Abstract
Background
It has been shown that a large variation is present and exploitable from wild Solanum species but most of it is still untapped. Considering the thousands of Solanum accessions in different gene banks and probably even more that are still untouched in the Andes, it is a challenge to exploit the diversity of tomato. What have we gained from tomato domestication and breeding and what can we gain in the future?
Scope
This review summarizes progress on tomato domestication and breeding and current efforts in tomato genome research. Also, it points out potential challenges in exploiting tomato biodiversity and depicts future perspectives in tomato breeding with the emerging knowledge from tomato-omics.
Conclusions
From first domestication to modern breeding, the tomato has been continually subjected to human selection for a wide array of applications in both science and commerce. Current efforts in tomato breeding are focused on discovering and exploiting genes for the most important traits in tomato germplasm. In the future, breeders will design cultivars by a process named ‘breeding by design’ based on the combination of science and technologies from the genomic era as well as their practical skills.
Key words: Breeding, domestication, genomics, Solanum lycopersicum
BACKGROUND
Tomato (Solanum lycopersicum) originated from the Andean region now encompassed by part of Chile, Boliva, Ecuador, Colombia and Peru. The time and place of domestication of tomato are not known with certainty. The tomato had reached a fairly advanced stage of domestication before being taken to Europe in the 15th century and further domestication on a much more intense level occurred throughout Europe in the 18th and 19th centuries (Sims, 1980). Since the 20th century, human beings have created a huge array of morphologically different cultivars and forms from the single species S. lycopersicum via plant breeding (see below). Through domestication, research and breeding activities that were implemented by scientists and breeders worldwide, modern tomato varieties (mostly hybrids) have been developed with all shapes, colours and sizes.
The advent of genomics has brought a real boost to the generation of data, knowledge and tools that can be applied in breeding, which has transformed breeding from a rather individually based activity to a multidisciplinary teamwork that is most suited to exploit genes from tomato germplasm in an efficient way. As a result, it is expected that the improvement in tomato cultivars will continue in the future. In this review, we look at the domestication and breeding of tomato and the new insights that have come from recent developments in tomato genome research. This is followed by a discussion on what was gained and what can be gained in the future during the domestication and breeding of tomato.
TOMATO BIODIVERSITY
Diversity among wild tomato relatives and within domesticated tomato
Tomato belongs to the Solanaceae family, which includes >3000 species with origins in both the Old (eggplant in China and India) and New World (pepper/potato/tomato in Central and South America; Knapp, 2002). The phylogenetic classification of the Solanaceae has been recently revised and the genus Lycopersicon re-integrated into the Solanum genus with its new nomenclature. Solanum section Lycopersicon includes the cultivated tomato (S. lycopersicum) and 12 additional wild relatives. Solanum lycopersicum is the only domesticated species (Peralta et al., 2006).
Wild tomatoes have a large genetic diversity, especially within the self-incompatible species like S. chilense and S. peruvianum (Rick, 1988). Tremendous variation has been revealed by molecular markers and it is striking that more genetic variation was observed within a single accession of the self-incompatible species than in all accessions of any of the self-compatible species (Miller and Tanksley, 1990; Bretó et al., 1993; Sacks et al., 1997; Villand et al., 1998; Egashira et al., 2000). The genetic variation present in the wild species has been investigated intensively for specific traits and is being exploited in tomato breeding (e.g. Walter, 1967; Rick and Chetelat, 1995; Larry and Joanne, 2007).
Compared with the rich reservoir in wild species, the cultivated tomato is genetically poor. It is estimated that the genomes of tomato cultivars contain <5 % of the genetic variation of their wild relatives (Miller and Tanksley, 1990). The lack of diversity in the cultivated tomato can be visualized using DNA technologies. Very few polymorphisms within the cultivated tomato genepool have been identified, even using sensitive molecular markers (Van der Beek et al., 1992; Villand et al., 1998; Park et al., 2004; Garcia-Martinez et al., 2005; Tam et al., 2005). Tomato domestication experienced a severe genetic bottleneck as the crop was carried from the Andes to Central America and from there to Europe. The initial domestication process was, in part, reached by selecting preferred genotypes in the existing germplasm. Selection of a horticultural crop like tomato is usually done on a single plant basis and with small numbers of selected plants. In a predominantly inbreeding species, genetic variation tends to decrease, even without selection. As a consequence, genetic drift is a major process that reduces genetic variation.
Most likely, no exchange of genetic information with the wild germplasm took place until 1940. Up till then the renowned geneticist and plant breeder Charlie Rick (University of California, Davis) observed that crosses between wild and cultivated species generated a wild array of novel genetic variation in the offspring. Since then, breeding from wild species via interspecific crosses has lead to great utilization of the favourable attributes hidden in tomato exotics in the 20th century (see below).
Collection and preservation of genetic resources
The collection, description, propagation and distribution of genetic materials are of the utmost importance in tomato breeding. It is noteworthy to mention Dr Charlie Rick who organized numerous expeditions to the Andes to hunt for wild tomato species. Over the years in the later half of the 20th century, thousands of accessions of the wild Solanum species have been collected and maintained at the Tomato Genetics Resource Center in Davis, California (TGRC, http://tgrc.ucdavis.edu/). In addition to the natural collections, the TGRC produced a large proportion of monogenic mutants and miscellaneous genetic stocks of tomato. In the Netherlands, the Botanical and Experimental Garden (http://www.bgard.science.ru.nl/) pays special attention to the Solanaceae germplasm collection and maintains the most extensive ex situ plant collections of non-tuberous Solanaceae species in the world. In addition to preservation of the wild species accessions, a comprehensive mutant population has been developed (Menda et al., 2004) (http://zamir.sgn.cornell.edu/mutants/), which is an isogenic tomato ‘mutation library’ containing a total of 13 000 M(2) families derived from treatment with EMS (ethyl methane sulfonate) and fast-neutron mutagenesis. All the mutants serve as basic resources for exploring gene function to discover the ‘genes that make tomatoes’.
In addition to the genetic resources maintained in the genebanks, wild tomato relatives still grow in the centre of origin of tomato from the northern part of Chile to Colombia. In 2005, two new species of wild tomatoes were identified from Peru (Peralta et al., 2005). In Loja province, Ecuador, a large collection of wild tomatoes of different species has been sampled and is currently being maintained by Dr Morales at the University of Loja (pers. comm.). Most wild tomatoes are endemic in narrow geographical regions and also have very small populations, making them vulnerable to extinction. Therefore, to discover and maintain species diversity in biodiverse regions is extremely important.
