Abstract
Ions derived from nano-electrospray ionization (nano-ESI) of α-synuclein, a 14.5 kDa, 140 amino acid residue protein that is a major component of the Lewy bodies associated with Parkinson’s disease, have been subjected to ion trap and beam-type collisional activation. The former samples products from fragmentation at rates generally lower than 100 s−1 whereas the latter samples products from fragmentation at rates generally greater than 103 s−1. A wide range of protein charge states spanning from as high as [M+17H]17+ to as low as [M+4H]4+ have been formed either directly from nano-ESI or via ion/ion proton transfer reactions involving the initially formed protein cations and have been subjected to both forms of collision-induced dissociation (CID). The extent of sequence information (i.e., number of distinct amide bond cleavages) available from either CID method was found to be highly sensitive to protein precursor ion charge state. Furthermore, the relative contributions of the various competing dissociation channels were also dependent upon precursor ion charge state. The qualitative trends in the changes in extent of amide bond cleavages and identities of bonds cleaved with precursor ion charge state were similar for two forms of CID. However, for every charge state examined, roughly twice the primary sequence information resulted from beam-type CID relative to ion trap CID. For example, evidence for cleavage of 86% of the protein amide bonds was observed for the [M+9H]9+ precursor ion using beam-type CID whereas 41% of the bonds were cleaved for the same precursor ion using ion trap CID. The higher energies required to drive fragmentation reactions at rates necessary to observe products in the beam experiment access more of the structurally informative fragmentation channels, which has important implications for whole protein tandem mass spectrometry.
1. Introduction
The identification and characterization of proteins is a major activity in proteomics. The most widely used approaches to identify proteins rely on chemical or enzymatic protein digestion followed by mass spectrometry or tandem mass spectrometry (MS/MS) of the digestion products [1–5]. Experimental procedures based on the analysis of peptides from protein digestion are commonly referred to as ‘bottom up” approaches. While bottom up approaches are quite powerful, it is also widely recognized that potentially important information can be lost in the process, such as the identities and locations of post-translational modification, due to the complexity of the mixture generated via digestion, poor sequence representation in the peptides observed, lack of intact protein mass information, etc. An alternative approach to protein identification and characterization, which is referred to as the ‘top down’ approach, involves the tandem mass spectrometry of intact protein ions [6–9]. A key advantage to any top down approach is the direct connection between the mass of the intact protein and the structural information derived from its dissociation. This connection is lost with bottom up approaches.
Despite the potential advantages associated with top down approaches, they are not as mature as bottom up approaches. Further development is needed in areas such as protein separations, ionization of whole protein mixtures, and whole protein dissociation, particularly for proteins in excess of 200 kDa [10]. From the standpoint of tandem mass spectrometry, the approach to dissociation is particularly important because it plays a key role in determining the nature and extent of the structural information that can be generated from an isolated gaseous protein ion. A variety of dissociation approaches have been examined for whole protein dissociation, including slow heating methods [11] such as collisional activation by sustained off-resonance irradiation [12] in Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometry [13], infra-red multi-photon dissociation (IRMPD) [14] and ion trap collisional activation [15] in electrodynamic ion traps [16]. Slow heating methods are characterized by ion activation and deactivation processes occurring in parallel with dissociation such that a roughly steady-state precursor ion internal energy distribution is achieved during the ion activation period [17,18]. A variety of faster activation methods that do not achieve a condition of comparable ion activation and deactivation rates have also been applied to whole protein ions including UV photodissociation [19], surface-induced dissociation [20], and beam-type collisional activation at relatively low collisions energies (i.e. 10–500 eV) [21–23] and high collision energies (i.e. > 1 keV) [24]. In terms of time-frames associated with the activation step, slow heating methods generally employ activation periods of tens to hundreds of milliseconds, whereas, in the case of low energy beam-type CID (10–1000 eV laboratory collision energies), tens of microseconds are more typical for the activation period. Electron capture dissociation [25] (ECD) and electron transfer dissociation [26] (ETD) have also been applied to protein ions and have been shown to be complementary to the vibrational excitation techniques mentioned above, particularly in the characterization of post-translational modifications.
The factors that play significant roles in the nature of the structural information that is forthcoming from the dissociation products of a whole protein ion include inter alia the ion activation approach and, often, the specific conditions used, the polarity [27] and charge state of the ion, and the modification state of the ion (e.g., the presence or absence of disulfide linkages [28]). Most reported systematic investigations of the charge state dependent dissociation behavior of whole protein ions have been conducted under ion trap collisional activation conditions [29–32]. These studies have shown that the favored dissociation channels are highly dependent upon precursor ion charge state with intermediate charge states providing the greatest sequence coverage. Favored cleavages N-terminal to proline residues and C-terminal to aspartic acid residues, the latter being particularly prominent at relatively low charge states, are also noted, in analogy with peptide ion dissociation behavior [33]. Preferred cleavage channels are expected to be particularly prominent under the slow heating conditions of ion trap collisional activation. Far fewer studies with ion activation techniques that give rise to faster dissociation than ion trap collisional activation, IRMPD, or SORI-CID have been reported that describe dissociation as a function of precursor ion charge state. In this work, we describe the charge state dependent dissociation behavior of a model protein, α-synuclein, under both ion trap and beam-type collisional activation conditions. These approaches are both accessible with hybrid tandem mass spectrometers that combine both beam and ion trapping functions. In this case, the experiments are performed on a hybrid quadrupole/time-of-flight tandem mass spectrometer and constitute the only systematic comparison of these commonly used CID approaches as applied to a wide range of charge states of whole protein of this size (14,461 Da, 140 amino acid residues). The only other protein examined over a wide charge state range under both activation conditions is bovine ubiquitin [34] (8,451 Da, [M+12H]12+-[M+6H]6+). In that work, the maximum sequence coverage obtained from any charge state under any condition was 70% and the total sequence coverage obtained was ~77%.
Alpha-synuclein belongs to a natively unfolded class of proteins at physiological pH. Its structure, as well as its interactions with various ligands, has been extensively studied [35–38]. These studies have included a recent report of the top-down tandem mass spectrometry of the complex of α-synuclein with spermine, with particular emphasis placed on ECD as the dissociation technique [39]. The primary structure (see below for complete sequence) consists of 3 main regions. The N-terminal region, which includes residues 1–60, contains four imperfect KTKEGV repeat units as well as the A30P, A53T, and E46K mutation sites affiliated with Parkinson’s disease [40]. The hydrophobic region, comprised of residues 61–95, is responsible for the fibrillation properties of α-synuclein. The insoluble fibrillar form of α-synuclein is a major component of the Lewy bodies, which are hallmark lesions of Parkinson’s disease (PD) [41,42]. It has been reported that misfolding and aggregation of α-synuclein play a crucial role in PD [43,44]. The C-terminal region (residues 96–140) is acidic due to the presence of many acidic amino acid residues. In this study, we have undertaken an examination of the charge state dependent fragmentation of protonated α-synuclein from [M+17H]17+ to [M+4H]4+ as a function of activation conditions to examine the relative merits of two widely accessible sets of collisional activation conditions mentioned above (i.e., ion trap collisional activation and low energy beam-type collisional activation).
