Abstract
Background
Archaeological evidence has revealed that barley (Hordeum vulgare) is one of the oldest crops used by ancient farmers. Studies of the time and place of barley domestication may help in understanding ancient human civilization.
Scope
The studies of domesticated genes in crops have uncovered the mechanisms which converted wild and unpromising wild species to the most important food for humans. In addition to archaeological studies, molecular studies are finding new insights into the process of domestication. Throughout the process of barley domestication human selection on wild species resulted in plants with more harvestable seeds. One of the remarkable changes during barley domestications was the appearance of six-rowed barley. The gene associated with this trait results in three times more seed per spike compared with ancestral wild barley. This increase in number of seed resulted in a major dichotomy in the evolution of barley. The identification of the six-rowed spike gene provided a framework for understanding how this character was evolved. Some important barley domestication genes have been discovered and many are currently being investigated.
Conclusions
Identification of domestication genes in crops revealed that most of the drastic changes during domestication are the result of functional impairments in transcription factor genes, and creation of new functions is rare. Isolation of the six-rowed spike gene revealed that this trait was domesticated more than once in the domestication history of barley. Six-rowed barley is derived from two-rowed ancestral forms. Isolation of photoperiod-response genes in barley and rice revealed that different genes belonging to similar genetic networks partially control this trait.
Key words: Barley, Hordeum, domestication gene, genome evolution, common ancestor, vrs1
INTRODUCTION
Some 10 000 years before present (bp) ancient farmers selected wild species leading to domestication of crops on which humans are dependent today. During this agricultural revolution, people saved seeds from plants with favoured traits for the next generation, and over time they converted seemingly unpromising wild species into reliable and bountiful crops (Doebley, 2004). Modification included increase in the number of seeds, improved seed fertility, change in plant architecture, change in seed size and shape, adaptation of flowering time to different areas, and loss of seed shattering.
Hordeum, Triticum and Secale belong to the tribe Triticeae, the Poaceae family. Poaceae is considered to be monophyletic; therefore all grasses belonging to this family may have evolved from a single ancestor (Devos, 2005). The genus Hordeum consists of 32 species and 45 taxa including diploid (2n = 2x = 14), tetraploid (2n = 4x = 28) and hexaploid (2n = 6x = 42) cytotypes (Bothmer et al., 1995). The majority of Hordeum species are perennials and different species have different reproductive systems (Bothmer et al., 2003). Cultivated barley (H. vulgare ssp. vulgare L.) and its wild progenitor (H. vulgare ssp. spontaneum C. Koch.) belong to a single biological species, which is an annual and is diploid. No crossing barriers have been developed between the wild and cultivated forms, therefore spontaneous and artificial crosses are easily obtained (Asfaw and Bothmer, 1990). There has been a high frequency of introgression in areas where the wild and cultivated forms are in close contact.
The immediate ancestor of cultivated barley was first discovered in Turkey by the German botanist Carl Koch, and described by him as a separate species, H. spontaneum. However, based on the biological species concept (Bothmer et al., 1995), the progenitor form is nowadays regarded as a subspecies [ssp. spontaneum (C. Koch) Thell.] within the same major species, H. vulgare, as cultivated barely (ssp. vulgare) (Bothmer et al., 2003). The first definite sign of barley cultivation has been recorded from the Middle East ‘arc’ more than 10 000 bp (Zohary and Hopf, 2000). To investigate the time and place of barley domestication, the study of genes related to key steps in domestication can give valuable insights. In this review, recent progress in understanding the transition of wild barley to domesticated forms is described by focusing on genetics and biological functions of important traits that have been induced during the domestication of barley cultivars.
IMPORTANT TRAITS FOR BARLEY DOMESTICATION AND MIGRATION
During the process of domestication, barley has gradually accumulated traits that facilitated agricultural production. Selection may have been unconscious, i.e. as a result of environmental selection or conscious as a result of deliberate choice by man (Bothmer et al., 2003). Three key traits – selection for non-brittle rachis, six-rowed spike and naked caryopsis – were involved in barley domestication (Salamini et al., 2002). These mutations are associated with the transition of wild barley to cultivated barley. Migration of barley to regions outside its place of origin was accelerated through mutations to develop reduced vernalization requirement and photoperiod insensitivity (Bothmer et al., 2003). Barley was spread to different geographic areas by the accumulation of diversity for these traits.
Non-brittle rachis
The most important trait for barley domestication is probably non-brittle rachis. Non-brittle rachis results in efficient harvest without loss of grains. Spikes of the non-brittle mutant remain longer on the plant in the field after maturation, so spikes with this mutant were harvested with higher frequency than spikes with brittle rachis by ancient farmers (Bothmer et al., 1990). Seed dispersal systems are designed to enable wild plants to survive in nature, but the loss of natural dispersal mechanisms was essential for agriculture. The earliest archaeological clue for non-brittle barley comes from Tell Abu Hureyra from 9500 bp (Hillman et al., 1989). Seed shattering in barley has two forms: brittle rachis and weak rachis (Kandemir et al., 2000, and references cited therein).
In Hordeum, spikes disarticulate immediately above each rachis node to form typical wedge-shaped spikelets (Bothmer et al., 1995). Disarticulation scars in wild barley are smooth which helps in seed dispersal, whereas in cultivated barley threshing produces rough dehiscence scars on grains detached from rachis segments. Anatomically, the rachis nodes are clearly constricted in brittle spikes, but are not constricted in non-brittle spikes (Ubisch, 1915). The brittleness of the rachis in barley promotes seed dispersal together with the rough awn, which can become attached to animals for effective dispersal (Bothmer et al., 1995).
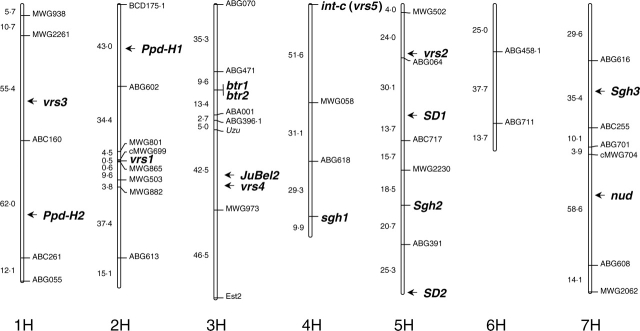
The most important non-brittle rachis genes for barley domestication are btr1 and btr2. Takahashi and Yamamoto (1949) clarified the monogenic recessive inheritance of non-brittle rachis of cultivated barley based on crosses between cultivars and ssp. spontaneum. Two-way test crosses revealed that two independent recessive genes (btr1 and btr2) cause non-brittle rachis (Takahashi and Hayashi, 1964). Btr1Btr2 (double dominant genotypes) strongly constrict the rachis node, whereas one recessive allele btr1Btr2 or Btr1btr2 does not result in constriction of the rachis node (Ubisch, 1915). These recessive genes have been independently established by natural mutations from wild progenitors, which have a brittle rachis (Fig. 1; Takahashi, 1987). The two genes are tightly linked (Takahashi and Hayashi, 1964) and located on the short arm of barley chromosome 3HS (Fig. 2; Komatsuda and Mano, 2002). The recessive nature of non-brittle rachis suggests a mutation for loss of function in Btr1 and Btr2. Phylogenetic studies using markers closely linked to btr1/btr2 determined that cultivated barley consists of two geographic types, western and eastern (Komatsuda et al., 2004; Azhaguvel and Komatsuda, 2007). These results support two independent domestication hypotheses of barley as proposed by Takahashi (1955).
