Abstract
During the past decade elevated phospholipase D (PLD) activity has been reported in virtually all cancers where it has been examined. PLD catalyzes the hydrolysis of phosphatidylcholine to generate the lipid second messenger phosphatidic acid (PA). While many targets of PA signaling have been identified, the most critical target of PA in cancer cells is likely to be mTOR – the mammalian target of rapamycin. mTOR has been widely implicated in signals that suppress apoptotic programs in cancer cells – frequently referred to as survival signals. mTOR exists as two multi-component complexes known as mTORC1 and mTORC2. Recent data has revealed that PA is required for the stability of both mTORC1 and mTORC2 complexes – and therefore also required for the kinase activity of both mTORC1 and mTORC2. PA interacts with mTOR in a manner that is competitive with rapamycin, and as a consequence, elevated PLD activity confers rapamycin resistance – a point that has been largely overlooked in clinical trials involving rapamycin-based strategies. The earliest genetic changes occurring in an emerging tumor are generally ones that suppress default apoptotic programs that likely represent the first line of defense of cancer. Targeting survival signals in human cancers represents a rational anti-cancer therapeutic strategy. Therefore, understanding the signals that regulate PA levels and how PA impacts upon mTOR could be important for developing strategies to de-repress the survival signals that suppress apoptosis. This review summarizes the role of PA in regulating the mTOR-mediated signals that promote cancer cell survival.
Keywords: Phospholipase D, Phosphatidic acid, mTOR, Rapamycin, Survival signals, Cancer
1. Introduction
During the past fifteen years many studies have shown that in response to mitogenic signals and activated oncoproteins there is an increase in phospholipase D (PLD) activity. PLD activity is elevated in response to several growth factors including epidermal growth factor (EGF) [1], platelet-derived growth factor [2], fibroblast growth factor [3,4], insulin [5], insulin-like growth factor 1 [6], and vascular endothelial growth factor [7]. In addition, PLD activity is elevated in response to the oncoproteins v-Src [8], v-Ras [9, 10], v-Fps [11], and v-Raf [12]. The elevated PLD activity in cells transformed by the Ras oncoprotein is required for cell transformation [13]. In addition, elevated expression of either c-Src or the EGF receptor, in combination with elevated expression of PLD, transforms rat fibroblasts [14–16]. PLD has also been reported to induce anchorage-independent growth and enhance cell cycle progression of mouse fibroblasts [17]. These studies reveal elevated PLD activity is correlated with mitogenic and oncogenic signals.
Elevated PLD expression has also been shown to prevent cell cycle arrest and apoptosis. High intensity Raf signaling induces cell senescence [18, 19] or, if cells are deprived of serum, apoptosis [20]. Elevated expression of PLD suppressed the apoptosis induced by high intensity Raf signals [20]. Similarly, rat fibroblasts overexpressing c-Src undergo apoptosis in response to growth factor deprivation, and elevated PLD expression suppressed this apoptosis [21]. These early studies in rodent fibroblasts indicate that in addition to enhancing cell proliferation, PLD is required for the “survival signals” that suppress default apoptotic programs.
The studies implicating PLD in mitogenic signaling and the suppression of apoptosis suggest that PLD activity might be a factor in human cancer where mitogenic signals are constitutively active and suppression of apoptotic programs is critical. Consistent with this hypothesis, elevated PLD activity and expression has been reported in a variety of human cancer tissues including breast, gastric, kidney, and colon [22–25]. In addition, elevated PLD activity has been reported in several human cancer cell lines including those derived from breast, lung, bladder, pancreatic, and kidney cancers [26–30]. Importantly, the elevated PLD activity in these cells was shown to be critical for suppressing apoptosis in these cell lines. Thus, elevated PLD activity in human cancers is likely a critical aspect of tumorigenesis that promotes cell proliferation and suppresses the default apoptotic programs that prevent cancer.
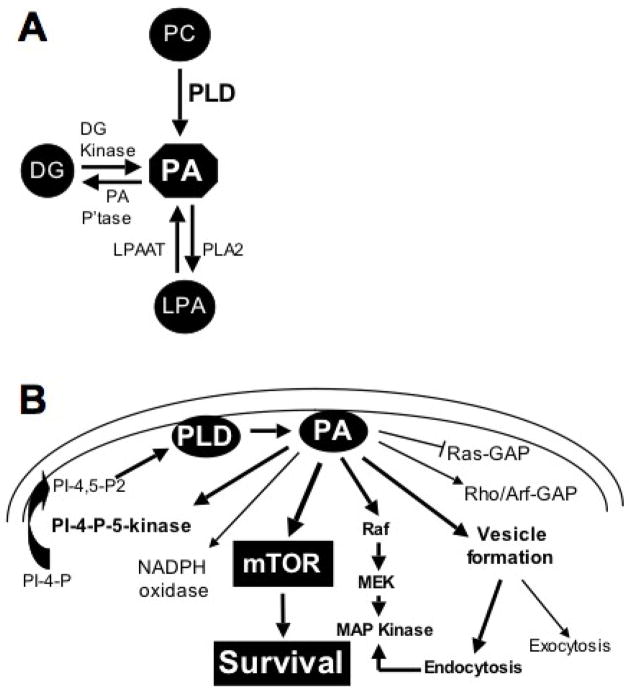
There are two mammalian PLD isoforms – PLD1 and PLD2 – the distinct functions of which are poorly understood [31]. Both catalyze the hydrolysis of phosphatidylcholine to phosphatidic acid (PA) and choline. PA is a central node for lipid signaling and can be generated from lysophosphatidic acid (LPA) and diacylglycerol (DG) as well from phosphatidylcholine (Fig. 1A) – although it is likely that the most relevant source in cancer cells is via the PLD-mediated hydrolysis of phosphatidylcholine [31]. PA is also metabolically converted to the lipid second messengers LPA and DG (Fig. 1A). However, while DG and LPA have important second messenger function, there are several targets of PA have been identified (Fig. 1B) and it is believed that the most significant effects of elevated PLD activity are mediated by targets of PA. Significantly, these include Raf [32] and the mammalian target of rapamycin (mTOR) [33] – both of which are commonly dysregulated in human cancers. mTOR, like PLD, has been implicated in cancer cell survival signals [34–36]. While the role of PA in regulating mTOR has been controversial [37], recent reports have strongly implicated PLD and its metabolite PA in the regulation of both mTOR complexes – mTORC1 and mTORC2 [38–40]. This review summarizes the role of PLD in the regulation of mTOR and the potential of combining strategies for targeting PLD and mTOR signals in human cancer.
Fig. 1.
