Abstract
Background
The objective of this study was to dissect into quantitative trait loci (QTLs) the large morphological and physiological differences between cultivated azuki bean (Vigna angularis) and a wild relative and to infer the commonalities of the QTLs for domestication-related traits across the Asian Vigna and with other warm-season legumes.
Methods
Two linkage maps, for the BC1F1 and F2 populations, respectively, from the same cross between azuki bean and V. nepalensis were developed. Using these linkage maps QTLs for 33 domestication-related traits were analysed and mapped. The location of mapped QTLs was compared with locations of similar QTLs in other warm-season legumes.
Key Results
QTLs were detected for seed-, pod-, stem- and leaf-related traits. Most traits were controlled by between two and nine QTLs but several traits, such as pod dehiscence, were controlled by single genes. QTLs for domestication-related traits were restricted to particular regions of the azuki bean genome, especially linkage groups 1, 2, 4, 7 and 9. Linkage groups 1 and 2 had QTLs for a suite of traits including pod size, germination, seed size and lower stem length. QTLs on linkage groups 7 and 9 were associated with upper stem length, maximum leaf size and pod and seed size. Pleiotropy or close linkage of genes for domestication-related traits is suggested in these regions. While some QTLs are common to azuki bean and other warm-season legumes, many are recorded for the first time in azuki bean.
Conclusions
QTLs for a large number of domestication-related traits have been mapped for the first time in azuki bean. QTLs with unexpected effect and new QTLs for traits such as seed size have been found. The results provide a foundation that will be useful for improvement of azuki bean and related legumes.
Key words: Azuki bean, Vigna angularis var. angularis, Vigna nepalensis, wild species, QTL, microsatellite
INTRODUCTION
Most domesticated legumes belong to two distinct phylogenetic clades, the inverted-repeat-lacking clade that includes many of the temperate or cool season legume crops, and the Phaseoleae clade that includes many of the tropical or warm-season legume crops (Wojciechowski et al., 2004). In the Phaseoleae a large group of closely related domesticated species is present in the genera Phaseolus (five species domesticated in the Americas) and Vigna (eight species domesticated in Africa and Asia). In addition, the Phaseoleae includes the most important domesticated legume in terms of production, the soybean [Glycine max (L.) Merr.]. All of the domesticated species in the Phaseolus–Vigna group are diploid except one, the tetraploid species Vigna reflexo-pilosa Hayata. The close phylogenetic relationship of the Phaseolus–Vigna group suggests that at the genome level there is likely to be a high degree of synteny among the domesticates of this group.
The genus Vigna is naturally distributed in the New World, Africa, Asia, Australia and the Pacific and has been divided into six subgenera. Most of the Vigna species that occur in Asia belong to the subgenus Ceratotropis and are known as the Asian Vigna. In the subgenus Ceratotropis six species have been domesticated. These six species are azuki bean [Vigna angularis var. angularis (Willd.) Ohwi and Ohashi], mungbean [V. radiata (L.) Wilczek], black gram [V. mungo (L.) Hepper], rice bean [V. umbellata (Thunb.) Ohwi and Ohashi], moth bean [V. aconitifolia (Jacq.) Maréchal] and creole bean [V. reflexo-pilosa var. glabra]. Among them mungbean, azuki bean and black gram are the most important in terms of production (Tomooka et al., 2002).
Azuki bean (also known as adzuki) is a traditional legume grown across East Asia and northern South Asia (Zong et al., 2003). The presumed wild ancestor of cultivated azuki bean is V. angularis var. nipponensis (Yamaguchi, 1992). This wild species is distributed across a wide area of Japan, the Korean peninsula and China, including Taiwan, Nepal and Bhutan (Vaughan et al., 2004). Recently, a new species, V. nepalensis Tateishi and Maxted, from Nepal and Bhutan has been described (Tateishi and Maxted, 2002). Vigna nepalensis resembles V. angularis var. nipponensis in morphological characters and is genetically closely related to cultivated azuki bean with which it is cross compatible. They are considered to belong to the same species complex (Tomooka et al., 2006). Since V. nepalensis is within the primary genepool of azuki bean it was used in a cross with azuki bean to develop a molecular linkage map of azuki bean. Azuki bean was the first Asian Vigna with a molecular linkage map that has resolved the 11 linkage groups corresponding to the haploid chromosome number of the diploid Asian Vigna (Han et al., 2005).
Azuki bean shows numerous differences in morphological and physiological traits associated with domestication compared with its closely related wild relatives. Domestication of azuki bean has resulted in a conspicuous increase in seed and pod size, non-twining growth habit, and loss of seed dormancy and seed dispersal ability. In addition, seed colour variation that is not found in its wild relatives is present in azuki bean cultivars. Among the domesticated Asian Vigna and their presumed wild ancestors, seed size, seed colour and life history traits differ markedly. For example, cultivated mungbean generally has green seeds that are about five times the size of wild mungbean, while cultivated azuki bean usually has red seeds that are more than eight times the size of the wild azuki bean (Tomooka et al., 2000).
Since the Phaseolus–Vigna group includes many domesticated species, comparative genome studies of the domestication syndrome in this group may shed light on cryptic species-specific genes for morphological and physiological variation (Hawkes, 1983; Hammer, 1984). The genetic control of some domestication-related traits has been characterized in some of the warm-season legumes including mungbean of the Asian Vigna and New World Phaseolus (Boutin et al., 1995; Koinange et al., 1996; Menacio-Hautea et al., 1993; Papa et al., 2005).
The cross between V. nepalensis and V. angularis that was used to develop the molecular linkage map of azuki bean (Han et al., 2005) was the basis for the research presented here. The overall objectives of this study were to provide a foundation for comparative analysis of the domestication syndrome across the Asian Vigna and other warm-season legumes. The specific objectives in this study were (a) to develop a molecular linkage map based on the F2 population of a cross between V. nepalensis and V. angularis, the same cross for which a molecular linkage map of azuki bean based on the BC1F1 population has been published (Han et al., 2005); (b) to dissect into QTLs the morphological and physiological traits associated with the domestication of azuki bean; and (c) to infer commonality of QTLs for domestication-related traits based on information available from other crosses involving warm-season legumes.
MATERIALS AND METHODS
Mapping population
The segregating populations studied were derived from the cross between the wild species Vigna nepalensis (female) and the cultivated azuki bean (male and recurrent parent for backcross) and used to construct molecular linkage maps and for trait measurement. Vigna nepalensis (JP107881) was collected in Nepal and the cultivated azuki bean (JP81481) is a landrace from Tokushima Prefecture, Japan. These accessions were obtained from the Ministry of Agriculture, Forestry and Fisheries, Gene Bank, Tsukuba, Japan.
The BC1F1 population, consisting of 187 individuals (from 200 seeds), was planted in pots in a vinyl greenhouse of the National Institute of Agrobiological Sciences (NIAS), Tsukuba, Japan, 36°2′N 140°8′E, from June to November 2002, and used for DNA extraction to construct the molecular linkage map. The F2 population, consisting of 141 individuals (from 144 seeds), was grown from June to November 2003 under the same conditions as the BC1F1, used for DNA extraction to construct a molecular linkage map and evaluated for domestication-related traits (Table 1). BC1F1:2 lines each consisting of ten individuals per line were planted in the field in June 2003 and 2004 at NIAS and were evaluated based on mean value for each trait per line. The F2:3 lines each consisting of ten individuals per line were planted in the field in June 2004 at NIAS and were evaluated in a similar way. Ten plants of each parent were grown together with these populations. In total, four sets of data for seedling and vegetative domestication-related traits were obtained from BC1F1:2 (2003, 2004), F2 and F3 generations except for seed and pod size traits.
Table 1.
Domestication-related traits examined, their trait and QTL abbreviations and evaluation method
| Organ | General attribute | Trait | Trait abbreviation | QTL/gene | Evaluation method | Evaluated population |
|---|---|---|---|---|---|---|
| Pod | Pod size | Podlength (cm) | PDL | Pdl | Length of straight pod | BC1F1, F2 |
| Podwidth (mm) | PDW | Pdw | Width of pod at widest part | BC1F1, F2 | ||
| Pod dehiscence | Pod –Number of twist (count) | PDT | Pdt | Number of twists along the length of the shattered pod | BC1F1, F2 | |
| Seed | Seed size | Seedweight (g) | SD100WT | Sd100wt | Weight of 100 seeds | BC1F1, F2 |
| Seedlength (mm) | SDL | Sdl | Maximum distance from top to bottom of the seed | BC1F1, F2 | ||
| Seedwidth (mm) | SDW | Sdw | Maximum distance from hilum to its opposite side | BC1F1, F2 | ||
| Seedthickness (mm) | SDT | Sdt | Maximum distance between both sides of the hilum | BC1F1, F2 | ||
| Seed colour | Seed coat colour | SDC | Sdc | Tan or red | BC1F1, F2 | |
| Seed coat colour black mottle color | SDCBM | Sdcbm | Present or absent | BC1F1, F2 | ||
| Seed germination | Seed – Germination in field (%) | SDG | Sdg | Germination percentage at 21 d after sowing in field | BC1F1:2, F2:3 | |
| Seed – Coat permeable (%) | SDP | Sdp | Percent of imbibed seeds at 21 d after sowing at 25 °C in incubator | BC1F1:2, F2:3 | ||
| Stem | Epicotyl colour | Epicotyl colour | ECC | Ecc | Red or green | F2 |
| Stem size | Epicotyl length (cm) | ECL | Ecl | Length from cotyledon to primary leaf | BC1F1:2, F2, F2:3 | |
| Stem – Internode length (1st to 10th) (cm) | ST1-10I | St1-10i | Length from primary leaf node to each node | BC1F1:2, F2, F2:3 | ||
| Stem length (cm) | STL | Stl | Length from primary leaf node to 10th trifoliate leaf node | BC1F1:2, F2, F2:3 | ||
| Stem thickness (mm) | STT | Stt | Stem diameter below the primary leaf | BC1F1:2, F2, F2:3 | ||
| Growth habit | Stem – twining (%) | STTW | Sttw | Stem twining beyond the main stem 10th internode | BC1F1:2, F2:3 | |
| Leaf | Leaf size | Leaf – Primary leaf length (mm) | LFPL | Lfpl | Distance from pulvinus to leaf tip | BC1F1:2, F2, F2:3 |
| Leaf – Primary leaf width (mm) | LFPW | Lfpw | Widest part of primary leaf | BC1F1:2, F2, F2:3 | ||
| Leaf – maximum leaflet length (mm) | LFML | Lfml | Length of the largest terminal leaflet on a leaf between 1st trifoliate leaf node and 10th trifoliate leaf node | BC1F1:2, F2, F2:3 | ||
| Leaf – maximum leaflet width (mm) | LFMW | Lfmw | Width of the largest terminal leaflet on a leaf between 1st trifoliate leaf node and 10th trifoliate leaf node | BC1F1:2, F2, F2:3 | ||
| Branch | Number | Branch number (count) | BRN | Brn | Number of branches on main stem from 1st trifoliate leaf node to 10th trifoliate leaf node | BC1F1:2, F2, F2:3 |
| Position | Branch – Position of 1st branch (ith node) | BRP | Brp | Position of the first branch on main stem from 1st trifoliate leaf node to 10th trifoliate leaf node | BC1F1:2, F2, F2:3 | |
| Flower | Flowering time | Flower – Days to first flower (days) | FLD | Fld | Number of days from sowing to 1st flowering | F2 |
Information on climate (monthly average temperature, rainfall and daylight hours) for 2002–2004 when the segregating populations were grown is provided (see Supplementary Information 1, available at online).
Trait measurement
Thirty-three domestication-related traits were evaluated. These traits related to the size of various plant parts (seeds, pods, leaf and stem), plant habit (stem and branch), life cycle (flowering time), pod dehiscence, seed and epicotyl colour and seed germination (Table 1). These traits are all known to differ between wild and cultivated azuki bean and, therefore, are considered to be directly or indirectly the result of changes associated with domestication. Of these, 30 were treated as quantitative traits and three – epicotyl colour, seed coat colour and black mottle of seed coat – were treated as qualitative traits. The seedling traits (epicotyl length, primary leaf length and width) were recorded when the 1st trifoliate leaf opened. The vegetative traits (1st to 10th internode length, stem diameter, leaf length and width, number of branches and growth habit) were recorded when the 10th trifoliate expanded completely. When the uppermost internode showed twining extension, the growth habit characteristic was recorded as ‘1’; when the uppermost internode was non-twining it was recorded as ‘0’.