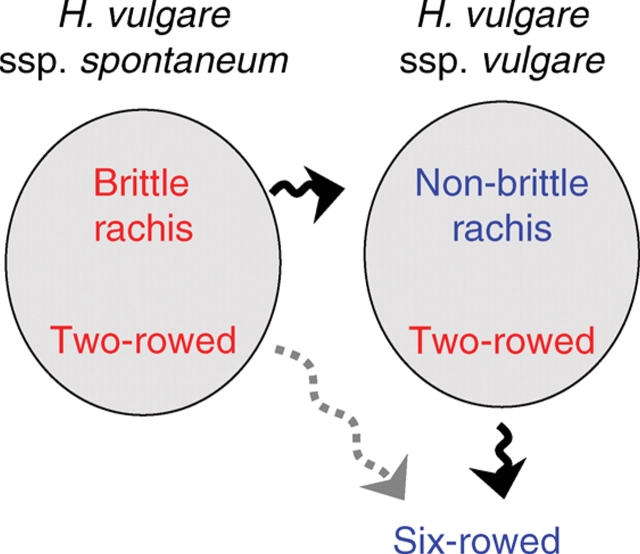
Fig. 1.
A schematic diagram of the domestication process in barley regarding brittle rachis and row-type.
Fig. 2.
Consensus map of barley domestication-related genes. RFLP markers from the cross Azumamugi × Kanto Nakate Gold (Mano et al., 2001) were considered. Distances between markers are given in centimorgans. Genes were imposed on the genetic map by comparative mapping approach.
Allelic variation was implied from Btr1 and Btr2. Alleles of Btr1.a and Btr2.k complementary produced brittle rachis in the presence of the dominant D gene (chromosome 7H), whereas Btr1.h and Btr2.h of wild barley do not need the D factor to produce brittle rachis (Komatsuda and Mano, 2002; Senthil and Komatsuda, 2005). Komatsuda et al. (2004) showed two QTLs on chromosomes 5H and 7H (the D gene) for brittle rachis in addition to Btr1 and Btr2.
Brittle-rachis genes are located on homeologous group 3 chromosomes in Hordeum, Triticum, Aegilops, Dasypyrum and Thinopyrum (Watanabe and Ikebata, 2000, and references cited therein; Li and Gill, 2006). It remains to be determined whether they are orthologous or not. Konishi et al. (2006) identified rice shattering gene qSH1 which encodes a BEL1-type homeobox gene. The qSH1 gene is located on the long arm of rice chromosome 1. Barley chromosome 3H and rice chromosome 1 are syntenous (Devos, 2005), and JuBel2 (the barley orthologue of qSH1) (Li and Gill, 2006) was mapped on the long arm of barley chromosome 3H (Fig. 2; Müller et al., 2001; Castiglioni et al., 1998). As JuBel2 and btr1/btr2 are located in different arms of barley chromosome 3H, JuBel2 does not correspond to barley btr1/btr2. Therefore, the btr1 and btr2 genes remain to be cloned. To elucidate the origin of cultivated barley, cloning of non-brittle rachis genes will be necessary. The btr1 gene has been fine-mapped and is now delimited to a 0·84-cM region using AFLP-derived STS markers (Azhaguvel et al., 2006).
Six-rowed spike
During the process of cereal domestication, humans have selected in wild species toward the general direction of increased yield (Harlan et al., 1973). One of the most conspicuous selections for increased seeds was the appearance of a six-rowed spike during barley domestication in the Middle East. Six-rowed barley produces three times as many seeds per spike as two-rowed barley and is a change of dramatic agronomic importance. Various theories have been proposed to the evolutionary pathway of six-rowed cultivars. Åberg (1938) assumed that six-rowed cultivated barley derived from a six-rowed form with a brittle rachis known as H. agriocriton found in the early 1930s in western China. Another theory assumed a single evolutionary line from ssp. spontaneum to ssp. vulgare (Fig. 1) which is nowadays in favour with barley scientists. Remains of six-rowed barley appear very early in the aceramic Neolithic beds in Tell Abu Hureyra from 8800 bp onwards (Helbaek, 1959; Zohary and Hopf, 2000). Archaeological evidence comes from Ali Kosh (Helbaek, 1969) where remains of two-rowed barley are dated at about 9000 bp with sporadic six-rowed elements among the two-rowed materials. This is a sign that the six-rowed character in barley was derived from two-rowed barley during domestication.
The spike architecture of Hordeum species is unique among the Triticeae, which possess three spikelets at each rachis node (Bothmer and Jacobsen, 1985; Bothmer et al., 1995). Exhibition of the two-rowed phenotype in wild barley suggests that the two-rowed spike is the ancestral form, which was changed to a six-rowed spike in cultivated barley by mutation during domestication. In wild and cultivated barley the central spikelet is fertile and goes on to develop into a grain. The two lateral spikelets are sterile in the two-rowed type (wild and cultivated barley), but are fertile in six-rowed type (only cultivated barley). The fertile lateral spikelets that develop into grain appeared during barley domestication (Zohary, 1963; Bothmer et al., 1995). In wild barley, the three spikelets form a light, arrowhead-like dispersal unit that both facilitates seed dispersal by animals and aids seed burial. The numerous upward-oriented barbs on the lemma and awn are also part of the dispersal and self-planting mechanisms. Reduced awn barbing seems important in its utilization as animal feed which is found in two-rowed cultivars. Six-rowed spontaneous mutants are not preferred for survival in the wild and they are eliminated naturally and rapidly from wild barley populations (Zohary, 1963; Bothmer et al., 1995).
There are at least five independent loci controlling the six-rowed spike phenotype in barley. Six-rowed spike 1 (vrs1), a recessive gene located on chromosome 2HL (Fig. 2), is observed in all six-rowed cultivars. Wild barleys have dominant alleles for Vrs1, whereas cultivated barleys have a dominant Vrs1 (two-rowed) allele or a recessive vrs1 (six-rowed) allele depending on their phenotypic row-type. Cultivated barleys with recessive allele at vrs1 are completely six-rowed over the whole spike (Lundqvist et al., 1997). More than 90 mutant lines have been induced for this locus from two-rowed barley (Lundqvist et al., 1997), which supports the hypothesis that six-rowed barley was derived from two-rowed barley by mutation. Allelic variation based on awn length development of lateral spikelets was observed among six-rowed barleys as vrs1.a and vrs1.c. In the vrs1.a allele, which exists in most of the six-rowed cultivars, awn lengths of lateral spikelets are nearly the same as the central spikelets (Fig. 3E; Lundqvist et al., 1997). The vrs1.c allele is a six-rowed allele with awn-like appendages on the lemma of lateral spikelets (Lundqvist et al., 1997). Two-rowed barleys also have different alleles of the Vrs1.p with pointed-tip lateral spikelets (Fig. 3D), Vrs1.b with round-tip lateral spikelets (Fig. 3B, C) and Vrs1.t with extremely rudimentary lateral spikelets (Fig. 3A). Analysis of a closely linked marker to vrs1 suggested a hypothesis that six-rowed barley originated more than once in the history of barley domestication (Tanno et al., 2002).