TOMATO DOMESTICATION
Tomatoes were domesticated in America; however, the original site of domestication and the early events of domestication are largely obscure (Peralta and Spooner, 2007). Two hypotheses have been advanced for the original place of tomato domestication, one Peruvian and the other Mexican. Although definite proof for the time and place of domestication is lacking, Mexico is presumed to be the most probable region of domestication, with Peru as the centre of diversity for wild relatives (Larry and Joanne, 2007). Solanum lycopersicum cerasiforme is thought to be the ancestor of cultivated tomato, based on its wide presence in Central America and the presence of a shorten style length in the flower (Cox, 2000). However, recent genetic investigations have shown that the plants known as ‘cerasiforme’ are a mixture of wild and cultivated tomatoes rather than being ‘ancestral’ to the cultivated tomatoes (Nesbitt and Tanksley, 2002).
Tomato domestication syndrome
Domestication has triggered a wide range of morphological and physiological traits that distinguish domesticated crops from their wild ancestors. These characteristics are collectively referred to as the domestication syndrome (Frary and Doganlar, 2003). The exact trait composition of domestication syndrome varies among crops. Generally, syndrome characteristics include a more compact growth habit, increased earliness, reduction/loss of seed dispersal and dormancy, gigantism and increased morphological diversity in the consumed portion of the plant (Frary and Doganlar, 2003). With advances in genome mapping and quantitative genetic analyses, the genetic basis is being dissected for traits that are related to domestication in many crops (Poncet et al., 2004). Studies on the domestication syndrome and domestication process have revealed that numerous traits that distinguish crop plants from their wild relatives are often controlled genetically by a relatively small number of loci with effects of unequal magnitude (reviewed in Frary and Doganlar, 2003).
In tomato, domestication syndrome traits have been studied for growth habit (self-pruning, plant height and earliness) and fruit traits (set, size, shape, colour and morphology) and the qualitative genes and quantitative trait loci (QTLs) underlying these syndrome characteristics have been identified (Grandillo and Tanksley, 1996; Doganlar et al., 2000; Frary and Doganlar, 2003; Tanksley, 2004). Comparative genetics revealed that many loci associated with similar traits are co-localized in tomato, pepper and eggplant of the Solanaceous family (Frary et al., 2000; Donganlar et al., 2002; Van der Knaap and Tanksley, 2002; Frary and Doganlar, 2003). The fact that tomato and other Solanaceous crops share common QTLs despite their independent domestication on different continents may indicate that relatively few loci are implicated overall in the drastic phenotypic changes observed in domestication. Below, we focus on a few domestication features and processes that are related to tomato fruit and seed.
Fruit size
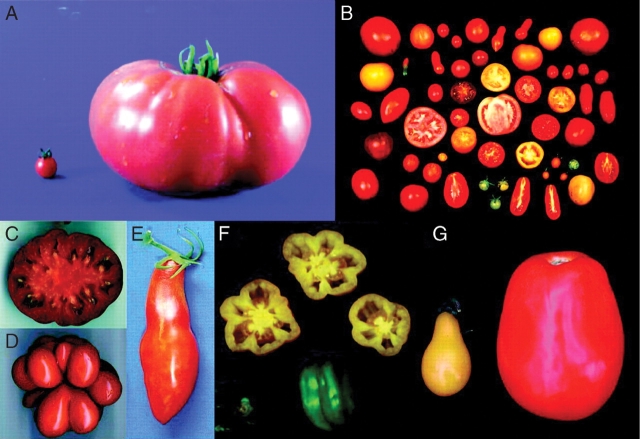
An obvious feature of tomato domestication is the massive increase in fruit size. Wild tomato species have tiny fruits made to propagate the species and not to feed human beings. Domestication has transformed the once small wild tomato into the present-day cultivars. Because domestication occurred in prehistoric times, the evolution pathway related to the transition in tomato fruit size is unknown. Most likely, mutations associated with larger fruit were selected and accumulated during selection by early humans. By crossing a wild and a cultivated tomato, mutations in about six QTLs seem to have been responsible for transforming the small berries of wild tomatoes to the extremely large fruit now associated with modern cultivars (Fig. 1, reviewed in Tanksley, 2004). One of these QTLs is fruit weight 2·2 (fw2·2) that changes fruit weight by up to 30%. It is believed that mutation(s) in the fw2·2 locus was the first step on the road to domestication and responsible for a key transition during tomato domestication (Alpert et al., 1995; Frary et al., 2000). Cloning of fw2·2 has shown that this locus codes for a negative repressor of cell division, and the changes from small to large fruit are caused by mutations in the promoter sequence. These mutations are associated with a lower total transcript level of fw2·2 during the cell-division phase of fruit development as well as a shift in the timing of expression (Cong et al., 2002). Molecular clock-based estimates indicate that the large fruit allele arose long before the tomato was domesticated (Nesbitt and Tanksley, 2002).
Fig. 1.
Collage depicting wide variation in size and shape of tomato fruit. (A) The large-fruited tomato ‘Giant Heirloom’ common to modern agriculture (right) and the typical fruit of a related wild species (L. pimpinellifolium). (B) The range of fruit size and shape variation in tomato. (C) Cross-section of the fruit from a plant homozygous for a mutation at the fasciated locus causing multiple locules. (D) Alternate allele of fasciated associated with unfused carpels. (E) Fruit from ‘Long John’, which carries mutations at both the sun and ovate loci causing extremely long, narrow fruit. (F) Bell pepper-type fruit produced by ‘Yellow Stuffer’. (G) Fruit from two different cultivars homozygous for a mutation at the ovate locus. In the variety on the left, the ovate mutation results in the production of fruit that are both elongated and constricted at the stem end of the fruit (hence, the pear shape). However, in the processing variety on the right, the ovate mutation causes elongated fruit but has a much reduced effect on neck constriction. This figure is from Tanksley SD. 2004. The Plant Cell 16: S181–S189.