The seed and pod traits were investigated using the seeds and pods from BC1F1 and F2 plants that were harvested in 2002 and 2003, respectively. Length, width and thickness of five seeds were measured using digital callipers. Seed germination was investigated under two different conditions. The first experiment was carried out in the field from 15 June in 2003 and 2004 using seeds from BC1F1 and F2 plants, respectively, stored at room temperature after harvest. Ten unscarified seeds were sown in the field and the number and date of germinated seeds up to 21 d after planting were recorded. Germination was recorded as the proportion of germinated seeds to total surviving seeds, including germinated seeds and seeds recovered from the field soil. The second experiment was carried out in the laboratory. Ten unscarified seeds were placed on wet filter paper and incubated in the dark at 25 °C for 21 d. The number of seeds that expanded after imbibing water was recorded as a percentage. For pod traits, number of twists along the length of the naturally shattered pod dried at 32 °C for 2 weeks was recorded on ten pods, and then their length and width were measured after soaking in water to reconstitute their shape. As an index of pod dehiscence, the ratio of twist number to length of pod was used.
Map construction and QTL analysis
A molecular linkage map based on the F2 population was constructed using framework SSR markers that were distributed throughout the BC1F1 genetic linkage map (Han et al., 2005). Total genomic DNA from F2 individuals was extracted from 100 mg of fresh leaf tissue using the EZ1 DNA Tissue kit (QIAGEN, Valencia, CA, USA). The SSR analysis and linkage analysis of SSR markers in the F2 population were carried out according to the method of Han et al. (2005).
Prior to QTL analysis, the frequency distributions of phenotypes in BC1F1, BC1F1:2, F2 and F2:3 populations were examined for each trait (see Supplementary Information 2, available online). The proportional data for traits related to seed germination and growth habit was transformed to arcsine data. Before conducting the QTL analysis, outliers with a value greater than ± 3 s.d. of the trait average were removed to reduce the effect of strong deviation from normality.
QTL analysis was conducted by using the software package MultiQTL, ver. 2·5 (http://www.multiqtl.com/) according to the procedure described by Peng et al. (2003). In brief, the entire genome was scanned for each of the domestication-related traits using general interval mapping with the following approach. First, a single QTL model was fitted for each trait-chromosome (linkage group) combination. A permutation test (Churchill and Doerge, 1994) was used to obtain chromosome-wise statistical significance of the putative QTL when the single-QTL model was fitted. Estimates of the main parameters (QTL effect, its position, and the proportion of explained phenotypic variance explained) were evaluated at the thresholds of the test statistics obtained from bootstrap analysis (Lebreton and Visscher, 1998) based on 10 000 runs per linkage group. When the LOD graph indicated the possibility of two QTLs, a two-linked-QTL model was fitted for each trait-chromosome combination (Korol et al., 1998). A permutation test was used to obtain chromosome-wise statistical significance of the putative QTL when the two-linked-QTL model was fitted. The hypothesis of two linked QTLs (H2) was compared with two alternatives – H0 (the chromosome has no effect) and H1 (only one QTL in the chromosome) – at P < 0·01 level. For the traits evaluated over 2 years on the same population, the single- or two-linked-QTL model for multiple-environments (Jansen et al., 1995) was fitted and tested using the same procedure as described above. In order to correct for multiple comparisons, experimental-wise significance level for all QTLs was declared at P < 0·001 and estimated based on method of Benjamini and Hochberg (1995) and QTLs significant at FDR (false discovery rate) = 5 % are reported in the present study.
To test for randomness of the genomic distribution of QTLs for domestication-related traits identified, χ2 tests were calculated under the assumption of independent gene action. To test whether or not QTLs were randomly distributed along a linkage group, a Poisson distribution function P(x) = e−μμx/x!, where x is the number of QTL per 10-cM interval and μ is the resulting QTL density on linkage group, was calculated.
QTL nomenclature followed the style of Somta et al. (2006). The trait abbreviation is followed by the population number [BC1F1 or BC1F1:2 (1) and F2 or F2:3 (2)], linkage group and then QTL number. QTLs were first assigned to the BC1F1 or BC1F1:2 populations and then only new QTLs were assigned in the F2 or F2:3 populations.
RESULTS
Establishment of the linkage map
The BC1F1 map constructed by Han et al. (2005) consisted of 381 markers (205 SSR, 94 RFLP and 187 AFLP markers) across 11 linkage groups. The linked markers span a total distance of 832·1 cM.
The F2 map constructed here used 74 SSR markers covering 649·7 cM. This was equivalent to 78·1 % of the BC1F1 map. For the F2 map, segregation distortion was observed in 28 % (21 out of 74) of markers. All 21 markers were distorted in favour of alleles from the wild female parent and 14 of these were found on linkage groups 4, 6, 8 and 11. The order of the SSR markers on each linkage group was consistent in the two maps, BC1F1 and F2, although there were small differences in the distance between the markers (see Supplementary Information 3, available online).
Field data analysis
The means, standard deviations and broad sense heritabilities in 2003 and 2004 of traits in the parental lines and F2 and F2:3 populations are shown (Table 2). These populations showed a high degree of morphological and physiological variation. The means of lines (or plants) in BC1F1, BC1F1:2, F2 and F2:3 populations generally fell between the means of cultivated and wild parents for all traits except the position of the first branch. The seed germination (%), pod dehiscence and size of organs such as leaf, stem, seed and pod differed markedly between the parents. For internode lengths, the 1st to 3rd internodes in the cultivated parent were longer than those in the wild parent, whereas the 4th to 10th internodes in the cultivated parent were shorter than those in the wild parent. For qualitative traits, in the cultivated parent seed coat colour was red and the epicotyl was green, while the wild parent had a black mottled over tan seed coat and red epicotyl.
Table 2.
Trait mean, standard deviation and heritability values for parents, the BC1F1:2 (2003, 2004), F2 and F2:3 populations of the cross between V. nepalensis and cultivated azuki bean
| BC1F1:2 population (2003) |
BC1F1:2 population (2004) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Vigna nepalensis |
V. angularis |
BC1F1:2 |
Heritability (%) |
V. nepalensis |
V. angularis |
BC1F1:2 |
Heritability (%) | |||||||
| Trait† | Mean | s.d.§ | Mean | s.d. | Mean | s.d. | Mean | s.d. | Mean | s.d. | Mean | s.d. | ||
| PDL (cm)‡ | 7·2 | 0·85 | 9·2 | 1·03 | 9·0 | 1·21 | 39·7 | No investigation | ||||||
| PDW (mm)‡ | 4·8 | 0·15 | 10·2 | 0·30 | 8·5 | 0·59 | 84·2 | No investigation | ||||||
| PDT (count)‡ | 3·0 | 0·12 | 0·7 | 0·19 | 2·6 | 2·10 | 99·4 | No investigation | ||||||
| SD100WT (g)‡ | 1·7 | 0·18 | 16·3 | 0·14 | 9·1 | 1·61 | 99·0 | No investigation | ||||||
| SDL (mm)‡ | 3·7 | 0·17 | 7·3 | 0·14 | 6·1 | 0·44 | 87·3 | No investigation | ||||||
| SDW (mm)‡ | 2·5 | 0·11 | 5·6 | 0·07 | 4·6 | 0·32 | 91·4 | No investigation | ||||||
| SDT (mm)‡ | 2·0 | 0·06 | 5·2 | 0·04 | 4·2 | 0·33 | 97·7 | No investigation | ||||||
| SDC‡ | Tan | Red | T : R = 94 : 91 (χ2 = 0·049) | No investigation | ||||||||||
| SDCBM‡ | Present | Absent | P : A = 96 : 89 (χ2 = 0·265) | No investigation | ||||||||||
| SDG (%) | 2·5 | 5·59 | 70·1 | 5·40 | 50·1 | 34·07 | 97·4 | 0·0 | 0·00 | 76·7 | 5·77 | 32·5 | 30·19 | 98·2 |
| SDP (%) | 0·0 | 0·00 | 75·0 | 7·07 | 31·1 | 32·53 | 97·6 | No investigation | ||||||
| ECC | No investigation | No investigation | ||||||||||||
| ECL (cm) | No investigation | 0·7 | 0·11 | 7·9 | 0·08 | 4·9 | 1·12 | 99·3 | ||||||
| ST1I (cm) | 0·1 | 0·06 | 2·1 | 0·40 | 1·3 | 0·46 | 60·5 | 0·2 | 0·00 | 1·9 | 0·66 | 1·7 | 0·43 | – |
| ST2I (cm) | 0·2 | 0·06 | 2·0 | 0·59 | 1·6 | 0·58 | 49·1 | 0·5 | 0·03 | 2·0 | 0·29 | 1·8 | 0·43 | 76·5 |
| ST3I (cm) | 0·4 | 0·10 | 2·4 | 0·87 | 2·1 | 0·75 | 31·8 | 1·6 | 0·18 | 2·1 | 0·29 | 2·2 | 0·48 | 75·0 |
| ST4I (cm) | 0·6 | 0·15 | 3·0 | 0·43 | 2·7 | 0·93 | 88·2 | 3·6 | 0·22 | 2·8 | 0·50 | 3·2 | 0·82 | 77·7 |
