Abstract
Fanconi anemia (FA) is an autosomal recessive disorder characterized by congenital abnormalities, bone marrow failure, chromosome fragility, and cancer susceptibility. At least eleven members of the FA gene family have been identified using complementation experiments. Ubiquitin-proteasome has been shown to be a key regulator of FA proteins and their involvement in the repair of DNA damage. Here, we identified a novel functional link between the FA/BRCA pathway and E2F-mediated cell cycle regulome. In silico mining of a transcriptome database and promoter analyses revealed that a significant number of FA gene members were regulated by E2F transcription factors, known to be pivotal regulators of cell cycle progression – as previously described for BRCA1. Our findings suggest that E2Fs partly determine cell fate through the FA/BRCA pathway.
Keywords: E2F, Fanconi anemia, cell cycle, DNA damage/repair, transcriptional regulation, bioinformatics
Introduction
Fanconi anemia (FA) is an autosomal recessive disorder characterized by congenital abnormalities, bone marrow failure, chromosome fragility, and cancer susceptibility (Joenje and Patel, 2001). To date, at least eleven members of this family, including FANCA, FANCB, FANCC, FANCD1 (alias BRCA2), FANCD2, FANCE, FANCF, FANCG (alias XRCC9), FANCJ, FANCL, and FANCM (Joenje and Patel 2001), have been identified using complementation experiments. Among them, eight FA-associated genes have been identified so far (FANCA, FANCC, FANCD1, FANCD2, FANCE, FANCF, FANCG, and FANCL (Strathdee et al. 1992; Joenje and Patel, 2001; Timmers et al. 2001; Howlett et al. 2002; Meetei et al. 2003). The FANCA, FANCC, FANCE, FANCF, FANCG, and FANCL proteins are part of a nuclear multiprotein core complex that triggers the monoubiquitination of the FANCD2 protein during S phase of the cell cycle and after exposure to DNA crosslinking agents (Garcia-Higuera et al. 2001; Meetei et al. 2003; Soulier et al. 2005). Monoubiquitinated FANCD2 colocalizes in nuclear foci with BRCA1, FANCD1, and NBS1 (Garcia-Higuera et al. 2001; Howlett et al. 2002; Nakanishi et al. 2002). This evidence suggests a link between the FA protein complex and the cellular BRCA1 repair machinery. Therefore, the FA/BRCA pathway is thought to be involved in the repair of DNA damage to maintain genomic integrity (D’Andrea and Grompe, 2003). The cellular targets of the FA/BRCA pathway remain unknown.
A remarkably high clinical variability exists among FA patients (Auerbach et al. 2001; Alter, 2003). Besides the ubiquitination of FA proteins, other regulatory mechanisms may affect the expression level of FA proteins; such regulatory mechanisms may contribute to the clinical variability observed among FA patients. Bioinformatics is a useful tool for integrating complex gene functions. It also allows the establishment of biological frameworks or “system biology”. It is believed to connect between gene regulation and phenotypes. Our current standard concept for gene regulation can be roughly divided into two aspects; namely, gene transcription and protein degradation. If the significant regulators of gene transcription or protein degradation for each component of complex gene networks could be revealed, one could easily imagine the gene network and extract the phenotypes of cells or model organisms. As a logical consequence, system biology views are an unavoidable necessity for current biology studies.
We have focused on the E2F transcription factor-regulated transcriptome because E2F family members integrate the upstream signals to the downstream target genes during the monitoring of proper cell cycle progression. Indeed, the downstream target genes of E2F1 can be divided into several categories including DNA replication, DNA damage/repair, apoptosis, differentiation, and development (Bracken et al. 2004). Evidence that E2F family members regulate BRCA1 expression and interact with BRCA1 protein (Wang et al. 2000; Oberley et al. 2003; Bindra and Glazer, 2006) prompted us to investigate comprehensively the relationship between E2Fs and FA genes.