2. Experimental Section
2.1 Materials
Acetic acid was obtained from Mallinckrodt Baker (Phillipsburg, NJ), 2,2,3,3,4,4,5,5,6,6,7,7,8,8,8-pentadecafluoro-1-octanol (PFO) from Sigma-Aldrich (St. Louis, MO), and α-synuclein from rPeptide (Bogart, GA). PFO was prepared in a methanol solution containing 1% ammonium hydroxide with a final concentration of 400 µM.
2.2 Purification of α-synuclein by RP-HPLC
An α-synuclein stock solution with a concentration of 1 mg/mL was prepared in H2O and was purified by reversed-phase high-performance liquid chromatography on an Agilent (Palo Alto, CA) model 1100 system, using an Aquapore RP-300 (7 µm particle size, 100×4.6 mm i.d.) column (Perkin-Elmer, Wellesley, MA) operated at a flow rate of 1 ml/min. A linear 60 min gradient from 0 to 100% buffer B was used, where buffer A was 0.1% (v) aqueous TFA and buffer B was (60:40) acetonitrile: water containing 0.09% TFA. The column was maintained at room temperature and the absorbance was monitored at 215 nm. The α-synuclein was eluted after ~43 min. The collected fractions were lyophilized and dissolved in 100 µl of water to a concentration of 1.5 mg/ml. Samples were prepared in aqueous 1% acetic acid and adjusted to a final concentration of 0.75 mg/ml for positive nano-ESI. The solution was then stored in a freezer at −20 °C until mass spectrometric analysis.
2.3 Ionization Methods
The nano-ESI emitters were made by a Sutter Instruments (Novato, CA, USA) micropipette puller model P-87. For the proton transfer ion/ion reactions, a home-built pulsed dual ESI source was used for the formation of positive ions of proteins and for the formation of deprotonated PFO, as fully described elsewhere [45].
2.4 Mass Spectrometry
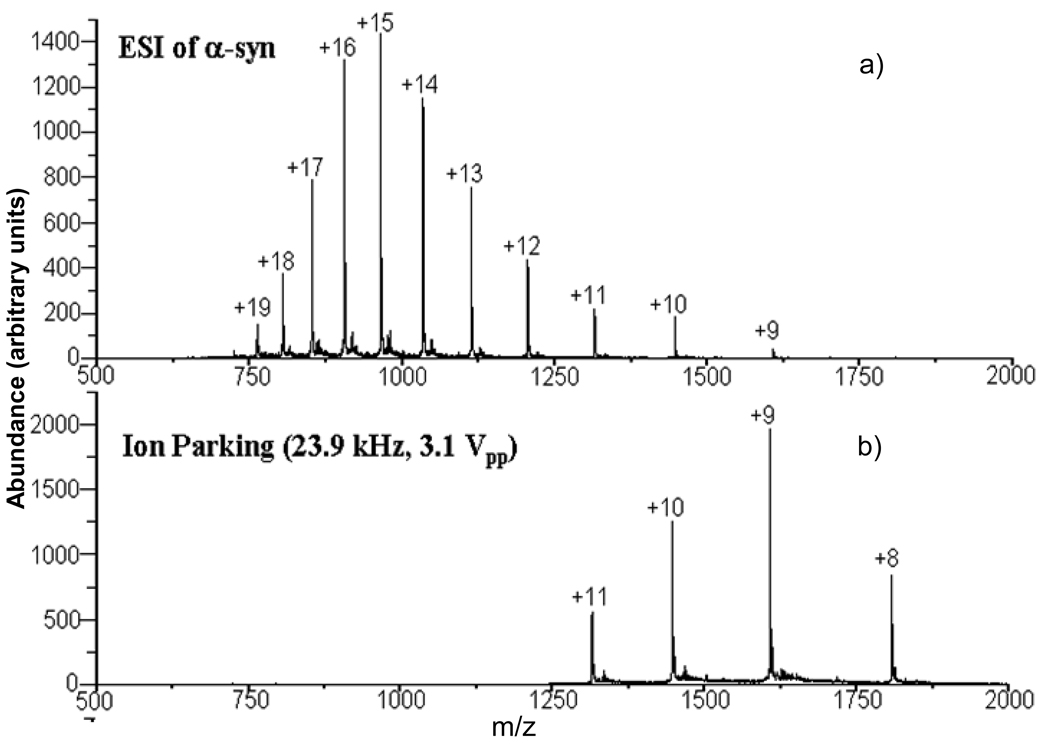
All experiments were performed using a commercial quadrupole/ time-of-flight (QqTOF) tandem mass spectrometer (QSTAR XL, Applied Biosystems/MDS Sciex) [46] modified for ion/ion reaction studies [47]. For ion trap experiments, there were up to six experimental steps used to collect charge state dependent fragmentation data: cation accumulation, transmission mode ion/ion reaction [48] in Q0 whereby anions were transmitted through Q0 while the protein ions were stored, precursor ion isolation via Q1, collision induced dissociation (CID) in Q2 using resonance excitation, proton transfer ion-ion reaction in Q2, and mass analysis via orthogonal acceleration TOF. The ion-ion reaction step prior to mass analysis was used to reduce the charge states of the CID products largely to singly-charged species in order to simplify the product ion assignment. When the charge state of interest was already present in relatively high abundance from the initial charge state distribution, no ion/ion reactions were conducted in Q0. When the charge state of interest was a low charge state and not available directly from nano-ESI, the ion/ion proton transfer reaction in Q0 was performed to form the charge state of interest from higher charge states formed directly via nano-ESI. Parallel ion parking [49] was sometimes used to concentrate precursor ions into a limited number of charge states for subsequent mass-selection via Q1. Figure 1 provides an example of results from a transmission-mode ion/ion reaction in Q0 with parallel ion parking used to concentrate charte into the 8+-11+ charge states. Beam-type CID experiments were performed using the same instrument. The experimental procedures for ion parking and beam-type CID used here have been reported in detail elsewhere [50,51]. In brief, the following experimental sequence was used: 1) cation accumulation (200–250 ms) in Q0; 2) ion parking in Q0 (to obtain low charge state species); 3) precursor ion isolation via Q1; 4) beam-type CID in Q2 with the injection energy determined by the DC rod offset difference between Q0 and Q2; 5) mutual storage mode ion-ion reactions in Q2; and 6) mass analysis of ions using a reflectron time-of-flight mass analyzer. The MS/MS spectra shown here were typically the average of 500–1000 individual scans.