| ST5I (cm) | 0·7 | 0·31 | 3·1 | 0·51 | 3·0 | 1·19 | 87·7 | 5·8 | 0·09 | 3·1 | 0·41 | 4·3 | 1·02 | 91·3 |
| ST6I (cm) | 1·0 | 0·15 | 3·3 | 0·77 | 3·5 | 1·72 | 89·6 | 11·9 | 0·31 | 3·5 | 0·42 | 5·3 | 1·25 | 91·2 |
| ST7I (cm) | 1·3 | 0·20 | 3·3 | 0·68 | 3·7 | 2·18 | 94·7 | 17·2 | 0·14 | 3·5 | 0·41 | 6·3 | 1·65 | 96·5 |
| ST8I (cm) | No investigation | 18·5 | 0·71 | 3·9 | 0·25 | 7·3 | 2·03 | 93·2 | ||||||
| ST9I (cm) | No investigation | 21·8 | 0·80 | 4·2 | 0·31 | 8·3 | 2·40 | 93·7 | ||||||
| ST10I (cm) | No investigation | 23·0 | 1·50 | 4·5 | 0·46 | 9·5 | 2·85 | 85·0 | ||||||
| STL (cm) | 4·3 | 0·76 | 19·2 | 3·22 | 17·9 | 6·67 | 87·7 | 104·8 | 3·06 | 39·4 | 3·78 | 54·9 | 11·93 | 91·7 |
| STTW (%) | 100·0 | 0·00 | 0·0 | 0·00 | 26·0 | 31·71 | 100·0 | 100·0 | 0·00 | 0·0 | 0·00 | 56·6 | 30·89 | 100·0 |
| STT (mm) | 2·9 | 0·47 | 5·0 | 0·36 | 4·4 | 0·68 | 62·7 | 2·1 | 0·06 | 6·9 | 0·46 | 5·1 | 0·65 | 74·9 |
| LFPL (mm) | 22·4 | 1·93 | 40·8 | 3·08 | 38·3 | 4·72 | 70·3 | 21·3 | 1·27 | 34·7 | 3·25 | 35·7 | 3·94 | 60·8 |
| LFPW (mm) | 17·5 | 1·32 | 40·7 | 1·97 | 35·7 | 4·07 | 83·1 | 17·4 | 1·41 | 35·0 | 2·75 | 33·8 | 3·53 | 61·7 |
| FLML (mm) | No investigation | 91·2 | 1·37 | 104·5 | 2·45 | 104·5 | 7·54 | 93·0 | ||||||
| FLMW (mm) | No investigation | 62·4 | 0·81 | 90·2 | 1·86 | 80·6 | 6·35 | 94·9 | ||||||
| BRN (count) | 3·7 | 0·58 | 2·3 | 0·47 | 2·2 | 0·74 | 49·7 | 5·1 | 0·20 | 3·0 | 0·54 | 2·2 | 0·72 | 68·4 |
| BRP (ith) | 1·0 | 0·00 | 1·7 | 0·20 | 1·8 | 0·58 | 94·0 | 1·1 | 0·20 | 2·9 | 0·33 | 4·0 | 0·78 | 87·6 |
| FLD (day) | No investigation | No investigation | ||||||||||||
| F2 population (2003) | F2:3 population (2004) | |||||||||||||
| V. nepalensis | V. angularis | F2 | Heritability (%) | V. nepalensis | V. angularis | F2:3 | Heritability (%) | |||||||
| Trait† | Mean | s.d. | Mean | s.d. | Mean | s.d. | Mean | s.d. | Mean | s.d. | Mean | s.d. | ||
| PDL (cm)‡ | 7·3 | 0·11 | 10·1 | 0·37 | 8·0 | 1·05 | 93·3 | No investigation | ||||||
| PDW (mm)‡ | 5·2 | 0·26 | 9·5 | 0·29 | 7·0 | 0·62 | 80·1 | No investigation | ||||||
| PDT (count)‡ | 5·9 | 0·47 | 0·5 | 0·04 | 3·4 | 1·80 | 96·5 | No investigation | ||||||
| SD100WT (g)‡ | 1·8 | 0·07 | 10·7 | 0·23 | 5·0 | 1·20 | 97·9 | No investigation | ||||||
| SDL (mm)‡ | 3·4 | 0·10 | 6·4 | 0·18 | 5·0 | 0·47 | 90·7 | No investigation | ||||||
| SDW (mm)‡ | 2·5 | 0·06 | 4·9 | 0·08 | 3·7 | 0·32 | 95·6 | No investigation | ||||||
| SDT (mm)‡ | 2·2 | 0·05 | 4·5 | 0·11 | 3·2 | 0·34 | 93·9 | No investigation | ||||||
| SDC‡ | Tan | Red | T : R = 116 : 25 (χ2 = 3·974) | No investigation | ||||||||||
| SDCBM‡ | Present | Absent | P : A = 104 : 37 (χ2 = 0·116) | No investigation | ||||||||||
| SDG (%) | No investigation | 3·3 | 5·77 | 76·7 | 11·55 | 25·8 | 26·46 | 88·1 | ||||||
| SDP (%) | No investigation | 0·0 | 0·00 | 80·0 | 14·14 | 14·0 | 23·47 | 81·8 | ||||||
| ECC | Red | Green : G = 108 : 33 (χ2 = 0·191) | No investigation | |||||||||||
| ECL (cm) | 1·7 | 0·18 | 8·0 | 1·07 | 4·1 | 0·94 | 32·5 | 0·7 | 0·16 | 7·7 | 0·16 | 2·4 | 0·88 | 96·7 |
| ST1I (cm) | 0·2 | 0·03 | 1·7 | 0·27 | 0·6 | 0·20 | 4·8 | 0·3 | 0·02 | 2·0 | 0·42 | 0·8 | 0·29 | – |
| ST2I (cm) | 0·2 | 0·06 | 2·4 | 0·53 | 1·1 | 0·51 | 43·8 | 0·6 | 0·07 | 1·9 | 0·16 | 1·1 | 0·33 | 85·5 |
| ST3I (cm) | 0·8 | 0·24 | 2·8 | 0·52 | 2·0 | 1·05 | 85·3 | 1·5 | 0·20 | 1·9 | 0·22 | 1·6 | 0·46 | 78·4 |
| ST4I (cm) | 1·7 | 0·31 | 3·4 | 0·40 | 4·4 | 3·67 | 99·1 | 3·8 | 0·28 | 2·6 | 0·24 | 2·3 | 0·71 | 86·0 |
| ST5I (cm) | 3·5 | 0·77 | 3·9 | 0·70 | 9·2 | 6·75 | 98·8 | 5·9 | 0·15 | 2·9 | 0·30 | 3·0 | 0·85 | 92·2 |
| ST6I (cm) | 8·5 | 2·04 | 5·2 | 0·96 | 14·0 | 8·51 | 96·5 | 10·8 | 0·33 | 3·3 | 0·37 | 3·7 | 1·17 | 90·8 |
| ST7I (cm) | 15·3 | 2·48 | 6·7 | 1·08 | 19·0 | 8·43 | 94·8 | 12·9 | 0·38 | 3·5 | 0·26 | 4·6 | 1·58 | 95·8 |
| ST8I (cm) | 22·8 | 2·72 | 10·1 | 3·06 | 22·0 | 7·04 | 83·1 | 14·6 | 0·55 | 3·6 | 0·52 | 5·6 | 2·10 | 93·5 |
| ST9I (cm) | 25·2 | 2·59 | 14·6 | 3·05 | 22·7 | 6·68 | 82·0 | 15·5 | 0·61 | 3·8 | 0·67 | 6·8 | 2·57 | 93·8 |
| ST10I (cm) | 25·1 | 1·92 | 16·9 | 2·88 | 22·6 | 6·62 | 86·3 | 17·2 | 0·35 | 4·0 | 0·74 | 8·2 | 3·26 | 96·8 |
| STL (cm) | 103·2 | 8·60 | 67·7 | 8·60 | 117·5 | 38·16 | 94·9 | 83·8 | 1·75 | 37·3 | 3·54 | 40·0 | 12·07 | 94·6 |
| STTW (%) | No investigation | 100·0 | 0·00 | 0·0 | 0·00 | 73·5 | 33·91 | 100·0 | ||||||
| STT (mm) | 3·7 | 0·18 | 4·6 | 0·32 | 3·7 | 0·45 | 68·3 | 2·2 | 0·04 | 6·9 | 0·17 | 5·2 | 0·90 | 98·1 |
| LFPL (mm) | 28·2 | 2·20 | 57·6 | 6·12 | 47·3 | 4·56 | – | 21·5 | 1·13 | 33·3 | 2·66 | 30·7 | 3·36 | 63·1 |
| LFPW (mm) | 22·5 | 1·75 | 56·1 | 5·33 | 42·9 | 3·81 | – | 17·7 | 0·26 | 34·7 | 1·72 | 27·7 | 3·48 | 87·5 |
| FLML (mm) | 125·2 | 6·81 | 131·0 | 9·56 | 135·0 | 14·88 | 68·9 | 86·8 | 4·31 | 105·7 | 4·02 | 99·2 | 10·54 | 84·3 |
| FLMW (mm) | 80·7 | 6·07 | 103·4 | 6·25 | 92·6 | 13·43 | 79·0 | 62·8 | 3·86 | 87·8 | 1·69 | 79·0 | 9·15 | 89·4 |
| BRN (count) | 9·2 | 0·42 | 1·9 | 0·74 | 6·8 | 2·02 | 91·1 | 5·4 | 0·51 | 3·9 | 0·28 | 2·1 | 0·81 | 74·3 |
| BRP (ith) | 1·8 | 0·42 | 1·1 | 0·32 | 2·8 | 1·35 | 92·4 | 1·0 | 0·00 | 2·2 | 0·33 | 4·3 | 1·21 | 96·2 |
| FLD (day) | 96·3 | 1·06 | 49·1 | 0·99 | 79·2 | 7·30 | 98·0 | No investigation | ||||||
† Trait abbreviations are shown in Table 1.
‡ The BC1F1 generation was evaluated for these traits.
§ s.d., Standard deviation.
* Significant at 5 % level.
The traits analysed showed nearly normal distributions among lines (or plants) in these populations. In 2004, each internode length showed a normal distribution in both the BC1F1:2 and F2:3 populations (see Supplementary Information 2, available online). However, for some internodes mean values were distorted [BC1F1:2 (2004) internodes 1–3, F2:3 internodes 1 and 4–7] and some lines showed transgressive segregation [BC1F1:2 (2004) internodes 4 and 5, F2:3 internode 1].
The F2 mean for seed germination percentge in the field showed a distribution distorted towards the wild parent type. A binomial distribution was observed for twist number in the pod suggesting that a single gene controls this trait. A binomial distribution was also observed for twining versus non-twining growth in the BC1F1:2 population in 2003. However, in the BC1F1:2 (2004) and F3 populations the mean number of lines with twining growth was distorted towards the wild parent. Generally the traits measured showed high broad sense heritability (exceeding 0·6; Table 2). The segregation ratios for the three qualitative traits, seed coat colour, black mottle on the seed coat and epicotyl colour, matched the ratios expected for control by a single gene in both populations except that seed coat colour in the F2 population showed distorted segregation significant at the 5 % level. Red seed coat colour and green epicotyl colour of the cultivated parent are recessive. Black mottle and tan seed coat colour, and red epicotyl colour are dominant.
In general, similar or related traits showed significant positive correlations at P ≤ 0·05 in all populations (see Supplementary information 4, available online). For example, there were significant positive correlations between stem length and each internode length and also significant positive correlations between lengths of each internode. Traits related to seed size were positively correlated with pod length and stem thickness but negatively correlated with the days to flowering in the F2 population. Seed germination percentage was positively correlated with seed coat colour but not with traits related to seed size.
Detection of putative QTL
The results of the QTL analysis for each trait in each population are shown in Table 3. Only QTLs with a probability of ≤ 0·001 were considered. Although many other QTLs were detected when a probability level of ≤ 0·01 was used, these are not reported to ensure a high level of confidence in the QTLs named. One to nine QTLs were detected for each trait except for branch position for which no QTL was detected. For 22 out of the 30 quantitative traits two to six QTLs were detected.
Table 3.
QTLs detected in the backcrossed (BC1F1:2) and self-pollinated (F2 and F2:3) populations
| Trait | QTL | LG | LOD | P value | Loci position (cM) | BC1F1:2 (2003) |
BC1F1:2 (2004) |
LG | LOD | P value | Loci position (cM) | F2 (2003) |
F2:3 (2004) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEV | Additive effect | PEV | Additive effect | PEV | Additive effect | Dominant effect | D/[A] | PEV | Additive effect | Dominant effect | D/[A] | ||||||||||
| PDL* | Pdl2·1·1 + | 1 | NI† | 1 | 5·36 | 0·00009 | 88·5 | 0·177 | 1·16 | 0·17 | 0·15 | NI | |||||||||