In the present study, we evaluated the possible link between E2F transcription factors and FA genes by analyzing the FA gene promoters in silico followed by a luciferase-based promoter assay. These analyses, combined with a public microarray data search, allowed us to identify a novel aspect of the FA pathway that is partially regulated in a cell cycle-dependent manner via E2Fs. The discovery that both FA genes and BRCA1 are under the control of E2Fs suggests that the FA/BRCA pathway is an effector of E2F-regulated cell cycle progression and DNA damage/repair signaling. Comprehensive regulome analyses of the FA gene using cell cycle-associated transcriptional factors may enable us to open a new window onto the system biology of FA genes.
Materials & Methods
Bioinformatics
The E2F1 gene expression profile identified using adenovirus-mediated gene transfer in SKMEL-2 melanoma cells (Affymetrix GeneChip analysis (Jamshidi-Parsian et al. 2005)) was deposited in the Gene Expression Omnibus (GEO), which is maintained by The National Center for Biotechnology Information (NCBI, NIH) (http://www.ncbi.nlm.nih.gov/). By searching the database, FANCA (Probe No. 33145), FANCC (Probe Nos. 35713, 1982_s, and 160034_s), FANCD1 (Probe Nos. 1503, 1989, and 1990_g), FANCG (Probe No. 37584), and FANCL (Probe No. 33125) mRNAs were identified (Accession No. GDS1078; Platform No. GPL91). The expression profile suggested by an analysis of the EST (expressed sequence tag) counts in human tissues and organs was searched using the UniGene database (EST Profile Viewer, NCBI, NIH). The data sets used for the FA genes are shown in Table 1. The number of transcripts per million was calculated based on the gene EST/total EST in the pool, and this value was exported to an Excel file. Transfac software (http://motif.genome.jp/) was then used to determine the E2F family of transcription factors binding-elements.
Table 1.
| Gene | UniGene | Mapping | Genomic clone | Primer sequences |
|---|---|---|---|---|
| FANCA | Hs.567267 | 16q24.3 | AC005565 | F: 5′-GCTTGGTTGGCCAGGTGGT-3′, R: 5′-ATGGCCTTGGCGCCTACAG-3′ |
| FANCB | Hs.554740 | Xp22.2 | AC140846 | F: 5′-AGGCCCTCAGCCTAGGTCC-3′, R: 5′-CAGCGGCAACATACCGGAG-3′ |
| FANCC | Hs.494529 | 9q22.3 | AL157384 | F: 5′-AAGAAGCCAGCGCCCCTTC-3′, R: 5′-TGGAATTTTCCCGCGGTCG-3′ |
| FANCD1 | Hs.34012 | 13q12.3 | AL445212 | F: 5′-CCACCCAAACATGAGCTGG-3′, R: 5′-CTCTGCCGCCTAGTTTCAG-3′ |
| FANCD2 | Hs.208388 | 3p26 | AC007999 | F: 5′-TGGGCGAGCTTCTCTTCAC-3′, R: 5′-ACTTTCCCGCCAGGCCCGA-3′ |
| FANCE | Hs.302003 | 6p22-p21 | AL022721 | F: 5′-CCCGACATCTCCCTTGAAAT-3′, R: 5′-TTGGGAGACCGGAGAAACCC-3′ |
| FANCF | Hs.632151 | 11p15 | AC103801 | F: 5′-AAGCGCGGAGACGTTCATG-3′, R: 5′-GCGATCCAGGTGCTGCAGA-3′ |
| FANCG | Hs.591084 | 9p13 | AC004472 | F: 5′-GTTAGTTAGGCTGCTTTAC-3′, R: 5′-TGGGTTCCCGCTTCCACCGA-3′ |
| FANCJ | Hs.532799 | 17q22-q24 | AC060798 | F: 5′-TGGATGCCGAAGTTCTCGCC-3′, R: 5′-GAAAGGGCACGAGCCCTTCC-3′ |
| FANCL | Hs.646789 | 2p16.1 | AC007250 | F: 5′-GGTCAATAAAGATGGGTAGG-3′, R: 5′-TGGGTCCTGCACATGCGCAG-3′ |
| FANCM | Hs.509229 | 14q21.3 | AL121809 | F: 5′-GAATGAGGCACGTTAGACGC-3′, R: 5′-AGCTCAACCGCTACGGTTCC-3′ |
Plasmids
