Abstract
Background and Aims
Barley (Hordeum vulgare ssp. vulgare) cultivation started between 9500 and 8400 years ago, and was a major part of ancient agriculture in the Near East. The brittle rachis is a critical trait in the domestication process.
Methods
A DNA sequence closely linked to the brittle rachis complex was amplified and resequenced in a collection of cultivated barleys, wild barleys (H. vulgare ssp. spontaneum) and weedy brittle rachis varieties (H. vulgare ssp. vulgare var. agriocrithon). The sequence was used to construct a phylogenetic tree.
Key Results
The phylogeny separated the W- (btr1-carrying) from the E- (btr2-carrying) cultivars. The wild barleys had a high sequence diversity and were distributed throughout the W- and E-clades. Some of the Tibetan var. agriocrithon lines were closely related to the E-type and others to the W-type cultivated barleys, but an Israeli var. agriocrithon line has a complex origin.
Conclusions
The results are consistent with a diphyletic origin of barley. The W- and E-type cultivars are assumed to have evolved from previously diverged wild barley via independent mutations at Btr1 and Btr2.
Key words: Hordeum vulgare, cultivated barley, wild barley, weedy barley, var. agriocrithon, btr1, btr2, domestication, evolution
INTRODUCTION
Barley (Hordeum vulgare ssp. vulgare) was one of the first crop species to be developed in the ‘Fertile Crescent’ (Zohary and Hopf, 1993). Archaeological remains of non-brittle barley grains indicate that selection by man of tough rachis forms of wild barley (H. vulgare ssp. spontaneum) was probably the initial stage of the domestication process (Harlan, 1992). In addition to the Fertile Crescent, Tibet, Ethiopia and Morocco have all been proposed as alternative candidate regions for the site of barley domestication (Åberg, 1938; Xu, 1982; Bekele, 1983; Molina-Cano et al., 1987, 2005; Zohary, 1996). The six-rowed brittle Tibetan barley H. agriocrithon was identified by Åberg (1938), and was considered to be the progenitor of cultivated six-rowed barley (Åberg, 1940; Friesleben, 1943). However, other authorities have suggested that it was derived from a hybrid between wild barley and six-rowed cultivated barley (Zohary, 1963; Konishi, 2001; Tanno and Takeda, 2004), even though the presence in Tibet of a true ssp. spontaneum has yet to be established. Some further alternatives are that H. agriocrithon arose from a secondary mutant, or that it descends from a weedy hybridized segregant out of a hybrid between oriental and occidental-type cultivated barleys, which have diverged substantially from one another (Bothmer et al., 1995). As a result, the latter authors have suggested H. vulgare ssp. vulgare var. agriocrithon (hereafter var. agriocrithon) as the proper taxonomic classification of this subspecies.
The origin of barley remains to be resolved. Evidence has been presented that the mutation from brittle to non-brittle rachis must have occurred on at least two independent occasions (Takahashi, 1955). Supporting the hypothesis of a polyphyletic origin, Zohary (1996) opined that domestication was a multiple event. However, Badr et al. (2000) have suggested that the Israel–Jordan area section of the Fertile Crescent was the only place where wild barley was domesticated, proposing instead a monophyletic origin. Molecular studies of the key traits implicated in the domestication process should provide better objective evidence than studies of genes or markers which are genetically independent of the critical domestication genes for resolving the domestication question (Komatsuda et al., 2004).
The brittle rachis is one of the most critical traits in the evolution and domestication of barley. In wild barley, this character is determined by two complementary genes, Btr1 and Btr2, tightly linked to one another on chromosome 3H (Takahashi and Hayashi, 1964). In cultivated barleys, one or other of these has been lost by mutation. Most occidental cultivars are of genotype btr1Btr2 and are referred to hereafter as W-type, while most oriental ones are Btr1btr2 (E-type) (Takahashi, 1955). Using markers derived from a high-density AFLP-based genetic map based on an E-type × W-type cross, a phylogenetic analysis showed a clear separation between the E- and W-clades (Komatsuda et al., 2004). The AFLP marker e09m25-08, which co-segregated with btr1/btr2 (Komatsuda et al., 2004; Senthil and Komatsuda, 2006), was converted to an STS (sequence-tagged site) format, and high-resolution mapping using this assay demonstrated a low level of recombination with btr1 (0·21 cM; Azhaguvel et al., 2006; Vidya Saraswathi et al., 2006). The definition of sequence polymorphism in a closely linked marker (0·1 cM) was used to infer the multiple origin of six-rowed barley (Tanno et al., 2002), and these conclusions have recently been verified following the isolation of the six-rowed spike gene vrs1 (Komatsuda et al., 2007). Since the non-brittle rachis genes have yet to be cloned, we have adopted a similar approach to track the genealogy of the brittle rachis gene complex in the evolution from wild to cultivated barley.