| Pdl1·7·1 + | 7 | 18·63 | 0·00009 | 8·5 | 0·391 | 1·50 | 7 | 4·70 | 0·00018 | 1·6 | 0·170 | 0·96 | −0·48 | −0·50 | |||||||
| Pdl1·8·1 + | 8 | 9·18 | 0·00010 | 48·1 | 0·221 | 1·12 | 8 | 4·80 | 0·00027 | 59·7 | 0·194 | 1·24 | −0·15 | −0·12 | |||||||
| Pdl1·9·1 + | 9 | 3·19 | 0·00050 | 5·5 | 0·096 | 0·73 | 9 | ||||||||||||||
| Pdl1·10·1− | 10 | 3·45 | 0·00050 | 42·2 | 0·107 | −0·78 | 10 | ||||||||||||||
| PDW* | Pdw2·2·1. + | 2 | NI | 2 | 4·05 | 0·00045 | 41·9 | 0·194 | 0·68 | −0·03 | −0·05 | NI | |||||||||
| Pdw1·7·1 + | 7 | 14·96 | 0·00009 | 8·7 | 0·338 | 0·68 | 7 | ||||||||||||||
| Pdw1·9·1 + | 9 | 3·74 | 0·00040 | 10·6 | 0·107 | 0·37 | 9 | 7·18 | 0·00010 | 10·2 | 0·233 | 0·43 | −0·02 | −0·06 | |||||||
| Pdw2·9·2 + | 9 | 9 | 48·0 | 0·53 | −0·14 | −0·27 | |||||||||||||||
| Pdw1·11·1 + | 11 | 3·91 | 0·00010 | 27·2 | 0·105 | 0·37 | 11 | ||||||||||||||
| PDT* | Pdt1·7·1− | 7 | 81·47 | 0·00010 | 11·3 | 0·905 | −3·99 | NI | 7 | 27·42 | 0·00009 | 0·1 | 0·610 | −3·35 | 1·54 | 0·46 | NI | ||||
| SD100WT* | Sd100wt2·1·1 + | 1 | 1 | 6·35 | 0·00100 | 28·5 | 0·279 | 1·15 | −0·16 | −0·14 | NI | ||||||||||
| Sd100wt1·1·2 + | 1 | 4·61 | 0·00010 | 91·2 | 0·133 | 1·14 | NI | 1 | 88·2 | 0·74 | 0·25 | 0·33 | |||||||||
| Sd100wt1·2·1 + | 2 | 11·63 | 0·00010 | 33·5 | 0·268 | 1·65 | 2 | 8·16 | 0·00009 | 45·5 | 0·313 | 1·85 | −0·14 | −0·08 | |||||||
| Sd100wt2·5·1 + | 5 | 5 | 3·78 | 0·00091 | 2·3 | 0·136 | 1·11 | −0·26 | −0·23 | ||||||||||||
| Sd100wt2·9·1 + | 9 | 9 | 7·47 | 0·00010 | 6·8 | 0·252 | 0·99 | −0·14 | −0·14 | ||||||||||||
| Sd100wt2·9·2 + | 9 | 9 | 44·7 | 0·84 | −0·32 | −0·38 | |||||||||||||||
| SDL* | Sdl2·1·1 + | 1 | NI | 1 | 7·76 | 0·00010 | 20·6 | 0·311 | 0·39 | −0·06 | −0·16 | NI | |||||||||
| Sdl1·1·2 + | 1 | 4·96 | 0·00010 | 96·6 | 0·132 | 0·31 | 1 | 88·4 | 0·49 | 0·01 | 0·01 | ||||||||||
| Sdl1·2·1 + | 2 | 6·26 | 0·00010 | 32·8 | 0·162 | 0·35 | 2 | 5·86 | 0·00009 | 43·9 | 0·245 | 0·62 | −0·05 | −0·07 | |||||||
| Sdl2·3·1 + | 3 | 3 | 4·57 | 0·00018 | 38·2 | 0·202 | 0·49 | −0·19 | −0·39 | ||||||||||||
| Sdl1·7·1 + | 7 | 4·23 | 0·00018 | 21·7 | 0·117 | 0·30 | 7 | ||||||||||||||
| Sdl1·8·1 + | 8 | 3·98 | 0·00030 | 41·1 | 0·118 | 0·29 | 8 | 5·50 | 0·00009 | 43·4 | 0·195 | 0·52 | 0·09 | 0·18 | |||||||
| Sdl1·9·1 + | 9 | 3·40 | 0·00050 | 10·3 | 0·100 | 0·27 | 9 | ||||||||||||||
| Sdl1·11·1 + | 11 | 4·36 | 0·00010 | 20·6 | 0·114 | 0·29 | 11 | ||||||||||||||
| SDW* | Sdw1·1·1 + | 1 | 3·34 | 0·00100 | 93·1 | 0·093 | 0·18 | NI | 1 | NI | |||||||||||
| Sdw1·2·1 + | 2 | 8·11 | 0·00010 | 31·0 | 0·212 | 0·28 | 2 | 7·86 | 0·00009 | 44·7 | 0·297 | 0·48 | −0·01 | −0·02 | |||||||
| Sdw2·3·1 + | 3 | 3 | 4·12 | 0·00100 | 44·4 | 0·182 | 0·32 | −0·11 | −0·33 | ||||||||||||
| Sdw2·9·1 + | 9 | 9 | 5·69 | 0·00030 | 8·3 | 0·212 | 0·20 | 0·00 | 0·02 | ||||||||||||
| Sdw2·9·2 + | 9 | 9 | 46·9 | 0·21 | −0·13 | −0·60 | |||||||||||||||
| SDT* | Sdt1·1·1 + | 1 | 5·79 | 0·00010 | 90·3 | 0·159 | 0·25 | NI | 1 | ||||||||||||
| Sdt1·2·1 + | 2 | 10·22 | 0·00010 | 29·4 | 0·260 | 0·32 | 2 | 7·17 | 0·00100 | 47·9 | 0·281 | 0·49 | 0·04 | 0·07 | NI | ||||||
| Sdt2·5·1 + | 5 | 5 | 4·23 | 0·00055 | 3·0 | 0·154 | 0·35 | −0·04 | −0·11 | ||||||||||||
| Sdt2·9·1 + | 9 | 9 | 6·38 | 0·00040 | 8·8 | 0·231 | 0·24 | −0·02 | −0·07 | ||||||||||||
| Sdt2·9·2 + | 9 | 9 | 46·3 | 0·22 | −0·13 | −0·60 | |||||||||||||||
| SDG | Sdg1·1·1 + | 1 | 21·98 | 0·00009 | 37·9 | 0·304 | 15·16 | 0·264 | 11·00 | 1 | NI | ND‡ | |||||||||
| Sdg1·1·2 + | 1 | 99·0 | 8·56 | 5·60 | 1 | ||||||||||||||||
| Sdg1·2·1− | 2 | 7·95 | 0·00009 | 41·9 | 0·137 | −10·87 | 0·104 | −6·76 | 2 | ||||||||||||
| Sdg1·4·1− | 4 | 8·14 | 0·00009 | 7·8 | 0·094 | −4·93 | 0·149 | −2·60 | 4 | ||||||||||||
| Sdg1·4·2− | 48·0 | −1·39 | −4·68 | ||||||||||||||||||
| Sdg1·9·1− | 9 | 6·35 | 0·00009 | 3·5 | 0·065 | −5·02 | 0·103 | −6·71 | 9 | ||||||||||||
| SDP | Sdp1·1·1 + | 1 | 15·02 | 0·00010 | 36·1 | 0·374 | 13·83 | NI | 1 | 10·87 | 0·00100 | 54·7 | NI | 0·400 | 12·48 | −0·94 | −0·08 | ||||
| Sdp1·1·2 + | 1 | 94·9 | 14·74 | 1 | 90·1 | 12·34 | −0·94 | −0·08 | |||||||||||||
| Sdp1·4·1− | 4 | 5·69 | 0·00010 | 11·8 | 0·176 | −1·10 | 4 | ||||||||||||||
| Sdp1·4·2− | 46·8 | −9·90 | |||||||||||||||||||
| Sdp1·9·1− | 9 | 4·03 | 0·00010 | 2·6 | 0·106 | −8·12 | 9 | ||||||||||||||
| ECL | Ecl2·1·1 + | 1 | NI | 1 | 12·17 | 0·00010 | 5·1 | 0·225 | 1·03 | 0·03 | 0·03 | 0·201 | 0·91 | −0·10 | −0·11 | ||||||
| Ecl2·1·2 + | 1 | 1 | 85·7 | 0·49 | 0·14 | 0·28 | 0·75 | 0·39 | 0·52 | ||||||||||||
| Ecl1·2·1 + | 2 | 13·11 | 0·00010 | 27·6 | 0·319 | 1·23 | 2 | 31·74 | 0·00010 | 45·0 | 0·579 | 2·07 | 0·12 | 0·06 | 0·575 | 1·72 | −0·13 | −0·07 | |||
| ST1I | St1i2·1·1 + | 1 | 1 | 11·47 | 0·00100 | 8·8 | 0·198 | 0·86 | 0·08 | 0·09 | 0·160 | 0·84 | 0·37 | 0·44 | |||||||
| St1i1·1·2 + | 12·64 | 0·00009 | 93·0 | 0·138 | 0·31 | 0·210 | 0·37 | 86·5 | 0·46 | 0·02 | 0·05 | 0·94 | 0·04 | 0·05 | |||||||
| St1i1·2·1 + | 2 | 13·89 | 0·00009 | 35·0 | 0·045 | 0·17 | 0·294 | 0·44 | 2 | 22·07 | 0·00010 | 44·0 | 0·386 | 1·67 | −0·28 | −0·17 | 0·371 | 1·80 | −0·15 | −0·08 | |
| St1i1·11·1 + − | 11 | 5·53 | 0·00046 | 8·3 | 0·068 | 0·02 | 0·075 | −0·01 | 11 | ||||||||||||
| St1i1·11·2 + | 11 | 24·2 | 0·20 | 0·23 | 11 | ||||||||||||||||
| ST2I | St2i1·1·1 + | 1 | 11·91 | 0·00009 | 91·5 | 0·101 | 0·35 | 0·229 | 0·38 | 1 | |||||||||||
| St2i1·2·1 + | 2 | 13·23 | 0·00009 | 36·2 | 0·026 | 0·16 | 0·304 | 0·45 | 2 | 15·43 | 0·00010 | 46·3 | 0·341 | 1·61 | −0·04 | −0·02 | 0·333 | 1·43 | 0·08 | 0·06 | |
| ST3I | St3i1·1·1 + | 1 | 11·05 | 0·00009 | 92·6 | 0·114 | 0·48 | 0·199 | 0·41 | 1 | |||||||||||
| St3i1·2·1 + | 2 | 9·92 | 0·00009 | 37·8 | 0·016 | 0·14 | 0·251 | 0·48 | 2 | 14·30 | 0·00010 | 46·9 | 0·386 | 1·70 | −0·19 | −0·11 | 0·376 | 1·29 | 0·02 | 0·01 | |
| ST4I | St4i1·1·1 + | 1 | 15·16 | 0·00009 | 90·4 | 0·188 | 0·80 | 0·221 | 0·73 | 1 | 7·42 | 0·00010 | 90·0 | 0·063 | 0·17 | −0·36 | −2·12 | 0·020 | 1·16 | 0·07 | 0·06 |
| St4i1·2·1 + | 2 | 4·68 | 0·00009 | 29·3 | 0·016 | 0·15 | 0·131 | 0·57 | 2 | 10·51 | 0·00010 | 45·9 | 0·255 | 1·30 | −0·04 | −0·03 | 0·244 | 1·15 | 0·05 | 0·04 | |
| ST5I | St5i1·1·1 + | 1 | 13·78 | 0·00009 | 89·4 | 0·154 | 0·87 | 0·210 | 0·87 | 1 | 6·79 | 0·00010 | 86·9 | 0·100 | 0·22 | −0·48 | −2·22 | 0·032 | 0·96 | −0·12 | −0·12 |
| St5i2·2·1 + | 2 | 2 | 12·26 | 0·00010 | 32·3 | 0·371 | 0·53 | −0·07 | −0·14 | 0·313 | 0·66 | −0·13 | −0·20 | ||||||||
| St5i2·2·2. + | 2 | 2 | 52·3 | 0·84 | 0·17 | 0·20 | 0·37 | 0·13 | 0·35 | ||||||||||||
| ST6I | St6i1·1·1 + | 1 | 9·09 | 0·00009 | 88·1 | 0·124 | 1·06 | 0·130 | 0·81 | 1 | |||||||||||
| St6i2·2·1 + | 2 | 2 | 11·39 | 0·00010 | 20·9 | 0·296 | 0·24 | −0·02 | −0·09 | 0·238 | 0·39 | −0·16 | −0·40 | ||||||||
| St6i2·2·2 + | 2 | 2 | 45·7 | 0·96 | 0·32 | 0·33 | 0·68 | 0·09 | 0·14 | ||||||||||||
| St6i1·5·1 + | 5 | 5·35 | 0·00009 | 17·1 | 0·103 | 0·97 | 0·068 | 0·59 | |||||||||||||
| St6i2·7·1− | 7 | 7 | 5·25 | 0·00030 | 18·8 | 0·090 | −0·74 | 0·04 | 0·05 | 0·079 | −0·84 | −0·10 | −0·12 | ||||||||
| St6i1·9·1− | 9 | 6·10 | 0·00009 | 6·3 | 0·094 | −0·93 | 0·085 | −0·66 | 9 | ||||||||||||
| ST7I | St7i1·1·1 + | 1 | 6·17 | 0·00009 | 83·1 | 0·117 | 1·19 | 0·077 | 0·80 | 1 | |||||||||||
| St7i2·2·1 + | 2 | 2 | 9·35 | 0·00010 | 26·5 | 0·258 | 0·42 | 0·01 | 0·01 | 0·195 | 0·45 | −0·17 | −0·38 | ||||||||
| St7i2·2·2 + | 2 | 2 | 49·5 | 0·45 | 0·35 | 0·78 | 0·45 | 0·11 | 0·25 | ||||||||||||
| St7i1·3·1− | 3 | 4·05 | 0·00091 | 11·0 | 0·043 | −0·68 | 0·093 | −0·93 | 3 | ||||||||||||
| St7i1·4·1− | 4 | 4·16 | 0·00073 | 53·4 | 0·065 | −0·88 | 0·058 | −0·72 | 4 | 5·74 | 0·00050 | 64·5 | 0·187 | −1·15 | −0·06 | −0·05 | 0·179 | −0·68 | −0·08 | −0·12 | |
| St7i1·5·1 + | 5 | 4·03 | 0·00055 | 14·6 | 0·084 | 1·00 | 0·056 | 0·71 | 5 | ||||||||||||
| St7i1·7·1− | 7 | 6·88 | 0·00010 | 13·8 | 0·119 | −0·73 | 0·067 | −0·05 | 7 | ||||||||||||
| St7i1·7·2− | 7 | 39·4 | −0·69 | −0·70 | 7 | 8·33 | 0·00010 | 18·7 | 0·118 | −0·88 | 0·01 | 0·01 | 0·109 | −1·11 | −0·15 | −0·14 | |||||
| St7i1·9·1− | 9 | 9·37 | 0·00009 | 4·5 | 0·163 | −1·43 | 0·100 | −0·97 | 9 | 7·85 | 0·00010 | 14·7 | 0·195 | −0·97 | 0·45 | 0·47 | 0·138 | −0·65 | −0·04 | −0·06 | |
| ST8I | St8i2·7·1− | 7 | NI | 7 | 10·21 | 0·00010 | 18·8 | 0·139 | −0·95 | −0·07 | −0·08 | 0·130 | −1·23 | −0·17 | −0·14 | ||||||
| St8i1·9·1− | 9 | 3·15 | 0·00060 | 9·1 | 0·099 | −1·21 | 9 | 5·42 | 0·00050 | 14·2 | 0·099 | −0·57 | 0·37 | 0·65 | 0·055 | −0·76 | 0·04 | 0·05 | |||
| ST9I | St9i1·7·1− | 7 | 4·71 | 0·00009 | 38·9 | NI | 0·138 | −1·75 | 7 | 14·92 | 0·00010 | 20·5 | 0·248 | −1·20 | −0·36 | −0·30 | 0·214 | −1·30 | −0·10 | −0·08 | |
| ST10I | St10i1·4·1− | 4 | 4·02 | 0·00010 | 52·7 | NI | 0·111 | −1·84 | 4 | ||||||||||||