Fruit shape
In addition to an increase in fruit size, the domestication of fruit-bearing species often has resulted in tremendous shape variation. Wild tomatoes bear fruits that are almost invariably round, while cultivars available today show a wide variety of shapes: round, oblate, pear-, torpedo- and bell-shaped (Fig. 1, reviewed in Tanksley, 2004). Selection for increased fruit size may have lead to phenotypic changes in fruit shape by pleiotropic effects of the fruit size loci or expression of ‘hidden’ fruit shape alleles that have little or no visible effect on fruit shape in a small-fruited background (Tanksley et al., 1996; Grandillo et al., 1999). Other factors contributable to changes in fruit shape are demands of machine-harvestable tomatoes (starting in 1960s) and human inclination towards valuing novelty. Recent genetic studies have revealed several loci that cause phenotypic differences in fruit shape that distinguish the domesticated tomato from its wild relatives (reviewed in Tanksley, 2004). The ovate gene determines a change from round to elongated or pear-shaped tomato fruit (Liu et al., 2002). Allelic variation in sun and fs8·1 loci can cause elongated and square fruit shape, respectively, mainly for processing tomatoes (Ku et al., 2000; Van der Knaap and Tanksley, 2001).
Seeds size and weight
The seeds of domesticated plants are normally much larger than those of their wild counterparts. Tomato seeds are not the primary food product and do not belong to the tomato domestication syndrome; tomato seeds, however, are important for breeding. Compared with their wild relatives, the seeds of cultivated tomato have become several-fold larger and the change in seed weight was most likely in response to the selection pressure for uniform germination and seedling vigour (Doganlar et al., 2000). QTLs for seed weight have been identified that are often in close proximity to loci for fruit weight and soluble-solids content (Doganlar et al., 2000; Tanksley, 2004). Seed weight is positively correlated to fruit weight but negatively correlated to soluble-solids content (Goldman et al., 1995; Grandillo and Tanksley, 1996). One QTL, sw4·1, contributes significantly to seed weight variation and it is likely that it differentiates large-seeded cultivated tomato from its small-seeded wild relatives (Doganlar et al., 2000).
TOMATO BREEDING IN THE 20TH CENTURY
History of tomato breeding
At the end of the 19th century, numerous cultivars of tomato were available in different colours and for different purposes. These cultivars could be considered as landraces and products of domestication and some early breeding. Most of these cultivars require open pollination and the propagation was done by the farmers and growers who could easily obtain seeds from the fruits for the next generation. Because tomatoes do not naturally out-cross very often, seeds of a tomato produce plants resembling the parent. Due to this property, earlier tomato cultivars were selected and inherited in a family or community, thus earning the name heirloom. Heirloom tomato varieties are open-pollinated and are unique in size, shape and colour (Watson, 1996). Available in varieties with diverse characteristics, heirloom tomatoes tend to be prized for their distinctive flavours. At the beginning of the 20th century public institutes, mainly in the USA, became more involved in tomato breeding. Meanwhile, private companies were formed and commercial breeding has shifted open-pollinated cultivars to hybrids. Hybrids combine good characters from both parents that will segregate in the progeny, discouraging seed propagation by growers. The advantages of hybrid varieties over true breeding varieties were so great that the growers would buy hybrid seeds at higher prices. In 1946, the first hybrid tomato cultivar ‘Single Cross’ was released (Dorst, 1946). Eventually, nearly all tomato cultivars for the fresh market and an increasing number of cultivars using in processing are hybrids.
Although the process of plant breeding is theoretically simple, it does create novelty. The art of tomato breeding is identifying and combining the specific traits for each market. The fresh product is sold in a wide range of shapes and sizes, from the small cherry tomato to very large beef tomatoes. Typically breeders are continuously improving their breeding lines, either by making new crosses with their own material or by using the cultivars of their competitors, which is allowed by breeders' law (UPOV Breeders Rights, 1961). Breeders in this fashion use naturally occurring recombinations to produce cultivars that combine favourable traits. New traits are rarely introduced from wild germplasm as it may take many generations to remove the deleterious genes that go along with the introduced genes due to linkage drag. When the parental lines are more fixed (F4 to F6), crosses are made to produce test hybrids. After several generations of testing at the breeders' site and eventually at the farmers' sites, the best hybrids are selected for commercial usage. Hybrids of tomato show some heterosis, but this is only selected for at the latest stage of the breeding programme, when test hybrids are generated. In earlier generations the parent lines are selected at a single plant basis but not for combining abilities or heterosis. So, recurrent selection programmes to select parents with the best combining abilities, like that used in field crops, is not a common practice in tomato breeding.
Nowadays, there are a dozen tomato-breeding companies which are the main players in the world market. In order to survive, seed companies must continuously develop new cultivars with added value and hence commercial tomato breeding is very innovative. The average turnover time of commercial tomato cultivars is approx. 5 years. This system results in a very competitive breeding practice and breeding companies can only get return on their investments if they can get high prices for their seeds. This is certainly true for the fresh tomato market, and less so for the tomato-processing industry where seed (and tomato fruit) prices are much lower and the volumes much larger. The annual value of the worldwide tomato seed market is about half a billion euros with the majority being for fresh market tomatoes.
Traits of modern tomato cultivars
The goals of public and private tomato breeding programmes vary widely depending on location, need and resources. In general, breeding goals in tomato have gone through four phases: breeding for yield in the 1970s, for shelf-life in the 1980s, for taste in the 1990s and for nutritional quality currently. To be successful, growers must produce a high yield of high-quality fruit, while holding production costs as low as possible. Therefore, many of the breeding goals focus on characteristics that reduce production costs or ensure reliable production of high yields with high-quality fruits.
Resistance to biotic and abiotic stresses
One of the most prominent issues in tomato breeding is breeding for resistance to the most destructive pests and pathogens. The tomato hosts >200 species of a wide variety of pests and pathogens that can cause significant economic losses. Often, these pests and pathogens have to be controlled by using chemical compounds like fungicides or pesticides. These methods may not be fully effective, raise production costs and require compliance with chemical-use laws. They also cause concern regarding potential risk for the growers, the consumers and for the environment. Nature has provided a great wealth of resistances that are available in the wild species (Fig. 2). Many of the resistances are simply inherited, and remarkable successes have been made in transferring disease-resistance genes into cultivated tomato. One of the first examples was the exploitation of Cladosporium fulvum resistance from S. pimpinellifolium in 1934 (Walter, 1967). Since tomato is subtropical in origin, tomato production is sub-optimal over large parts of the tomato crop-growing areas, due to unfavourable environmental conditions, caused by abiotic factors including high or low temperatures, excessive water or drought, and soil salinity or alkalinity. In tomato breeding germplasm much genetic variation for such stress tolerance exists and is used for breeding (e.g. Rick and Chetelat, 1995; Wang et al., 2003; Venema et al., 2005).