MATERIALS AND METHODS
Plant material
Twenty-three barley Hordeum vulgare ssp. vulgare L. cultivars, three accessions of H. vulgare ssp. vulgare var. agriocrithon (Åberg) Bowd., and 18 of wild barley H. vulgare ssp. spontaneum C. Koch. were obtained from several sources (Table 1). In addition, one line of H. bulbosum and one of H. murinum were included as potential paraphyletic outgroups for the phylogenetic study. The taxonomic treatment follows Bothmer et al. (1995). Genotype with respect to btr1/btr2 was taken from the work of Takahashi et al. (1983), Komatsuda and Mano (2002) and Komatsuda et al. (2004). Exploratory DNA amplification and sequencing were carried out on DNA templates from ‘Azumamugi’ (AZ, E-type), ‘Kanto Nakate Gold’ (KNG, W-type) and OUH602 (wild barley).
Table 1.
Plant materials used for the phylogenetic analysis (E, Btr1Btr1btr2btr2; W, btr1btr1Btr2Btr2)
| Taxon | Name/accession number | Origin | Phenotype | Genotype | Row type | Source* |
|---|---|---|---|---|---|---|
| ssp. vulgare | Azumamugi | Japan | Non-brittle | E | 6 | 1 |
| Bonus | Sweden | Non-brittle | W | 2 | 2 | |
| Cairo 1 (OUB369) | Egypt, Cairo | Non-brittle | E | 6 | 3 | |
| Caveda | Spain | Non-brittle | W | 6 | 3 | |
| Chevalier | UK | Non-brittle | W | 2 | 4 | |
| Debre Zeit 29 | Ethiopia | Non-brittle | W | 2 | 4 | |
| Dissa | Germany | Non-brittle | W | 6 | 5 | |
| Esfahan 1 (OUI032) | Iran, Esfahan | Non-brittle | E | 6 | 3 | |
| Goheung Covered 1 (OUK001) | South Korea, Goheung | Non-brittle | E | 6 | 3 | |
| Golden Promise | UK | Non-brittle | W | 2 | 4 | |
| Hanna | Czechoslovakia | Non-brittle | W | 2 | 4 | |
| Haruna Nijo | Japan | Non-brittle | W | 2 | 3 | |
| Hayakiso 2 | Japan | Non-brittle | E | 6 | 3 | |
| Kanto Nakate Gold | Japan | Non-brittle | W | 2 | 1 | |
| Kristina | Sweden | Non-brittle | W | 2 | 2 | |
| Misato Golden | Japan | Non-brittle | W | 2 | 1 | |
| Morex | USA | Non-brittle | Unknown | 6 | 6 | |
| Natsudaikon Mugi | Korea | Non-brittle | W | 6 | 4 | |
| New Golden | Japan | Non-brittle | W | 2 | 1 | |
| Pukou 1 (OUC018) | China, Pukou | Non-brittle | E | 6 | 3 | |
| Sama 1 (OUN005) | Nepal, Sama | Non-brittle | E | 6 | 3 | |
| Soren Oumugi 19329 | Former USSR | Non-brittle | E | 6 | 1 | |
| Tayeh 1 (OUC331) | China, Tayeh | Non-brittle | E | 6 | 3 | |
| var. agriocrithon | OUH786 | Tibet, Tsela Dzong | Brittle | 6 | 3 | |
| OUH797 | Tibet, Tsela Dzong | Brittle | 6 | 3 | ||
| OUH802 | Isreal, N.Negev | Brittle | 6 | 3 | ||
| ssp. spontaneum | H3140A | Cyprus | Brittle | 2 | 7 | |
| OUH602 | Caspian Sea Reagion | Brittle | 2 | 3 | ||
| OUH624 | Afghanistan, Heart | Brittle | 2 | 3 | ||
| OUH630 | Afghanistan, Kandahar | Brittle | 2 | 3 | ||
| OUH638 | Jordan | Brittle | 2 | 3 | ||
| OUH644 | Turkmenistan, Sumbar | Brittle | 2 | 3 | ||
| OUH707 | Iraq, Karkuk | Brittle | 2 | 3 | ||
| OUH725 | Turkey, Mardin | Brittle | 2 | 3 | ||
| OUH726 | Turkey, Silvan | Brittle | 2 | 3 | ||
| OUH728 | Iran, Kermanshah | Brittle | 2 | 3 | ||
| OUH729 | Iran, Karand | Brittle | 2 | 3 | ||
| OUH730 | Turkmenistan, Karakala | Brittle | 2 | 3 | ||
| OUH742 | Iraq, Jarmo | Brittle | 2 | 3 | ||
| OUH743 | Iraq, Karkuk | Brittle | 2 | 3 | ||
| OUH776 | Morocco, Djebel | Brittle | 2 | 3 | ||
| OUH777 | Morocco, Djebel | Brittle | 2 | 3 | ||
| OUH783 | Libya, Taknis | Brittle | 2 | 3 | ||