Figure 1.
a) Nano-ESI mass spectrum of α-synuclein. b) Post-ion/ion reaction data from a parallel ion parking transmission mode reaction involving α-synuclein cations and PFO anions in Q0.
The protein’s molecular weight and CID fragments of α-synuclein were calculated by using PROWL– Protein Information Retrieval Online World Wide Web Lab (http://prowl.rockefeller.edu/prowl/proteininfo.html) and Protein Prospector (http://prospector.ucsf.edu). A signal-to-noise ratio of 3 was used as the abundance threshold for assigning a peak and product ion assignments are within +/− 1 Da of the predicted mass.
3. Results and Discussion
3.1 Ion Trap Collision-Induced Dissociation
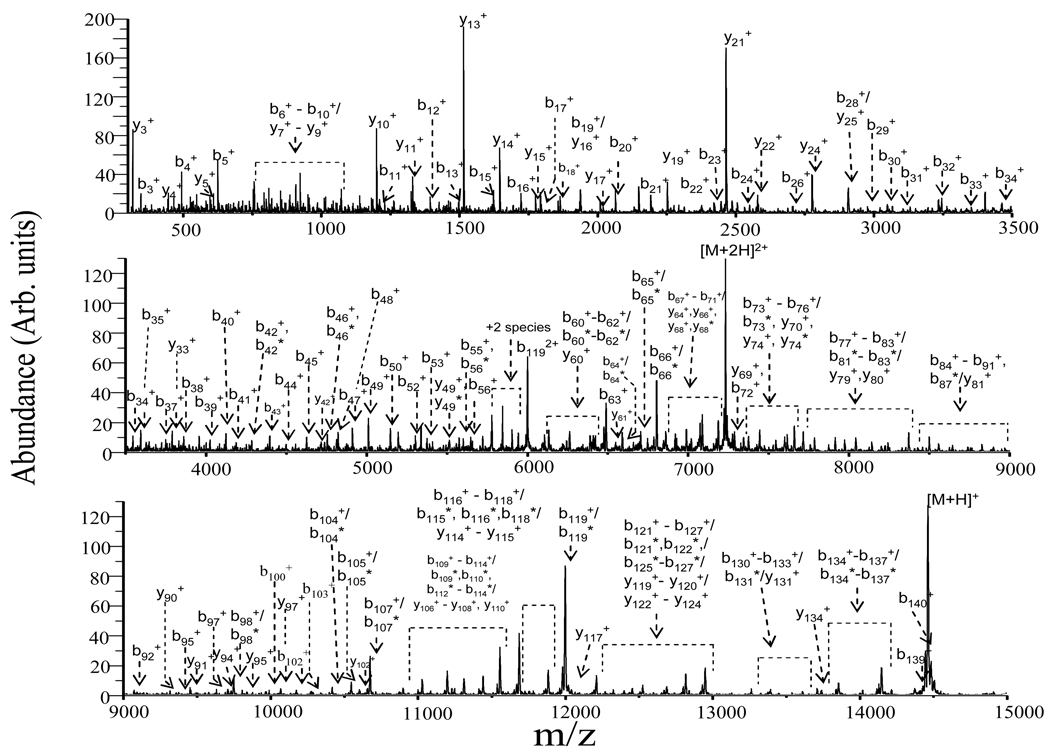
Protonated α-synuclein ions ranging in charge sate from +4 to +17 were subjected to ion trap collisional activation in the Q2 linear ion trap (LIT), followed by ion/ion proton transfer reaction with deprotonated PFO in order to reduce product ions to largely the +1 charge state. The relative abundances of the product ions resulting from ion trap CID were found to be insensitive to the amplitude of the dipolar excitation voltage over the range of amplitudes that gave rise to >50% conversion of precursor ions to products while leading to minimal precursor ion ejection. Activation periods of 20–250 ms were examined as well. These results, as well as those observed in previous ion trap CID studies of protein cations [28–32], suggest that relatively little change in protein structural information is noted over the rate range of 5–50 s−1 typically accessed by the ion trap experiment. Post-ion/ion MS/MS results for the [M+9H]9+ ion of α-synuclein are shown in Figure 2, which illustrates the products resulting from ion trap activation conditions (dipolar excitation at 81.06 kHz, 600 mVp-p, 20 ms, nitrogen as a collision gas at ~ 5 mTorr) in the Q2 LIT after a 200-ms mutual storage ion/ion reaction period. Since most of the ions are reduced to singly charged species via ion/ion reactions, most of product ions are readily assigned by comparing observed m/z values with those of the expected singly charged product masses of α-synuclein from amide bond cleavages. Products from cleavage of 57 of the protein amide bonds are identified from the fragmentation of the [M+9H]9+ ion under these conditions with cleavage of the D119/P120 bond, which leads to the b119/y21 complementary pair, being most prolific. Cleavages C-terminal to aspartic acid and N-terminal to proline each tend to be frequently observed and cleavages at D-P sites are particularly favored.
Figure 2.
Ion trap CID post-ion/ion reaction MS/MS spectrum of the [M + 9H]9+ ion of α-synuclein via activation at 81.06 kHz, 600 mVp-p, nitrogen as a collision gas at ~ 5 mTorr. The loss of water from the corresponding b or y ions is labeled with an asterisk.