Fig. 2.

Overview of mapped resistance genes and QTLs (quantitative trait loci) on the tomato genome. Marker loci are taken from the core FRLP map of Tanksley et al. (1992). Resistance genes and QTLs are printed in bold and italics with QTLs underlined. This figure is from Bai and Lindhout (2005).
Yield and heterosis
Heterosis is a biological phenomenon manifesting itself in hybrids that are more vital, adaptive and productive than their parents. Heterosis has been explained by over-dominance and by additive effects. However, it is still unclear how much each of these effects contributes to the total heterosis effect (Birchler et al., 2006; Semel et al., 2006). Breeders prefer to develop F1 hybrids, not only for heterosis, but also for their uniformity and protection against illegal reproduction.
Fruit quality
Fruit quality is a combination of visual stimuli like size, shape and colour, and sensory factors like sugar, acidity and taste. Moreover, the consumers' perception of quality is also heavily influenced by the products' appearance and information like sun-ripe, biological, transgenic, etc. Research on the genetic control of fruit quality traits has been dominated by studies of the ripening process and determination of soluble solid content (Rick and Chetelat, 1995).
Ripening is of interest to tomato breeders, since ripening affects several quality traits, like colour, flavour and soluble solids content. Another factor that is especially important for fresh-market tomatoes is their shelf-life. During ripening, several processes occur that negatively affect the storage of the fruit. Some genes involved in ripening like polygalacturonase and ethylene synthase have been cloned (DellaPenna et al., 1986). Tomato cultivars with a long shelf-life have been obtained mainly by using mutants non-ripening (nor) and ripening inhibitor (rin), e.g. the ‘Daniela’ hybrid that was released >15 years ago by the BonTom tomato breeding group (Faculty of Agriculture, Hebrew University of Jerusalem, Israel). The genetic make-up of Daniela combines the rin gene with some selected polygenes for firmness and slow ripening, together with other genes generating high yields of large, quality fruit.
The red colour of tomato is determined by the colour of the skin and of the flesh. The skin colour varies from yellow to colourless, while the flesh colour varies between green and red. During ripening, there is a 500-fold increase in the level of lycopene in the tomato fruit. Increased lycopene has proven nutritional value as an antioxidant that is associated with a low incidence of certain forms of human cancer (Miller et al., 2002). Recently, high-lycopene tomatoes have been sold as specialities in the fresh market.
Flavour is the sum of the interaction between sugars, acids and a set of approx. 30 volatile compounds (Tieman et al., 2006). Some prediction of flavour can be made by measuring the acidity and refraction index, which is equivalent to the soluble solids content. Although flavour is a complicated trait, it has been shown that a significant improvement in tomato flavour could be attained by increasing sugar and acid contents in tomato fruits by genetic manipulation (Jones and Scott, 1983). However, breeding for volatiles has not yet been performed intensively, as little is known about the relationships between flavour, aroma and volatiles. Recently, via genomics and targeted metabolite approaches, many QTLs have been identified that reproducibly alter the composition of volatile and non-volatile chemicals important to tomato flavour (Tikunov et al., 2005; Tieman et al., 2006).
Biotechnology and transgenic or genetically modified organisms
The basic technologies that help breeders extend their germplasm with new sources or genes are applicable in tomato: in vitro techniques to regenerate tomato plants from tissue or single cells, embryo-rescue and cell fusion techniques to combine cells of non-crossable species and transformation technologies to introduce genes from other organisms. The potential for breeding applicants is evident. Tomato was the first food crop for which transgenic fruits were commercially available. The first transgenic cultivar ‘Flavr-Savr’™ was relased in 1994 (NBIAP News Report, July 1994). The consumers were excited and readily bought and consumed the transgenic tomatoes. However, this success did not last long due to a combination of bad performance of other agricultural traits, like yield (a classic example that one gene or one trait does not make a successful variety) and some marketing problems. Another transgenic tomato was released in the UK and used for canned products. The transgenic tomato showed a reduction in the expression of the polygalacturonase gene that is involved in fruit softening (http://www.ncbe.reading.ac.uk/NCBE/GMFOOD/firstfruit.html). Again, consumers were enthusiastic. Since tomato is a relatively simple system for transformation with Agrobacterium tumefaciens, tomatoes harbouring other transgenes have also been made, although not yet exploited commercially. These traits include other biosynthesis genes affecting ripening or fruit quality, and constructs for herbicide, virus and insect resistance that were hitherto difficult or not possible to breed using a more traditional approach (Schuch et al., 1991).
The release of transgenic tomatoes came to a complete stop at the end of the 20th century. The reasons are complicated and include expensive patent filing that is associated with transgenic crops and consumer concerns, especially in the EU. In addition, most commercially important traits are too complex to benefit from the incorporation of a single transgene (Strauss, 2003).
Marker-assisted breeding
The advent of molecular markers and linkage maps has made it possible to find associations between markers and phenotypes. Breeders can use a known association of molecular markers with a trait or a chromosome segment to select the presence of molecular markers rather than the phenotype. This process is known as marker-assisted selection. Originally, the tomato morphological map was generated using visually distinguishable morphological mutants and, later, isozymes were added to the ‘classical’ map. These isozymes were the first generation of molecular markers. With the advent of DNA-based molecular markers like RFLPs and AFLPs, complete genetic maps have been generated in tomato (Tanksley et al., 1992; Haanstra et al., 1999). The generation of simple PCR markers in the form of CAPS (cleaved amplified polymorphic sequence) and SCAR (sequence characterized amplified region) has promoted the application of DNA markers in the breeding programme for marker-assisted selection, a starting point for molecular breeding (Bai et al., 2004). Nowadays, dozens of genes, important for tomato breeding, have been mapped and molecular markers have been made available online (http://sgn.cornell.edu). Breeders use these markers to a great extent with the main aim of increasing the efficiency of breeding programmes. Via marker-assisted selection, the paradigm of plant breeding has changed from selection of phenotypes towards selection of genes, either directly or indirectly.