| PI282597 | Israel, C. Israel | Brittle | 2 | 8 | ||
| H. bulbosum | H3878 | Italy | Brittle | 2 | 3 | |
| H. murinum | H74 | Egypt | Brittle | 2 | 3 |
* 1, National Institute of Crop Science, Tsukuba, Japan; 2, Nordic Gene Bank, Alnarp, Sweden; 3, Research Institute for Bioresources, Okayama University, Kurashiki, Japan; 4, National Institute of Agrobiological Sciences, Tsukuba, Japan; 5, Sapporo Breweries, Nitta, Japan; 6, School of Biosciences, Washington State University, Pullman, USA; 7, Swedish University of Agricultural Sciences, Alnarp, Sweden; 8, USDA-ARS, Aberdeen, Idaho, USA.
DNA isolation, amplification and sequencing
Genomic DNA was extracted from young leaves following procedures described by Komatsuda et al. (1998). The e09m25-08STS-Ext sequence was amplified by the primers M679M06a620U037 (5′-AGAAGCTCACAGGGTTAGAAT-3′) or M679M06a990U073 (5′-TTGTGAAGGCTCTCCAGAGTC-3′) in combination with M679M06a990L643 (5′-TACGAGGAGCTGGTCAAGGAA-3′) (Fig. 1). The 10-μL PCRs contained 20 ng genomic DNA, 300 nm each primer, 200 µm dNTP, 25 mm TAPS (N-Tris(hydroxymethyl)methyl-3-amino-propanesulphonic acid, pH 9·3), 50 mm KCl, 1 mm 2-mercaptoethanol, 2·5 mm MgCl2 and 0·25 U ExTaq DNA polymerase (Takara, Tokyo). Reactions were denatured (94 °C/5 min), amplified for 30 cycles of 94°C/30 s, 62 °C/30 s and 72 °C/30 s, and finally incubated at 72 °C for 7 min. Amplicons were separated by 1·8 % (w/v) agarose (Iwai Kagaku, Tokyo) gel electrophoresis, eluted from the gel and purified using the Qiaquick gel purification kit (Qiagen, USA). The purified DNAs were sequenced using the Bigdye Terminator version 3·1 (ABI, Tokyo) system.
Fig. 1.
Genomic sequence of the e09m25-08STS-Ext amplicon in Morex (BAC M679M06), AZ, KNG and OUH602. The MseI and EcoRI restriction sites responsible for the AFLP fragment e09m25-08 are indicated in Morex and KNG. The fragment sequence is extended in both directions in the Morex BAC clone. Primers M679M06a620U037 and M679M06a990L643 were used to amplify the full sequences from the four lines. Later M679M06a990U073, instead of M679M06a620U037, was used for phylogenetic study because it generated more strong and stable PCR products than did M679M06a620U037. The sequences flanked by the arrows (552 nts) were considered for the phylogenetic analysis. Bases that are identical in all four sequences are indicated by asterisks (*).
Phylogenetic analysis
Sequence alignment was performed using CLUSTAL W (Thompson et al., 1994) with manual refining. Indels (insertions and deletions) shared by two or more taxa were included as ‘01’ codes for the analysis, in addition to all nucleotide substitutions. Phylogenetic trees were constructed by the neighbor-joining method of Saitou and Nei (1987). Trees were computed with PAUP* 4·0b10 (Swofford 1998). The confidence of each clade was estimated by bootstrap analysis using 1000 pseudo-replicates.
Recombination analyses
Two methods were used to search for intragenic recombination. The first employed the program GENECONV 1·81 (http://www.math.wustl.edu/~sawyer/geneconv/index.html) developed by Sawyer (1998). The global permutation P values are based on BLAST like global scores (10 000 replicates). The second involved a search for recombination using DnaSP program version 4·10·7 (Rozas et al., 2003).