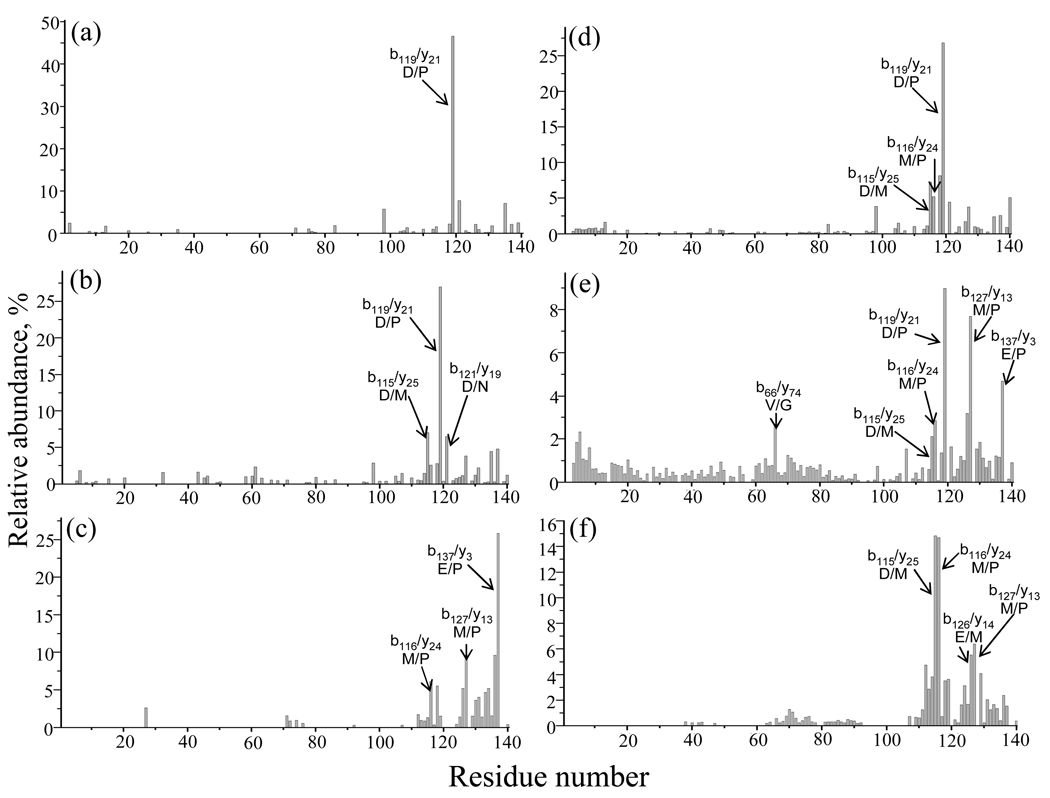
The ion trap fragmentation of α-synuclein ions ranging from [M+4H]4+ to [M+17H]17+ has parallels with that of other proteins observed previously under ion trap CID conditions. For example, preferential cleavage C-terminal to aspartic acid residues and loss of H2O molecules from protonated precursor ions at low charge states (e.g., charge states ≤ +5 in the case of α-synuclein) and relatively extensive non-specific amide bond fragmentation at intermediate charge states (e.g., +9) is common to all proteins studied to date. Generalizations regarding the highest charge states (e.g., +15 and higher for α-synuclein) are more difficult to draw. However, fragmentation is often limited to a few channels and cleavage N-terminal to proline residues is often observed. The fragmentation behaviors of the +5, +9, and +15 charge states under ion trap CID conditions are summarized in Figure 3 panels a–c, respectively, which show the summed complementary b/y ion abundances as a function of residue number. For the ion trap CID of the [M+5H]5+ parent ion, ~ 67% of the total product ion abundance was accounted for by fragmentation C-terminal to aspartic acid residues. However, the contribution from C-terminal aspartic acid cleavages decreased to ~ 48% in the product ion abundances of the intermediate charge state, [M+9H]9+, with the appearance of more evenly weighted (52%) non-specific fragmentation channels (11% are from the contribution of N-terminal cleavages to proline). For the high charge state ion, [M+15H]15+, cleavage N-terminal to proline residues is the only clearly favored fragmentation channel, which accounted for 42% of the total product ion abundance. Since proline and aspartic acid residues are adjacent at residues 119 and 120, cleavage at D119/P120 is particularly prominent at the low and intermediate charge states (see Figure 3a and 3b). However, the abundances of the product ions from this fragmentation channel decrease dramatically for the [M+15H]15+ ion, and the dominant fragmentation channel is replaced by cleavage at E137/P138.
Figure 3.
Summed b and y abundances for complementary ion pairs as a function of residue number resulting from cleavages of the α-synuclein backbone with ion trap CID for (a) [M+5H]5+, (b) [M+9H]9+, and (c) [M+15H]15+ ions; and with beam-type CID for (d) [M+5H]5+, (e) [M+9H]9+, and (f) [M+15H]15+ ions.
Proton mobility [52,53,54] appears to play a particularly important role in determining the number of amide bond dissociation channels that compete. At very low charge states, in which protons are likely to be of relatively low mobility due to location at particularly basic sites and high degrees of intramolecular charge solvation, low energy and low frequency processes (i.e., those with entropic constraints in achieving the transition state, such as neutral losses that arise from rearrangement reactions) tend to dominate. These include the C-terminal aspartic acid cleavage and losses of small molecules such as water and ammonia. The highest degrees of proton mobility are likely to be found at intermediate charge states in which electrostatic repulsion tends to minimize intramolecular charge solvation but is not so strong so as to prevent proton mobility in the activated ion population. As amide bond cleavages are facilitated by the presence of a proton [55], this situation gives rise to the greatest diversity of amide bond cleavages. At high charge states, intramolecular electrostatic repulsion can limit proton mobility thereby resulting in the contribution of a limited number of dissociation channels.
3.2 Beam-Type Collision Induced Dissociation
All beam-type CID experiments were performed using nitrogen as the collision gas at a pressure of 5 mTorr. This pressure has been shown to provide both relatively high CID product yields and relatively high ion/ion reaction rates [44]. To study the effect of injection energy in beam-type CID of α-synuclein, the percent sequence coverage (% Sequence), as defined in equation 1:
| (1) |
was determined as a function of injection energy (IE) of the precursor ion into Q2 in the laboratory frame of reference. Note that at the pressures used here, the protein precursor ions can undergo several hundred collisions with nitrogen during passage through Q2. Hence, we avoid use of the term “collision energy” by use of the term “injection energy” because the energies of successive collisions are reduced by momentum transfer in previous collisions. In the present apparatus, injection energy is determined by the difference between the DC off-set potentials of Q0 and Q2 multiplied by the charge state of the precursor ion, as indicated in equation 2:
| (2) |
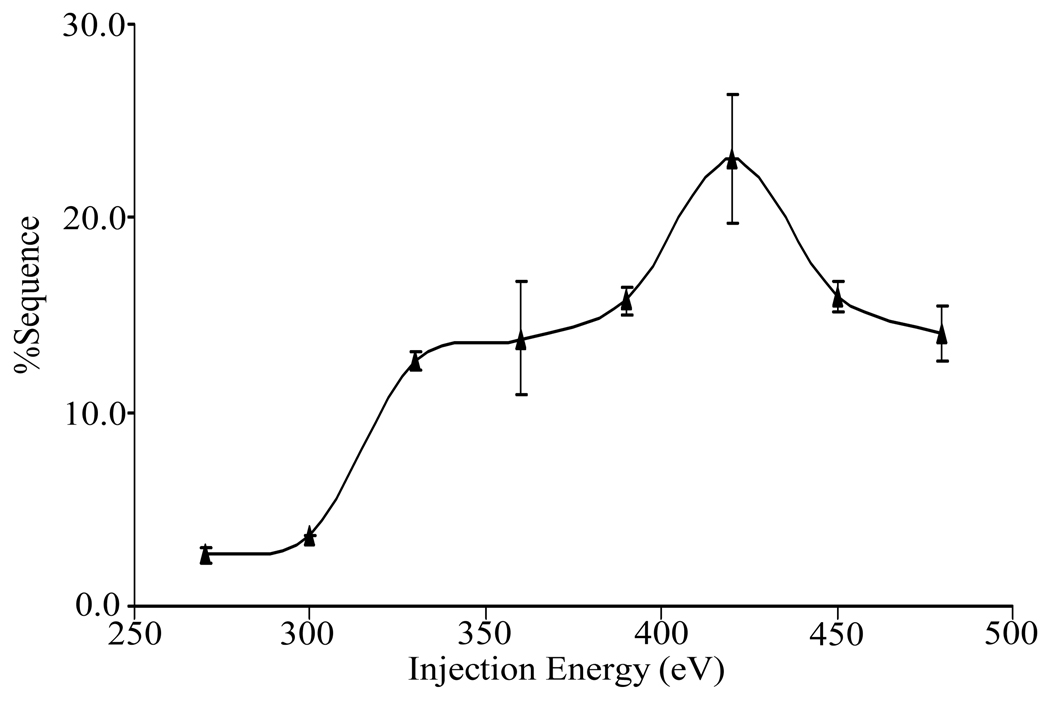
where DCQ0 is the offset DC potential applied to Q0, DCQ2 is the offset DC potential applied to Q2, and Z is the charge of precursor ion. Figure 4 provides an example of the dependence of %Sequence on collision energy for the [M+15H]15+ ion. As the injection energy increases, %Sequence initially increases, reaches a maximum, and then decreases. The initial increase presumably arises from greater internal energy deposition into the precursor, which allows a larger number of competitive dissociation channels to be accessed. As the injection energy increases further, however, sequential fragmentation processes give rise to products that are difficult to identify and deplete the abundances of first generation products thereby making relatively low level sequence-informative products more difficult to identify or detect. For the [M+15H]15+ ion, the injection energy giving high sequence coverage with minimal secondary fragmentation falls in the range of 380 eV to 430 eV. This range proved to be useful for all of the relatively high charge states (e.g., +12 to +17). The optimal range of collision energies for lower charge states was found to be higher, probably due to lower internal electrostatic repulsion, which results in higher kinetic stability. The optimum injection energy range for intermediate charge states (e.g., +7 to +11) was found to be 600 to 700 eV and that for the lowest charge states examined (+4 and +5) was 750 eV to 800 eV.