FUTURE TOMATO BREEDING
QTLs and introgression lines (ILs)
The genetics of a quantitative trait is hard to study, since the effect of each gene is small and often influenced by environment or by the interaction with other genes (epistasis). Many important tomato traits as described above are genetically controlled by a combined action of QTLs with favourable alleles often present in the wild species (e.g. Fulton et al., 2000; Bai et al., 2003; Gur and Zamir, 2004). To introgress the wild favourable allele into cultivated tomato, marker-assisted selection plays an important role and the map positions and markers linked to the QTLs provide a basis for breeders to design optimal breeding strategies.
To map QTLs in tomato, interspecific populations have been extensively used. However, in an interspecific cross, multiple segregating QTL at the whole genome level often tend to mask the effects of one another (e.g. Tanksley et al., 1996; Grandillo et al., 1999). Compared with interspecific crosses, ILs are more powerful in QTL identification. ILs carry a single introgressed region, and are otherwise identical for the rest of their genome. As a result, the phenotypic variation in these lines can be associated with individual introgression segments. In tomato, several sets of ILs have been developed for wild relatives of tomato. These include S. pennellii (Eshed and Zamir, 1995), and S. habrochaites (Monforte and Tanksley, 2000). Introgressions have also been made with S. lycopersicoides and S. sitiens (Chetelat and Meglic, 2000; Michael et al., 2005; Canady et al., 2006). These sets of ILs can be requested from the Tomato Genetics Resource Centre in Davis, California (http://tgrc.ucdavis.edu). These introgression libraries have potential in breeding for quantitative traits, as different genomic regions of wild tomato species can be pyramided into new breeding lines (Fig. 3; Fridman et al., 2004). Thus, ILs (also called prebreds) will offer tomato breeders a powerful tool to optimize the uses of genetic variation in nature by bringing together in one genotype alleles that maximize yield, resistance to biotic and abiotic stress, etc.
Fig. 3.
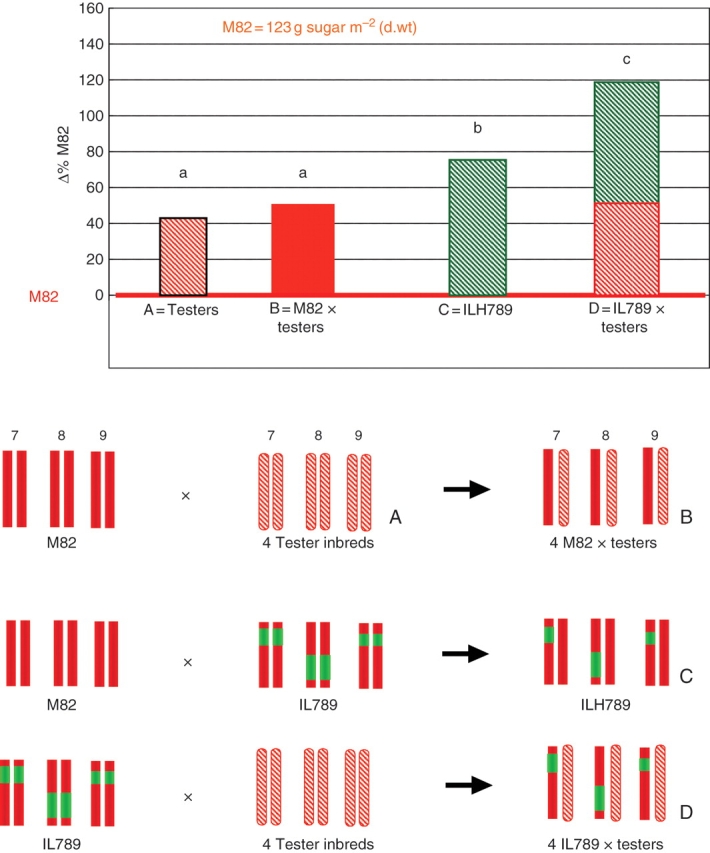
Pyramiding introgression lines (ILs) into breeding lines to explore the genetic variation in Brix Yield (BY, measured in g m–2). The upper panel shows the BY differences among different genotypes (shown as ▵ % from M82; base line represents M82) and the lower panel shows the combinations of tomato chromosomes 7, 8 and 9 in these genotypes. Yield differences between M82 (red chromosomes 7, 8 and 9), the four tomato tester inbreds (A = testers, pink chromosomes 7, 8 and 9), and their hybrids (B = M82 × testers) result from allelic variation present in the cultivated tomato gene pool. Since M82, the multiple-introgression line (IL789), and their hybrid IL789 × M82 (C = ILH789) differ only in three S. pennellii segments (green chromosomes), any BY difference between them is associated with the exotic allelic variation. The yield of the hybrids of IL789 with the four testers (D = IL789 × testers) results from both cultivated and exotic variation. This figure is from Gur and Zamir (2004) with modification.
Exploiting the tomato genome
New technologies, initially based on DNA polymorphisms (genetic markers), but nowadays based on much wider technologies, designated as ‘genomics’, have enabled the international community to explore the plant genome with the aim of understanding how organisms function and how genes make an organism. In tomato, the International Solanaceae Genomics Project (SOL) was initiated in 2003 with sequencing the tomato genome as it first cornerstone. The genome of tomato is being sequenced by an international consortium of ten countries (Mueller et al., 2005b). Under the umbrella of SOL, there are two additional projects, EU-SOL and Lat-SOL. The EU-SOL project focuses on the development of high quality and healthy tomato and potato varieties with improved consumer-, processor- and producer-directed traits. The Lat-SOL aims at joining efforts in Latin America and promoting information and resource flow between laboratories working in basic and applied aspects of Solanaceae species, establishing post-genome techniques and integrating researches with the existing SOL and EU-SOL programmes. These genomics projects are generating a huge amount of data, such as tomato gene databases, the gene expression database, the tomato metabolite database, genome annotations, etc, which are maintained in the SOL Genomics Network (SGN; http://sgn.cornell.edu, a genomics information resource for the Solanaceae family and related families of Asterids). It is expected that these developments will result in an important step forward in our knowledge and understanding of the structure and function of the tomato genome. The tomato genome sequence will provide the foundation for a new sequenced-based, comparative resource that will aid in linking the Solanaceae to each other and outward to other families (Mueller et al., 2005a).