| St10i1·7·1− | 7 | 4·29 | 0·00009 | 39·5 | 0·130 | −2·01 | 7 | 13·38 | 0·00010 | 22·4 | 0·171 | −0·96 | −0·32 | −0·34 | 0·140 | −1·37 | −0·07 | −0·05 | |||
| St10i1·9·1− | 9 | 4·22 | 0·00020 | 8·5 | 0·135 | −2·04 | 9 | ||||||||||||||
| STL | Stl1·1·1 + | 1 | 10·82 | 0·00009 | 88·0 | 0·178 | 5·19 | 0·130 | 8·19 | 1 | |||||||||||
| Stl2·2·1 + | 2 | 2 | 10·97 | 0·00010 | 25·9 | 0·246 | 0·41 | −0·03 | −0·07 | 0·201 | 0·68 | −0·20 | −0·29 | ||||||||
| Stl2·2·2 + | 50·4 | 0·77 | 0·27 | 0·35 | 0·50 | 0·25 | 0·49 | ||||||||||||||
| Stl2·4·1− | 4 | 4 | 5·90 | 0·00050 | 63·8 | 0·176 | −1·11 | −0·09 | −0·08 | 0·168 | −0·79 | 0·00 | 0·00 | ||||||||
| Stl1·5·1 + | 5 | 4·00 | 0·00073 | 14·2 | 0·084 | 3·44 | 0·049 | 4·82 | 5 | ||||||||||||
| Stl1·7·1− | 7 | 7·29 | 0·00010 | 15·2 | 0·114 | −3·01 | 0·082 | −0·41 | 7 | ||||||||||||
| Stl1·7·2− | 7 | 39·7 | −1·67 | −5·43 | 7 | 10·59 | 0·00010 | 19·2 | 0·196 | −1·14 | −0·16 | −0·14 | 0·184 | −1·07 | −0·11 | −0·10 | |||||
| Stl1·9·1− | 9 | 7·14 | 0·00009 | 6·0 | 0·109 | −4·03 | 0·105 | −7·29 | |||||||||||||
| STTW | Sttw1·4·1− | 4 | 7·51 | 0·00009 | 52·3 | 0·080 | −6·56 | 0·126 | −8·07 | 4 | NI | ||||||||||
| Sttw1·7·1− | 7 | 5·21 | 0·00009 | 33·6 | 0·108 | −8·79 | 0·059 | −3·42 | 7 | 6·09 | 0·00009 | 21·7 | 0·231 | −42·67 | 6·36 | 0·15 | |||||
| Sttw1·9·1− | 9 | 15·77 | 0·00009 | 5·1 | 0·274 | −22·40 | 0·147 | −9·42 | 9 | ||||||||||||
| STT | Stt1·4·1 + | 4 | 7·71 | 0·00009 | 52·2 | 0·048 | 0·27 | 0·159 | 0·51 | 4 | |||||||||||
| Stt1·6·1 + | 6 | 4·12 | 0·00055 | 0·9 | 0·020 | 0·16 | 0·100 | 0·40 | 6 | ||||||||||||
| Stt1·9·1 + | 9 | 5·26 | 0·00018 | 6·5 | 0·072 | 0·34 | 0·085 | 0·37 | 9 | 7·16 | 0·00010 | 22·1 | 0·124 | 0·42 | −0·44 | −1·05 | 0·044 | 0·91 | −0·19 | −0·20 | |
| LFPL | Lfpl1·1·1 + | 1 | 5·26 | 0·00018 | 85·0 | 0·124 | 3·09 | 0·052 | 1·62 | 1 | ND | ND | |||||||||
| Lfpl1·7·1 + − | 7 | 6·67 | 0·00030 | 12·6 | 0·054 | −1·15 | 0·123 | 1·82 | 7 | ||||||||||||
| Lfpl1·7·2 + | 7 | 36·1 | 2·73 | 1·14 | 7 | ||||||||||||||||
| LFPW | Lfpw2·2·1 + | 2 | 2 | 5·77 | 0·00040 | 45·1 | 0·043 | 0·19 | 0·12 | 0·60 | 0·022 | 1·13 | −0·08 | −0·07 | |||||||
| Lfpw1·7·1 + − | 7 | 5·68 | 0·00060 | 11·8 | 0·047 | −0·94 | 0·104 | 1·26 | 7 | ||||||||||||
| Lfpw1·7·2 + | 7 | 37·6 | 2·29 | 1·11 | 7 | ||||||||||||||||
| Lfpw1·8·1 + | 8 | 4·00 | 0·00100 | 37·3 | 0·086 | 1·95 | 0·043 | 1·21 | 8 | ||||||||||||
| Lfpw1·9·1 + | 9 | 4·24 | 0·00073 | 14·4 | 0·061 | 1·77 | 0·064 | 1·58 | 9 | 5·96 | 0·00020 | 18·3 | 0·054 | 0·30 | −0·15 | −0·52 | 0·030 | 0·93 | −0·10 | −0·11 | |
| Lfpw2·11·1 + | 11 | 11 | 5·61 | 0·00030 | 10·9 | 0·086 | 0·43 | 0·14 | 0·33 | 0·059 | 1·02 | 0·10 | 0·10 | ||||||||
| Lfpw1·11·1 + | 11 | 5·18 | 0·00009 | 20·6 | 0·068 | 1·86 | 0·087 | 1·88 | 11 | ||||||||||||
| LFML | Lfml1·5·1 + | 5 | 3·62 | 0·00020 | 22·2 | NI | 0·112 | 4·92 | 5 | ||||||||||||
| Lfml1·6·1− | 6 | 2·92 | 0·00100 | 29·2 | 0·088 | −4·31 | 6 | ||||||||||||||
| Lfml1·7·1 + | 7 | 4·93 | 0·00010 | 17·4 | 0·147 | 5·65 | 7 | ||||||||||||||
| Lfml2·8·1 + | 8 | 8 | 5·52 | 0·00070 | 46·6 | 0·170 | 0·82 | −0·02 | −0·03 | 0·152 | 0·38 | −0·01 | −0·02 | ||||||||
| LFMW | Lfmw1·5·1 + | 5 | 4·51 | 0·00010 | 14·8 | NI | 0·123 | 4·34 | 5 | ||||||||||||
| Lfmw1·6·1− | 6 | 4·31 | 0·00010 | 29·1 | 0·110 | −4·08 | 6 | ||||||||||||||
| Lfmw1·7·1 + | 7 | 3·44 | 0·00050 | 26·3 | 0·115 | 4·18 | 7 | ||||||||||||||
| Lfmw2·8·1 + | 8 | 8 | 5·81 | 0·00040 | 39·0 | 0·172 | 0·83 | 0·18 | 0·22 | 0·140 | 0·60 | 0·02 | 0·03 | ||||||||
| Lfmw1·9·1 + | 9 | 6·07 | 0·00010 | 7·1 | 0·166 | 5·06 | 9 | ||||||||||||||
| BRN | Brn1·10·1− | 10 | 5·90 | 0·00009 | 36·6 | 0·154 | −0·55 | 0·029 | −0·21 | 10 | ND | ND | |||||||||
| BRP | ND | ND | ND | ND | |||||||||||||||||
| FLD | Fld2·4·1− | NI | NI | 4 | 4·99 | 0·00009 | 57·6 | 0·239 | −9·94 | 1·03 | 0·10 | NI | |||||||||
* The BC1F1 generation was evaluated for these traits.
† NI, Not investigated.
‡ ND, Not detected.
Seed germination (SDG and SDP)
In azuki bean domestication has resulted in loss of dormancy (or high percentage seed germination). For seed germination in the field, six QTLs were detected on linkage groups 1, 2, 4 and 9. Alleles from the cultivated parent at two of these QTLs on linkage group 1 involved a strong positive effect on germination percentage. Unexpectedly, the four remaining QTLs from the cultivated parent had a negative effect on germination percentage in the field. Based on the laboratory test for permeability of the seed coat to water, five QTLs were detected on linkage groups 1, 4 and 9. At two QTLs on linkage group 1, alleles of the cultivated parent had the effect of increasing the percentage of seeds with a permeable seed coat. Based on their location, these two QTLs are probably the same as those found on linkage group 1 in the field test and are the only QTLs detected from the cultivated parent that promote water absorption by seeds and germination.
Pod dehiscence (PDT)
Azuki bean domestication has involved a loss of pod dehiscence reflected in reduced ability for the pod wall to twist. The frequency distribution for pod dehiscence (reflected by the number of twists on the pod after opening) suggests that a single recessive gene controls loss of pod dehiscence. One QTL with a high heritability (above 96%) was located on linkage group 7.
Increase in organ size
Seed size, pod length and width and stem thickness were considered.
Seed size (SD100WT, SDL, SDW and SDT): five to eight QTLs for traits related to seed size were located on linkage groups 1, 2, 3, 5, 7, 8, 9 and 11 (Table 3). At all QTLs, alleles from the cultivated parent increase the size of each trait. The QTLs with the largest phenotypic contribution for all four traits were located on linkage group 2 (from 16 % for seed length to 31 % for seed weight).
Pod length and width (PDL, PDW): five QTLs were detected for pod length, on linkage groups 1, 7, 8, 9 and 10. The QTLs located on linkage group 7 had the largest effect. This region on linkage group 7 coincided with the QTL region for pod dehiscence. Of the five QTLs detected for pod width, the QTL with largest effect was also located close to the QTL region for pod dehiscence.
Stem thickness (STT): three QTLs for stem thickness were detected on linkage groups 4, 6 and 9.
Growth habit
Despite very different weather conditions in 2003 and 2004 (see Supplementary Information 1, available online) many common QTLs were detected for stem length, internode length and twining habit. The QTLs with large effects were located on linkage groups 1, 2, 7 and 9. QTLs affecting length of stem and lower internodes were located on linkage groups 1 and 2. Alleles from the cultivated parent result in longer lower internodes and shorter upper internodes.
For twining habit (STTW) three QTLs were detected, on linkage groups 4, 7 and 9. The two regions on linkage groups 7 and 9 with QTLs for twining habit are the same as those having QTLs of the cultivated parent for shorter stem and internodes.
Leaf shape
Nine QTL for maximum leaf length and width were found. However, different QTLs were detected in the BC1F1:2 population from those found in the F2 and F2:3 populations.
Flowering time
One QTL was detected on linkage group 4, with the allele of the cultivated parent resulting in earlier flowering.
Epicotyl traits (ECL and ECC)
Three QTLs having alleles of the cultivated parent for increased epicotyl length were detected on linkage groups 1 and 2. Location of two of these QTLs for epicotyl length is in a similar position to those for stem length and lower internode length.
The gene that controlled epicotyl colour was mapped on linkage group 4.
Seed colour
Seed colour traits were recorded as (a) seed colour either red or tan (SDC) and (b) presence or absence of black mottle (SDCBM) (Table 2). The genes that controlled the seed coat colour and black mottle on the seed were mapped to linkage groups 1 and 4, respectively. The gene for seed colour is closely linked to a QTL for seed germination on linkage group 1 and that for black mottle seed coat was closely linked to epicotyl colour on linkage group 4.
The independence and distribution of QTL related to domestication traits across the azuki bean genome and linkage groups
Assuming independent gene action, 65–85 QTLs for domestication-related traits were identified and, assuming pleiotropy or close linkage at all loci, 16 or 17 positions with possible pleiotropic effects were identified (Table 4). Overall χ2 values were 39·4–53·1 for independent gene action and 5·2 and 4·7 for pleiotropy or close linkage (Table 4). Thus the assumption of independent gene action was rejected and the assumption of pleiotropy or close linkage could not be rejected.
Table 4.