Signaling through phosphatidic acid. A. PA is a central node in lipid second messenger signaling. It is generated primarily from phosphatidylcholine (PC) by a hydrolysis reaction that releases the choline head group. PA can also be generated from diacylglycerol (DG) through by a phosphorylation reaction catalyzed by DG kinase; and from lysophosphatidic acid (LPA) via an acylation reaction catalyzed by LPA acyl-transferase (LPAAT). PA can also be converted to both DG and LPA by PA phosphatase (PA P’tase) and a type 2 phospholipase (PLA2). B. PA has many downstream targets. PA has been reported to regulate GTPase activating proteins (GAPs) for Ras, Rho and Arf GTPases; PA also activates an NADPH oxidase [31]. PA activates phosphatidylinositol-4-phosphate (PI-4-P)-5-kinase [31], which may create a positive feedback loop to generate the PI-4,5-bis-phosphate (PI-4,5-P2) required for both PLD1 and PLD2 activity. The likely relevant targets for survival signals provided by PLD and PA are Raf and mTOR, which regulate progression through the cell cycle and suppress apoptosis [32,37]. PA is also required for endocytosis, which along with Raf, contributes to the activation of MAP kinase [73].
2. The PLD-mTOR connection
2.1. PA competes with rapamycin for binding to mTOR
The first report directly linking PLD activity with mTOR was the discovery that PA is required for mTOR activity [33]. PA was shown to bind the FKBP12-rapamycin binding (FRB) domain of mTOR in a manner that is competitive with rapamycin-FKBP12 [33]. Consistent with a competition between PA and rapamycin-FKBP12 for mTOR, it was subsequently demonstrated that elevated levels of PLD activity conferred rapamycin resistance in human breast cancer cell lines [42]. The recently reported NMR structure of the mTOR FRB domain bound to PA revealed an interaction between Arg2109 in the FRB domain and the phosphate group on PA [43] and was consistent with the model proposed by Chen and colleagues [33, 44]. Arg2109 is at the end of an α-helical section of the FRB domain that is adjacent to another α-helix. The groove in between the two α-helices forms a hydrophobic pocket where rapamycin binds mTOR [43]. The glycerol backbone and proximal aliphatic components of PA are proposed to insert into this pocket. Thus, there is an emerging theory that the mechanism by which rapamycin inhibits mTOR is by preventing the interaction between mTOR and PA.
2.2. PA is required for stabilization of mTOR complexes
mTOR exists in two complexes – mTORC1 and mTORC2 [45, 46]. While a role for PA in the regulation of mTORC1 has been widely reported [37], a role for PA in the regulation of mTORC2 is only emerging. First, Sabatini and colleagues reported that prolonged treatment with rapamycin suppressed mTORC2, and significantly, that rapamycin disrupted the interaction between mTOR and Rictor, which is a component of mTORC2 [47–48]. Thus, if rapamycin interferes with the interaction between mTOR and Rictor, and if rapamycin inhibits mTOR by interfering with the interaction between mTOR and PA, then it is likely that PA is critical for the association between mTOR and Rictor to form or stabilize mTORC2. Consistent with this hypothesis, we recently reported that suppression of cellular PA level inhibited the association between mTOR and Rictor and suppressed phosphorylation of Akt at the mTORC2 site at Ser473 [40, 41]. Suppression of PA levels also prevented the association between mTOR and Raptor to form mTORC1 [40]. Thus, the emerging mechanism for the role of PA in the regulation of mTOR is that PA is required for the formation and/or stabilization of mTOR complexes.
2.3 Differential stability of mTORC1 and mTORC2 and sensitivity to rapamycin
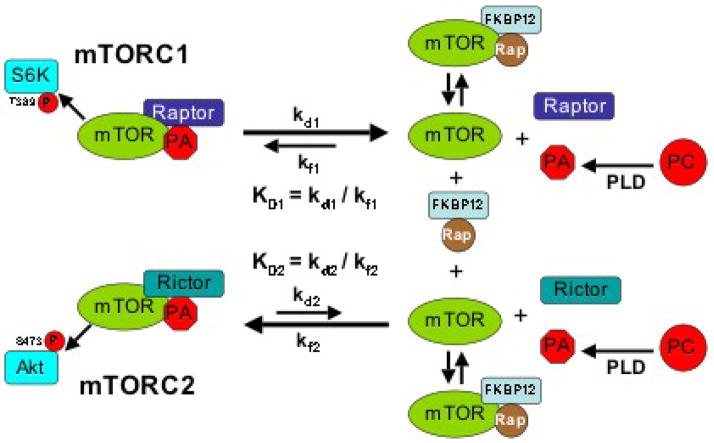
Although mTORC2 was originally described as a rapamycin resistant form of mTOR [46], more recently it was reported that mTORC2 is suppressed with prolonged rapamycin treatment [47, 48]. These findings could be explained if mTORC2 is a much more stable complex than mTORC1. Since rapamycin binds mTOR is a manner that it is competitive with PA [33, 42], and PA is required for the stability of mTOR complexes [40], then the differential sensitivity of mTORC1 and mTORC2 could be explained by a differential affinity of mTORC1 and mTORC2 for PA. A model for the differential stability of mTOR complexes is shown in Fig. 2. The greater sensitivity of mTORC1 to rapamycin is consistent with a lower affinity of PA for mTORC1 relative to mTORC2. The weaker association between PA and mTORC1 means that PA and mTORC1 will dissociate more often and thus, lower concentrations of rapamycin would be needed to replace PA on mTORC1 when it dissociates. In contrast, mTORC2 binds PA more strongly and therefore dissociations are rare – meaning that very high concentrations of rapamycin would be needed to bind the low concentrations of mTOR obtained when PA dissociates from mTORC2. To put it in terms of physical constants, the dissociation constant for PA and mTORC2 (KD2) would be less than the dissociation constant for mTORC1 and PA (KD1) (Fig. 2). The rate constants described in the model are only for the interaction between mTOR and PA, and therefore, the model represents an oversimplification – in that the involvement of Rictor and Raptor along with other components of mTORC1 and mTORC2 complexes have been neglected. However, the model does provide a first approximation for the differential stability of mTORC1 and mTORC2 complexes that is consistent the observed differential sensitivities to rapamycin.
Fig. 2.