Super domestication: breeding in the genomics era
With the advance of tomato genome sequences and genomics, the genetic basis of plant growth and development are expected to be better understood. Knowing the candidate genes for important traits and having the knowledge of exact functional nucleotide polymorphism within the gene, breeders can easily identify useful alleles in the wild germplasm and create novel genotypes by introgressing and pyramiding favourite unused natural alleles and/or even by shuffling and re-organization of genomic sequences.
Also, genome sequences and genomics offer exciting new perspectives and opportunities to clone and study the origin of domestication-related genes (or gene families), track rates of sequence divergence over time, and provide hints about how genes evolve and generate products. Up till now, two domestication-related genes in tomato have been cloned, ovate and fw2·2. As discussed above, mutation in the locus ovate resulted in a loss of wild-type function, while mutation(s) in the locus fw2·2 was associated with gene regulation. In addition to ovate and fw2·2, five other domestication-related genes have been cloned in rice and maize and a remarkable feature of the domesticated alleles of all the five genes is that they are functional and encode transcription factors that regulate other genes by directly binding to their DNA (reviewed in Doebley, 2006). Learning from domestication and with more and more available knowledge on genomics, plant breeders might consider manipulating transcription and regulation factors in the genome to generate a pool of new trait variation. Very recently, Davuluri et al. (2005) demonstrated that manipulation of a plant regulatory gene can influence the production of several phytonutrients generated from independent biosynthetic pathways and lead to a novel genotype that cannot be achieved by a conventional breeding approach.
CONCLUSIONS
Tomato domestication and breeding: what have we gained and what can we gain in the future?
In this review, we have looked into the domestication and breeding processes in tomato and the new insights from recent developments in tomato genome research. We show that large variation is present and exploitable in the wild Solanum species. As a consequence of inbreeding during tomato domestication, the genetic diversity in cultivated tomato is now very narrow. Much of the genetic variation was due to spontaneously occurring mutations that were rapidly introduced into new cultivars if these had added value. Sooner or later, a ceiling of the potential of tomato breeding by only using the cultivated germplasm will be reached. To explore tomato biodiversity, ILs that carry small introgressed chromosome fragments from related wild species in a cultivated tomato background are most useful. However, it is virtually impossible to exploit all individual wild accessions by generating genetic libraries of ILs, considering >75 000 Solanum accessions are conserved in genebanks around the world (Larry and Joanne, 2007). It is expected that in the next decade the genome sequence of tomato, at least for the gene-dense regions, will be determined. With techniques like Eco-tilling, allele mining will greatly facilitate the identification of useful genes in the wild tomato germplasm (Comai et al., 2004). Thanks to the synteny among the Solanaceous crops and even the microsynteny with Arabidopsis it is expected that our knowledge about the function of tomato genes will gradually increase. The increasing knowledge from tomato -omics will challenge traditional tomato breeding methodology. With all the genomics, expression and metabolite databases, breeders' capital will shift from the field to the computer. The breeder will select the best combinations of genotypes and design programmes to combine traits in new cultivars in a ‘breeding by design’ process (Peleman and Van der Voort, 2003).
ACKNOWLEDGEMENTS
Funding to pay the Open Access publication charges for this article was provided by the OECD.
LITERATURE CITED
- Alpert KB, Grandillo S, Tanksley SD. fw 2·2: a major QTL controlling fruit weight is common to both red- and green-fruited tomato species. Theoretical and Applied Genetics. 1995;91:994–1000. doi: 10.1007/BF00223911. [DOI] [PubMed] [Google Scholar]
- Bai Y, Lindhout P. New Challenges for Durable Resistance Breeding in Tomato. Proceedings of the XVth meeting of the Eucarpia Tomato Working Group; 20–23 September, 2005; Bari, Italy. 2005. [Google Scholar]
- Bai Y, Huang CC, Van der Hulst R, Meijer-Dekens F, Bonnema G, Lindhout P. QTLs for tomato powdery mildew resistance (Oidium lycopersici) in Lycopersicon parviflorum G1·1601 co-localize with two qualitative powdery mildew resistance genes. Molecular Plant Microbe Interaction. 2003;16:169–176. doi: 10.1094/MPMI.2003.16.2.169. [DOI] [PubMed] [Google Scholar]
- Bai Y, Feng X, Van der Hulst R, Lindhout P. A set of simple PCR markers converted from sequence specific RFLP markers on tomato chromosomes 9 to 12. Molecular Breeding. 2004;13:281–287. [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S. Unraveling the genetic basis of hybrid vigor. Proceedings of the National Academy of Sciences of the USA. 2006;103:12957–12958. doi: 10.1073/pnas.0605627103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breto MP, Asins MJ, Carbonell EA. Genetic variability in Lycopersicon species and their genetic relationship. Theoretical and Applied Genetics. 1993;86:113–120. doi: 10.1007/BF00223815. [DOI] [PubMed] [Google Scholar]
- Canady MA, Ji YF, Chetelat RT. Homeologous recombination in Solanum lycopersicoides introgression lines of cultivated tomato. Genetics. 2006;174:1775–1778. doi: 10.1534/genetics.106.065144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetelat RT, Meglic V. Molecular mapping of chromosome segments introgressed from Solanum lycopersicoides into cultivated tomato (Lycopersicon esculentum) Theoretical and Applied Genetics. 2000;100:232–241. [Google Scholar]
- Comai L, Till BJ, Reynolds SH, Greene EA, Codomo C, Enns L, et al. Large-scale discovery of natural polymorphisms by Ecotilling. The Plant Journal. 2004;37:778–786. doi: 10.1111/j.0960-7412.2003.01999.x. [DOI] [PubMed] [Google Scholar]
- Cong B, Liu J, Tanksley SD. Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proceedings of the National Academy of Sciences of the USA. 2002;99:13606–13611. doi: 10.1073/pnas.172520999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox S. I Say Tomayto, You Say Tomahto. 2000. http://lamar.colostate.edu/~samcox/Tomato.html .