The human FA promoter fragments were generated using polymerase chain reaction (PCR) from genomic DNA, and ligated into pGL3-Basic vectors (Promega, Madison, WI, U.S.A). PCR primers were designed to amplify 318-bp (−274/+44), 333-bp (−263/+70), 298-bp (−232/+66), 318-bp (−286/+32), 224-bp (−208/+16), 1330-bp (−1287/+43), 328-bp (−373/+37), 442-bp (−441/+1), 256-bp (−249/+7), 860-bp (−835/+25), and 410-bp (−732/−128) fragments of the human FANCA, FANCB, FANCC, FANCD1, FANCD2, FANCE, FANCF, FANCG, FANCJ, FANCL, and FANCM promoter sequences, respectively; the above numbering is relative to the transcription initiation site at +1, described in the UniGene Database (Genome View, NCBI, NIH). The GenBank Accession numbers of the genomic clones used for the designation of the PCR primers are shown in Table 1. A KpnI site was added to the forward primer and a BglII site was added to the reverse primer to facilitate the subcloning of the promoter regions of FANCA, FANCB, FANCC, FANCD1, FANCD2, FANCF, FANCG, FANCJ, FANCL, and FANCM, respectively. A KpnI site was added to the forward primer and a HindIII site was added to the reverse primer to facilitate the subcloning of the promoter region of FANCE. The following plasmids that were used have been previously described: pcDNA3-E2F1, E2F2, E2F3, E2F4, E2F5, and E2F6 (Yoshida and Inoue, 2004; Hayashi et al. 2006).
Cell culture and luciferase assay
HeLa and WI-38 cells were cultured in Earle’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, U.S.A) supplemented with 10% fetal bovine serum and antibiotics. Preparation of the adenovirus, the virus infection procedure, and the Western blot analysis were as described previously (Goto et al. 2006). For the promoter assay, 2 × 104 HeLa cells were transfected with FuGENE6, in accordance with the manufacturer’s instructions (Roche Diagnostic, Basel, Switzerland). Briefly, 400 ng of the expression plasmid, 200 ng of the firefly luciferase reporter plasmid (pGL3, Promega, Madison, WI, U.S.A.), and 0.6 ng of the Renilla luciferase reporter plasmid (pRL-TK, Promega, Madison, WI, U.S.A.) per 24-well dish were used for each transfection. The cells were harvested 24 hours after the transfection, and a luciferase assay was performed using the Dual-Luciferase Reporter Assay System, in accordance with the manufacturer’s protocol (Promega, Madison, WI, U.S.A.). As a control for transfection efficiency, the firefly luciferase activity values were normalized to the Renilla luciferase activity values. Data are presented as the mean values ± standard deviation.
RT (reverse transcriptase)-PCR
Total cellular RNA was extracted from the WI-38 cells using an RNeasy Mini Kit (Qiagen, Valencia, CA, U.S.A.), in accordance with the manufacturer’s instructions. The RT step was also performed according to the manufacturer’s directions (Invitrogen, Carlsbad, CA, U.S.A.). Briefly, 500 ng of extracted RNA, oligo(dT) primer, and 1 × annealing buffer were diluted in 8 μL of RNase/DNase-free water, heated to 65 °C for 5 minutes, and then chilled on ice. For the first-strand cDNA synthesis, a heat-denatured RNA solution, along with 2 × first-strand reaction mix and the SuperScript III/RNase OUT enzyme mix, was added to make up 20 μL of the reaction mixture, followed by incubation at 50 °C for 50 minutes, and then heating at 85 °C for 5 minutes and cooling on ice. The following primers were designed to amplify 300-bp long FANCC (NM_000136) cDNA: sense, 5′-gggcctctctcctgttctga-3′ and antisense, 5′-gaggtcagggcttccaggct-3′. PCR was then performed as follows: denaturation for 2 minutes at 94 °C, followed by 25–30 cycles at 94 °C for 15 seconds, 55 °C for 30 seconds, and 68 °C for 30 seconds. As a control, a GAPDH primer set was used (R&D Systems, Minneapolis, MN, U.S.A).