Model for the differential effects of rapamycin on mTORC1 and mTORC2. The dissociation constants (KD) for mTORC1 and mTORC2 with PA represent the ratios for the rate constants for dissociation (kd) and formation (kf). Recent studies [40] are consistent with a model whereby the rate constant for the dissociation of mTORC1 to PA and mTOR (kd1) is greater than rate constant for the dissociation of mTORC2 to PA and mTOR (kd2). Thus, there are fewer dissociations of PA from mTORC2. The ability of rapamycin-FKBP12 to suppress mTORC1 and mTORC2 is dependent on how frequently mTOR becomes available to bind rapamycin-FKBP12. There would be more mTOR generated from mTORC1 than from mTORC2 and therefore less rapamycin would be required to compete with PA for binding to the mTOR derived from mTORC1. In contrast, the rare dissociations of mTORC2 would require much more rapamycin-FKBP12 to compete with PA to capture the rare mTOR proteins derived from mTORC2. Reducing PA levels would shift the equilibrium in favor of dissociation of the mTOR complexes therefore reduce the concentration of rapamycin-FKBP12 needed to bind to and suppress mTOR.
2.4 Effect of PA on mTORC1 and mTORC2 signals and rapamycin sensitivity
The differential effect of rapamycin on mTORC1 and mTORC2 may explain previous reports where rapamycin had different effects at different concentrations. Rapamycin suppressed the proliferation of MCF7 breast cancer cells with an IC50 of 20 nM, whereas rapamycin suppressed S6 kinase phosphorylation with an IC50 of 1 nM [42]. These data are consistent with rapamycin inhibiting mTORC1, which is required for the phosphorylation of S6 kinase, at 1 nM, and suppressing mTORC2, which is likely required for cell proliferation, at 20 nM. MCF7 cells have low levels of PLD activity and therefore low levels of PA – and consistent with a competition between PA and rapamycin for mTOR, if PLD activity was increased in the MCF7 cells, there was a corresponding increase in the IC50s for the suppression of both S6 kinase phosphorylation and cell proliferation [42]. This effect was also observed in skeletal muscle where mechanical stimulation elevated PLD activity and increased the IC50 for the effect of rapamycin [49]. MDA-MB-231 breast cancer cells have high levels of PLD activity and also exhibited a differential sensitivity of S6 kinase phosphorylation and cell proliferation to rapamycin. However, the IC50s were much higher at 20 nM and 10 μM respectively for S6 kinase phosphorylation and cell proliferation. Importantly, if PLD activity was suppressed, the IC50s for suppression of both cell proliferation and S6 kinase phosphorylation went down [42]. We just reported that reducing the PLD activity in 786-O kidney cancer cells increases sensitivity of both mTORC1 and mTORC2 complex stability to rapamycin [40]. This effect was observed for both S6 kinase phosphorylation and Akt phosphorylation at Ser473. Importantly, reducing PLD activity makes the relatively rapamycin resistant mTORC2 sensitive to concentrations that could be used therapeutically. This could be important because the suppression of cell proliferation [42] and the induction of apoptosis [26, 29] required higher levels of rapamycin – implying the involvement of mTORC2. Similarly, in 786-O kidney cancer cells, higher levels of rapamycin were required to suppress the expression of HIF2α than was required to suppress HIF1α [41]. It turned out that the expression of HIF2α was dependent on mTORC2, whereas HIF1α was dependent on both mTORC1 and mTORC2 [41] – explaining the relative insensitivity of HIF2α to rapamycin. Importantly, HIF2α expression is more critical than HIF1α for tumorigenesis [50, 51]. Thus, targeting mTORC2 could be more important than targeting mTORC1 for cancer therapeutics. The ability to increase rapamycin sensitivity of mTORC2 by suppressing PA levels may be a viable strategy for targeting mTORC2 in cancers such as kidney cancer where HIF2α and mTORC2 are implicated.
3. Other signals regulating mTOR
3.1 PA is necessary, but not sufficient to activate mTOR
While recent studies clearly demonstrate a PA requirement for the activation of both mTORC1 and mTORC2, there is also evidence that PA is not sufficient to activate either mTORC1 or mTORC2. Chen and colleagues demonstrated that exogenously provided PA stimulated the activation of the mTORC1 as indicated by increased phosphorylation of the mTORC1 substrates S6 kinase eukaryotic initiation factor 4E binding protein-1 in HEK293 cells, however the effect was dependent on the presence of amino acids [33]. Similarly, we have found that in MDA-MB-231 cells, where there is a high level of PLD activity, there is very little phosphorylation of Akt at the mTORC2 site at Ser473 [26]. However, insulin stimulates Akt phosphorylation at Ser473 in a PLD-dependent manner [40]. Thus, while PA was necessary for activating both mTORC1 and mTORC2, elevated PLD activity and PA levels are not sufficient.
3.2 Signals mediated by Akt
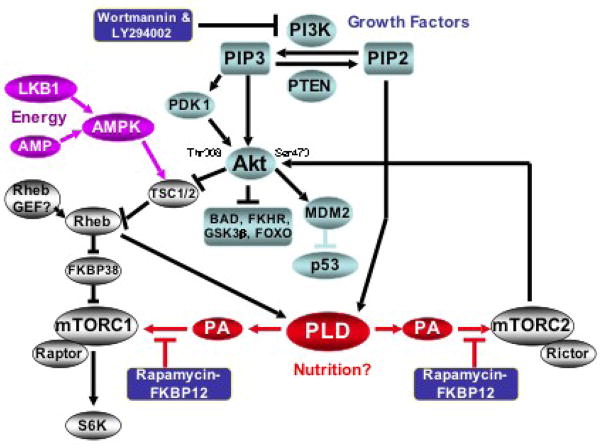
mTORC1 is an indirect target of survival signals generated by phosphatidylinositol (PI)-3-kinase (PI3K). PI3K generates the lipid second messenger (PI)-3,4,5-tris-phosphate (PIP3), which leads to the recruitment of Akt - a kinase that phosphorylates several substrates critical for cell cycle progression and the suppression of apoptotic programs [52]. PIP3 generation also leads to the recruitment of the phosphoinositide-dependent kinase 1 (PDK1) which phosphorylates Akt at Ser308 [52]. Akt is then able to phosphorylate several substrates – one of which is the tuberous sclerosis complex (TSC1/2) – a GTPase activating protein complex that suppresses Rheb, a GTPase that contributes directly to the activation of mTOR [53]. Akt suppresses TSC1/2 leading to the activation of Rheb and mTOR. TSC1/2 can also be targeted and activated by LKB1 tumor suppressor protein, which along with high levels of AMP activates the AMP-dependent kinase [54], which then phosphorylates and activates TSC1/2 resulting in the down-regulation of Rheb and inhibition of mTOR. Thus, there are signals that both activate and suppress mTOR through the TSC1/2 complex and Rheb. Importantly, very recent data reveals that Rheb directly interacts with and activates PLD1 [38, 39]. It was demonstrated that suppression of TSC2 elevates PLD activity and S6 kinase phosphorylation and that this increase was dependent on PLD1. Thus, PLD activity may regulate mTORC1 in cooperation with PI3K and other signals that regulate the GTPase activating potential of TSC1/2.