Fig. 3.
Barley spikelets in one rachis node. (A) Ethiopian landrace var. deficiens; rudimentary lateral spikelets (Vrs1.t). (B) Wild barley var. spontaneum; sterile lateral spikelets (Vrs1.b). (C) Two-rowed cultivar var. distichon; sterile lateral spikelets (Vrs1.b). (D) Wild barley var. proskowetzii; short-awned or tip-pointed lateral spikelets (Vrs1.p). (E) Six-rowed cultivar convar. vulgare; fully fertile and awned lateral spikelets (vrs1.a).
Six-rowed spike 2 (vrs2), six-rowed spike 3 (vrs3) and six-rowed spike 4 (vrs4) are independent recessive genes, which are located on chromosome 5HL, 1HL and 3HL, respectively (Fig. 2; Lundqvist et al., 1997). These loci are detected only in induced mutant lines and there are no reports of mutation in these loci in cultivars. Alleles at these loci enhance the development of lateral spikelets to various degrees depending on their position in the spike (Lundqvist et al., 1997).
Six-rowed spike 5 (vrs5 or int-c), a recessive gene located on chromosome 4HS, is detected in many two-rowed barley cultivars and more than 20 induced mutant lines (Lundqvist et al., 1997). Alleles at the int-c locus modify the degree of fertility in lateral spikelets and produce an intermediate spike type. Of two alleles of this locus observed in cultivars, the int-c.b allele prevents anther development in lateral spikelets, whereas the Int-c.h allele allows the development of anthers and promotes occasional seed set in lateral spikelets (Leonard, 1942; Woodward, 1947; Lundqvist and Lundqvist, 1987). It remains to be determined whether the wild ancestor of domesticated barley had a dominant allele for Int-c or a dominant mutation created Int-c during domestication.
Among the five six-rowed spike loci only the natural mutation on vrs1 and vrs5/int-c are observed in barley cultivars. Development of six-rowed barley is highly dependent on the evolution in these two genes. The effects of these two genes are opposite to each other in the sense that the dominant Vrs1 allele suppresses the development of lateral spikelets but the dominant Int-c.h allele promotes development of lateral spikelets and occasional seed set. Because recessive vrs1 is observed in all cultivated six-rowed barley cultivars, recessive mutation of this gene during barley domestication is the key point for the origin of six-rowed barley.
Naked caryopsis
The hulled or naked caryopsis character of barley is an important agronomic trait because of its direct link to dietary use. Hulled barley has caryopses with the husk cemented to the grain, while naked barley grows with easily separable husks upon threshing. The remains of naked kernels have been found in Ali Kosh about 8000 bp which means a mutation for this trait occurred early in the domestication of barley (Helbaek, 1969). Harlan (1995) suggested that the change to non-brittle rachis preceded the emergence of naked caryopsis during the barley domestication. A single recessive gene, nud, located on chromosome 7HL (Fig. 2; Scholz, 1955; Fedak et al., 1972), controls the naked caryopsis character, suggesting that easy separation of the husk results from a mutation that damaged gene function. Naked barley is distributed widely in the world, but there is a higher preference for naked barleys in East Asian countries such as China, Korea and Japan, and it is especially common in Tibet and the northern parts of Nepal, India and Pakistan (Bothmer et al., 2003). Since the frequency is low in the Western countries, Vavilov (1926) considered southern Asia to be a centre of origin for naked barley. It has, however, become clear that naked barley was grown in Anatolia (Turkey) and in northern Europe in ancient times (Helbaek, 1969). The Naked gene has multiple effects on many traits of barley, like yield reduction and lower seed weight (Choo et al., 2001).
Molecular analyses of a closely linked marker to the nud gene support the hypothesis of a monophyletic origin of this gene (Taketa et al., 2004). This analysis clearly separated hulled, naked and wild barley into four alleles. One hundred naked cultivars have the same allele (monophyletic), and only one wild barley (OUH625, among 53 wild lines) from south-western Iran showed the same allele as the naked barley group. No hulled barley (among 106 cultivars) that carries same allele as a naked barley group. Taketa et al. (2004) hypothesized that naked barley originated from wild barley directly or it originated from hulled domesticated barley, which is now extinct. High-density and high-resolution mapping has delimited the nud gene to a 0·66-cM region using AFLP markers (Kikuchi et al., 2003; Taketa et al., 2006). A BAC contig spanning the hulled or naked caryopsis locus (Nud/nud) with a length of 240 kb was made by chromosome walking (Amano et al., 2006).
Reduced dormancy
Seed dormancy is defined as the temporary inability of a viable seed to germinate under favourable environmental conditions (Simpson, 1990). This trait enables seeds to survive adverse conditions and allows seed dispersal to a wider region (Snape et al., 2001b). A high level of seed dormancy is a problem in cultivars when rapid seed germination on planting is needed. In the malthouse, seeds must germinate rapidly and completely upon imbibition of water, and a high level of dormancy after harvest is economically undesirable (Carn, 1980; Ullrich et al., 1993, 1997). Nevertheless, stringent phenotypic selection against seed dormancy can lead to the development of barley cultivars susceptible to pre-harvest sprouting, which is also highly undesirable (Prada et al., 2004). Generally, a moderate level of seed dormancy is thought to be appropriate for barley cultivars (Han et al., 1999; Romagosa et al., 1999).
Barley seed dormancy is a quantitative trait that is affected by several genes, environmental factors, and by gene × environment interactions (Ullrich et al., 1996). Many seed dormancy QTLs have been identified in barley (Han et al., 1996; Edney and Mather, 2004; Prada et al., 2004; Zhang et al., 2005; Vanhala and Stam, 2006). Cultivars of different pedigrees may have different dormancy genes, which explain the various QTL analysis results from different mapping populations. However, it is important to note that common major QTLs (SD1, SD2) have been identified in various studies (Li et al., 2004).
SD1 and SD2 located at different loci on chromosome 5H (Fig. 2) are important in the study of seed dormancy. SD1 has been detected as a major QTL near the centromere region on the long arm of chromosome 5H in most of the barley QTL mapping projects (Han et al., 1996; Edney and Mather, 2004; Prada et al., 2004; Zhang et al., 2005). A single Mendelian gene controls the allele at SD1 in which the dormancy allele is dominant. SD1 shows the largest and most consistent effect on dormancy (Han et al., 1999). This gene has been delimited to a 4·4-cM region in chromosome 5H (Han et al., 1996, 1999). SD2, a region near the distal end of the long arm of chromosome 5H, has been detected as a QTL for seed dormancy (Takeda, 1996; Ullrich et al., 1996). This gene controls a moderate level of seed dormancy, which makes it a promising candidate gene for utilization in barley breeding (Gao et al., 2003). Fine mapping has resolved the SD2 QTL to a 0·8-cM interval (Gao et al., 2003). A gene coding for GA20-oxidase was identified as a candidate gene of SD2 using barley–rice synteny (Li et al., 2004), but its gene identity remains to be proven. SD1 is epistatic to SD2 at early-ripening stages, but they seem to act additively at later ripening stages (Romagosa et al., 1999).