Distribution of QTLs for domestication-related traits (P ≤ 0·001) under hypotheses of pleiotropy or independent gene action
| Pleiotropy or close linkage |
Independent gene action |
||||||
|---|---|---|---|---|---|---|---|
| No. of QTL |
No. of QTL |
||||||
| Length (cM) | Observed | Expected | χ2 | Observed | Expected | χ2 | |
| BC1F1:2 | |||||||
| LG1 | 104 | 2 | 3·0 | 0·31 | 17 | 14·8 | 0·33 |
| LG2 | 60 | 1 | 1·7 | 0·29 | 10 | 8·5 | 0·25 |
| LG3 | 50 | 1 | 1·4 | 0·12 | 1 | 7·1 | 5·25* |
| LG4 | 58 | 2 | 1·6 | 0·07 | 8 | 8·2 | 0·01 |
| LG5 | 49 | 1 | 1·4 | 0·11 | 5 | 7·0 | 0·55 |
| LG6 | 40 | 2 | 1·1 | 0·65 | 3 | 5·7 | 1·27 |
| LG7 | 50 | 3 | 1·4 | 1·75 | 17 | 7·1 | 13·77*** |
| LG8 | 55 | 1 | 1·6 | 0·20 | 3 | 7·8 | 2·97 |
| LG9 | 43 | 1 | 1·2 | 0·04 | 14 | 6·1 | 10·18** |
| LG10 | 58 | 1 | 1·6 | 0·26 | 2 | 8·2 | 4·73* |
| LG11 | 31 | 2 | 0·9 | 1·42 | 5 | 4·4 | 0·08 |
| Total | 598 | 17 | 17·0 | 5·24 | 85 | 85 | 39·39*** |
| F2 (F2:3) | |||||||
| LG1 | 117 | 4 | 2·9 | 0·40 | 13 | 11·9 | 0·11 |
| LG2 | 71 | 2 | 1·8 | 0·03 | 19 | 7·2 | 19·34** |
| LG3 | 58 | 1 | 1·4 | 0·14 | 2 | 5·9 | 2·56 |
| LG4 | 66 | 1 | 1·6 | 0·25 | 3 | 6·7 | 2·04 |
| LG5 | 43 | 1 | 1·1 | 0·01 | 2 | 4·4 | 1·28 |
| LG6 | 37 | 0 | 0·9 | 0·92 | 0 | 3·8 | 3·75 |
| LG7 | 39 | 2 | 1·0 | 1·08 | 9 | 4·0 | 6·44* |
| LG8 | 72 | 2 | 1·8 | 0·02 | 4 | 7·3 | 1·49 |
| LG9 | 51 | 2 | 1·3 | 0·42 | 12 | 5·2 | 9·02** |
| LG10 | 55 | 0 | 1·4 | 1·37 | 0 | 5·6 | 5·58** |
| LG11 | 32 | 1 | 0·8 | 0·05 | 1 | 3·2 | 1·55 |
| Total | 641 | 16 | 16·0 | 4·69 | 65 | 65 | 53·15*** |
* P < 0·05; ** P < 0·01, *** P < 0·001.
The observed number of QTLs in each 10-cM interval for each linkage group was compared with the expected number of QTLs. The null hypothesis that QTLs were randomly distributed was rejected for many linkage groups (Table 5). The uneven distribution of QTLs on linkage groups 1 and 2 was pronounced for both the BC1F1:2 and F2 (F2:3).
Table 5.
Distribution of QTLs on each linkage group based on (A) the null hypothesis for Poisson goodness of fit for the BCF1 (F1:2) and (B) on each linkage group based on the null hypothesis for Poisson goodness of fit F2 (F2:3)
| Observed no. of QTL per 10-cM interval |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LG | 0–10 | 11–20 | 21–30 | 31–40 | 41–50 | 51–60 | 61–70 | 71–80 | 81–90 | 91–100 | 101–110 | 111–120 | Expected† | χ2 |
| (A) | ||||||||||||||
| 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 5 | 10 | 0 | – | 1·55 | 19928·7*** |
| 2 | 0 | 0 | 3 | 6 | 1 | 0 | 0 | – | – | – | – | – | 1·43 | 54·7*** |
| 3 | 0 | 1 | 0 | 0 | 0 | 0 | – | – | – | – | – | – | 0·17 | 0·0 |
| 4 | 1 | 1 | 0 | 0 | 2 | 4 | – | – | – | – | – | – | 0·67 | 38·1*** |
| 5 | 0 | 4 | 1 | 0 | 0 | – | – | – | – | – | – | – | 1·00 | 13·5** |
| 6 | 1 | 0 | 2 | 0 | 0 | – | – | – | – | – | – | – | 0·60 | 0·8 |
| 7 | 2 | 6 | 2 | 7 | 0 | 0 | – | – | – | – | – | – | 2·83 | 21·8*** |
| 8 | 0 | 0 | 0 | 1 | 2 | 0 | – | – | – | – | – | – | 0·50 | 1·1 |
| 9 | 11 | 3 | 0 | 0 | 0 | – | – | – | – | – | – | – | 2·80 | 1608·5*** |
| 10 | 1 | 1 | 0 | 0 | 0 | 0 | – | – | – | – | – | – | 0·33 | 0·2 |
| 11 | 1 | 0 | 4 | 0 | – | – | – | – | – | – | – | – | 1·25 | 8·7* |
| (B) | ||||||||||||||
| 1 | 2 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 6 | 2 | 0 | 0 | 1·08 | 113·7*** |
| 2 | 0 | 0 | 3 | 1 | 13 | 2 | 0 | 0 | – | – | – | – | 2·38 | 109414·0*** |
| 3 | 0 | 0 | 0 | 1 | 1 | 0 | – | – | – | – | – | – | 0·33 | 0·2 |
| 4 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | – | – | – | – | – | 0·43 | 1·3 |
| 5 | 2 | 0 | 0 | 0 | 0 | – | – | – | – | – | – | – | 0·40 | 3·5 |
| 6 | 0 | 0 | 0 | 0 | – | – | – | – | – | – | – | – | 0·00 | – |
| 7 | 2 | 4 | 3 | 0 | – | – | – | – | – | – | – | – | 2·25 | 2·6 |
| 8 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 0 | – | – | – | – | 0·50 | 0·3 |
| 9 | 3 | 4 | 1 | 0 | 4 | 0 | – | – | – | – | – | – | 2·00 | 7·5 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | – | – | – | – | – | – | 0·00 | – |
| 11 | 0 | 1 | 0 | 0 | – | – | – | – | – | – | – | – | 0·25 | 0·1 |
† Expected number of QTL per 10-cM interval.
* P < 0·05; ** P < 0·01, *** P < 0·001.
DISCUSSION
Reproducibility of QTLs
Despite the use of a stringent threshold (P ≤ 0·001) for detecting QTLs in the two populations, about one-third are common (Table 3). For example, one-third of the QTLs (eight out of 24) for traits related to seed size were detected in both the BC1F2 (2003) and F3 populations, although the seeds of these populations were harvested in different years. For pod length, two QTLs detected in the F2 population were also observed in the BC1F2 (2003) although the heritability in the BC1F2 (2003) was lower than in the F2 population, suggesting that environmental factors may influence pod length. Growth habit traits are sensitive to the growing environment (Koinange et al., 1996). Even though temperature, rainfall and solar radiation received varied greatly in 2003 and 2004, analysis of stem internode length, stem length and stem twining revealed 15 QTLs common to the BC1F2 and F2/F3 out of a total of 45 QTLs detected.
The genetics of domestication-related traits
In azuki bean a few domestication-related traits are controlled by a single major gene and most of them are controlled by a small number of QTLs. Results from this study reveal that azuki bean differs from related legumes in the genetic control of several simply inherited traits. For example, a few genes control pod dehiscence in common bean (P. vulgaris L.) (Koinange et al., 1996) but in azuki bean a single gene controls this trait. While a single gene controls twining plant habit in common bean (Koinange et al., 1996), in this study three QTLs for twining habit (evaluated in a different way) were detected.
For the majority of traits measured, between two and nine QTLs on two or more linkage groups were detected. QTLs for many related traits such as pod size, seed size and leaf shape and stem-related traits co-occur in the same location on the same linkage groups. This can be explained by developmental allometry (Smartt, 1976).
QTLs for internode length suggest gene association across the genome resulting in an effect that is best described as a cascade as they act at sequential stages in the plant life cycle. Most QTLs for lengths of the lower internodes (internodes 1–5) are located on linkage groups 1 and 2. For lengths of upper internodes (internodes 6–10) most QTLs are on linkage groups 4, 7 and 9.
Seed germination in azuki bean
Unexpectedly, at four QTLs, alleles of the cultivated parent were associated with lower seed germination. Koinange et al. (1996) reported that in common bean, at one QTL the allele of the cultivated parent was associated with a higher level of dormancy than the allele of the wild parent. There are several possible explanations for why cultivated azuki bean may have QTLs for lower germination. These may include prevention of premature germination in the field or the tendency of local farmers in Japan to use seeds from good harvests over several planting seasons in their home garden plots of azuki bean. Alternatively, some alleles that promote seed germination may be linked to traits not suitable for cultivated azuki, or QTLs for delayed germination in the wild parent used here may differ from QTLs in other wild germplasm of the azuki bean complex. Currently, hard-seededness in cultivated azuki bean is an occasional problem for azuki bean paste makers (Lumpkin and McClary, 1994). Azuki breeding which targets cultivated alleles that cause a lower germination percentage may help overcome this problem.
Distribution of QTL for domestication-related traits
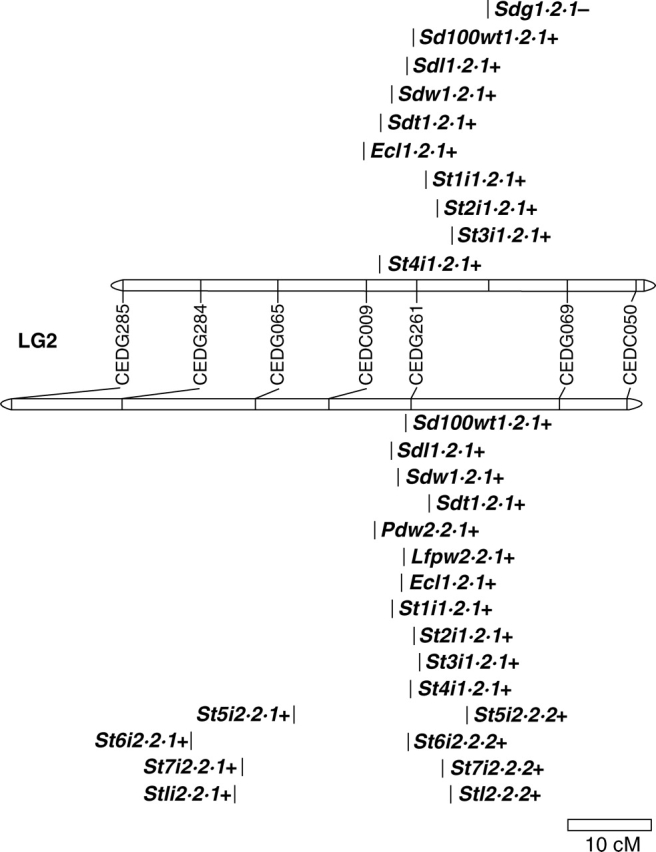
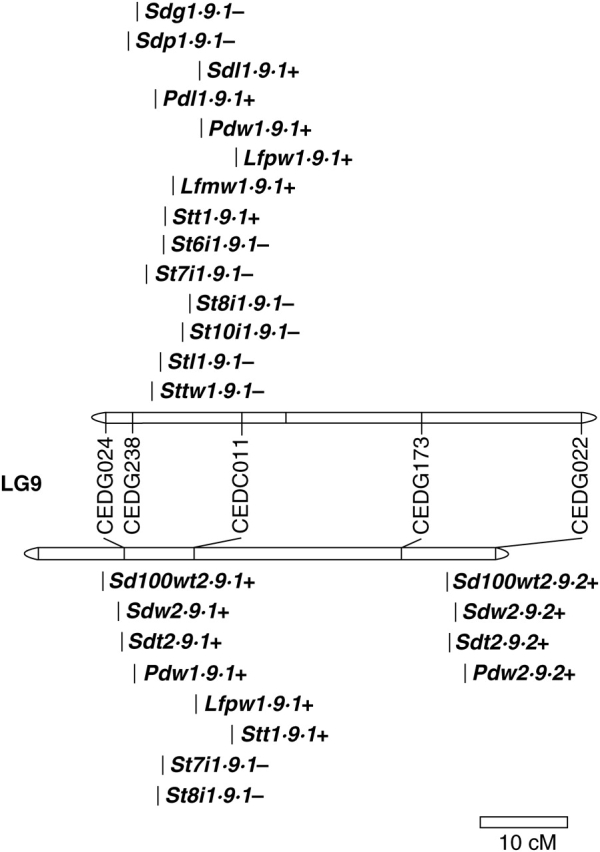
The genes controlling domestication-related traits are not randomly distributed across crop genomes (Poncet et al., 2000; Doebley and Stec, 1991, 1993). For example, in common bean (Phaseolus vulgaris L.) three out of the 11 linkage groups, D1, D2 and D7, play an important role in growth habit, phenology, seed dispersal, dormancy, pod length and seed size (Koinange et al., 1996). The distribution of domestication syndrome QTLs departed significantly from random across the azuki bean genome (Table 4). Particularly important linkage groups with major QTL for domestication-related traits were linkage groups 1, 2, 4, 7 and 9 and are discussed below. For the linkage groups 1 and 2, domestication syndrome QTLs were non-randomly distributed across the linkage group in both populations studied (Table 5). The locations of QTLs on linkage groups 2 and 9 are shown in Figs 1 and 2 and figures for other linkage groups are also provided online (see Supplementary information 3).