Figure 4.
The percent sequence coverage of the fragments that result from beam-type CID as a function of ion injection energy for +15 charge state of α-synuclein. Three replicate measurements were performed to establish the error bars.
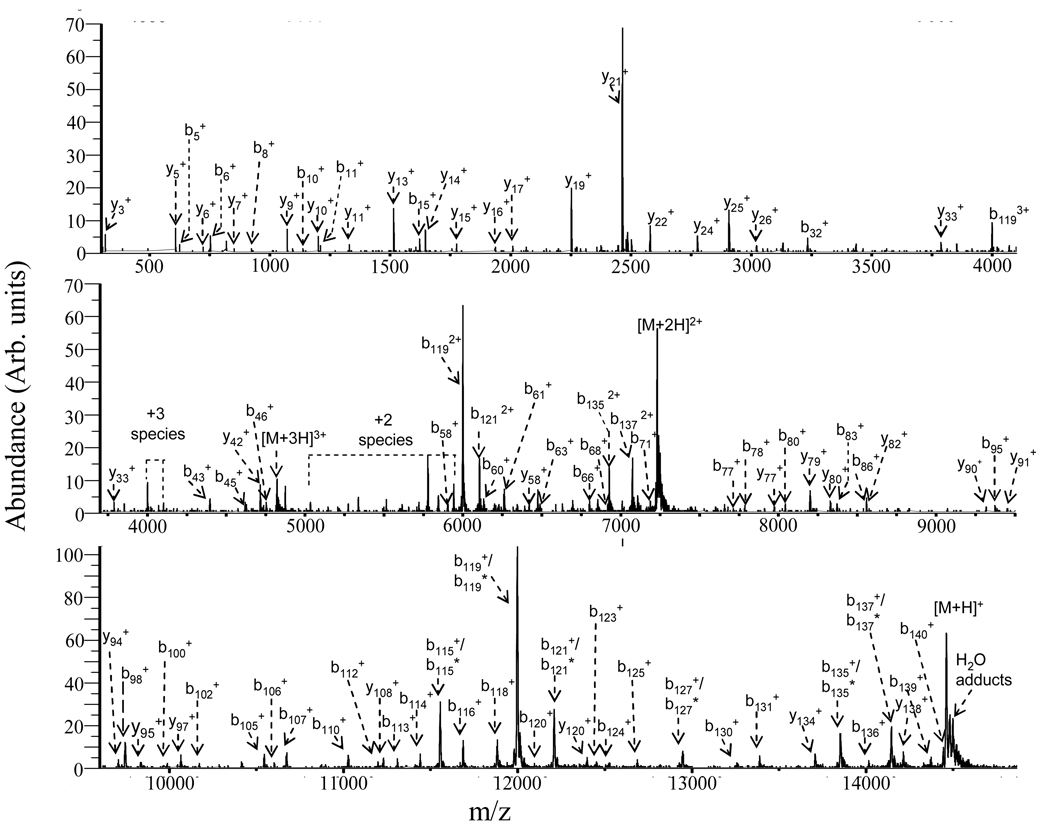
Post ion/ion reaction product ion spectra were obtained from the [M+4H]4+ to [M+17H]17+ precursor ions of α-synuclein within the respective injection energy ranges to maximize %Sequence for each charge state. The post-ion/ion reaction product ion data for the [M+9H]9+ ion collected under beam-type CID conditions (pressure = ~5 mTorr N2; laboratory injection energy = 630 eV) is shown in Figure 5 for illustration. Extensive fragmentation was observed as evidenced by the fragmentation in b-, y-type ions, and by the loss of water molecules from the corresponding b- and y-type ions, which are labeled with asterisks. These data can be compared directly with the data of Figure 2, which shows the results from ion trap CID.
Figure 5.
Post-ion/ion reaction MS/MS spectrum of the [M+9H]9+ ion of α-synuclein: injection energy, 630 eV; collision gas, nitrogen at ~ 5mTorr. The loss of water from the corresponding b or y ions is labeled with an asterisk.