Reduced vernalization requirement
Vernalization is the requirement for a period of low temperature for a plant to make the transition from a vegetative to a reproductive state. The genes for the vernalization pathway prevent flower development during the winter, providing protection for floral organs against cold. One of the prerequisites for the expansion of barley production must have been development of a spring growth habit (no or reduced requirement for vernalization). Almost all wild barleys have a winter growth habit with the exception of a few strains which are regarded as hybrids with spring cultivars (Takahashi et al., 1963, 1968). Both winter and spring barley are cultivated in mid-latitudinal regions including North Africa, southern Europe and Asia according to climate conditions. The development of barley lines lacking a vernalization requirement expanded barley cultivation to areas where spring sowing is necessary to avoid winter injury.
Three genes sgh1, Sgh2 and Sgh3 are detected in spring growth habit barleys and their allelic genes (Sgh1, sgh2 and sgh3) are required for winter growth habit. Regarding the epistatic effects among these genes, only one genotype (Sgh1sgh2sgh3) showed winter growth habit. The multiple alleles of Sgh2I and Sgh2II, account for a gradation of vernalization requirements (Takahashi and Yasuda, 1956). Linkage analysis revealed the location of these spring growth habit genes on barley chromosomes 4H, 5H and 7H, respectively (Fig. 2; Takahashi and Yasuda, 1956, 1958; Yasuda, 1969; Laurie et al., 1995; Yan et al., 2006).
The first domesticated barleys are likely to have had a winter growth habit. Later a dominant mutation occurred in the sgh2 locus, resulting in spring barley of Sgh1Sgh2sgh3 (type I) (Takahashi et al., 1963, 1968). The Sgh2 mutation occurred many times independently based on molecular analysis of the gene region (Ohmura et al., 2006). Subsequently, mutations from Sgh1 and sgh3 might have occurred independently in type I, because both sgh1 and Sgh3 spring genes are mostly associated with Sgh2. Other genotypes are supposed to be hybrids (Takahashi et al., 1963, 1968; Bothmer et al., 2003), but the multiple origin of Sgh2 implies a more complex story. These three genes have also been called in barley Vrn-H1 (corresponding to Sgh2), Vrn-H2 (corresponding to Sgh1) and Vrn-H3 (corresponding to Sgh3). Similar epistatic interactions and map locations indicate that barley and wheat vernalization genes are orthologous (Laurie et al., 1995; Dubcovsky et al., 1998; Yan et al., 2004), and wheat VRN1 (orthologue of barley Sgh2), VRN2 (orthologue of barley Sgh1) and VRN3 (orthologue of barley Sgh3) were isolated by positional cloning (Yan et al., 2003, 2004, 2006).
Both Sgh2 and VRN1 are dominant for spring growth habit. The wheat VRN1 gene is an APETALA1 gene of the MADS-box gene family, which initiates the transition from the vegetative to reproductive apex (Yan et al., 2003). Both Sgh1 and VRN2 are dominant for winter growth habit. Wheat VRN2 encodes zinc finger in the first exon and the CCT domain in the second exon (called ZCCT1), which inhibits the transition of plants from the vegetative to reproductive stage (Yan et al., 2004). Transcription of this gene is gradually down-regulated by vernalization. Therefore wheat VRN1 and VRN2 have opposite transcription profiles. Southern blot analysis of barley genomic DNA using wheat ZCCT1 as a probe in 85 barley cultivars showed the presence of this gene in 23 winter barley lines and deletion of this gene in 62 spring barley lines (Yan et al., 2004). Both barley Sgh3 and wheat VRN3 are dominant for spring growth habit. These genes are FLOWERING LOCUS T (FT) orthologues and were isolated from rice and arabidopsis related to flowering time (Yan et al., 2006, and references cited therein). The dominant allele with a high level of transcript shows early flowering in barley and wheat. Based on the tentative model present by Yan et al. (2006), VRN2 negatively regulates VRN1 and VRN3 and VRN2 is down-regulated by vernalization. First intron of Vrn-H1 may include an intronic regulatory element for the flowering repressor mediated by Vrn-H2 gene products of barley (Fu et al., 2005).
In barley, photoperiod has an important main effect and interactive role with vernalization in determining flowering time (Karsai et al., 2005; Trevaskis et al., 2006). Vernalization and photoperiod sensitivity may contribute to low temperature tolerance by maintaining plants in a vegetative state and expression of low temperature tolerance genes (Karsai et al., 2001; Mahfoozi et al., 2001).
Photoperiod insensitivity
Plants have evolved to ensure that flowering occurs when there is the greatest chance of pollination, seed development and seed dispersal. These constraints apply to wild ancestors, but the modification of flowering time by human selection has been essential to the spread of barley worldwide.
In barley, flowering time is a highly variable phenotypic trait with major implications for adaptation to geographic regions (Calder, 1965). This physiological trait is controlled by many genes including photoperiod response genes (Laurie, 1997). Plant growth and development, including photoperiod-dependent flowering, is regulated by the products of the red/far-red light phytochrome and the blue/UV-A light cryptochrome photoreceptor gene families (Cashmore et al., 1999; Lin, 2000; Quail, 2002). Wild barley has been classified as a quantitative long-day (LD) species, implying that the heading time is advanced by increasing day length (Boyd et al., 2003). Spring barley accumulated LD-insensitive mutants to allow an extended vegetative growth period under LD. This is likely to have favoured expansion of the barley production area into higher latitudes.
The major determinant of LD response in barley is the Ppd-H1 locus located on chromosome 2HS (Fig. 2; Laurie et al., 1995; Karsai et al., 1997; Decousset et al., 2000). Wild barleys have a dominant allele at the Ppd-H1 locus, while cultivated barleys can be divided into two groups either having or lacking the dominant allele. Ppd-H1 plants head about 20 d earlier than ppd-H1 plants under LD conditions (16 h of light) (Turner et al., 2005). The recessive nature of photoperiod-insensitive, ppd-H1, suggests that reduced response results from a mutation that impairs gene function. Cross-hybridizing markers show that Ppd-H1 can be homoeologous to the wheat Ppd-1 series of genes on chromosome group 2 of wheat (Snape et al., 2001a). Cloning of Ppd-H1 (Turner et al., 2005) revealed a pseudo-response regulator (PRR) which is different from the major rice photoperiod response genes, Hd1 and Hd3a (Yano et al., 2000; Kojima et al., 2002) but is likely to be orthologous to rice Hd2 (OsPRR37; Murakami et al., 2005). Significant pleiotropic effects of the Ppd-H1 locus under LD on plant height, plant yield, tiller yield, spike length and grain number per tiller were detected (Laurie et al., 1994; Sameri et al., 2006). In all cases, presence of the photoperiod-insensitive allele resulted in higher values. A second major photoperiod response gene, Ppd-H2, has been mapped on the long arm of the chromosome 1H (Fig. 2). Explicit differences in flowering time under short days (10 h) were observed, but Ppd-H2 has little effect under long days (13–16 h) (Laurie et al., 1995; Szücs et al., 2006). Genes and mutants at >14 other loci have been associated with earliness in barley (Lundqvist et al., 1997). These genes in various combinations permit plant breeders to adapt barley for production in many parts of the world.
ISOLATION OF SIX-ROWED SPIKE 1, VRS1 GENE
Elucidation of the origin of six-rowed barley has been a long-term goal of barley scientists. Helbaek (1959) hypothesized that six-rowed barley originated from two-rowed cultivated barley based on archaeological evidence. Tanno et al. (2002) proposed that six-rowed barley had two different origins based on sequence analysis of a closely linked DNA marker to vrs1. Isolation of the vrs1 gene is crucial for testing these hypotheses.