RESULTS
The e09m25-08STS-Ext locus is highly variable
The AZ, KNG and OUH602 e09m25-08STS-Ext sequences were highly variable (Fig. 1). The AZ fragment was 554 bp in length, the KNG one 589 bp and the OUH602 one 562 bp. The AZ and KNG sequences differed from one another by 26 single nucleotide substitutions and ten indels, and KNG and OUH602 by 30 single nucleotide substitutions and eight indels; but AZ and OUH602 differed by just four single nucleotide substitutions and four indels (Fig. 1). The sequences from ‘Morex’ and KNG were identical to one another. Both the MseI and EcoRI recognition sites, which are responsible for the AFLP fragment e09m25-08, were present in the KNG sequence (Fig. 1), while the absence of the fragment in AZ and OUH was due to the loss of both of these sites (Komatsuda et al., 2004; Senthil and Komatsuda 2006). An STS marker (e50m21-01STS) which maps 0·63 cM proximal to btr1/btr2 (Azhaguvel et al., 2006) was also considered but, despite an amplicon size of > 1 kb, only limited polymorphism existed between the wild barley OUH602 and the cultivars AZ and KNG (data not shown).
Phylogenetic analysis based on the e09m25-08STS-Ext locus
When the original forward primer (M679M06a620U037) was replaced by M679M06a990U073, a better level of amplification efficiency and stability was achieved (Fig. 1). A single fragment was amplified from all accessions of cultivated barley, var. agriocrithon and wild barley (data not shown). The multiple sequence alignment generated a matrix consisting of 44 taxonomic entities and 552 nucleotide sites, of which 490 were invariant, 25 variable but parsimony-uninformative, and 37 variable and parsimony-informative. Ten phylogenetically informative indels were added to the data matrix. Although an attempt was made to use either H. bulbosum and H. murinum to provide an outgroup(s) to root the phylogenetic tree, this was not possible, because neither of these templates amplified a single species amplicon. As a result, an un-rooted tree was constructed. The resulting un-rooted tree consisted of two major clades (Fig. 2), separated with a bootstrap value of 100.
Fig. 2.
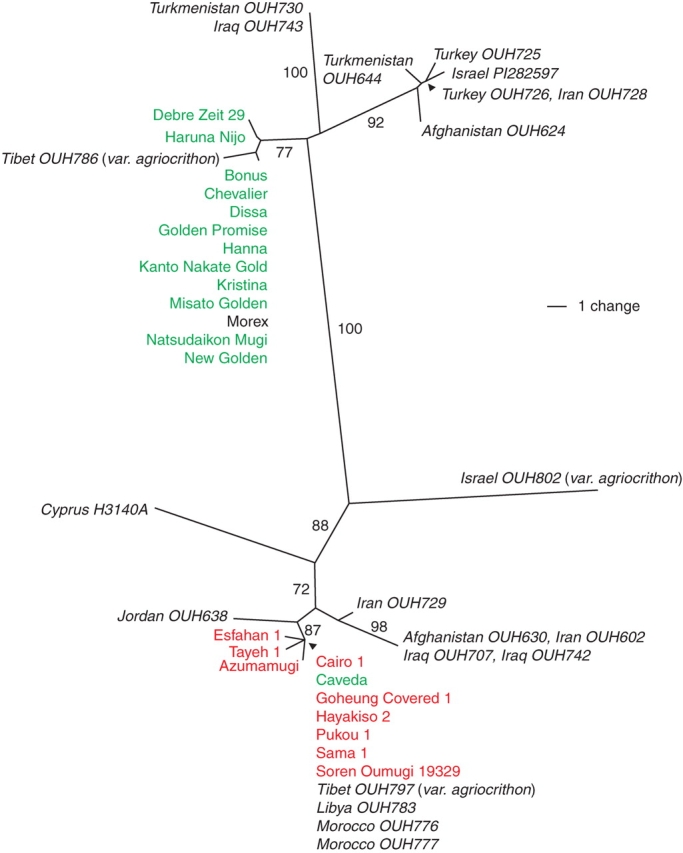
Neighbor-joining tree obtained from the sequence analysis of e09m25-08STS-Ext. Wild barley lines are represented by country of origin, followed by accession numbers in italics. The three six-rowed var. agriocrithon lines have brittle-rachis, and are classified as H. vulgare ssp. vulgare (Bothmer and Jacobsen, 1985). Cultivated barley lines are represented in plain text. Cultivars in the upper clade shown in green are W-types (btr1) except for ‘Morex’ (unknown btr status). Cultivars in the lower clade shown in red are E-types (btr2) except for the W-type ‘Caveda’. Bootstrap values with 1000 replicates > 60 % are shown.