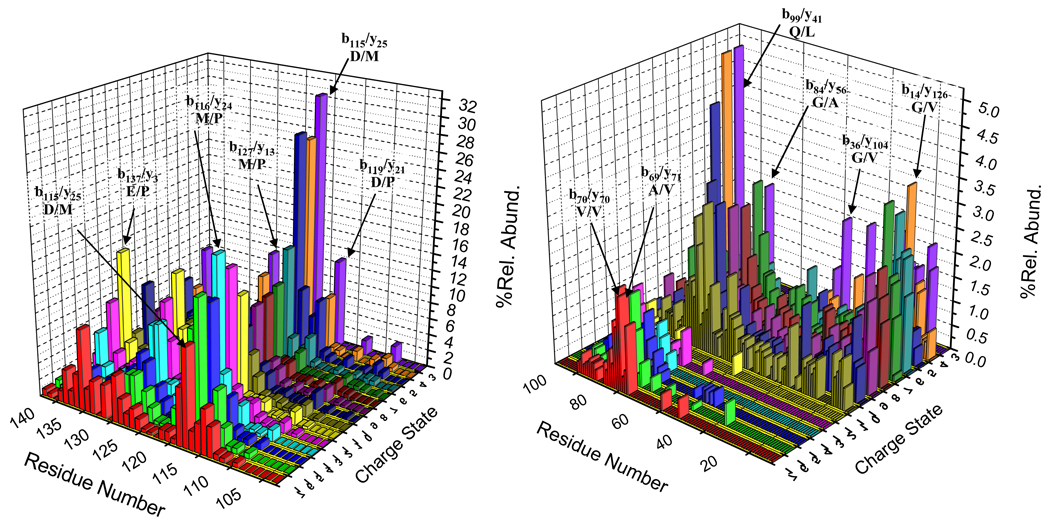
The normalized summed complementary b- and y-ions from each fragmentation channel plotted as a function of residue number for all charge states examined by beam-type CID is shown in Figure 6. The fragmentation behavior of precursor ions in the highest charge state grouping, [M+12H]12+-[M+17H]17+, shows dominant cleavages C-terminal to acidic residues (Asp, and Glu) and N-terminal to the proline residue. Moreover, small molecule losses (e.g., H2O losses) are also observed. All charge states in this group show evidence for cleavage at D115/M116 (b115), M116/P117 (b116), V118/D119 (b118), D119/P120 (b119), E126/M127 (b126/y14), M127/P128 (b127/y13), and E137/P138 (b137). In the charge states of +15 to +17, the D115/M116 cleavage channel gives rise to the most abundant product ion (viz., b115). while the b116 product ion from cleavage at M116/P117 gives rise to the base peak for the +12 to +14 charge states. The %Sequence values determined from the +12 to +17 charge states are 32%, 34%, 35%, 44%, 44% and 38%, respectively.
Figure 6.
Summed abundances of b- and y-type complementary product ions derived from beam-type CID are plotted as a function of residue number and parent ion charge state for all parent ion charge states examined. The abundance scale is normalized by dividing the summed b- and y- type ions by the total product ion signal for the relevant charge state.
Many of the major fragmentation channels noted for the high charge states also contribute significantly in the dissociation of the precursor ions of the intermediate charge state group, [M+6H]6+ to [M+11H]11+. However, this group of precursor ions tends to also show extensive non-specific fragmentation of the protein backbone. The b- and y-type product ions produced from this charge state group reflect from 69% (+11 charge state) to 86% (+9 charge state) of the sequence. This charge state group shows evidence for major cleavages at A107/P108 (b107/y33), D115/P116 (b115/y25), D119/P120 (b119/y21), E126/M127 (b126/y14), M127/P128 (b127/y13) and E137/P138 (b137/y3). At charge states below [M+6H]6+, higher injection energy is needed in order to yield significant degrees of fragmentation. Small molecule loss is particularly prominent, as has also been noted previously for low charge state ions under ion trap CID conditions. Multiple water molecule losses are also commonly observed. The preferential amide bond fragmentation is observed at D119/P120 (b119/y21) for both of the lowest charge states examined.
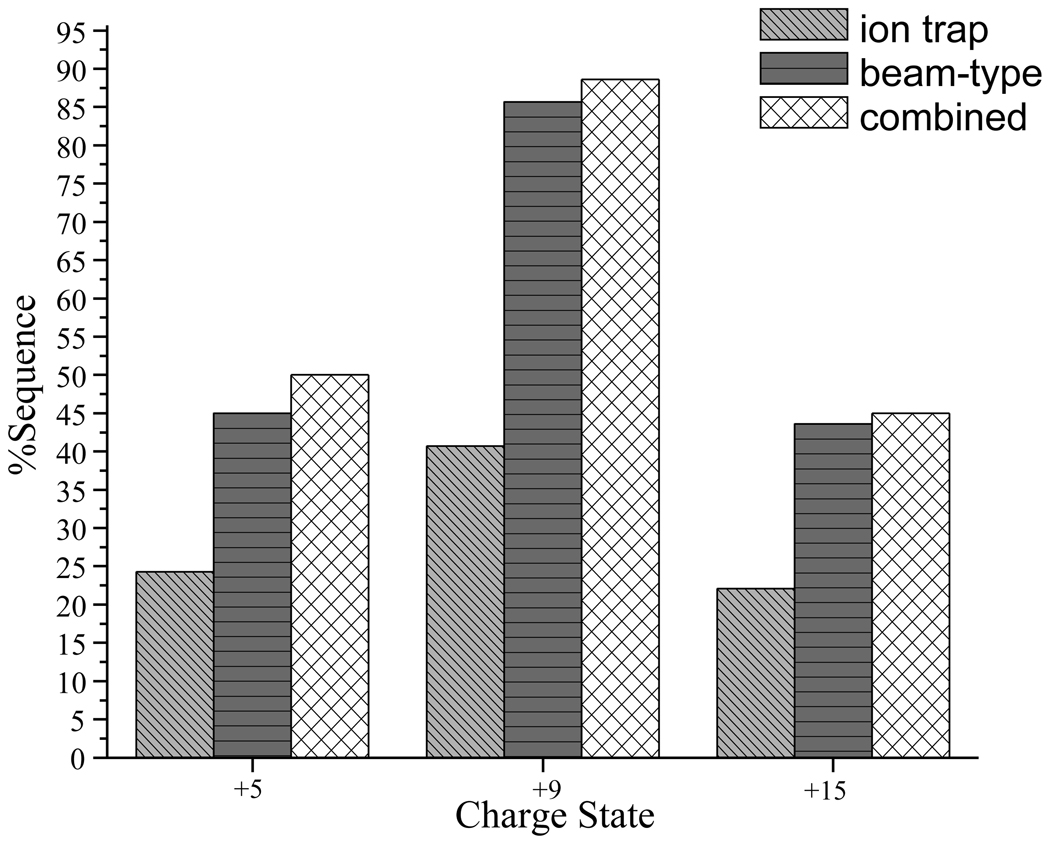
The normalized summed complementary b- and y-ion abundances from the [M+5H]5+, [M+9H]9+, and [M+15H]15+ precursor ions from ion trap and beam-type CID are compared in Figure 3. Beam-type CID of [M+5H]5+, [M+9H]9+, and [M+15H]15+ ions (see Figure 3 panels d–f) yielded %Sequence values of 45%, 86%, and 44%, respectively, while ion trap CID yielded values of 27%, 41%, and 22%, respectively. (A signal-to-noise ratio of ≥ 3 was used as the abundance criterion for assigning a label to a product ion.) The sequence information available from ion trap CID is largely reproduced in the beam-type CID data while the latter contains additional sequence information, as summarized in Figure 6. For both ion trap CID and beam-type CID data, the highest %Sequence (~ 40% from ion trap CID, ~ 80% for beam-type CID) was noted for intermediate charge states (+6 to +11). Generally, approximately twice as much sequence coverage was found for each of the charge states in the beam-type activation experiments, with the highest (~86%) found for the [M+9H]9+ ion. Fewer cleavages were found for the high charge state and low charge state groups (~ 40–45%), presumably due to Coulombic repulsion in high charge states and proton sequestration in low charge states, as mentioned above.