To clone vrs1, the six-rowed spike gene was delimited to 1 cM using STSs derived from RFLP and AFLP markers (Komatsuda et al., 1998, 1999; He et al., 2004). Comparison of six high-resolution maps using different parents revealed that marker order around the vrs1 gene is constant and there is no drastic rearrangement among barley cultivars in this region (Komatsuda and Tanno, 2004). After long-term gene mapping efforts, a barley–rice synteny approach was used taking advantage of the complete rice genomic sequence (International Rice Genome Sequencing Project, 2005) and extensive available barley ESTs in public databases for marker enrichment. This strategy enabled vrs1 to be delimited to 0·06 cM (Pourkheirandish et al., 2007). The vrs1 gene was isolated by means of positional cloning (Komatsuda et al., 2007). A BAC contig spanning the vrs1 locus with a length of 518 kb was made by chromosome walking. Cloning of the vrs1 gene revealed that Vrs1 encodes a member of the homeodomain-leucine zipper (HD-ZIP) I class of transcription factors. The dominant nature of Vrs1 suggests VRS1 is a repressor protein that binds to the DNA of genes and regulates the development of lateral spikelets as a transcription factor. The result agrees with the theory of Doebley (2006) that most domestication genes involve changes to transcription factors.
Analysis of induced mutants from two-rowed barley (Komatsuda et al., 2007) revealed that loss of function of Vrs1 caused six-rowed or intermediate phenotypes in mutant spikes. The functional character of Vrs1 in two-rowed barley agrees with the hypothesis of Helbaek (1969) based on archaeological evidence that six-rowed barley originated from two-rowed barley. This result also confirmed that two-rowed barley is the ancestral form of barley. In situ hybridization of Vrs1 revealed that this gene is expressed only in lateral spikelets at the immature stage (Komatsuda et al., 2007), which is the reason that this gene only affects the development of lateral spikelets. Sequence analysis of the vrs1 gene among a worldwide collection of six-rowed cultivars revealed three independent origins for six-rowed barley. Two of them (alleles vrs1.a2 and vrs1.a3) were derived from their immediate ancestor two-rowed cultivated barley (alleles Vrs1.b2 and Vrs1.b3 Fig. 3C) by a single nucleotide mutation. This result agrees with the hypothesis of Tanno et al. (2002) that six-rowed barley originated more than once during barley domestication. The origin of vrs1.a1, the most widespread allele and probably the first allele of six-rowed barley, was not found among two-rowed cultivars tested in the study (Komatsuda et al., 2007). It remains to be determined whether this six-rowed allele was derived from extinct two-rowed cultivated barley or it is directly derived from wild barley and then outcrossing with non-brittle lines resulted in six-rowed cultivars with the vrs1.a1 allele. To answer this question, sequence analysis of vrs1 among a worldwide collection of wild barleys would be useful.
Characterization of a 518-kb barley contig harbouring the vrs1 gene revealed that the six-rowed spike 1 gene is located in a gene poor-region (Pourkheirandish et al., 2007). Recombination was highly suppressed around the vrs1 gene in this contig. Comparison of the barley vrs1 contig and rice genome sequence revealed that the rice orthologue of vrs1 (Oshox14) is located in rice chromosome 7 instead of a collinear region in rice chromosome 4. This result suggests that a translocation occurred in barley or rice during evolution.
CONCLUSIONS
The above review shows the importance of understanding on genes related to crop domestication and migration. Crop domestication and migration made agriculture efficient, which was the critical stage in development of human civilizations (Salamini et al., 2002). It is hypothesized that changing climate might have lead to communities cultivating wild plants in a limited area to survive and that was the starting point of human agriculture (Salamini et al., 2002). The precise time/s when and region/s where barley domestication occurred are not entirely known. The vrs1 gene affects the visual yield of barley because plants have three times as many seeds per spike as two-rowed plants. The study of isolated alleles at the six-rowed spike locus has resulted in a leap in the understanding of barley domestication. To further elucidate the story of barley domestication, it will be necessary to isolate and study critical genes such as non-brittle rachis (btr1 and btr2) and naked caryopsis (nud).
Each cereal crop is derived from a single common ancestor (Moore, 1995). Based on the assumption of Moore (1995) different crops should carry the same genes derived from their common ancestor. The grass family emerged approx. 60 million years ago and the Triticeae diverged approx. 12 million years ago (Devos, 2005). Traits are controlled by a number of genes located in different genetic loci, acting as a network within the cell. These networks are inherited from a common ancestor of crops. The ancestors of crops diverged from each other millions of years ago, but domestication was initiated about 10 000 bp. Thus, different mutations during domestication may have targeted different genes from the same network corresponding to a single trait in different crops. For example, isolation of photoperiod-response genes in barley and rice revealed that the mechanism of photoperiod response was inherited from a common ancestor in barley and rice, but different mutations targeted different genes during evolution after separation of barley and rice (Turner et al., 2005). Most of the domestication genes isolated so far are transcription factors (Doebley, 2006). The regulatory genes, which control the expression of structural genes in target tissue at specific developmental stages, may evolve to control different genes downstream from these regulatory genes in different crops. Even though some regulatory elements are orthologues in different crops, it is possible that these orthologue elements interfere with different responses. Most of the domestication traits are the result of a mutation that leads to a loss of function (Doebley, 2006). The vrs1 locus has a unique function in barley that controls the development of lateral spikelets. This function may not exist in the other cereals like wheat or rice because of their inflorescence structure. Comparison of barley and rice for vrs1 revealed that, even though an vrs1 orthologue exists in rice, it is not in a collinear location with barley, which revealed an evolutionary rearrangement. This rearrangement may relate to a special function of this gene in barley.
ACKNOWLEDGMENTS
We thank Dr S. Taketa, Dr S. Nakamura, Dr K. Kato and Dr D. Vaughan for comments on the manuscript. The financial support from the Ministry of Agriculture, Forestry and Fisheries of Japan (Rice Genome Project MP1113b and Green Techno Project GD3006) and research grant of CREST program of Japan Science and Technology Agency (JST) to T. Komatsuda are gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by the OECD.
