerson DA. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res. 1998;26:967–973. doi: 10.1093/nar/26.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe A, Ma DP, Wilson RK, Wong JF. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- Sue CM, Tanji K, Hadjigeorgious G, Andreu AL, Nishino I, Krishna S, Bruno C, Hirano M, Shanske S, Bonilla E, Fischel-Ghodsian N, DiMauro S, Friedman R. Maternally inherited hearing loss in a large kindred with a novel T7511C mutation in the mitochondrial DNA tRNASer(UCN) gene. Neurology. 1999;52:1905–1908. doi: 10.1212/wnl.52.9.1905. [DOI] [PubMed] [Google Scholar]
- Thyagarajan D, Bressman S, Bruno C, Przedborski S, Shanske S, Lynch T, Fahn S, DiMauro S. A novel mitochondrial 12SrRNA point mutation in Parkinsonism, deafness and neuropathy. Ann Neurol. 2001;48:730–736. [PubMed] [Google Scholar]
- Verhoeven K, Ensink RJ, Tiranti V, Huygen PL, Johnson DF, Schatteman I, Van Lae L, Verstreken M, Van de Heyning P, Fischel-Ghodsian N, Zeviani M, Cremers CW, Willems PJ, Van Camp G. Hearing impairment and neurological dysfunction associated with a mutation in the mitochondrial tRNASer(UCN) gene. Eur J Hum Genet. 1999;7:45–51. doi: 10.1038/sj.ejhg.5200247. [DOI] [PubMed] [Google Scholar]
- Yao YG, Kong OP, Bandelt HJ, Kivisild T, Zhang YP. Phylogeographic diffeentiation of motochondrial DNA in Han Chinese. Am J Hum Genet. 2002;70:635–651. doi: 10.1086/338999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young WY, Zhao L, Qian Y, Wang Q, Li N, Greinwld JH, Guan MX. Extremely low penetrance of hearing loss in four Chinese families with the mitochondrial 12S rRNA A1555G mutation. Biochem Biophys Res Commun. 2005;328:1244–1251. doi: 10.1016/j.bbrc.2005.01.085. [DOI] [PubMed] [Google Scholar]
- Zhao H, Li R, Wang Q, Yan Q, Deng JH, Han D, Bai Y, Young WY, Guan MX. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am J Hum Genet. 2004a;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Young WY, Yan Q, Li R, Cao J, Wang Q, Li X, Peters JL, Han D, Guan MX. Functional characterization of the mitochondrial 12S rRNA C1494T mutation associated with aminoglycoside-induced and non-syndromic hearing loss. Nucleic Acids Res. 2005;33:1132–1139. doi: 10.1093/nar/gki262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Young WY, Li R, Wang Q, Qian Y, Guan MX. Clinical evaluation and sequence analysis of the complete mitochondrial genome of three Chinese patients with hearing impairment associated with the 12S rRNA T1095C mutation. Biochem Biophys Res Commun. 2004b;325:1503–1508. doi: 10.1016/j.bbrc.2004.10.199. [DOI] [PubMed] [Google Scholar]