Rheb has also been implicated in regulating an mTOR inhibitor known as FKBP38 [55]. Of interest with regard to the regulation of mTOR by PLD and PA is that FKBP38 binds to the FRB domain of mTOR, which also binds PA and rapamycin. Thus, it is possible that Rheb stimulates the dissociation of FKBP38 from mTOR by activating PLD1 and generating PA. PA could then compete with FKBP38 for the FRB domain and displace FKBP38 from mTOR. While this is an attractive hypothesis, the role that FKBP38 has in regulating mTORC1 has been challenged [56] and FKBP38 may not regulate mTORC1 in all contexts.
A further complication in the regulation of mTOR has emerged over the past few years involving the role of Akt in the regulation of mTOR. The complication revolves around the phosphorylation of Akt at Thr308 and Ser473 in response to cellular signals. Complete Akt activation not only involves the phosphorylation of Akt at the PDK1 site at Thr308, but also at the mTORC2 site at Ser473 [57]. While the kinase activity of Akt is significantly higher when doubly phosphorylated at both Thr308 and Ser473, TSC1/2 phosphorylation by Akt was not affected in cells where mTORC2 kinase activity and Akt phosphorylation at Ser473 was suppressed [58, 59]. In contrast, the phosphorylation of FOXO by Akt was suppressed by mTORC2 disruption [58, 59]. This could imply that the phosphorylation of Akt at Ser 473 impacts on substrate specificity. With regard to the phosphorylation of FOXO, mTORC1 is functioning downstream of Akt and mTORC2 is functioning upstream of Akt. An additional complication is that inhibitors of PI3K suppress phosphorylation of Akt at both Thr308 and Ser473, and ironically, phosphorylation of Akt at the mTORC2 site at Ser473 has been widely used as an indicator of PI3K activity in spite of no direct evidence linking the activation of mTORC2 to PI3K. Furthermore, PI3K and mTOR are related kinases and inhibitors of PI3K also inhibit mTOR [60]. Thus, at present, there is considerable confusion as to how mTORC2 is activated and Akt gets phosphorylated at Ser473 in response to growth factors such as insulin. But it is clear that insulin-induced increases in Akt phosphorylation are dependent on PLD [40]. The signals regulating mTOR and Akt are shown in Fig. 3. In this model, PLD is apparently downstream of Akt for the activation of mTORC1 and upstream of Akt for the activation of mTORC2. It has been reported that PLD activity is elevated in response to amino acids [38] and PLD may therefore be critical for the nutritional sensing that mTOR is known to regulate [61].
Fig. 3.
Survival signals generated by PI3K and PLD that target mTOR. In the PI3K pathway, PIP3 recruits Akt and PDK1, a kinase that phosphorylates Akt at Thr308. Akt is also phosphorylated at Ser 473 by mTORC2. Phosphorylation of Akt at Ser473 elevates Akt kinase activity [57] and likely impacts substrate specificity [52]. Akt then phosphorylates several proteins that suppress cell cycle progression – including the TSC1/2 complex, a GTPase activating protein complex that suppresses the GTPase Rheb [53]. Rheb has been reported to stimulate dissociation of FKBP38 from FRB domain of mTOR [55], which contributes to the activation of mTORC1. Rheb was also recently reported to activate PLD1 [38]. PLD-generated PA competes with rapamycin to bind mTORC1 – and possibly FKBP38 – and is required for the mTOR activation of S6 kinase. PLD is also required for the activation of mTORC2, which phosphorylates Akt at Ser473. mTOR is targeted by growth factors, nutrition and energy sensing signals – indicated by different colors.
4. Differential roles for PLD1 and PLD2 in regulating mTOR
4.1 Regulation of PLD1 and PLD2
There are two mammalian PLD isoforms – PLD1 and PLD2 – that can be distinguished by different mechanisms of regulation and sub-cellular distribution [31]. PLD1 has a predominantly peri-nuclear localization and is regulated by members of the Ras family of GTPases including ARF [62], Rho [63], and Ral [64]. Importantly, it was recently reported that the GTPase Rheb activates PLD1 [38]. PLD2 is largely restricted to lipid raft fractions on the plasma membrane and its mode of regulation is not well understood [31]. Both PLD1 and PLD2 have a stringent requirement for PI-4,5-bis-phosphate (PIP2) and therefore presumably on the kinases that generate PIP2 [31]. In this regard, it is of interest that PA also activates PI-4-phosphate-5-kinase [65], which generates PIP2 from PIP and may represent a positive feedback mechanism and a means for PLD1 to stimulate PLD2 [66].
4.2 Regulation of mTORC1 by PLD1 and PLD2
Several studies have investigated the dependence of S6 kinase activation on PLD1. Exogenously expressed PLD1 increased S6 kinase phosphorylation in rat fibroblasts [67]. LPA-induced S6 kinase phosphorylation was shown to be dependent on PLD1 [68]. The activation of S6 kinase by Cdc42 was also dependent on PLD1 [69]. Suppression of PLD1 expression blocked S6 kinase phosphorylation in B16 melanoma cells [70]. And the recent finding that Rheb, which is implicated in the activation of mTORC1, directly activates PLD1 [38] strongly supports a role for PLD1 in the activation of mTORC1. Thus, there is a clear connection between PLD1 and mTORC1. However, there is also evidence that PLD2 contributes to the activation of mTORC1. Exogenously expressed PLD2 increased S6 kinase phosphorylation in MCF7 breast cancer cells [67]. We recently reported that catalytically inactive dominant negative mutants of both PLD1 and PLD2 could suppress S6 kinase phosphorylation. However, a combination of both PLD1 and PLD2 dominant negative mutants was more effective in suppressing S6 kinase phosphorylation [40]. PLD2 was reported to form a functional complex with mTOR and Raptor through a TOS (TOR signaling) motif in PLD2, and this interaction was essential for mitogen stimulation of S6 kinase phosphorylation [71]. This group was unable to suppress S6 kinase phosphorylation with PLD1 siRNA. Similarly, Sun et al. [38] were unable to suppress S6 kinase phosphorylation with PLD2 siRNA. These apparent conflicting results may reflect the perils of working with PLD siRNAs. We have found that the half-lives of PLD1 and especially PLD2 are quite long – making the down-regulation of PLD protein using the siRNA approach difficult [30, 40]. Attesting to the axiom that one positive result is worth ten negative results, current studies suggest that both PLD1 and PLD2 are required for the activation of mTORC1. Although it is not clear how both PLD1 and PLD2 contribute to the activation of mTOR, we have proposed that elevated PLD1 leads to the activation of PLD2 by increasing levels of PIP2 required for the activity of PLD2 [66]. This could explain the apparent involvement of both PLD1 and PLD2 in the activation of mTORC1. Both PLD1 and PLD2 were able to suppress protein phosphatase 2A in a rapamycin-dependent manner [72] and dominant negative mutants for both PLD1 and PLD2 suppressed receptor-mediated endocytosis [73] – further supporting a model whereby PLD1 and PLD2 work together to generate PA.