Each clade contained a mixture of wild and domesticated types. The upper clade included all (bar one) of the W-type cultivars, together with a group of wild barleys of diverse geographical origin (e.g. OUH624 from Afghanistan, OUH728 from Iran, OUH725 and OUH726 from Turkey, OUH644 from Turkmenistan, and PI282597 from Israel), while a small sub-clade linked two accessions from Iraq (OUH743) and Turkmenistan (OUH730). The Japanese cultivars fell within the W-type cluster, as expected, given that they were bred from European germplasm. The Ethiopian ‘Debre Zeit 29’ (a variety classified deficiens in some cases) also belonged to this cluster, along with one var. agriocrithon accession from Tibet (OUH786) (Fig. 2). The other major clade included wild barleys from Jordan (OUH638), Iran (OUH729), the Caspian Sea Region (OUH602), Iraq (OUH707 and OUH742) and Afghanistan (OUH630). A wild barley from Cyprus (H3140A) and a var. agriocrithon line from Israel (OUH802) were also grouped in this clade but were distantly separated from the other members. The E-type cultivars all clustered within this clade, which also contained the Spanish ‘Caveda’ (a W-type cultivar) and one Tibetan var. agriocrithon line (OUH797). The sequence of the Moroccan (OUH776 and OUH777) and Libyan (OUH783) wild barleys was identical with that of the major E-type cultivars (Fig. 2).
Sequence analysis of var. agriocrithon lines with cultivars
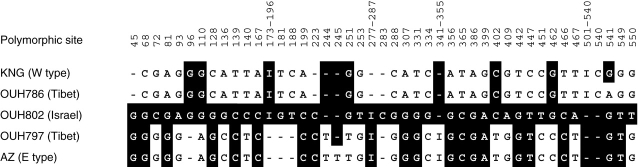
In all, 45 SNPs (single nucleotide polymorphisms) and indels were recognized in the comparison between the sequences of the var. agriocrithon lines and representative E- and W-type cultivars (Fig. 3). OUH802 has a unique sequence and its origin could not be explained by recombination events between E- and W-type versions of the sequence. No recombination was detected by Geneconv with any of the sequence pairs based on Bonferroni-corrected Karlin–Altschul P values (P > 0·05). Also no recombination was detected by DnaSP.
Fig. 3.
Sequence comparison of three var. agricrithon (OUH identifiers) lines with ‘Azumamugi’ (E-type) and ‘Kanto Nakate Gold’ (W-type). The polymorphic sites in the e09m25-08STS-Ext sequence were extracted from a multiple sequence alignment. Single nucleotide substitutions and indels shared with OUH802 are indicated by shading.
DISCUSSION
Since there are no biological barriers to hybridization between wild and cultivated barley, all wild and cultivated barleys, including var. agriocrithon, are deemed to belong to a single biological species (Bothmer et al., 1995). Thus, as a result of gene flow, any rigorous taxonomic distinction between wild and cultivated barleys is problematical. Of the traits that differentiate the wild from the cultivated form, the foremost is brittle versus non-brittle rachis. The e09m25-08STS-ext sequence, which is tightly linked with btr1/btr2, exhibits a high amount of sequence diversity in wild barley, but is less polymorphic within either the E- or the W-type groups (Figs 1 and 2). This marker thus provides a legitimate handle for inferring the history of barley domestication. Phylogenetic analysis based on the marker sequence separates the E- from the W-type barley cultivars with only a single exception (Fig. 2). A phylogeny based on AFLP markers linked to btr1/btr2 was similarly able to differentiate between the E- and W-types, while a tree constructed from AFLP loci unlinked with btr1/btr2 showed no clear separation between them (Komatsuda et al., 2004). Neither of these phylogenetic trees could resolve the wild barleys, as most clustered in the middle of the tree. In contrast, the present approach has grouped the wild lines into two clades, indicating that this marker region is probably strongly correlated with the divergence of btr1/btr2 in the E- and W-type cultivars. If the origin of domesticated barley was a multiple event, then the E- and W-type cultivars would have derived independently from widely diverged wild barleys.