Results
Expression profiles of human FA mRNAs
To investigate the common expression patterns of the FA mRNA, we initially compared the abundance of ESTs among major human tissues and organs including the brain, heart, lung, liver, stomach, small intestine, colon, kidney, pancreas, testis, and ovary. In silico analysis revealed that the expression patterns of the FA mRNA differed considerably in human tissues and organs (Fig. 1). One characteristic feature of the expression patterns of FA genes was that FA genes were less expressed in the heart but were relatively well expressed in the stomach, colon, testis, and ovary. We speculated that the expression of FA genes tends to be enriched in proliferative tissues.
Figure 1.
Expression profiles of FA. A search was conducted based on the EST counts in human tissues and organs in the UniGene database (NCBI, NIH). The data sets used for the FA genes are listed in Table 1. The number of transcripts per million was calculated from the gene EST/total EST in the pool.
Next, we asked which transcription factor regulates the FA/BRCA pathway. The E2F family of transcription factors plays pivotal roles in the cell cycle progression and DNA damage repair pathways (Bracken et al. 2004). Therefore, we searched for FA mRNAs in public databases where the E2F family of transcription factors was either overexpressed or knocked down. A GEO database search revealed the deposition of microarray data for FANCA, FANCC, FANCD1, FANCG, and FANCL (see Materials & Methods). FANCA and FANCL mRNAs were clearly upregulated in E2F1 overexpressed SKMEL-2 melanoma cells, whereas FANCD1 and FANCG mRNAs were unchanged after E2F1 overexpression. On the other hand, the upregulation of FANCC mRNA by E2F1 was shown by one probe used in a microarray experiment but not by other probes. To substantiate the E2F1-regulated FANCC mRNA expression, human normal lung fibroblast WI-38 cells were infected with an adenovirus expressing E2F1. A Western blot analysis revealed that a faint band of endogenous E2F1 protein was detected in the control adenovirus-infected cell lysates, whereas significant amounts of exogenous E2F1 protein were detected in the E2F1 overexpressed cell lysates (Fig. 2, left panel). Twenty-four hours after the virus infection, the FANCC mRNA level was found to be upregulated, while the GAPDH mRNA level remained unchanged (Fig. 2, right panel). This result suggests that FANCC could be a downstream target gene of E2F1.
Figure 2.
E2F1 activates FANCC expression in WI-38 cells. (A) Cultures were infected with adenovirus encoding E2F1 or mock (control), and the E2F1 protein level was determined using a Western blot analysis with an anti-E2F1 antibody. Molecular weights (kDa) are shown on the left. The position of the E2F1 protein is indicated by the arrow on the right. RNA was extracted from WI-38 cells 24 hours after infection and analyzed using RT-PCR for FANCC and GAPDH.
Transcriptional regulation of human FA genes
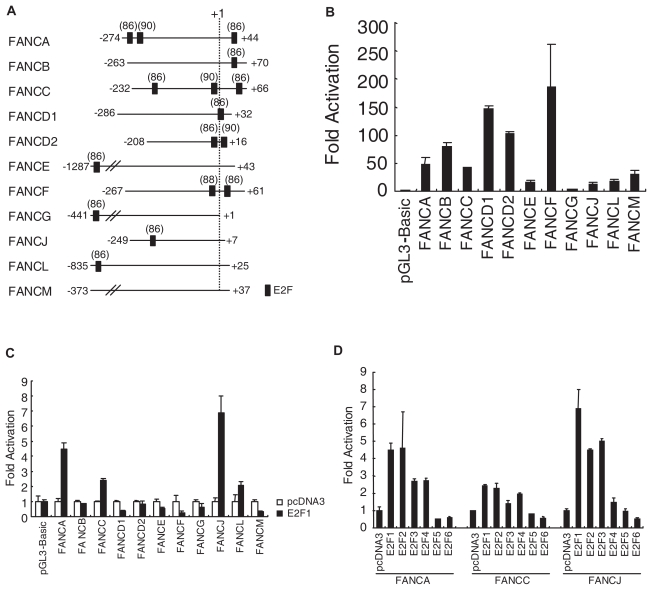
To gain more insight into the E2F-regulation of FA genes, we searched the proximal region of the transcription start site for potential cis-elements using Transfac software (ver. 4.0, cut off 85). We focused on the identification of E2F-binding consensus binding sequences within 1.5-kbp upstream and 0.5-kbp downstream of the transcription start site. At least one or up to three E2F-binding consensus sequences were identified for all FA genes except the FANCM gene (Fig. 3A).