4.3 Regulation of mTORC2 by PLD1 and PLD2
A role for PLD in the regulation of mTORC2 has only recently been established, but there is evidence that both PLD1 and PLD2 contribute to the activation of mTORC2 as well. Akt phosphorylation at Ser473 is constitutively elevated in 786-O kidney cancer cells and is dependent on the mTORC2 component Rictor. Dominant negative mutants for both PLD1 and PLD2 suppressed phosphorylation of Akt at Ser473 in the 786-O cells [40]. Similarly, both PLD1 and PLD2 dominant negative mutants suppressed insulin-induced Akt phosphorylation at Ser473 in MDA-MB-231 breast cancer cells, which have low levels of Akt phosphorylation [40]. Thus it appears likely that PLD1 and PLD2 are working together to activate both mTORC1 and mTORC2.
5. Targeting PLD-mTOR signals and rapamycin resistance in cancer cells
5.1 Targeting PLD-mTOR survival signals
Targeting mTOR in anti-cancer therapies has attracted much attention in recent years largely due to a link between mTOR and survival signals in human cancer cells [34–36]. Much has been written about the potential of targeting mTOR because, in principle, the suppression of survival signals in cancer cells should result in apoptotic cell death and tumor regression. While the principle of targeting mTOR-mediated survival signals in human cancer offers an attractive therapeutic option, early clinical trials with rapamycin and rapamycin analogues have been largely disappointing [34]. One of the problems with targeting mTOR is that mTORC2 may be more important for the survival signals. HIF2α, which is critical for tumor formation by kidney cancer cells [50, 51], is dependent on mTORC2 [40] and higher concentrations of rapamycin are required to suppress mTORC2. Thus, while rapamycin based therapies may be able to suppress mTORC1, current dosages are unlikely to affect mTORC2. However, it was recently demonstrated that suppressing PA levels renders mTORC2 sensitive to rapamycin concentrations in nano-molar ranges that are clinically achievable [40]. Therefore, combination therapies that include rapamycin and strategies that suppress PLD activity could be used to target mTORC2. In this regard, it was recently reported that the natural product honokiol suppresses PLD activity in human cancer cells [75]. While honokiol does not suppress PLD directly, it does suppress the PLD activity that is elevated in response to the stress of serum withdrawal [75], and may therefore target the PLD signals that are protecting the cancer cells from apoptosis. Importantly, honokiol was well tolerated in mouse xenograft studies [76]. Targeting PLD directly has intrinsic problems in that PLD1 has been implicated in protein trafficking in the Golgi [31], and therefore suppression of PLD activity in general would likely be toxic to normal cells. However, targeting PLD2 specifically could be useful in that suppression of PLD2 expression is tolerated in cells much better than suppression of PLD1 (our unpublished observations). To this end, Alex Brown’s group has just reported the generation of isoform-specific PLD inhibitors from halopemide [77], which was shown previously to suppress PLD activity directly [78]. Similarly, Mike Frohman’s group has identified a compound that suppresses PLD2, but inhibits only a subset of cellular activities that are blocked by 1-butanol, which suppresses effects mediated by both PLD1 and PLD2 [79]. This compound, known as FIPI suppressed cell migration, which has been correlated previously with PLD-mediated survival signals in cancer cells [27]. Thus, it may be possible to suppress PLD2 specifically, which may have less harmful side effects that could occur with inhibition of PLD1 and PLD2. And since FIPI block cell migration, it is possible that this compound could be specific for the elevated PLD activity in cancer cells where PLD activity is critical for cell migration.
5.2 Rapamycin resistance
Another problem with rapamycin based therapeutic strategies is that the ability of rapamycin to suppress either mTORC1 or mTORC2 is dependent on the level of PLD activity and PA. As described above, PA contributes to the activation of mTOR in a manner that is competitive with rapamycin, and elevated PLD activity increases the concentration of rapamycin needed to suppress the growth of human cancer cells - conferring rapamycin resistance [42]. Since PLD activity is elevated in a large number of human cancers [35, 74], high levels of PA in cancer cells may represent a serious obstacle for successful treatment with rapamycin or rapamycin derivatives that are acting in a manner that is competitive with PA. Moreover, rapamycin treatment could actually select for cells with elevated PLD activity and, since elevated PLD activity also promotes metastatic phenotypes [27], rapamycin could actually accelerate tumor progression and metastasis. Increases in PA could also occur by increases in diacylglycerol kinase and lysophosphatidic acyl-transferase, which also increase PA levels (Fig. 1A). The key point is that elevated levels of PA confers rapamycin resistance and therefore rapamycin-based therapeutic strategies must take into account mechanisms that generate PA, which increases the concentration of rapamycin needed to suppress either mTORC1 or mTORC2. Strategies that suppress PLD activity and reduce PA levels offer the potential to increase sensitivity to rapamycin and rapamycin derivatives. Suppressing PLD activity would also avoid the problem of selecting for cells with elevated PLD activity in the presence of rapamycin.
6. Conclusions
Elevated PLD activity has been observed in a large number of human cancers and has been shown to suppress apoptosis [74]. PLD activity is commonly elevated in cancer cell lines in response to the stress of serum withdrawal [27]. It has now become apparent that a key target of PLD survival signals is mTOR. mTOR has been implicated as a key regulator of stress responses by shutting down under conditions of poor nutrition or hypoxia [36, 61]. In order for a cancer cell to survive and proliferate, it must overcome normal cellular responses to stress in order to continue dividing in an emerging tumor where there is poor vascularization. Activating mTOR under these conditions allows cells to continue proliferating and avoid apoptosis. Although PA is necessary for mTOR complex stability and activity, it is not sufficient to fully activate mTORC1 or mTORC2. While there is much known about the additional signals that are needed to activate mTORC1, there is little known about how mTORC2 is activated. Further studies on these signals will be important for developing strategies to suppress the survivals signals mediated by PLD and mTOR.
The apparent mechanism for the suppression of mTOR by rapamycin is to prevent the interaction between mTOR and PA, which facilitates the formation and/or stability of mTOR complexes [40]. Consistent with this hypothesis, reducing the level of PA, like rapamycin, disrupts the association between mTOR and Raptor and between mTOR and Rictor [40]. Importantly, elevated PLD activity confers resistance to rapamycin [42, 49], which could have important unintended clinical consequences – in that rapamycin treatment could actually select for cancer cells with elevated PLD activity. The elevated PLD activity would not only generate rapamycin resistance, it could also make cells more malignant because elevated PLD activity also stimulates the metastatic phenotypes of increased cell migration and invasion [27]. Thus, the potential for PLD to impact the targeting mTOR in multiple ways in anti-cancer therapies argues strongly that serious attention be given to the role of PLD and PA in the regulation of mTOR – especially with regard to targeting mTOR in anti-cancer therapies.