There has been a long-standing debate concerning the origin of the six-rowed, brittle var. agriocrithon ever since its discovery in Tibet. Interestingly, the marker sequence of one of the var. agriocrithon lines (OUH786) was similar to that of W-type cultivars, whereas another (OUH797) shared its sequence with some of the E-type cultivars. The cultivar × ssp. spontaneum origin hypothesis requires both a recombination event between e09m25-08STS-ext and btr1/btr2 and the existence of wild barley in the vicinity of where cultivars are grown. Therefore, we propose a rather simpler model, based on a back mutation in the btr1/btr2 region from cultivar to var. agriocrithon forms in Tibet. This is a likely origin for OUH797 because E-type cultivars are six-rowed, not only in the present sample but also in general (Takahashi, 1955). The origin of OUH786 may be more complicated.
Var. agriocrithon has frequently been also found in Israel, Cyprus and Libya (for a review, see Bothmer and Jacobsen, 1985). The Israeli var. agriocrithon line used here (OUH802) was well separated from both E- and W-type cultivars, as well as from all the other wild barleys and var. agriocrithon form (Fig. 2). Its marker sequence could not have been the outcome of a hybridization event, followed by recombination (Fig. 3). Recombination analysis by Geneconv and DnaSP did not reveal any recombination between any pairs of the marker sequences. Furthermore, it is unlikely that the brittle rachis of this line originated from a double recombination event between btr1Btr2 and Btr1btr2 to generate a brittle Btr1Btr2 genotype, because these two loci are tightly linked (Takahashi and Hayashi, 1964; Komatsuda et al., 2004). The introgression of a six-rowed spike gene (vrs1) from cultivated to wild barley by outcrossing or by spontaneous mutation of Vrs1 in wild barley may therefore have been responsible for the six-rowed spike phenotype of this form. Thus the line may represent an example of the wide ranging genetic diversity of ssp. spontaneum, from which it was derived. Considerable molecular diversity within var. agriocrithon lines has been reported (Tanno and Takeda, 2004). At the least, however, the present study supports the view that var. agriocrithon lines from Tibet and Israel must have different origins (Komatsuda et al., 2004).
The wild barleys as a group do not cluster with either the E- or the W-types, and there is no clear association between geographic origin and placement within the phylogenetic tree. An exception to this generality is that the Libyan and Moroccan wild accessions share complete homology with some of the E-type cultivars. This was not surprising, given that a considerable number of North African cultivated barleys are of E-type (Takahashi et al., 1983). Although a close relationship appears to hold between Oriental and North African barley, it is unclear as to whether either the E-type cultivars originated from North African wild barley (Molina-Cano et al., 1982, 1999) or whether the two forms share the same sequence as a result of gene flow from E-type cultivars to wild barley. However, this former scenario seems improbable, given that North Africa is so geographically distant from East Asia. It is therefore hard to argue that North African wild barley could have been the immediate ancestor of the modern E-type cultivars. Morocco has not been considered as a secondary centre of barley origin (Blattner and Badani Méndez, 2001), and it has even been suggested that the Moroccan wild barley lines are weedy (Molina-Cano et al., 1982). We suppose that gene flow has resulted in ‘Caveda’ (and most of other Western–Mediterranean cultivars; Komatsuda et al., 2005) and North African wild barleys sharing alleles specific to these regions. These wild barley lines may be in a similar taxonomical situation as Tibetan var. agriocrithon.
Badr et al. (2000) excluded the possibility of a polyphyletic origin of barley, but notably this same research group has now moved to favour a diphyletic origin (Kilian et al., 2006). A polyphyletic origin is also favoured by a number of other authorities (Kolodinska et al., 2004; Komatsuda et al., 2004; Molina-Cano et al., 2005). On the basis of our comparative sequence-based study, we suggest that at least two independent brittle rachis wild populations were involved in barley domestication, and we therefore support the notion of a diphyletic origin for cultivated barley.
ACKNOWLEDGEMENTS
We thank the Research Institute for Bioresources, Okayama University, Kurashiki, Japan for providing the plant materials, D. Vidya Saraswathi for her technical assistance and Y. Turuspekove and M. Pourkheirandish for their discussions. We thank R. Koebner for linguistic assistance in the preparation of this manuscript. A research grant from Core Research for Evolutional Science and Technology (CREST) to T. Komatsuda is gratefully appreciated. P. Azhaguvel is a research fellow of Japan Society for the Promotion of Science (JSPS). Funding to pay the Open Access publication charges for this article was provided by the OECD.