Figure 3.
Transcriptional regulation of the human FA genes. (A) Schematic representation of the vicinity of the transcription start sites of the FA genes. The putative transcription factor E2F-binding sites (closed box) were searched within 1.5-kbp upstream and 0.5-kbp downstream of the transcription start sites, and are indicated with their scores (maximum score, 100) as calculated by the Transfac program. The transcription start sites (dotted lines) are indicated. The positions of the promoter constructs are numbered relative to the transcription initiation site at +1, as described in the NCBI UniGene database (NCBI, NIH). (B) FA promoter activities in asynchronously growing human cells. HeLa cells were transfected with 200 ng of reporter constructs and 400 ng of the expression vector for E2F1, together with 0.6 ng of pRL-TK. The pcDNA3 vector was used as a negative control. At 24 hours after the transfection, the cells were harvested and the extracts were prepared to measure the firefly and Renilla luciferase activities. Values are represented as the relative luciferase activities, with that of pGL3-Basic being defined as 1. (C) The E2F-binding motif(s) of the FANCA, FANCC, FANCJ, and FANCL promoters are sufficient to confer responses to ectopic E2F1 expression. The experiment was performed as described in (B). Values are represented as relative luciferase activities, with that of the control vector pcDNA3 being taken as 1. (D) Activation of the FA promoters by members of the E2F family. The experiment was performed as described in (B). Values are represented as the relative luciferase activities, with that of the control vector pcDNA3 being defined as 1.
To demonstrate the importance of the putative E2F-binding element for basal promoter activity, we generated promoter-luciferase constructs. These promoter constructs were used to study transient gene expression by transfecting them into HeLa cells and evaluating the firefly luciferase activities by measuring the chemiluminescence with a luminometer. The transfection efficiency was normalized by the dual luciferase assay, in which the corresponding Renilla luciferase activity upon co-transfection of the pRL-TK plasmid was also measured. As shown in Figure 3B, the FA promoter constructs used in this study showed various extents of increased activity, as determined by measuring the relative luciferase activities, when the activity of the control luciferase vector, pGL3-Basic, was defined as 1. The exogenous coexpression of E2F1 caused up to approximately 4.5-, 2.5-, and 7.0-fold increases in the FANCA, FANCC, and FANCJ promoter activities, respectively, compared to that of the pcDNA3 control vector (Fig. 3C). The promoter regions cloned for FANCL were slightly upregulated by the co-expression of E2F1. These results suggest that the E2F-binding motif(s) of the promoter constructs plays critical roles in the E2F1-mediated human FANCA, FANCC, and FANCJ promoter activities.
Next, we sought evidence to show that members of the E2F family transcriptionally regulated the FANCA, FANCC, and FANCJ genes. As shown in Figure 3D, the exogenous co-expression of E2F1 ~ E2F4 caused an increase in the human FANCA, FANCC, and FANCJ promoter constructs, whereas the co-expression of E2F5 or E2F6 was associated with no increase in promoter activity, compared to that in the pcDNA3 control vector. In contrast, the co-expression of E2F1 ~ E2F6 was associated with no increase in pGL3-Basic promoter activity (data not shown).
Discussion
Each FA has its own characteristic features, but their functions commonly belong to the same categories, such as DNA damage repair or S phase progression. In the present work, we investigated the common transcriptional regulatory factors that regulate FA genes. First of all, we examined the abundance of ESTs of FA in various human tissues and organs. This approach provided little information regarding the framework of the regulatory network of FA genes. Because we had been investigating the transcriptional network of E2F transcription factors, we noted that recent comprehensive gene expression profiling of the E2F transcriptome had pinpointed some FA genes under the E2F pathway (Fig. 4). Notably, a microarray approach revealed that FANCA could be regulated by E2Fs like E2F1, E2F2, and E2F3 (Vernell et al. 2003). By applying chromatin immunoprecipitation to isolate E2F4-bound sequences, the promoter regions of FANCD2, FANCG, and FANCL were shown to be potentially regulated by E2F4 (Weinmann et al. 2002; Cam et al. 2004). In addition, databases deposited in NCBI revealed that FANCA, FANCL, and probably FANCC genes were upregulated in E2F1-overexpressed SKMEL-2 melanoma cells. We demonstrated that the overexpression of E2F1 in human diploid primary fibroblast WI-38 cells upregulated FANCC mRNA expression. This evidence prompted us to examine the extent to which E2Fs contribute to the transcriptional regulation of FA genes.