Fig. 1.
QTLs detected on linkage group 2 for BC1F1 (upper) and F2 (lower) from the cross between V. nepalensis and azuki bean. The effect of the cultivated parent is indicated after each QTL name. For explanation of trait abbreviations, see Table 1.
Fig. 2.
QTLs detected on linkage group 9 for BC1F1 (upper) and F2 (lower) from the cross between V. nepalensis and azuki bean. The effect of the cultivated parent is indicated after each QTL name. For explanation of trait abbreviations, see Table 1.
Linkage group 1
On linkage group 1 the QTLs involved in loss of dormancy and increased size of organs such as seed, pod, stem and leaf were detected in a limited region of 16 cM. This suggests either close linkage or pleiotropy of these domestication-related traits (Table 4). Close linkage or pleiotropy for domestication traits is reported in several crops such as maize (Doebley et al., 1997), rice (Cai and Morishima, 2002), eggplant (Doganlar et al., 2002) and common bean (Koinange et al., 1996). The best studied case is the single locus tb1 (teosinte branched 1) of maize that controls multiple traits (Doebley et al., 1997).
The genes or QTLs for seed colour and loss of seed dormancy in azuki bean are closely linked and there is a significant correlation between these two traits. This helps explain why in wild and weedy populations, individuals with a red seed coat are not found, since lack of seed dormancy is a detrimental trait in the wild. Unlike seeds with a red seed coat, those with a tan seed coat can survive in the natural environment from year to year (Wang et al., 2004).
Linkage group 2 (Fig. 1)
QTLs found on this linkage group are all related to seed size and early stages of plant growth and affect many of the same traits as those of linkage group 1. QTLs for increased seed size (seed weight, seed length, seed width and seed thickness) and epicotyl length, stem and lower internode lengths were all detected in a 18-cM region on linkage group 2 (Fig. 1). The highest genetic contribution for all traits related to seed size occurred in this region.
Linkage group 4
On linkage group 4, the genes for epicotyl colour and black mottle on the seed coat are linked closely. The association between these two traits has been reported previously (Kaga et al., 1996). The only QTL detected for flowering date was located on this linkage group and is associated with the upper stem characteristics of upper internode length and stem twining.
Linkage group 7
Linkage group 7 appears to be the most important for changes in the pod during domestication as it is the only linkage group for which all three pod-related traits (pod length and width and pod twist number) were detected. In addition, several QTLs for growth habit such as stem length, upper internode length and stem twining were consistently detected in a narrow region of 6 cM.
Linkage group 9
QTLs for growth habit (stem length, internode length, stem thickness and determinancy) and leaf and seed size were detected mainly in a 10–15-cM region on linkage group 9 (Fig. 2). Cultivated parent alleles in this region shorten the stem and upper internode lengths and cause a non-twining plant type. This region has many QTLs in common with, and having the same influence as, cultivated azuki bean alleles on linkage group 7 discussed above. The QTLs in this region on linkage group 9 may also reflect pleiotropy or close linkage.
Comparison of QTLs and their location among warm-season legumes
Cross between azuki bean and V. angularis var. nipponensis
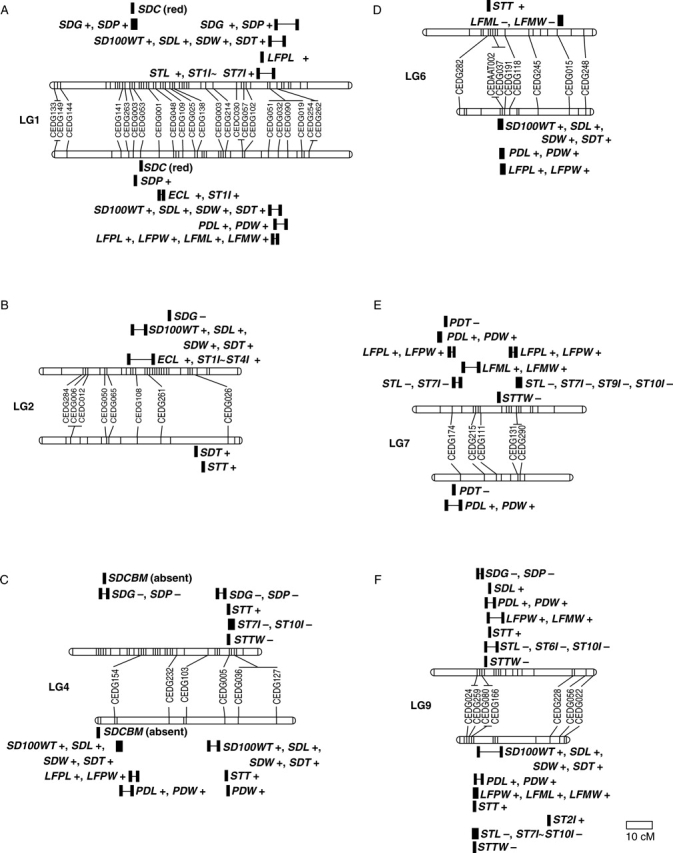
In Japan, cultivated and wild azuki bean exist as a crop complex as a result of gene flow (Wang et al., 2004). Many intermediate plant types can be found in many parts of Japan (Yamaguchi, 1992). To better understand the genetics of the azuki bean complex a parallel study (T. Isemura, unpubl. res.) was conducted involving a cross between a Japanese cultivar of azuki bean, ‘Kyoto Dainagon’, and an accession of its presumed ancestor from Japan, Vigna angularis var. nipponensis. Domestication-related traits were recorded in the F2 population in 2004. A summary of the location of QTLs for traits found in comparison with those reported here is shown in Fig. 3. This figure only shows linkage groups with the most QTLs from both studies. Many common QTLs are found on linkage groups 1 and 9 but this is not the case with the other linkage groups. Whereas linkage groups 2 and 7 have many QTLs for seed size and internode length in the cross studied here, in the cross involving V. angularis var. nipponensis they did not. For these two linkage groups only pod-related QTLs on linkage group 7 were common in both crosses. Similarly, linkage groups 4 and 6 had many QTLs for seed size and pod-related traits in the cross involving V. angularis var. nipponensis but QTLs for these traits were not found in the cross studied here. Only genes for black mottled seed coat and epicotyl colour on linkage group 4 were common on these linkage groups in the two crosses. If these differences in QTLs between these two crosses within the azuki bean complex reflect genetic differences, rather than genomic rearrangements, between the wild or cultivated parents azuki bean breeders may be able to use the different QTLs in their breeding programmes.
Fig. 3.
Comparative QTL maps of domestication-related traits found in the BC1F1:2 of the cross V. nepalensis and azuki bean (JP81481) and the F2 of the cross between V. angularis (‘Kyoto Dainagon’) and V. angularis var. nipponensis. For explanation of trait abbreviations, see Table 1.
Cross between azuki bean and V. nakashimae (Ohwi) Ohwi & Ohashi
The genes controlling red colour in the epicotyl and black mottle on seed coat were detected on linkage group 4 in this study, and these two loci are closely linked. Kaga et al. (1996, 2000) mapped the gene for red colour in epicotyl on linkage groups 4 and 1 using interspecific populations derived from the crosses between azuki bean and V. nakashimae and azuki bean and rice bean (V. umbellata), respectively. These two linkage groups were identified as the same on the basis of the order of three common RFLP markers (Han et al., 2005). The position of the locus for red epicotyl also coincides in the three maps.
Common bean (Phaseolus vulgaris L.)
One of the closest genera to Vigna is Phaseolus and these two genera have been taxonomically distinguished only since 1970 (Verdcourt, 1970). In order to compare the QTLs detected in this study with those in common bean (P. vulgaris) the relationships of the linkage groups were investigated on the basis of the distribution of common RFLP markers reported in the common bean maps (Vallejos et al., 1992; Koinange et al., 1996; Freyre et al., 1998; Yu et al., 2000; Blair et al., 2003) and azuki bean map (Han et al., 2005; this study). Although the number of common RFLP markers is low, the relationship between linkage groups in the two genera could be determined. Linkage groups 7 and 8 of azuki bean in this study corresponded to the linkage group B5 and B7 of common bean (Fig. 4). QTL for pod and growth habit traits were detected on linkage group 7 in this study, but no QTL for these traits were detected on linkage group B5 of common bean. However, Koinange et al. (1996) reported that linkage group B7 played an important role in pod length and seed size. QTLs for seed length and pod length were detected in almost the same region on linkage group 8 of azuki bean. It was not possible to clarify the relationship between linkage group 1 in this study and B1 of common bean where many QTL were detected in both crops. It will be necessary to determine if Phaseolus SSR markers can be placed on the genetic map of azuki bean and vice versa to clarify corresponding linkage group relationships between these two crops.
Fig. 4.
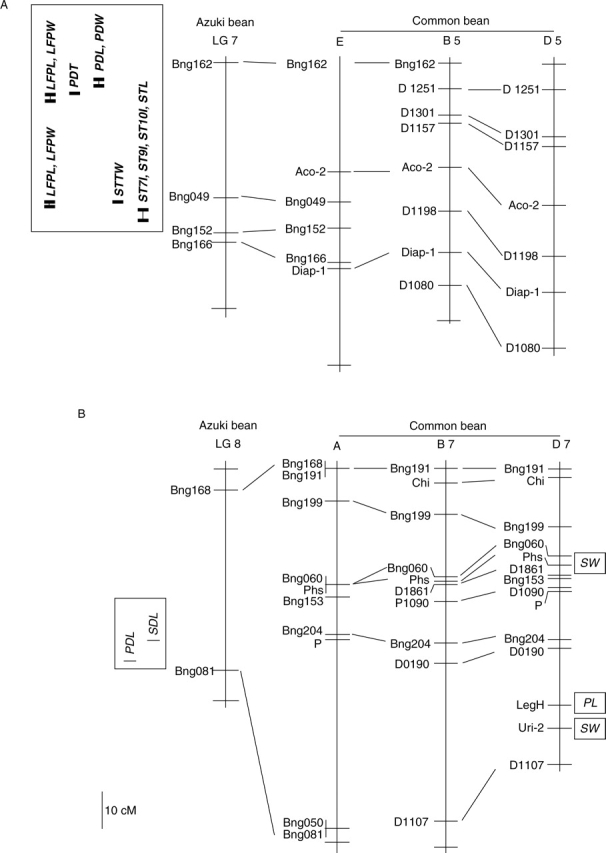
Comparative QTL maps for domestication-related traits between azuki bean and common bean (Phaseolus vulgaris). Linkage groups 7 and 8 of azuki bean in this study corresponded to linkage groups B5 and B7 on the current map of common bean, respectively. It was not possible to clarify the relationships for remaining linkage groups between azuki bean and common bean. Linkage groups of common bean, left to right, are after the maps of Vallejos et al. (1992), Freyre et al. (1998) and Koinange et al. (1996). The QTL for SW (seed weight) and PL (pod length) on linkage group D7 in common bean are after Koinange et al. (1996).
Cowpea [Vigna unguiculata (L.) Walp.] and mungbean [Vigna radiata (L.) Wilczek]
QTL analysis for seed weight was carried out in the Vigna domesticates cowpea and mungbean since this is an important trait related to yield (Fatokun et al., 1992). Using populations derived from crosses between cowpea and wild cowpea and mungbean and wild mungbean, two and four QTLs for seed weight, respectively, were reported (Fatokun et al., 1992). Linkage groups ii and vi in the cowpea map correspond to linkage groups 1 and 4, respectively, in the azuki bean map (Fig. 5). Linkage groups i, ii, iii and vi in the mungbean map correspond to linkage groups 9, 1, 8 and 10, respectively, in the azuki bean map (Fig. 5). In this study, a QTL for seed weight was detected on linkage group 1 at a location corresponding to that of a QTL for this trait on linkage group ii in cowpea and mungbean. This seed weight QTL appears to be conserved among these three species. QTLs for seed weight were also detected at similar locations on azuki bean linkage group 9 and mungbean linkage group i.
Fig. 5.