LITERATURE CITED
- Åberg E. Hordeum agriocrithon nova sp., a wild six-rowed barley. Annals of the Agricultural College of Sweden. 1938;6:159–216. [Google Scholar]
- Åberg E. The taxonomy and phylogeny of Hordeum L. sect. Cerealia Ands. Symbolae Botanicae Upsalienses. 1940;4:1–156. [Google Scholar]
- Azhaguvel P, Vidya Saraswathi D, Komatsuda T. High-resolution linkage mapping for the non-brittle rachis locus btr1 in cultivated × wild barley (Hordeum vulgare) Plant Science. 2006;170:1087–1094. [Google Scholar]
- Badr A, Müller K, Schäer-Pregl R, El Rabey H, Effgen S, Ibraim HH, et al. On the origin and domestication history of barley (Hordeum vulgare) Molecular Biology and Evolution. 2000;17:499–510. doi: 10.1093/oxfordjournals.molbev.a026330. [DOI] [PubMed] [Google Scholar]
- Bekele E. A differential rate of regional distribution of barley flavoniod patterns in Ethiopia, and a view on the centre of origin of barley. Heriditas. 1983;98:269–280. doi: 10.1111/j.1601-5223.1983.tb00605.x. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Badani Méndez A. RAPD data do not support a second centre of barley domestication in Morocco. Genetic Resources and Crop Evolution. 2001;48:13–19. [Google Scholar]
- Bothmer RV, Jacobsen N. Origin, taxonomy, and related species. In: Rasmusson D, editor. Barley ASA agronomy monograph. Vol. 26. Madison, WI: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America; 1985. pp. 19–56. [Google Scholar]
- Bothmer RV, Jacobsen N, Baden C, Jørgensen R, Linde-Laursen I. Systematic and ecogeographic studies on crop genepools. 2nd edn. Vol. 7. Rome: International Plant Genetic Resources Institute; 1995. An ecogeographical study of the genus Hordeum. [Google Scholar]
- Freisleben R. Ein neuer Fund von Hordeum agriocrithon Åberg. Zuchter. 1943;15:25–29. [Google Scholar]
- Harlan JR. Crops and man. Madison, WI: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America; 1992. [Google Scholar]
- Kilian B, Özkan H, Kohl J, von Haeseler A, Barale F, Deusch O, et al. Haplotype structure at seven barley genes: relevance to gene pool bottlenecks, phylogeny of ear type and site of barley domestication. Molecular Genetics and Genomics. 2006;276:230–241. doi: 10.1007/s00438-006-0136-6. [DOI] [PubMed] [Google Scholar]
- Kolodinska BA, Bothmer RV, Dayteg C, Rashal I, Tuvesson S, Weibull J. Inter simple sequence repeat analysis of genetic diversity and relationships in cultivated barley of Nordic and Baltic origin. Hereditas. 2004;141:186–192. doi: 10.1111/j.1601-5223.2004.01867.x. [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Mano Y. Molecular mapping of the intermedium spike-c (int-c) and non-brittle rachis 1 (btr1) loci in barley (Hordeum vulgare L.) Theoretical and Applied Genetics. 2002;105:85–90. doi: 10.1007/s00122-001-0858-0. [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Azhaguvel P, Vidya-Saraswathi D. Are North African barleys of type Eastern or Western? Czech Journal of Genetics and Plant Breeding. 2005;41(Special Issue):96. [Google Scholar]
- Komatsuda T, Maxim P, Senthil N, Mano Y. High-density AFLP map of nonbrittle rachis 1 (btr1) and 2 (btr2) genes in barley (Hordeum vulgare L.) Theoretical and Applied Genetics. 2004;109:986–995. doi: 10.1007/s00122-004-1710-0. [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Nakamura I, Takaiwa F, Oka S. Development of STS markers closely linked to the vrs1 locus in barley. Hordeum vulgare. Genome. 1998;41:680–685. [Google Scholar]
- Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proceedings of the National Academy of Sciences of the USA. 2007;104:1424–1429. doi: 10.1073/pnas.0608580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T. Genetic diversity in Hordeum agriocrithon E. Åberg, six-rowed barley with brittle rachis, from Tibet. Genetic Resources and Crop Evolution. 2001;48:27–34. [Google Scholar]
- Molina-Cano JL, Gómèz-Campo C, Conde J. Hordeum spontaneum C.Koch as a weed of barley fields in Morocco. Zeitschrift fur Pflanzenzuchtung. 1982;88:161–167. [Google Scholar]