Acknowledgments
Paige Yellen is acknowledged for thoughtful comments on the manuscript. The author of this work was supported by grants from the National Cancer Institute (CA46677) and a SCORE grant from the National Institutes of Health (GM60654). Research Centers in Minority Institutions (RCMI) award RR-03037 from the National Center for Research Resources of the National Institutes of Health, which supports infrastructure and instrumentation in the Biological Sciences Department at Hunter College, is also acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Song J, Jiang YW, Foster DA. EGF induces the production of biologically distinguishable diglyceride species from phosphatidylinositol and phosphatidylcholine: evidence for the independent activation of type C and type D phospholipases. Cell Growth Differ. 1994;5:79–85. [PubMed] [Google Scholar]
- 2.Plevin R, Cook SG, Palmer S, Wakelam MJ. Multiple sources of sn-1,2-diacylglycerol in platelet-derived-growth factor-stimulated swiss 3T3 fibroblasts. Biochem J. 1991;279:559–565. doi: 10.1042/bj2790559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motoike T, Bieger S, Wiegandt H, Unsicker K. Induction of phosphatidic acid by fibroblast growth factor in cultured baby hamster kidney fibroblasts. FEBS Lett. 1993;332:164–168. doi: 10.1016/0014-5793(93)80505-o. [DOI] [PubMed] [Google Scholar]
- 4.Sa G, Das T. Basic fibroblast growth factor stimulates cytosolic phospholipase A2, phospholipase C-γ1 and phospholipase D through distinguishable signaling mechanisms. Mol Cell Biochem. 1999;198:19–30. doi: 10.1023/a:1006970710298. [DOI] [PubMed] [Google Scholar]
- 5.Karnam P, Standaert ML, Galloway L, Farese RV. Activation and translocation of Rho and ADP ribosylation factor by insulin in rat adipocytes. Apparent involvement of phosphatidylinositol 3-kinase. J Biol Chem. 1997;272:6136–6140. doi: 10.1074/jbc.272.10.6136. [DOI] [PubMed] [Google Scholar]
- 6.Banno Y, Takuwa Y, Yamada M, Takuwa N, Ohguchi K, Hara A, Nozawa Y. Involvement of phospholipase D in insulin-like growth factor-I-induced activation of extracellular signal-regulated kinase, but not phosphatidylinositol 3-kinase or Akt, in Chinese hamster ovary cells. Biochem J. 2003;369:363–368. doi: 10.1042/BJ20021368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seymour LW, Shoaibi MA, Martin A, Ahmed A, Elvin P, Kerr DJ, Wakelam MJ. Vascular endothelial growth factor stimulates protein kinase C-dependent phospholipase D activity in endothelial cells. Lab Invest. 1996;75:427–437. [PubMed] [Google Scholar]
- 8.Song J, Pfeffer LM, Foster DA. v-Src increases diacylglycerol levels via a type D phospholipase-mediated hydrolysis of phosphatidylcholine. Mol Cell Biol. 1991;11:4903–4908. doi: 10.1128/mcb.11.10.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carnero A, Cuadrado A, del Peso L, Lacal JC. Activation of type D phospholipase by serum stimulation and ras-induced transformation in NIH3T3 cells. Oncogene. 1994;9:1387–1395. [PubMed] [Google Scholar]
- 10.Jiang H, Lu Z, Luo JQ, Wolfman A, Foster DA. Ras mediates the activation of phospholipase D by v-Src. J Biol Chem. 1995;270:6006–6009. doi: 10.1074/jbc.270.11.6006. [DOI] [PubMed] [Google Scholar]
- 11.Jiang YW, Song J, Zang Q, Foster DA. Phosphatidylcholine-specific phospholipase D activity is elevated in v-Fps-transformed cells. Biochem Biophys Res Comm. 1994;203:1195–1203. doi: 10.1006/bbrc.1994.2309. [DOI] [PubMed] [Google Scholar]
- 12.Frankel PA, Ramos M, Flom J, Bychenok S, Joseph T, Kerkhoff E, Rapp UR, Feig LA, Foster DA. Ral and Rho dependent activation of phospholipase D in v-Raf transformed cells. Biochem Biophys Res Comm. 1999;255:502–507. doi: 10.1006/bbrc.1999.0234. [DOI] [PubMed] [Google Scholar]
- 13.Buchanan FG, McReynolds M, Couvillon A, Kam Y, Holla VR, Dubois RN, Exton JH. Requirement of phospholipase D1 activity in H-RasV12-induced transformation. Proc Natl Acad Sci U S A. 2005;102:1638–1642. doi: 10.1073/pnas.0406698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Z, Hornia A, Joseph T, Sukezane T, Frankel P, Zhong M, Bychenok S, Xu L, Feig LA, Foster DA. Phospholipase D and RalA Cooperate with the EGF Receptor to Transform 3Y1 Rat Fibroblasts. Mol Cell Biol. 2000;20:462–467. doi: 10.1128/mcb.20.2.462-467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph T, Wooden R, Bryant A, Zhong M, Lu Z, Foster DA. Transformation of cells overexpressing a tyrosine kinase by phospholipase D1 and D2. Biochem Biophys Res Comm. 2001;289:1019–1024. doi: 10.1006/bbrc.2001.6118. [DOI] [PubMed] [Google Scholar]
- 16.Ahn BH, Kim SY, Kim EH, Choi KS, Kwon TK, Lee YH, Chang JS, Kim MS, Jo YH, Min DS. Transmodulation between phospholipase D and c-Src enhances cell proliferation. Mol Cell Biol. 2003;23:3103–3115. doi: 10.1128/MCB.23.9.3103-3115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min DS, Kwon TK, Park WS, Chang JS, Park SK, Ahn BH, Ryoo ZY, Lee YH, Lee YS, Rhie DJ, Yoon SH, Hahn SJ, Kim MS, Jo YH. Neoplastic transformation and tumorigenesis associated with overexpression of phospholipase D isozymes in cultured murine fibroblasts. Carcinogenesis. 2001;22:1641–1647. doi: 10.1093/carcin/22.10.1641. [DOI] [PubMed] [Google Scholar]
- 18.Samuels ML, McMahon M. Inhibition of platelet-derived growth factor- and epidermal growth factor-mediated mitogenesis and signaling in 3T3 cells expressing ΔRaf-1:ER, an estradiol-regulated form of Raf-1. Mol Cell Biol. 1994;14:7855–7866. doi: 10.1128/mcb.14.12.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerkhoff E, Rapp UR. High-intensity Raf signals convert mitotic cell cycling into cellular growth. Cancer Res. 1998;58:1636–1640. [PubMed] [Google Scholar]