- Davuluri GR, Van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, et al. Fruit-specific RNA-mediated suppression of DET1 enhances cartenoid and flavonoid content in tomatoes. Nature Biotechnology. 2005;23:890–895. doi: 10.1038/nbt1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Alexander DC, Bennett AB. Molecular cloning of tomato fruit polygalacturonase: analysis of polygalacturonase mRNA levels during ripening. Proceedings of the National Academy of Sciences of the USA. 1986;83:6420–6424. doi: 10.1073/pnas.83.17.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J. Unfallen grains: how ancient farmers turned weeds into crops. Science. 2006;312:1318–1319. doi: 10.1126/science.1128836. [DOI] [PubMed] [Google Scholar]
- Doganlar S, Frary A, Tanksley SD. The genetic basis of seed-weight variation: tomato as a model system. Theoretical and Applied Genetics. 2000;100:1267–1273. [Google Scholar]
- Doganlar S, Frary A, Daunay MG, Lester R, Tanksley S. Conservation of gene function in the Solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics. 2002;161:1713–1726. doi: 10.1093/genetics/161.4.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorst JCEA (VW) Een en twintigste beschrijvende rassenlijst voor landbouwgewassen. Wageningen, Rijkscommissie voor de samenstelling van de rassenlijst voor landbouwgewassen. 1946:221. [Google Scholar]
- Egashira H, Ishihara H, Takshina T, Imanishi S. Genetic diversity of the ‘peruvianum-complex’ (Lycopersicon peruvianum (L.) Mill. and L. chilense Dun.) revealed by RAPD analysis. Euphytica. 2000;116:23–31. [Google Scholar]
- Eshed Y, Zamir D. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield associated QTL. Genetics. 1995;141:1147–1162. doi: 10.1093/genetics/141.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A, Doganlar S. Comparative genetics of crop plant domestication and evolution. Turkish Journal of Agricultural Forestry. 2003;27:59–69. [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Van der Knaap E, Cong B, Liu JP. fw2·2: a quantitative trait locus key to evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D. Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science. 2004;305:1786–1789. doi: 10.1126/science.1101666. [DOI] [PubMed] [Google Scholar]
- Fulton TM, Grandillo S, Beck-Bunn T, Fridman E, Frampton A, Lopez J, et al. Advanced backcross QTL analysis of a Lycopersicon esculentum × Lycopersicon parviflorum cross. Theoretical and Applied Genetics. 2000;100:1025–1042. [Google Scholar]
- Garcia-Martinez S, Andreani L, Garcia-Gusano M, Geuna F, Ruiz JJ. Evolution of amplified length polymorphism and simple sequence repeats for tomato germplasm fingerprinting: utility for grouping closely related traditional cultivars. Genome. 2005;49:648–656. doi: 10.1139/g06-016. [DOI] [PubMed] [Google Scholar]
- Goldman IL, Paran I, Zamir D. Quantitative trait locus analysis of a recombinant inbred line population derived from a Lycopersicon esculentum × L. cheesmanii cross. Theoretical and Applied Genetics. 1995;90:925–932. doi: 10.1007/BF00222905. [DOI] [PubMed] [Google Scholar]
- Grandillo S, Tanksley SD. Analysis of horticultural traits differentiating the cultivated tomato from the closely related species Lycopersicon pimpinellifolium. Theoretical and Applied Genetics. 1996;92:935–951. doi: 10.1007/BF00224033. [DOI] [PubMed] [Google Scholar]
- Grandillo S, Ku HM, Tanksley SD. Identifying loci responsible for natural variation in fruit size and shape in tomato. Theoretical and Applied Genetics. 1999;99:978–987. [Google Scholar]
- Gur A, Zamir D. Unused natural variation can lift yield barriers in plant breeding. PLoS Biology. 2004;2:1610–1615. doi: 10.1371/journal.pbio.0020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haanstra JPW, Wye C, Verbakel H, Meijer-Dekens F, Van den Berg P, Odinot P, et al. An integrated high-density RFLP-AFLP map of tomato based on two Lycopersicon esculentum × L. pennellii F2 populations. Theoretical and Applied Genetics. 1999;99:254–271. [Google Scholar]
- Jones RA, Scott SJ. Improvement of tomato flavor by genetically increasing sugar and acid contents. Euphytica. 1983;32:845–855. [Google Scholar]
- Larry R, Joanne L. Genetic resources of tomato. In: Razdan MK, Mattoo AK, editors. Genetic improvement of solanaceous crops. Vol. 2. Enfield, NH: Science Publishers; 2007. Tomato. [Google Scholar]
- Liu J, Van Eck J, Cong B, Tanksley SD. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proceedings of the National Academy of Sciences of the USA. 2002;99:13302–13306. doi: 10.1073/pnas.162485999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S. Tobacco to tomatoes: a phylogenetic perspective on fruit diversity in the Solanaceae. Journal of Experimental Botany. 2002;53:2001–2022. doi: 10.1093/jxb/erf068. [DOI] [PubMed] [Google Scholar]
- Ku HM, Doganlar S, Tanksley SD. fs8·1, a major QTL, sets the pattern of tomato carpel shape well before anthesis. Theoretical and Applied Genetics. 2000;101:873–878. [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D. In silico screening of a saturated mutation library of tomato. The Plant Journal. 2004;38:861–872. doi: 10.1111/j.1365-313X.2004.02088.x. [DOI] [PubMed] [Google Scholar]
- Michael AC, Meglic V, Chetelat RT. A library of Solanum lycopersicoides introgression lines in cultivated tomato. Genome. 2005;48:685–697. doi: 10.1139/g05-032. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tanksley SD. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theoretical and Applied Genetics. 1990;80:437–448. doi: 10.1007/BF00226743. [DOI] [PubMed] [Google Scholar]
- Miller EC, Hadley CW, Schwartz SJ, Erdman JW, Boileau TWM, Clinton SK. Lycopene, tomato products, and prostate cancer prevention. Have we established causality? Pure and Applied Chemistry. 2002;74:1435–1441. [Google Scholar]