- Molina-Cano JL, Fra-Mon P, Salcedo G, Aragoncillo C, Roca de Togores F, García Olmedo F. Morocco as a possible domestication center for barley: biochemical and agromorphological evidence. Theoretical and Applied Genetics. 1987;73:531–536. doi: 10.1007/BF00289190. [DOI] [PubMed] [Google Scholar]
- Molina-Cano JL, Moralejo M, Igartua E, Romagosa I. Further evidence supporting Morocco as a center of origin of barley. Theoretical and Applied Genetics. 1999;98:913–918. [Google Scholar]
- Molina-Cano JL, Russell JR, Moralejo MA, Escacena JL, Arias G, Powell W. Chloroplast DNA microsatellite analysis supports a polyphyletic origin for barley. Theoretical and Applied Genetics. 2005;110:613–619. doi: 10.1007/s00122-004-1878-3. [DOI] [PubMed] [Google Scholar]
- Rozas JJ, Sánchez-DelBarrio Messeguer X, Rozas R. DnaSP, DNA polymorphism analysis by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The Neighbor-joining Method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sawyer SA. Statistical tests for detecting gene conversion. Molecular Biology and Evolution. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Senthil N, Komatsuda T. Comparative mapping of non-brittle rachis genes btr1 and btr2 using wild × occidental and wild × oriental cultivar populations in barley Hordeum vulgare L. Euphytica. 2006;145:215–220. [Google Scholar]
- Swofford D. PAUP*. Phylogenetic analysis using parsimony (*and other methods) Sunderland, MA: Sinauer Associates; 1998. ver 4. [Google Scholar]
- Takahashi R. The origin and evolution of cultivated barley. In: Demerec M, editor. Advances in genetics. Vol. 7. New York, NY: Academic Press; 1955. pp. 227–266. [Google Scholar]
- Takahashi R, Hayashi J. Linkage study of two complementary genes for brittle rachis in barley. Berichte des Ohara Institute für Landwirtschaftliche Biologie, Okayama University. 1964;12:99–105. [Google Scholar]
- Takahashi R, Yasuda S, Hayashi J, Fukuyama T, Moriya I, Konishi T. Catalogue of barley germplasm preserved in Okayama University. Kurashiki, Japan: Okayama University; 1983. [Google Scholar]
- Tanno K, Takeda K. On the origin of six-rowed barley with brittle rachis, agriocrithon (Hordeum vulgare ssp. f. agriocrithon (Åberg) Bowd.), based on a DNA marker closely linked to the vrs1 (six-row gene) locus. Theoretical and Applied Genetics. 2004;110:145–150. doi: 10.1007/s00122-004-1816-4. [DOI] [PubMed] [Google Scholar]
- Tanno K, Takaiwa F, Oka S, Komatsuda T. A nucleotide sequence linked to the vrs1 locus for studies of differentiation in cultivated barley (Hordeum vulgare L.) Hereditas. 1999;130:77–82. doi: 10.1111/j.1601-5223.1999.00077.x. [DOI] [PubMed] [Google Scholar]
- Tanno K, Taketa S, Takeda K, Komatsuda T. A DNA marker closely linked to the vrs1 locus (row-type gene) indicates multiple origins of six-rowed cultivated barley (Hordeum vulgare L.) Theoretical and Applied Genetics. 2002;104:54–60. doi: 10.1007/s001220200006. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acid Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidya Saraswathi D, Azhaguvel P, Senthil N, Koba T, Komatsuda T. Molecular mapping of non-brittle rachis genes btr1 and btr2 using STS markers in barley. Japan Agricultural Research Quarterly. 2006;40:239–242. [Google Scholar]
- Xu TW. Origin and evolution of cultivated barley in China. Acta Genetica Sinica. 1982;9:440–446. [Google Scholar]
- Yu Y, Tomkins JP, Waugh R, Frisch DA, Kudrna D, Kleinhofs A, et al. A bacterial artificial chromosome library for barley (Hordeum vulgare L.) and the identification of clones containing putative resistance genes. Theoretical and Applied Genetics. 2000;101:1093–1099. [Google Scholar]
- Zohary D. Spontaneous brittle six-rowed barley, their nature and origin. Proceedings of the 1st International Barley Genetics Symposium; 26–31; Wageningen, The Netherlands. Wageningen: 1963. Aug, pp. 27–31. [Google Scholar]
- Zohary D. The mode of domestication of the founder crops of Southwest Asian agriculture. In: Harris DR, editor. The origins and spread of agriculture and pastoralism in Eurasia. London: University College London Press; 1996. pp. 142–158. [Google Scholar]
- Zohary D, Hopf M. Domestication of plants in the old world. 2nd edn. Oxford: Oxford University Press; 1993. [Google Scholar]