- 20.Joseph T, Bryant A, Wooden R, Kerkhoff E, Rapp UR, Foster DA. Phospholipase D overcomes cell cycle arrest induced by high-intensity Raf signaling. Oncogene. 2002;21:3651–3658. doi: 10.1038/sj.onc.1205380. [DOI] [PubMed] [Google Scholar]
- 21.Zhong M, Shen Y, Zheng Y, Joseph T, Jackson D, Beychenok S, Foster DA. Phospholipase D prevents apoptosis in v-Src-transformed rat fibroblasts and MDA-MB-231 breast cancer cells. Biochem Biophys Res Comm. 2003;302:615–619. doi: 10.1016/s0006-291x(03)00229-8. [DOI] [PubMed] [Google Scholar]
- 22.Noh DY, Ahn SJ, Lee AR, Park IA, Kim JH, Suh PG, Ryu SH, Lee KH, Han JS. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer Lett. 2000;161:207–214. doi: 10.1016/s0304-3835(00)00612-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, Ehara H, Akao Y, Shamoto M, Nakagawa Y, Banno Y, Deguchi T, Ohishi N, Yagi K, Nozawa Y. Increased activity and intranuclear expression of phospholipase D2 in human renal cancer. Biochem Biophys Res Commun. 2000;278:140–143. doi: 10.1006/bbrc.2000.3719. [DOI] [PubMed] [Google Scholar]
- 24.Uchida N, Okamura S, Kuwano H. Phospholipase D activity in human gastric carcinoma. Anticancer Res. 1999;19:671–676. [PubMed] [Google Scholar]
- 25.Yamada Y, Hamajima N, Kato T, Iwata H, Yamamura Y, Shinoda M, Suyama M, Mitsudomi T, Tajima K, Kusakabe S, Yoshida H, Banno Y, Akao Y, Tanaka M, Nozawa Y. Association of a polymorphism of the phospholipase D(2) gene with the prevalence of colorectal cancer. J Mol Med. 2003;81:126–131. doi: 10.1007/s00109-002-0411-x. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Rodrik V, Foster DA. Alternative phospholipase D/mTOR survival signal in human breast cancer cells. Oncogene. 2005;24:672–679. doi: 10.1038/sj.onc.1208099. [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y, Rodrik V, Toschi A, Shi M, Hui L, Shen Y, Foster DA. Phospholipase D couples survival and migration signals in response to stress in human breast cancer cells. J Biol Chem. 2006;281:15862–15868. doi: 10.1074/jbc.M600660200. [DOI] [PubMed] [Google Scholar]
- 28.Shi M, Zheng Y, Garcia A, Foster DA. Phospholipase D provides a survival signal in human cancer cells with activated H-Ras or K-Ras. Cancer Lett. 2007;258:268–275. doi: 10.1016/j.canlet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gadir N, Jackson D, Lee E, Foster DA. Defective TGF-β signaling sensitizes human cancer cells to rapamycin. Oncogene. 2008;27:1055–1062. doi: 10.1038/sj.onc.1210721. [DOI] [PubMed] [Google Scholar]
- 30.Toschi A, Edelstein J, Rockwell P, Ohh M, Foster DA. HIFα expression in VHL-deficient renal cancer cells is dependent on phospholipase D. Oncogene. 2008;27:2746–2753. doi: 10.1038/sj.onc.1210927. [DOI] [PubMed] [Google Scholar]
- 31.Jenkins GM, Frohman MA. Phospholipase D: a lipid centric review. Cell Mol Life Sci. 2005;62:2305–2316. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rizzo MA, Shome K, Vasudevan C, Stolz DB, Sung TC, Frohman MA, Watkins SC, Romero G. Phospholipase D and its product, phosphatidic acid, mediate agonist-dependent raf-1 translocation to the plasma membrane and the activation of the mitogen-activated protein kinase pathway. J Biol Chem. 1999;274:1131–1139. doi: 10.1074/jbc.274.2.1131. [DOI] [PubMed] [Google Scholar]
- 33.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 34.Sawyers CL. Will mTOR inhibitors make it as cancer drugs? Cancer Cell. 2003;4:343–348. doi: 10.1016/s1535-6108(03)00275-7. [DOI] [PubMed] [Google Scholar]
- 35.Foster DA. Targeting mTOR-mediated survival signals in anticancer therapeutic strategies. Exp Rev Anticancer Ther. 2004;4:691–701. doi: 10.1586/14737140.4.4.691. [DOI] [PubMed] [Google Scholar]
- 36.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Foster DA. Regulation of mTOR by phosphatidic acid? Cancer Res. 2007;67:1–4. doi: 10.1158/0008-5472.CAN-06-3016. [DOI] [PubMed] [Google Scholar]
- 38.Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, Armstrong M, Brown HA, Chen J. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc Natl Acad Sci U S A. 2008;105:8286–8291. doi: 10.1073/pnas.0712268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, Chen J. mTOR signaling: PLD takes center stage. Cell Cycle. 2008;7:3118–3123. doi: 10.4161/cc.7.20.6881. [DOI] [PubMed] [Google Scholar]
- 40.Toschi A, Lee E, Xu L, Garcia A, Gadir N, Foster DA. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid – a competition with rapamycin. Mol Cell Biol. 2009;29 doi: 10.1128/MCB.00782-08. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toschi A, Lee E, Gadir N, Ohh M, Foster DA. Differential dependence of HIF1α and HIF2α on mTORC1 and mTORC2. J Biol Chem. 2008;283:34495 – 34499. doi: 10.1074/jbc.C800170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Zheng Y, Foster DA. Phospholipase D confers rapamycin resistance in human breast cancer cells. Oncogene. 2003;22:3937–3942. doi: 10.1038/sj.onc.1206565. [DOI] [PubMed] [Google Scholar]
- 43.Veverka V, Crabbe T, Bird I, Lennie G, Muskett FW, Taylor RJ, Carr MD. Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR. Oncogene. 2008;27:585–595. doi: 10.1038/sj.onc.1210693. [DOI] [PubMed] [Google Scholar]
- 44.Chen J, Fang Y. A novel pathway regulating the mammalian target of rapamycin (mTOR) signaling. Biochem Pharmacol. 2002;64:1071–1077. doi: 10.1016/s0006-2952(02)01263-7. [DOI] [PubMed] [Google Scholar]