Figure 4.
Model depicting the E2Fs-FA/BRCA axis in the control of cell fate. The model shown depicts the E2F-regulated expression of the FA/BRCA pathway as key determinants for cells entering into the DNA damage/repair pathway or the S phase of the cell cycle. Otherwise, cells go into apoptosis, which is exclusively regulated by E2F1 among the E2Fs. The promoter regions cloned for FANCA, FANCC, and FANCJ were identified as target sequences of E2Fs in the present study (surrounded by box). The regulation of FANCD2, FANCG, and FANCL as well as BRCA1 by E2Fs has also been previously reported or deposited in the public database (underlined). As posttranslational events, FANCA, FANCC, FANCE, FANCF, FANCG, and FANCL proteins trigger monoubiquitination of the FANCD2 protein during the S phase of the cell cycle and after DNA damage. Monoubiquitinated FANCD2 colocalizes in nuclear foci with BRCA1, FANCD1, and NBS1.
We prospectively analyzed the promoter regions of the eleven known FA genes using an in silico determination of the putative E2F1 consensus site and promoter analysis based on a luciferase reporter assay. From these studies, the promoter regions cloned for FANCA, FANCC, and FANCJ were found to be upregulated by the co-expression of E2F1; this evidence enabled us to propose a novel gene regulatory network that couples the E2F1 and FA/BRCA pathways (Fig. 4). FANCD2, FANCG, and FANCL may have E2F-responsive sites other than the region used in this study. Promoter analyses for mutations and the methylation status of FA genes have been well characterized; these statuses are known to affect the FA/BRCA pathway (Taniguchi et al. 2003). Among FA genes, the promoter of FANCC has been cloned and shown to be regulated by p53 (Savoia et al. 1995; Liebetrau et al. 1997). Once functional links between the E2F1 and FA/BRCA pathways have been established, the complete system biology of the FA/BRCA pathway could be analyzed by examining their promoter regulation by cell cycle-associated key transcriptional regulators, including p53.
BRCA1, a familial breast and ovarian cancer susceptibility gene, encodes nuclear phosphoproteins that function as tumor suppressors in human breast cancer cells. BRCA1 serves as an important negative regulator of the cell cycle through its interaction with E2F transcriptional factors and phosphorylation by cyclins/cdk (cyclin-dependent kinase) complexes (Wang et al. 1997). Moreover, the regulation of BRCA1 expression by the pRb (retinoblastoma protein)/E2Fs pathway has been extensively characterized (Wang et al. 2000; Oberley et al. 2003; Bindra and Glazer, 2006). The FA/BRCA pathway may be a pivotal effector regulated by activator-E2F signaling under the specific circumstance of DNA damage. Considering the predisposition to neoplasia, pRb mutations were expected to result in activator E2F overexpression; the subsequent expression of FA proteins might then compromise DNA damage, preventing the cells from progressing into a cancer phenotype.
In conclusion, we found that the FA/BRCA pathway is regulated by activator E2Fs responsible for the execution of the DNA damage/repair pathway. Most importantly, this pathway enables mechanistic links between E2F1 and FA genes, illuminating the molecular basis of DNA damage/repair and S phase progression. We propose that the present analysis might be used as a research working model to approach system biology, in combination with in silico and functional analyses, for a comprehensive characterization of cellular events in any given organism.
Acknowledgements
This study was supported in part by grants from The Ministry of Health, Labour and Welfare of Japan (Research on Toxicogenomics) and the Sato Memorial Foundation for Cancer Research.