As demonstrated above, the sequence information that can be obtained from different charge states is often complementary (see below). In the case of ion trap CID, the %Sequence value derived from information determined from all of the charge states examined was 72% (i.e., 101 backbone cleavages were represented when all precursor ion charge states were considered) while from beam-type CID, the %Sequence from all charge states was 96% (135 backbone cleavages represented). Figure 8 summarizes the specific backbone cleavages noted for each type of collisional activation, with the major channels indicated in bold. Clearly, more channels are accessed by beam-type CID, which is consistent with the fact that the beam experiment samples higher rate dissociation processes than the ion trap CID experiment and, therefore, samples ions of somewhat higher internal energies.
Figure 8.
Combined backbone cleavages resulting from (a) the ion trap CID of α-synuclein ions and from (b) the beam-type CID of α-synuclein ions. The specific cleavages (major peaks observed in spectra) are indicated by bold line for emphasis.
4. Conclusions
The charge state dependent fragmentation behavior of protonated α-synuclein ions from [M+17H]17+ to [M+4H]4+ has been examined by tandem mass spectrometry using both ion trap collisional activation and beam-type collisional activation in a quadrupole/time-of-flight (QqTOF) tandem mass spectrometer. The maximum number of amide bond cleavages is found for intermediate charge states with prominent contributions from cleavages C-terminal to aspartic acid residues and N-terminal to proline residues. Beam-type CID samples higher rate dissociation processes, which arise from higher internal energy ions. As a result, more amide bond cleavages can be accessed. Essentially for every charge state, evidence for the cleavage of twice as many amide bonds was noted from beam-type CID relative to ion trap CID. Remarkably, evidence for cleavage of 86% of the amide bonds is noted for beam-type CID of the [M+9H]9+ precursor ion. A similar diversity of cleavages was noted for the [M+8H]8+ ion as well. When information from beam-type CID of all charge states is considered, evidence for cleavage of 96% of the amide bonds is observed. These observations are qualitatively similar to those noted in the analogous comparison of activation conditions made for protonated ubiquitin [34]. However, greater sequence coverages were noted for the multiply protonated α-synuclein ions. These findings are particularly relevant for top-down protein ion characterization. Beam-type CID appears to be superior to slow heating methods for generating extensive sequence coverage for multiply protonated proteins, at least for proteins up to 14–15 kDa. The fact that moderate to low charge states provide the greatest extent of structurally informative fragmentation is also noteworthy in that this situation stands in contrast to ECD and ETD techniques, which generally provide more information from the highest charge states. The ion parking technique is well-suited for converting high charge states to lower charge states in a selective fashion and can therefore be useful in concentrating precursor ion signal into charge states most likely to provide the most extensive sequence information.
Figure 7.
Percent sequence coverage observed for α-synuclein ions from a representative of each charge state group ([M+5H]5+, [M+9H]9+, and [M+15H]15+) under ion trap CID and beam-type CID conditions as well as the sequence coverage associated with combining information from both activation conditions.
Acknowledgment
The authors acknowledge the support from the National Institutes of Health under Grant GM 45372.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aebersold R, Goodlett DR. Chem. Rev. 2001;101:269. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- 2.Hu L, Ye M, Jiang X, Feng S, Zou H. Anal. Chim. Acta. 2007;598:193. doi: 10.1016/j.aca.2007.07.046. [DOI] [PubMed] [Google Scholar]
- 3.Fournier ML, Gilmore JM, Martin-Brown SA, Washburn MP. Chem. Rev. 2007;107:3654. doi: 10.1021/cr068279a. [DOI] [PubMed] [Google Scholar]
- 4.Henzel WJ, Billeci TM, Stults JT, Wong SC, Grimley C, Watanabe C. Proc. Nat. Acad. Sci. USA. 1993;90:5011. doi: 10.1073/pnas.90.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yates JR., III Electrophoresis. 1998;19:893. doi: 10.1002/elps.1150190604. [DOI] [PubMed] [Google Scholar]
- 6.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. J. Am. Chem. Soc. 1999;121:806. [Google Scholar]
- 7.Kelleher NL. Anal. Chem. 2004;76:196A. [PubMed] [Google Scholar]
- 8.McLafferty FW, Breuker K, Jin M, Han XM, Infusini G, Jiang H, Kong XL, Begley TP. FEBS Journal. 2007;274:6256. doi: 10.1111/j.1742-4658.2007.06147.x. [DOI] [PubMed] [Google Scholar]
- 9.Reid GE, McLuckey SA. J. Mass Spectrom. 2002;37:663. doi: 10.1002/jms.346. [DOI] [PubMed] [Google Scholar]
- 0.Han XM, Jin M, Breuker K, McLafferty FW. Science. 2006;314:109. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 1.McLuckey SA, Goeringer DE. J. Mass Spectrom. 1997;32:461. [Google Scholar]
- 2.Senko MW, Speir JP, McLafferty FW. Anal. Chem. 1994;66:2801. doi: 10.1021/ac00090a003. [DOI] [PubMed] [Google Scholar]
- 3.Marshall AG, Hendrickson CL, Jackson GS. Mass Spectrom. Rev. 1998;17:1. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 4.Little DP, Speir JP, Senko MW, O’Connor PB, McLafferty FW. Anal. Chem. 1994;66:2809. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 5.Goeringer DE, McLuckey SA. J. Chem. Phys. 1996;104:2214. [Google Scholar]
- 6.March RE, Todd JFJ. Quadrupole ion trap mass spectrometry. 2nd ed. Hoboken, N.J: Wiley-Interscience; 2005. ISBN-13 978-0-471-48888-0. [Google Scholar]