LITERATURE CITED
- Åberg E. Hordeum agriocrithon nova sp., a wild six-round barley. The Annals of the Agricultural College of Sweden. 1938;6:159–216. [Google Scholar]
- Amano S, Awayama T, Saisho D, Sato K, Takeda K, Kawasaki S, et al. Construction of a BAC contig spanning the hulled or naked caryopsis locus (Nud/nud) in barley. Breeding Research. 2006;8:66. [Google Scholar]
- Asfaw Z, von Bothmer R. Hybridization between landrace varieties of Ethiopian barley (Hordeum vulgare ssp. vulgare) and the progenitor of barley (H. vulgare ssp. spontaneum) Hereditas. 1990;112:57–64. [Google Scholar]
- Azhaguvel P, Komatsuda T. A phylogenetic analysis based on nucleotide sequence of a marker linked to the brittle rachis locus indicates a diphyletic origin of barley. Annals of Botany. doi: 10.1093/aob/mcm129. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhaguvel P, Vidya-Saraswathi D, Komatsuda T. High-resolution linkage mapping for the non-brittle rachis locus btr1 in cultivated × wild barley (Hordeum vulgare) Plant Science. 2006;170:1087–1094. [Google Scholar]
- von Bothmer R, Jacobsen N. Origin, taxonomy, and related species. In: Rasmusson D, editor. Barley. Vol. 26. Madison, WI: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America; 1985. pp. 19–56. ASA Agronomy Monograph. [Google Scholar]
- von Bothmer R, Yen C, Yang JL. Does wild, six-rowed barley, Hordeum agriocrithon really exist? Plant Genetic Resources Newsletter. 1990;77:17–19. [Google Scholar]
- von Bothmer R, Jacobsen N, Baden C, Jørgensen RB, Linde-Laursen I. An ecogeographical study of the genus Hordeum. 2nd edn. Rome: International Plant Genetic Resources Institute; 1995. Systematic and Ecogeographic Studies on Crop Genepools 7. [Google Scholar]
- von Bothmer R, Sato K, Komatsuda T, Yasuda S, Fischbeck G. The domestication of cultivated barley. In: von Bothmer R, Hintum Tv, Knüpffer H, Sato K, editors. Diversity in barley (Hordeum vulgare) Amsterdam: Elsevier; 2003. pp. 9–27. [Google Scholar]
- Boyd WJR, Li CD, Grime CR, Cakir M, Potipibool S, Kaveeta L, et al. Conventional and molecular genetic analysis of factors contributing to variation in the timing of heading among spring barley (Hordeum vulgare L.) genotypes grown over a mild winter growing season. Australian Journal of Agricultural Research. 2003;54:1277–1301. [Google Scholar]
- Calder DM. Proceedings of the 12th Easter School in Agricultural Science. Butterworths; 1965. The growth of cereals and grasses. [Google Scholar]
- Carn JD. Detection of rain damaged barley at harvest (preharvest sprouting). Proceedings of the 30th annual Australian Cereal Chemistry Conference; Melbourne: Royal Australian Chemistry Institute; 1980. pp. 34–38. [Google Scholar]
- Cashmore AR, Jarillo JA, Wu Y-J, Liu D. Cryptochromes: blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- Castiglioni P, Pozzi C, Heun M, Terzi V, Müller KJ, Rohde W, Salamini F. An AFLP-based procedure for the efficient mapping of mutations and DNA probes in barley. Genetics. 1998;149:2039–2056. doi: 10.1093/genetics/149.4.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo T-M, Ho KM, Martin RA. Genetic analysis of a hulless × covered cross of barley using doubled-haploid lines. Crop Science. 2001;41:1021–1026. [Google Scholar]
- Decousset L, Griffiths S, Dunford RP, Pratchett N, Laurie DA. Development of STS markers closely linked to the Ppd-H1 photoperiod response gene of barley (Hordeum vulgare L.) Theoretical and Applied Genetics. 2000;101:1202–1206. [Google Scholar]
- Devos KM. Updating the ‘Crop Circle. Current Opinion in Plant Biology. 2005;8:155–162. doi: 10.1016/j.pbi.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Doebley J. The genetics of maize evolution. Annual Review of Genetics. 2004;38:37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- Doebley J. Unfallen grains: how ancient farmers turned weeds into crops. Science. 2006;312:1318–1319. doi: 10.1126/science.1128836. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theoretical and Applied Genetics. 1998;97:968–975. [Google Scholar]
- Edney MJ, Mather DE. Quantitative trait loci affecting germination traits and malt friability in a two-rowed by six-rowed barley cross. Journal of Cereal Science. 2004;39:283–290. [Google Scholar]
- Fedak G, Tsuchiya T, Helgason SB. Use of monotelotrisomics for linkage mapping in barley. Canadian Journal of Genetics and Cytology. 1972;14:949–957. [Google Scholar]
- Fu D, Szücs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Molecular Genetics and Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- Gao W, Clancy JA, Han F, Prada D, Kleinhofs A, Ullrich SE. Molecular dissection of a dormancy QTL region near the chromosome 7 (5H) L telomere in barley. Theoretical and Applied Genetics. 2003;107:552–559. doi: 10.1007/s00122-003-1281-5. [DOI] [PubMed] [Google Scholar]
- Han F, Ullrich SE, Clancy JA, Jitkov V, Kilian A, Romagosa I. Verification of barley seed dormancy loci via linked molecular markers. Theoretical and Applied Genetics. 1996;92:87–91. doi: 10.1007/BF00222956. [DOI] [PubMed] [Google Scholar]
- Han F, Ullrich SE, Clancy JA, Romagosa I. Inheritance and fine mapping of a major barley seed dormancy QTL. Plant Science. 1999;143:113–118. [Google Scholar]
- Harlan JR. Barley. In: Smartt J, Simmonds NW, editors. Evolution of crop plants. 2nd edn. London: Longman; 1995. pp. 140–147. [Google Scholar]
- Harlan JR, de Wet JMJ, Price EG. Comparative evolution of cereals. Evolution. 1973;27:311–325. doi: 10.1111/j.1558-5646.1973.tb00676.x. [DOI] [PubMed] [Google Scholar]
- He C, Sayed-Tabatabaei BE, Komatsuda T. AFLP targeting of the 1-cM region conferring the vrs1 gene for six-rowed spike in barley, Hordeum vulgare L. Genome. 2004;47:1122–1129. doi: 10.1139/g04-073. [DOI] [PubMed] [Google Scholar]
- Helbaek H. Domestication of food plants in Old World. Science. 1959;130:365–372. doi: 10.1126/science.130.3372.365. [DOI] [PubMed] [Google Scholar]
- Helbaek H. Plant-collecting, dry-farming and irrigation agriculture in prehistoric Deh Luran. In: Hole F, Flannery KV, Neely JA, editors. Prehistory and human ecology of the Deh Luran plain. An early village sequence from Khuzistan, Iran. University of Michigan: Memoirs of the Museum of Anthropology; 1969. pp. 383–426. [Google Scholar]
- Hillman GC, Colledge SM, Harris DR. Plant-food economy during the Epipalaeolithic period at Tell Abu Hureyra, Syria: dietary diversity, seasonality, and modes of exploitation. In: Harris DR, Hillman GH, editors. Foraging and farming: the evolution of plant exploitation. London: Unwin & Hyman; 1989. pp. 240–268. [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Kandemir N, Kudrna DA, Ullrich SE, Kleinhofs A. Molecular marker assisted genetic analysis of head shattering in six-rowed barley. Theoretical and Applied Genetics. 2000;101:203–210. [Google Scholar]