References
- Alter BP. Inherited bone marrow failure syndromes. Philadelphia: Saunders; 2003. [Google Scholar]
- Auerbach AD, Buchwald M, Joenje H. Fanconi anemia. New York: McGrawHill; 2001. [Google Scholar]
- Bindra RS, Glazer PM. Basal Repression of BRCA1 by Multiple E2Fs and Pocket Proteins at Adjacent E2F Sites. Cancer Biol Ther. 2006;5:1400–1407. doi: 10.4161/cbt.5.10.3454. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, et al. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Cam H, Balciunaite E, Blais A, et al. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- D’andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Goto Y, Hayashi R, Kang D, et al. Acute loss of transcription factor E2F1 induces mitochondrial biogenesis in HeLa cells. J Cell Physiol. 2006;209:923–934. doi: 10.1002/jcp.20802. [DOI] [PubMed] [Google Scholar]
- Hayashi R, Goto Y, Ikeda R, et al. CDCA4 is an E2F transcription factor family-induced nuclear factor that regulates E2F-dependent transcriptional activation and cell proliferation. J Biol Chem. 2006;281:35633–35648. doi: 10.1074/jbc.M603800200. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Jamshidi-Parsian A, Dong Y, Zheng X, et al. Gene expression profiling of E2F-1-induced apoptosis. Gene. 2005;344:67–77. doi: 10.1016/j.gene.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001;2:446–457. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- Liebetrau W, Budde A, Savoia A, et al. p53 activates Fanconi anemia group C gene expression. Hum Mol Genet. 1997;6:277–283. doi: 10.1093/hmg/6.2.277. [DOI] [PubMed] [Google Scholar]
- Meetei AR, DE Winter JP, Medhurst AL, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Taniguchi T, Ranganathan V, et al. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat Cell Biol. 2002;4:913–920. doi: 10.1038/ncb879. [DOI] [PubMed] [Google Scholar]
- Oberley MJ, Inman DR, Farnham PJ. E2F6 negatively regulates BRCA1 in human cancer cells without methylation of histone H3 on lysine 9. J Biol Chem. 2003;278:42466–42476. doi: 10.1074/jbc.M307733200. [DOI] [PubMed] [Google Scholar]
- Savoia A, Centra M, Ianzano L, et al. Characterization of the 5′ region of the Fanconi anaemia group C (FACC) gene. Hum Mol Genet. 1995;4:1321–1326. doi: 10.1093/hmg/4.8.1321. [DOI] [PubMed] [Google Scholar]
- Soulier J, Leblanc T, Larghero J, et al. Detection of somatic mosaicism and classification of Fanconi anemia patients by analysis of the FA/BRCA pathway. Blood. 2005;105:1329–1336. doi: 10.1182/blood-2004-05-1852. [DOI] [PubMed] [Google Scholar]
- Strathdee CA, Gavish H, Shannon WR, et al. Cloning of cDNAs for Fanconi’s anaemia by functional complementation. Nature. 1992;356:763–767. doi: 10.1038/356763a0. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Tischkowitz M, Ameziane N, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- Timmers C, Taniguchi T, Hejna J, et al. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell. 2001;7:241–248. doi: 10.1016/s1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- Vernell R, Helin K, Muller H. Identification of target genes of the p16INK4A-pRB-E2F pathway. J Biol Chem. 2003;278:46124–46137. doi: 10.1074/jbc.M304930200. [DOI] [PubMed] [Google Scholar]
- Wang A, Schneider-Broussard R, Kumar AP, et al. Regulation of BRCA1 expression by the Rb-E2F pathway. J Biol Chem. 2000;275:4532–4536. doi: 10.1074/jbc.275.6.4532. [DOI] [PubMed] [Google Scholar]
- Wang H, Shao N, Ding QM, et al. BRCA1 proteins are transported to the nucleus in the absence of serum and splice variants BRCA1a, BRCA1b are tyrosine phosphoproteins that associate with E2F, cyclins and cyclin dependent kinases. Oncogene. 1997;15:143–157. doi: 10.1038/sj.onc.1201252. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Yan PS, Oberley MJ, et al. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Inoue I. Regulation of Geminin and Cdt1 expression by E2F transcription factors. Oncogene. 2004;23:3802–3812. doi: 10.1038/sj.onc.1207488. [DOI] [PubMed] [Google Scholar]