- 7.McLuckey SA, Goeringer DE. J.Mass Spectrom. 1997;32:461. [Google Scholar]
- 18.De Hoffmann E, Stroobant V. Mass Spectrometry: Principles and Applications. 2nd ed. Hoboken, NJ: John Wiley & Sons Inc.; 2002. ISBN-10 0-471-48566-7. [Google Scholar]
- 9.Guan ZQ, Kelleher NL, O’Connor PB, Aaserud DJ, Little DP, McLafferty FW. Int. J. Mass Spectrom. Ion Processes. 1996;158:357. [Google Scholar]
- 20.Williams ER, Henry KD, McLafferty FW, Shabanowitz J, Hunt DF. J. Am. Soc. Mass Spectrom. 1990;1:413. doi: 10.1016/1044-0305(90)85022-E. [DOI] [PubMed] [Google Scholar]
- 21.Loo JA, Edmonds CG, Smith RD. Anal. Chem. 1991;63:2488. doi: 10.1021/ac00021a018. [DOI] [PubMed] [Google Scholar]
- 22.Loo JA, Edmonds CG, Smith RD. Anal. Chem. 1993;65:425. doi: 10.1021/ac00052a020. [DOI] [PubMed] [Google Scholar]
- 23.Loo JA, Edmonds CG, Smith RD. Science. 1990;248:201. doi: 10.1126/science.2326633. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Schey KL. J. Am. Soc. Mass Spectrom. 2008;19:231. doi: 10.1016/j.jasms.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zubarev RA, Kelleher NL, McLafferty FW. J. Am. Chem. Soc. 1998;120:3265. [Google Scholar]
- 26.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc. Nat. Acad. Sci. USA. 2004;101:9528. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chrisman PA, McLuckey SA. J. Proteome Res. 2002;1:549. doi: 10.1021/pr025561z. [DOI] [PubMed] [Google Scholar]
- 28.Stephenson JL, Jr, Cargile BJ, McLuckey SA. Rapid Commun. Mass Spectrom. 1999;13:2040. doi: 10.1002/(SICI)1097-0231(19991030)13:20<2040::AID-RCM754>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 29.Engel BJ, Pan P, Reid GE, Wells JM, McLuckey SA. Int. J. Mass Spectrom. 2002;219:171. doi: 10.1016/S1044-0305(01)00275-6. [DOI] [PubMed] [Google Scholar]
- 30.Hogan JM, McLuckey SA. J. Mass Spectrom. 2003;38:245. doi: 10.1002/jms.458. [DOI] [PubMed] [Google Scholar]
- 31.Watson DJ, McLuckey SA. Int. J. Mass Spectrom. 2006;255–256:53. [Google Scholar]
- 32.Newton KA, Chrisman PA, Wells JM, Reid GE, McLuckey SA. Int. J. Mass Spectrom. 2001;212:359. doi: 10.1002/rcm.512. [DOI] [PubMed] [Google Scholar]
- 33.Yu W, Vath JE, Huberty MC, Martin SA. Anal. Chem. 1993;65:3015. doi: 10.1021/ac00069a014. [DOI] [PubMed] [Google Scholar]
- 34.Xia Y, Liang X, McLuckey SA. Anal. Chem. 2006;78:1218. doi: 10.1021/ac051622b. [DOI] [PubMed] [Google Scholar]
- 35.Bortolus M, Tombolato F, Tessari I, Bisaglia M, Mammi S, Bubacco L, Ferrarini A, Maniero AL. J. Am. Chem. Soc. 2008;130:6690. doi: 10.1021/ja8010429. [DOI] [PubMed] [Google Scholar]
- 36.Carvajal P, Lanier T. The Unfolded Protein State Revisited. In: Pollack GH, Cameron IL, Wheatley DN, editors. Water and the Cell. The Netherlands: Springer; 2006. pp. 235–252. [Google Scholar]
- 37.Qin Z, Hu D, Han S, Hong D-P, Fink AL. Biochemistry. 2007;46:13322. doi: 10.1021/bi7014053. [DOI] [PubMed] [Google Scholar]
- 38.Rao JN, Dua V, Ulmer TS. Biochemistry. 2008;47:4651. doi: 10.1021/bi8002378. [DOI] [PubMed] [Google Scholar]
- 39.Xie Y, Zhang J, Yin S, Loo JA. J. Am. Chem. Soc. 2006;128:14432. doi: 10.1021/ja063197p. [DOI] [PubMed] [Google Scholar]
- 40.deLaureto PP, Tosatto L, Frare E, Marin O, Uversky VN, Fontana A. Biochemistry. 2006;45:11523. doi: 10.1021/bi052614s. [DOI] [PubMed] [Google Scholar]
- 41.Mirzaei H, Schieler JL, Rochet J-C, Regnier F. Anal. Chem. 2006;78:2422. doi: 10.1021/ac051978n. [DOI] [PubMed] [Google Scholar]
- 42.Ulmer TS, Bax A, Cole NB, Nussbaum RL. J. Biol. Chem. 2005;280:9595. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 43.Uversky VN, Fernández A, Fink AL. Structural and Conformational Prerequisites of Amyloidogenesis. In: Uversky VN, Fink AL, editors. Protein Reviews, Vol. 4 Protein Misfolding, Aggregation, and Conformational Diseases. Springer US; 2006. pp. 1–20. [Google Scholar]
- 44.Williams RB, Gutekunst WR, Joyner PM, Duan W, Li Q, Ross CA, Williams TD, Cichewicz RH. J. Agric. Food Chem. 2007;55:9450. doi: 10.1021/jf072241x. [DOI] [PubMed] [Google Scholar]
- 45.Xia Y, Liang X, McLuckey SA. J. Am. Soc. Mass Spectrom. 2005;16:1750. doi: 10.1016/j.jasms.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 46.Shevchenko A, Chernusevich I, Ens W, Standing KG, Thomson B, Wilm M, Mann M. Rapid Commun. Mass Spectrom. 1997;11:1015. doi: 10.1002/(SICI)1097-0231(19970615)11:9<1015::AID-RCM958>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 47.Xia Y, Chrisman PA, Erickson DE, Liu J, Liang X, Londry FA, Yang MJ, McLuckey SA. Anal. Chem. 2006;78:4146. doi: 10.1021/ac0606296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang X, McLuckey SA. J. Am. Soc. Mass Spectrom. 2007;18:882. doi: 10.1016/j.jasms.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chrisman PA, Pitteri SJ, McLuckey SA. Anal. Chem. 2006;78:310. doi: 10.1021/ac0515778. [DOI] [PubMed] [Google Scholar]
- 50.Xia Y, Thomson BA, McLuckey SA. Anal. Chem. 2007;79:8199. doi: 10.1021/ac071448m. [DOI] [PubMed] [Google Scholar]
- 51.Han H, Xia Y, McLuckey SA. Rapid Commun. Mass Spectrom. 2007;21:1567. doi: 10.1002/rcm.2994. [DOI] [PubMed] [Google Scholar]
- 52.Dongré AR, Jones JL, Somogyi A, Wysocki VH. J. Am. Chem. Soc. 1996;118:8365. [Google Scholar]
- 53.Nair H, Somogyi A, Wysocki VH. J. Mass Spectrom. 1996;31:1141. doi: 10.1002/(SICI)1096-9888(199610)31:10<1141::AID-JMS402>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 54.Wysocki VH, Tsaprailis G, Smith LL, Breci LA. J. Mass Spectrom. 2000;35:1399. doi: 10.1002/1096-9888(200012)35:12<1399::AID-JMS86>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 55.Baizs B, Suhai S. Mass Spectrom. Rev. 2005;24:508. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]