- Karsai I, Mészáros K, Hayes PM, Bedő Z. Effects of loci on chromosomes 2 (2H) and 7 (5H) on developmental patterns in barley (Hordeum vulgare L.) under different photoperiod regimes. Theoretical and Applied Genetics. 1997;94:612–618. [Google Scholar]
- Karsai I, Mészáros K, Láng L, Hayes PM, Bedő Z. Multivariate analysis of traits determining adaptation in cultivated barley. Plant Breeding. 2001;120:217–222. [Google Scholar]
- Karsai I, Szűcs P, Mészáros K, Filichkina T, Hayes PM, Skinner JS, et al. The Vrn-H2 locus is a major determinant of flowering time in a facultative × winter growth habit barley (Hordeum vulgare L.) mapping population. Theoretical and Applied Genetics. 2005;110:1458–1466. doi: 10.1007/s00122-005-1979-7. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Taketa S, Ichii M, Kawasaki S. Efficient fine mapping of the naked caryopsis gene (nud) by HEGS (high efficiency genome scanning)/AFLP in barley. Theoretical and Applied Genetics. 2003;108:73–78. doi: 10.1007/s00122-003-1413-y. [DOI] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiology. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Mano Y. Molecular mapping of the intermedium spike-c (int-c) and non-brittle rachis 1 (btr1) loci in barley (Hordeum vulgare L.) Theoretical and Applied Genetics. 2002;105:85–90. doi: 10.1007/s00122-001-0858-0. [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Tanno K. Comparative high resolution map of the six-rowed spike locus 1 (vrs1) in several populations of barley, Hordeum vulgare L. Hereditas. 2004;141:68–73. doi: 10.1111/j.1601-5223.2004.01820.x. [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Li W, Takaiwa F, Oka S. High resolution map around the vrs1 locus controlling two- and six-rowed spike in barley, Hordeum vulgare. Genome. 1999;42:248–253. [Google Scholar]
- Komatsuda T, Nakamura I, Takaiwa F, Oka S. Development of STS markers closely linked to the vrs1 locus in barley, Hordeum vulgare. Genome. 1998;41:680–685. [Google Scholar]
- Komatsuda T, Maxim P, Senthil N, Mano Y. High-density AFLP map of nonbrittle rachis 1 (btr1) and 2 (btr2) genes in barley (Hordeum vulgare L.) Theoretical and Applied Genetics. 2004;109:986–995. doi: 10.1007/s00122-004-1710-0. [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I–class homeobox gene. Proceedings of National Academy of Sciences of the USA. 2007;104:1424–1429. doi: 10.1073/pnas.0608580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, et al. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- Laurie DA. Comparative genetics of flowering time. Plant Molecular Biology. 1997;35:167–177. [PubMed] [Google Scholar]
- Laurie DA, Pratchett N, Bezant JH, Snape JW. Genetic analysis of a photoperiod response gene on the short arm of chromosome 2(2H) of Hordeum vulgare (barley) Heredity. 1994;72:619–627. [Google Scholar]
- Laurie DA, Pratchett N, Bezant JH, Snape JW. RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter × spring barley (Hordeum vulgare L.) cross. Genome. 1995;38:575–585. doi: 10.1139/g95-074. [DOI] [PubMed] [Google Scholar]
- Leonard WH. Inheritance of fertility in the lateral spikelets of barley. Genetics. 1942;27:299–316. doi: 10.1093/genetics/27.3.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Ni P, Francki M, Hunter A, Zhang Y, Schibeci D, et al. Genes controlling seed dormancy and pre-harvest sprouting in a rice-wheat-barley comparison. Functional & Integrative Genomics. 2004;4:84–93. doi: 10.1007/s10142-004-0104-3. [DOI] [PubMed] [Google Scholar]
- Li W, Gill BS. Multiple genetic pathways for seed shattering in the grasses. Functional & Integrative Genomics. 2006;6:300–309. doi: 10.1007/s10142-005-0015-y. [DOI] [PubMed] [Google Scholar]
- Lin C. Photoreceptors and regulation of flowering time. Plant Physiology. 2000;123:39–50. doi: 10.1104/pp.123.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist U, Lundqvist A. An intermedium gene present in a commercial six-row variety of barley. Hereditas. 1987;107:131–135. [Google Scholar]
- Lundqvist U, Franckowiak JD, Konishi T. New and revised description of barley genes. Barley Genetics Newsletter. 1997;26:22–516. [Google Scholar]
- Mahfoozi S, Limin AE, Fowler DB. Influence of vernalization and photoperiod responses on cold hardiness in winter cereals. Crop Science. 2001;41:1006–1011. [Google Scholar]
- Mano Y, Kawasaki S, Takaiwa F, Komatsuda T. Construction of a genetic map of barley (Hordeum vulgare L.) cross ‘Azumamugi’ × ‘Kanto Nakate Gold’ using a simple and efficient amplified fragment-length polymorphism system. Genome. 2001;44:284–292. [PubMed] [Google Scholar]
- Moore G. Cereal genome evolution: pastoral pursuits with ‘Lego’ genomes. Current Opinion in Genetics & Development. 1995;5:717–724. doi: 10.1016/0959-437x(95)80003-n. [DOI] [PubMed] [Google Scholar]
- Murakami M, Matsushika A, Ashikari M, Yamashino T, Mizuno T. Circadian-associated rice pseudo response regulators (OsPRRs): insight into the control of flowering time. Bioscience Biotechnology and Biochemistry. 2005;69:410–414. doi: 10.1271/bbb.69.410. [DOI] [PubMed] [Google Scholar]
- Müller J, Wang Y, Franzen R, Santi L, Salamini F, Rohde W. In vitro interactions between barley TALE homeodomain proteins suggest a role for protein–protein associations in the regulation of Knox gene function. The Plant Journal. 2001;27:13–23. doi: 10.1046/j.1365-313x.2001.01064.x. [DOI] [PubMed] [Google Scholar]
- Ohmura Y, Ishii M, Nishida H, Akashi Y, Kato K, Takeda K. Structural variation of vernalization response gene, Vrn-H1, in cultivated barley. Breeding Research. 2006;8:65. [Google Scholar]
- Pourkheirandish M, Wicker T, Stein N, Fujimura T, Komatsuda T. The barley six-rowed spike gene vrs1 reveals a breakdown of rice-barley micro collinearity by a transposition. Theoretical and Applied Genetics. 2007 doi: 10.1007/s00122-007-0522-4. doi: 10.1007/s00122-007-0522-4. [DOI] [PubMed] [Google Scholar]
- Prada D, Ullrich SE, Molina-Cano JL, Cistué L, Clancy JA, Romagosa I. Genetic control of dormancy in a Triumph/Morex cross in barley. Theoretical and Applied Genetics. 2004;109:62–70. doi: 10.1007/s00122-004-1608-x. [DOI] [PubMed] [Google Scholar]
- Quail PH. Photosensory perception and signalling in plant cells: new paradigms? Current Opinion in Cell Biology. 2002;14:180–188. doi: 10.1016/s0955-0674(02)00309-5. [DOI] [PubMed] [Google Scholar]
- Romagosa I, Han F, Clancy JA, Ullrich SE. Individual locus effects on dormancy during seed development and after ripening in barley. Crop Science. 1999;39:74–79. [Google Scholar]
- Salamini F, Özkan H, Brandolini A, Schäfer-Pregl R, Martin W. Genetics and geography of wild cereal domestication in the Near East. Nature Reviews Genetics. 2002;3::429–441. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- Sameri M, Takeda K, Komatsuda T. Quantitative trait loci controlling agronomic traits in recombinant inbred lines from a cross of oriental- and occidental-type barley cultivars. Breeding Science. 2006;56:243–252. [Google Scholar]
- Scholz F. Mutationsversuche an Kulturpflanzen. IV. Über den züchterischen Wert zwei röntgeninduzierter Gerstenmutanten. Kulturpflanze. 1955;3:69–89. [Google Scholar]