- Monforte A, Tanksley SD. Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: a tool for gene mapping and gene discovery. Genome. 2000;43:803–813. [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, et al. The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiology. (a) 2005;138:1310–1317. doi: 10.1104/pp.105.060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller L, Tanksley SD, Giovannoni JJ, Van Eck J, Stack S, Choi D, et al. The tomato sequencing project, the first cornerstone of the international Solanaceae project (SOL) Comparative and Functional Genomics. (b) 2005;6:153–158. doi: 10.1002/cfg.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt TC, Tanksley SD. Comparative sequencing in the genus Lycopersicon: implication for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics. 2002;162:365–379. doi: 10.1093/genetics/162.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YH, West MAL, St Clair DA. Evolution of AFLPs for germplasm fingerprinting and assessment of genetic diversity in cultivars of tomato (Lycopersicon esculentum L.) Genome. 2004;47:510–518. doi: 10.1139/g04-004. [DOI] [PubMed] [Google Scholar]
- Peleman JD, Van der Voort JR. Breeding by design. Trends in Plant Science. 2003;8:330–334. doi: 10.1016/S1360-1385(03)00134-1. [DOI] [PubMed] [Google Scholar]
- Peralta IE, Knapp S, Spooner DM. New species of wild tomatoes (Solannum Section Lycopersicon: Solanaceae) from Northern Peru. Systematic Botany. 2005;30:424–434. [Google Scholar]
- Peralta IE, Knapp S, Spooner DM. Nomenclature for wild and cultivated tomatoes. Tomato Genetics Cooperative Report. 2006;56:6–12. [Google Scholar]
- Peralta IE, Spooner DM. History, origin and early cultivation of tomato (Solanaceae) In: Razdan MK, Mattoo AK, editors. Genetic improvement of solanaceous crops. Vol. 2. Enfield, NH: Science Publishers; 2007. pp. 1–27. Tomato. [Google Scholar]
- Poncet V, Robert T, Sarr A, Gepts P. Quantitative trait loci analyses of the domestication syndrome and domestication process. In: Goodman R, editor. Encyclopedia of plant and crop science. New York, NY: Marcel Dekker; 2004. pp. 1069–1073. [Google Scholar]
- Rick CM. Tomato-like nightshades: affinities, auto-ecology, and breeders opportunities. Economic Botany. 1988;42:145–154. [Google Scholar]
- Rick CM, Chetelat RT. Utilization of related wild species for tomato improvement. Acta Horticulturae. 1995;412:21–38. [Google Scholar]
- Sacks EJ, Gerhardt LM, Graham EB, Thorrup TA, St Clair DA. Variation among 41 genotypes of tomato (Lycopersicon esculentum Mill.) for cross ability to L. peruvianum (L.) Mill. Annals of Botany. 1997;80:469–477. [Google Scholar]
- Schuch W, Kanczler J, Robertson D, Hobson G, Tucker G, Grierson D, et al. Fruit quality characteristics of transgenic tomato fruit with altered polygalacturonase activity. HortScience. 1991;26:1517–1520. [Google Scholar]
- Semel Y, Nissenbaum J, Menda N, Zinder M, Krieger U, Issman N, et al. Overdominant quantitative trait loci for yield and fitness in tomato. Proceedings of the National Academy of Sciences of the USA. 2006;103:12981–12986. doi: 10.1073/pnas.0604635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims WL. History of tomato production for industry around the world. Acta Horticulturae. 1980;100:25–26. [Google Scholar]
- Strauss SH. Regulation of biotechnology as though gene function mattered. BioScience. 2003;53:453–454. [Google Scholar]
- Tam SM, Mhiri C, Vogelaar A, Kerkveld M, Pearce SR, Grandbastien MA. Comparative analysis of genetic diversities within tomato and pepper collections detected by retrotransposon-based SSAP, AFLP and SSR. Theoretical and Applied Genetics. 2005;110:819–831. doi: 10.1007/s00122-004-1837-z. [DOI] [PubMed] [Google Scholar]
- Tanksley SD. The genetic, developmental and molecular bases of fruit size and shape variation in tomato. The Plant Cell. 2004;16:S181–S189. doi: 10.1105/tpc.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, De Vicente MC, Bonierbale MW, Broun P, et al. High density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Grandillo S, Fultom TM, Zamir D, Eshed Y, Petiard V, et al. Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theoretical and Applied Genetics. 1996;92:213–224. doi: 10.1007/BF00223378. [DOI] [PubMed] [Google Scholar]
- Tieman DM, Zeigler M, Schmelz EA, Taylor MG, Bliss P, Kirst M, et al. Identification of loci affecting flavour volatile emissions in tomato fruits. Journal of Experimental Botany. 2006;57:887–896. doi: 10.1093/jxb/erj074. [DOI] [PubMed] [Google Scholar]
- Tikunov Y, Lommen A, De Vos CHR, Verhoeven HA, Bino RJ, Hall RD, et al. A novel approach for nontargeted data analysis for metabolomics: large-scale profiling of tomato fruit volatiles. Plant Physiology. 2005;139:1125–1137. doi: 10.1104/pp.105.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UPOV Breeders Rights. International convention for the protection of new varieties in plants. 2 December 1961, as revised at Geneva on 10 November 1972, on 23 October 1978, and on 19 March 1991. 1961 [Google Scholar]
- Van der Beek JG, Verkerk R, Zabel P, Lindhout P. Mapping strategy for resistance genes in tomato based on RFLPs between cultivars: Cf9 (resistance to Cladosporium fulvum) on chromosome 1. Theoretical and Applied Genenetics. 1992;84:106–112. doi: 10.1007/BF00223988. [DOI] [PubMed] [Google Scholar]
- Van der Knaap E, Tanksley SD. Identification and characterization of a novel locus controlling early fruit development in tomato. Theoretical and Applied Genetics. 2001;103:353–358. [Google Scholar]
- Van der Knaap E, Tanksley SD. Extremely elongated tomato fruit controlled by four quantitative trait loci with epistatic interactions. Theoretical and Applied Genetics. 2002;104:241–247. doi: 10.1007/s00122-001-0776-1. [DOI] [PubMed] [Google Scholar]
- Venema JH, Linger P, Van Heusden AW, Van Hasselt PR, Brueggemann W. The inheritance of chilling tolerance in tomato (Lycopersicon spp.). Plant Biology. 2005;7:118–130. doi: 10.1055/s-2005-837495. [DOI] [PubMed] [Google Scholar]
- Villand J, Skroch PW, Lai T, Hanson P, Kuo CG, Nienhuis J. Genetic variation among tomato accessions from primary and secondary centers of diversity. Crop Science. 1998;38:1339–1347. [Google Scholar]
- Walter JM. Heredity resistance to disease in tomato. Annual Reviewers. 1967;5:131–160. [Google Scholar]
- Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- Watson B. Taylors guide to heirloom vegetables. New York, NY: Houghton Mifflin Co; 1996. [Google Scholar]