- 45.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 46.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 47.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 48.Zeng Z, Sarbassov DD, Samudio IJ, Yee KW, Munsell MF, Ellen Jackson C, Giles FJ, Sabatini DM, Andreeff M, Konopleva M. Rapamycin derivatives reduce mTORC2 signaling and inhibit AKT activation in AML. Blood. 2007;109:3509–3512. doi: 10.1182/blood-2006-06-030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hornberger TA, Chu WK, Mak YW, Hsiung JW, Huang SA, Chien S. The role of phospholipase D and phosphatidic acid in the mechanical activation of mTOR signaling in skeletal muscle. Proc Natl Acad Sci U S A. 2006;103:4741–4746. doi: 10.1073/pnas.0600678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 51.Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr Inhibition of HIF2α is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1:439–444. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tee AR, Blenis J. mTOR, translational control and human disease. Semin Cell Dev Biol. 2005;16:29–37. doi: 10.1016/j.semcdb.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Bai X, Ma D, Liu A, Shen X, Wang QJ, Liu Y, Jiang Y. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science. 2007;318:977–980. doi: 10.1126/science.1147379. [DOI] [PubMed] [Google Scholar]
- 56.Wang X, Fonseca BD, Tang H, Liu R, Elia A, Clemens MJ, Bommer UA, Proud CG. Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J Biol Chem. 2008;283:30482–30492. doi: 10.1074/jbc.M803348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 58.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 59.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 60.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 61.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 62.Xu L, Frankel P, Jackson D, Rotunda T, D’Souza-Schorey C, Foster DA. Elevated phospholipase D activity in H-Ras-, but not K-Ras-transformed cells by the synergistic action of RalA and ARF6. Mol Cell Biol. 2003;23:645–654. doi: 10.1128/MCB.23.2.645-654.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malcolm KC, Elliott CM, Exton JH. Evidence for Rho-mediated agonist stimulation of phospholipase D in rat1 fibroblasts. Effects of Clostridium botulinum C3 exoenzyme. J Biol Chem. 1996;271:13135–13139. doi: 10.1074/jbc.271.22.13135. [DOI] [PubMed] [Google Scholar]
- 64.Jiang H, Luo JQ, Urano T, Lu Z, Foster DA, Feig LA. Involvement of Ral GTPase in v-Src-induced phospholipase D activation. Nature. 1995;378:409–412. doi: 10.1038/378409a0. [DOI] [PubMed] [Google Scholar]
- 65.Moritz A, De Graan PN, Gispen WH, Wirtz KW. Phosphatidic acid is a specific activator of phosphatidylinositol-4- phosphate kinase. J Biol Chem. 1992;267:7207–7210. [PubMed] [Google Scholar]
- 66.Foster DA, Xu L. Phospholipase D in cell proliferation and cancer. Mol Cancer Res. 2003;1:789–800. [PubMed] [Google Scholar]
- 67.Hui L, Abbas T, Pielak R, Joseph T, Bargonetti J, Foster DA. Phospholipase D elevates the level of MDM2 and suppresses DNA damage-induced increases in p53. Mol Cell Biol. 2004;24:5677–5688. doi: 10.1128/MCB.24.13.5677-5686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kam Y, Exton JH. Role of phospholipase D1 in the regulation of mTOR activity by lysophosphatidic acid. FASEB J. 2004;18:311–319. doi: 10.1096/fj.03-0731com. [DOI] [PubMed] [Google Scholar]
- 69.Fang Y, Park IH, Wu AL, Du G, Huang P, Frohman MA, Walker SJ, Brown HA, Chen J. PLD1 regulates mTOR signaling and mediates Cdc42 activation of S6K1. Curr Biol. 2003;13:2037–44. doi: 10.1016/j.cub.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 70.Ohguchi K, Banno Y, Nakagawa Y, Akao Y, Nozawa Y. Negative regulation of melanogenesis by phospholipase D1 through mTOR/p70 S6 kinase 1 signaling in mouse B16 melanoma cells. J Cell Physiol. 2005;205:444–451. doi: 10.1002/jcp.20421. [DOI] [PubMed] [Google Scholar]
- 71.Ha SH, Kim DH, Kim IS, Kim JH, Lee MN, Lee HJ, Kim JH, Jang SK, Suh PG, Ryu SH. PLD2 forms a functional complex with mTOR/raptor to transduce mitogenic signals. Cell Signal. 2006;18:2283–2291. doi: 10.1016/j.cellsig.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 72.Hui L, Rodrik V, Pielak RM, Zheng Y, Foster DA. mTOR-dependent suppression of protein phosphatase 2A is critical for phospholipase D survival signals in human breast cancer cells. J Biol Chem. 2005;280:35829–35835. doi: 10.1074/jbc.M504192200. [DOI] [PubMed] [Google Scholar]
- 73.Shen Y, Xu L, Foster DA. Phospholipase D requirement for receptor-mediated endocytosis. Mol Cell Biol. 2001;21:595–602. doi: 10.1128/MCB.21.2.595-602.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Foster DA. Phospholipase D survival signals as a therapeutic target in cancer. Curr Signal Transduction Ther. 2006;1:295–303. [Google Scholar]
- 75.Garcia A, Zheng Y, Zhao C, Toschi A, Fan J, Shraibman N, Brown HA, Bar-Sagi D, Foster DA, Arbiser JL. Honokiol suppresses survival signals mediated by Ras-dependent phospholipase D activity in human cancer cells. Clin Cancer Res. 2008;14:4267–4274. doi: 10.1158/1078-0432.CCR-08-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bai X, Cerimele F, Ushio-Fukai M, Waqas M, Campbell PM, Govindaraja B, Der CJ, Battle T, Frank DA, Ye K, Murad E, Dubiel W, Soff G, Arbiser JL. Honokiol, a small molecular weight natural product, inhibits angiogenesis in vitro and tumor growth in vivo. J Biol Chem. 2003;278:35501–35507. doi: 10.1074/jbc.M302967200. [DOI] [PubMed] [Google Scholar]
- 77.Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, Armstrong MD, Arteaga CL, Lindsley CW, Brown HA. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat Chem Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Monovich L, Mugrage B, Quadros E, Toscano, Tommasi R, LaVoie S, Liu E, Du Z, LaSala D, Boyar W, Steed P. Optimization of halopemide for phospholipase D2 inhibition. Bioorg Med Chem Lett. 2007;17:2310–2311. doi: 10.1016/j.bmcl.2007.01.059. [DOI] [PubMed] [Google Scholar]
- 79.Su W, Yeku O, Olepu S, Genna A, Park JS, Ren H, Du G, Gelb M, Morris A, Frohman MA. FIPI, a Phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol Pharmacol. 2008 doi: 10.1124/mol.108.053298. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]