- Senthil N, Komatsuda T. Inter-subspecific maps of non-brittle rachis genes btr1/btr2 using occidental, oriental and wild barley lines. Euphytica. 2005;145:215–220. [Google Scholar]
- Simpson GM. Seed dormancy in grasses. New York, NY: Cambridge University Press; 1990. [Google Scholar]
- Snape JW, Butterworth K, Whitechurch E, Worland AJ. Waiting for fine times: genetics of flowering time in wheat. Euphytica. (a) 2001;119:185–190. [Google Scholar]
- Snape JW, Sarma R, Quarrie SA, Fish L, Galiba G, Sutka J. Mapping genes for flowering time and frost tolerance in cereals using precise genetic stocks. Euphytica. (a) 2001;120:309–315. [Google Scholar]
- Szücs P, Karsai I, von Zitzewitz J, Mészáros K, Cooper LLD, Gu YQ, et al. Positional relationships between photoperiod response QTL and photoreceptor and vernalization genes in barley. Theoretical and Applied Genetics. 2006;112:1277–1285. doi: 10.1007/s00122-006-0229-y. [DOI] [PubMed] [Google Scholar]
- Takahashi R. The origin and evolution of cultivated barley. In: Demerec M, editor. Advances in Genetics. Vol. 7. New York: Academic Press; 1955. pp. 227–266. [Google Scholar]
- Takahashi R. Barley Genetics V. Proceedings of the Fifth International Barley Genetics Symposium. Okayama: Academic Societies of Japan; 1987. Genetic features of east Asian barleys; pp. 7–20. [Google Scholar]
- Takahashi R, Hayashi J. Linkage study of two complementary genes for brittle rachis in barley. Bericht des Ohara Instituts für Landwirtschaftliche Biologie, Okayama. 1964;12:99–105. [Google Scholar]
- Takahashi R, Yamamoto J. Studies on the classification and the geographic distribution of barley varieties. 8. Nogaku Kenkyu. 1949;38:41–43. [Google Scholar]
- Takahashi R, Yasuda S. Genetic studies of spring and winter habit of growth in barley. Bericht des Ohara Instituts für Landwirtschaftliche Biologie, Okayama. 1956;10:245–308. [Google Scholar]
- Takahashi R, Yasuda S. Genetic studies on heading date in barley. In: Sakai K, Takahashi R, Akemine H, editors. Studies on the bulk method of plant breeding. Tokyo: Yokendo; 1958. pp. 44–64. [Google Scholar]
- Takahashi R, Hayashi S, Yasuda S, Hiura U. Chatacteristics of the wild and cultivated barleys from Afghanistan and its neighbouring regions. Bericht des Ohara Instituts für Landwirtschaftliche Biologie, Okayama. 1963;12:1–23. [Google Scholar]
- Takahashi R, Hayashi S, Hiura U, Yasuda S. A study of cultivated barleys from Nepal, Himalaya and North India with special reference to their phylogenetic differentiation. Bericht des Ohara Instituts für Landwirtschaftliche Biologie, Okayama. 1968;14:85–122. [Google Scholar]
- Takeda K. Varietal variation and inheritance of seed dormancy in barley. In: Noda K, Mares DJ, editors. Proceedings of the seventh international symposium on pre-harvest sprouting in cereals. Osaka: Centre for Academic Societies of Japan; 1996. pp. 205–212. [Google Scholar]
- Taketa S, Kikuchi S, Awayama T, Yamamoto S, Ichii M, Kawasaki S. Monophyletic origin of naked barley inferred from molecular analyses of a marker closely linked to the naked caryopsis gene (nud) Theoretical and Applied Genetics. 2004;108:1236–1242. doi: 10.1007/s00122-003-1560-1. [DOI] [PubMed] [Google Scholar]
- Taketa S, Awayama T, Amano S, Sakurai Y, Ichii M. High-resolution mapping of the nud locus controlling the naked caryopsis in barley. Plant Breeding. 2006;125:337–342. [Google Scholar]
- Tanno K, Taketa S, Takeda K, Komatsuda T. A DNA marker closely linked to the vrs1 locus (row-type gene) indicates multiple origins of six-rowed cultivated barley (Hordeum vulgare L.) Theoretical and Applied Genetics. 2002;104:54–60. doi: 10.1007/s001220200006. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES. HvVRN2 responds to day length, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiology. 2006;140:1397–405. doi: 10.1104/pp.105.073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- Von Ubisch G. Analyse eines Falles von Bastardatavismus und Faktorenkoppelung bei Gerste. Zeitschrift für induktive Abstammungs und Vererbungslehre. 1915;14:226–237. [Google Scholar]
- Ullrich SE, Hayes PM, Dyer WE, Blake TK, Clancy JA. Quantitative trait locus analysis of seed dormancy in ‘Steptoe’ barley. In: Walker-Simmons MK, Ried JL, editors. Pre-harvest sprouting in cereals 1992. St Paul, MN: American Association of Cereal Chemists; 1993. pp. 136–145. [Google Scholar]
- Ullrich SE, Han F, Blake TK, Oberthur LE, Dyer WE, Clancy JA. Seed dormancy in barley: genetic resolution and relationship to other traits. In: Noda K, Mares DJ, editors. Pre-harvest sprouting in cereals 1995. Osaka: Center for Academic Societies Japan; 1996. pp. 157–163. [Google Scholar]
- Ullrich SE, Han F, Jones BL. Genetic complexity of the malt extract trait in barley suggested by QTL analysis. American Society of Brewing Chemists. 1997;55:1–4. [Google Scholar]
- Vanhala TK, Stam P. Quantitative trait loci for seed dormancy in wild barley (Hordeum spontaneum C. Koch) Genetic Resources and Crop Evolution. 2006;53:1013–1019. [Google Scholar]
- Vavilov NI. Studies on the origin of cultivated plants. Bulletin of Applied Botany, Genetic, and Plant Breeding. 1926;16:1–248. [Google Scholar]
- Watanabe N, Ikebata N. The effects of homoeologous group 3 chromosomes on grain colour dependent seed dormancy and brittle rachis in tetraploid wheat. Euphytica. 2000;115:215–220. [Google Scholar]
- Woodward RW. The Ih, I, i alleles in Hordeum deficiens genotypes of barley. Journal of the American Society of Agronomy. 1947;39:474–482. [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proceedings of the National Academy of Sciences of the USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, et al. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, et al. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proceedings of the National Academy of Sciences of the USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. The Plant Cell. 2000;12:2473–2483. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S. Linkage and pleiotropic effects on agronomic characters of the genes for spring growth habit. Barley Newsletter. 1969;12:57–58. [Google Scholar]
- Zhang F, Chen G, Huang Q, Orion O, Krugman T, Fahima T, et al. Genetic basis of barley caryopsis dormancy and seedling desiccation tolerance at the germination stage. Theoretical and Applied Genetics. 2005;110:445–453. doi: 10.1007/s00122-004-1851-1. [DOI] [PubMed] [Google Scholar]
- Zohary D. Barley Genetics I. Proceeding of 1st International Barley Genetics Symposium. Wageningen: 1963. Spontaneous brittle six-rowed barley, their nature and origin; pp. 27–31. August 26–31. [Google Scholar]
- Zohary D, Hopf M. Domestication of plants in the old world: the origin and spread of cultivated plants in West Asia, Europe, and the Nile Valley. 3rd edn. New York, NY: Oxford University Press; 2000. [Google Scholar]