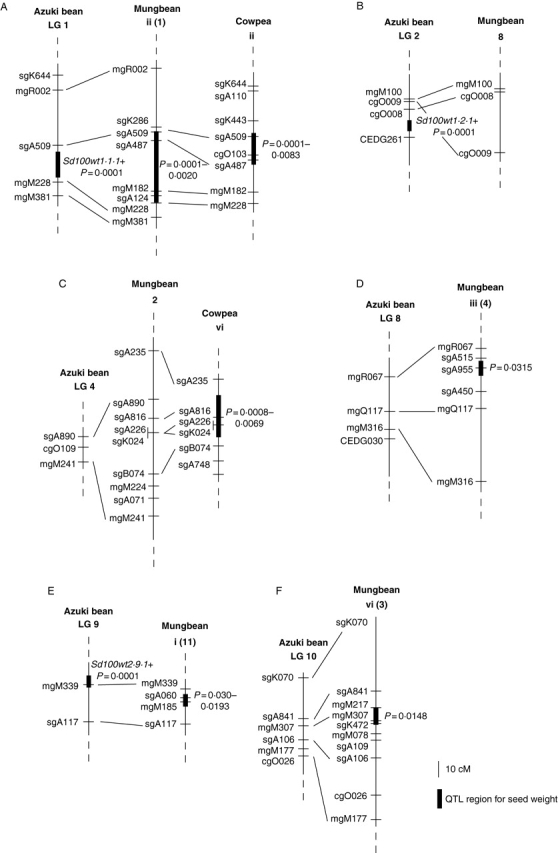
Comparative QTL maps for seed weight in three cultivated Vigna species, V. angularis (azuki bean), V. radiata (mungbean) and V. unguiculata (cowpea). Linkage groups of mungbean are after the maps of Fatokun et al. (1992) (Roman numerals) and Menancio-Hautea et al. (1993) (Arabic numerals). Linkage groups of cowpea are after the maps of Fatokun et al. (1992). The QTL regions for seed weight in mungbean and cowpea are after Fatokun et al. (1992).
Although the QTL with largest effect for seed weight was detected on the linkage group 2 in azuki bean, no QTL was detected on the linkage groups corresponding to this linkage group in cowpea and mungbean. QTLs on linkage group vi of cowpea, iii and vi of mungbean and 8 of azuki bean appear to be specific to these crops. These results suggest that the main genome regions related to increased seed weight under domestication do not correspond among these related species despite high homology between the linkage groups. In azuki bean, seed weight in cultivated taxa is about eight times that of the wild parent. In contrast, seed weight in cultivated and wild parents of crosses analysed for both cowpea and mungbean only exhibited a 5-fold difference (Fatokun et al., 1992). Azuki bean has among the largest seed for the cultivated Asian Vigna (Tomooka et al., 2000). It seems that increase in seed size in azuki bean compared with cowpea and mungbean involves some different loci.
Other legumes
In soybean (tribe Phaseoleae), a QTL for seed weight was detected near RFLP marker sgT153 on linkage group A (Maughan et al., 1996). This corresponds to linkage group 1 in azuki bean. However, this RFLP marker was well separated from the molecular markers associated with seed weight variation in azuki bean, mungbean and cowpea.
In pea (Pisum sativum L., tribe Vicieae) (Timmerman-Vaughan et al., 1996), a QTL for seed weight was also detected in the region that corresponds to the region with seed weight QTL on linkage group 1 of azuki bean and ii of cowpea and mungbean (tribe Phaseoleae) based on RFLP comparison. Therefore, it seems that this region has been conserved across the Leguminosae and plays an important role in increasing seed size.
CONCLUSIONS
A broad array of domestication-related traits in azuki bean have been analysed and their QTLs mapped on the azuki bean molecular linkage map. Most traits are controlled by two to nine QTLs that occur on different linkage groups. QTLs for domestication-related traits are not evenly distributed across the azuki bean linkage map and five out of the 11 linkage groups in azuki bean (linkage groups 1, 2, 4, 7 and 9) have 80 % of the QTLs detected. In addition, within a linkage group, QTLs are clustered.
Comparison of the positions of QTLs for domestication-related traits studied here with reports for related legumes has revealed that some regions with seed size QTLs are conserved among azuki bean, mungbean, cowpea, soybean and pea. However, there are also many QTLs that are not common either within the azuki bean complex or across the warm-season legumes. This study provides a foundation for use of molecular-assisted selection of domestication-related QTLs in azuki bean breeding programmes. The results presented here can be used to further enhance understanding of domestication in the Phaseolus–Vigna group.
SUPPLEMENTARY INFORMATION
Four sets of supplementary information are provided online at http://aob.oxfordjournals.org/. Supplementary Information 1 provides temperature, rainfall and sunlight hours at Tsukuba, Japan, for the months azuki populations were growing in 2002–2004. Supplementary information 2 shows the frequency distribution for all traits examined in all populations examined. Supplementary information 3 shows a comparison of QTLs found for BC1F1 and F2 molecular linkage maps. Supplementary information 4 shows the correlation coefficients among each trait for all populations examined.
ACKNOWLEDGEMENTS
This research was conducted while the first author (T.I.) was a recipient of a fellowship from the Japan Society for the Promotion of Science. The authors thank T. Nobori, N. Karino, S. Hirashima, K. Sugimoto, H. Uchiyama, T. Taguchi and H. Tomiyama for technical support. Funding to pay the Open Access publication charges for this article was provided by the OECD.
LITERATURE CITED
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B. 1995;57:289–300. [Google Scholar]
- Blair MW, Pedraza F, Buendia HF, Geitan-Solis E, Beebe SE, Gepts P, et al. Development of a genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.) Theoretical and Applied Genetics. 2003;107:1362–1374. doi: 10.1007/s00122-003-1398-6. [DOI] [PubMed] [Google Scholar]
- Boutin SR, Young ND, Olson TC, Yu ZH. Genome conservation among three legume genera detected with DNA markers. Genome. 1995;38:928–937. doi: 10.1139/g95-122. [DOI] [PubMed] [Google Scholar]
- Cai HW, Morishima H. QTL clusters reflect character associations in wild and cultivated rice. Theoretical and Applied Genetics. 2002;104:1217–1228. doi: 10.1007/s00122-001-0819-7. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A. Genetic analysis of the morphological differences between maize and teosinte. Genetics. 1991;129:285–295. doi: 10.1093/genetics/129.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A. Inheritance of morphological differences between maize and teosinte: comparison of results for two F2 populations. Genetics. 1993;134:559–570. doi: 10.1093/genetics/134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- Doganlar S, Frary A, Daunay MC, Lester RN, Tanksley SD. Conservation of gene function in the Solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics. 2002;161:1713–1726. doi: 10.1093/genetics/161.4.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatokun CA, Menancio-Hautea D, Danesh D, Young ND. Evidence for orthologous seed weight genes in cowpea and mungbean based on RFLP mapping. Genetics. 1992;132:841–846. doi: 10.1093/genetics/132.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyre R, Skroch PW, Geffroy V, Adam-Blondon AF, Shirmohamadali A, Johnson WC, et al. Towards an integrated linkage map of common bean. 4. Development of a core linkage map and alignment of RFLP maps. Theoretical and Applied Genetics. 1998;97:847–856. [Google Scholar]
- Hammer K. The domestication syndrome. Die Kulturpflanze. 1984;32:11–34. [in German with English summary] [Google Scholar]
- Han OK, Kaga A, Isemura T, Tomooka N, Vaughan DA. A genetic linkage map for azuki bean (Vigna angularis) Theoretical and Applied Genetics. 2005;111:1278–1287. doi: 10.1007/s00122-005-0046-8. [DOI] [PubMed] [Google Scholar]
- Hawkes J. The diversity of crop plants. Cambridge, MA: Harvard University Press; 1983. [Google Scholar]
- Jansen RC, Van Ooijen JM, Stam P, Lister C, Dean C. Genotype-by-environment interaction in genetic mapping of multiple quantitative trait loci. Theoretical and Applied Genetics. 1995;91:33–37. doi: 10.1007/BF00220855. [DOI] [PubMed] [Google Scholar]
- Kaga A, Ohnishi M, Ishii T, Kamijima O. A genetic linkage map of azuki bean constructed with molecular and morphological markers using an interspecific population (Vigna angularis × V. nakashimae) Theoretical and Applied Genetics. 1996;93:658–663. doi: 10.1007/BF00224059. [DOI] [PubMed] [Google Scholar]
- Kaga A, Ishii T, Tsukimoto K, Tokoro E, Kamijima O. Comparative molecular mapping in Ceratotropis species using an interspecific cross between azuki bean (Vigna angularis) and rice bean (V. umbellata) Theoretical and Applied Genetics. 2000;100:207–213. [Google Scholar]
- Koinange EMK, Singh SP, Gepts P. Genetic control of the domestication syndrome in common bean. Crop Science. 1996;36:1037–1045. [Google Scholar]
- Korol A, Ronin Y, Hayes P, Nevo E. Multi-interval mapping of correlated trait complexes: simulation analysis and evidence from barley. Heredity. 1998;80:273–284. [Google Scholar]
- Lebreton CM, Visscher PM. Empirical nonparametric bootstrap strategies in quantitative trait loci mapping: conditioning on the genetic model. Genetics. 1998;148:525–535. doi: 10.1093/genetics/148.1.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumpkin TA, McClary DC. Azuki bean: botany, production and uses. Wallingford: CAB International; 1994. [Google Scholar]
- Maughan PJ, Maroof MAS, Buss GR. Molecular-marker analysis of seed-weight: genomic locations, gene action, and evidence for orthologous evolution among three legume species. Theoretical and Applied Genetics. 1996;93:574–579. doi: 10.1007/BF00417950. [DOI] [PubMed] [Google Scholar]
- Menacio-Hautea D, Fatokun CA, Kumar L, Danesh D, Young ND. Comparative genome analysis of mungbean (Vigna radiata (L.) Wilczek) and cowpea (V. unguiculata (L.) Walpers) using RFLP mapping data. Theoretical and Applied Genetics. 1993;86:797–810. doi: 10.1007/BF00212605. [DOI] [PubMed] [Google Scholar]
- Papa R, Acosta J, Delgado-Salinas A, Gepts P. A genome-wide analysis of differentiation between wild and domesticated Phaseolus vulgaris from Mesoamerica. Theoretical and Applied Genetics. 2005;111:1147–1158. doi: 10.1007/s00122-005-0045-9. [DOI] [PubMed] [Google Scholar]
- Peng J, Ronin Y, Fahima T, Röder M, Li Y, Nevo E, Korol A. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proceedings of National Academy of Sciences of the USA. 2003;100:2489–2495. doi: 10.1073/pnas.252763199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet V, Lamy F, Devos KM, Gale MD, Sarr A, Robert T. Genetic control of domestication traits in pearl millet (Pennisetum glaucum L., Poaceae) Theoretical and Applied Genetics. 2000;100:147–159. doi: 10.1007/s00122-002-0889-1. [DOI] [PubMed] [Google Scholar]
- Smartt J. Tropical pulses. London: Longman; 1976. [Google Scholar]
- Somta P, Kaga A, Tomooka N, Kashiwaba K, Isemura T, Chaitieng B, et al. Development of an interspecific Vigna linkage map between Vigna umbellata (Thunb.) Ohwi & Ohashi and V. nakashimae (Ohwi) Ohwi & Ohashi and its use in analysis of bruchid resistance and comparative genomics. Plant Breeding. 2006;125:77–84. [Google Scholar]
- Tateishi Y, Maxted N. New species combinations in Vigna subgenus Ceratotropis (Piper) Verdcourt (Leguminosae, Phaseoleae) Kew Bulletin. 2002;57:625–633. [Google Scholar]
- Timmerman-Vaughan GM, McCallum JA, Frew TJ, Weeden NF, Russell AC. Linkage mapping of quantitative trait loci controlling seed weight in pea (Pisum sativum L.) Theoretical and Applied Genetics. 1996;93:431–439. doi: 10.1007/BF00223187. [DOI] [PubMed] [Google Scholar]
- Tomooka N, Kashiwaba K, Vaughan DA, Ishimoto M, Egawa Y. The effectiveness of evaluating wild species: searching for sources of resistance to bruchid beetle in the genus Vigna subgenus Ceratotropis. Euphytica. 2000;115:27–41. [Google Scholar]
- Tomooka N, Vaughan DA, Moss H, Maxted N. The Asian Vigna: genus Vigna subgenus Ceratotropis genetic resources. Dordrecht: Kluwer; 2002. [Google Scholar]
- Tomooka N, Kaga A, Vaughan DA. The Asian Vigna (Vigna subgenus Ceratotropis) biodiversity and evolution. In: Sharma AK, Sharma A, editors. Phanerogams (Angiosperm-Dicotyledons) Vol. 1. Enfield, NH: Science Publishers; 2006. pp. 87–126. Plant genome biodiversity and evolution Part C. [Google Scholar]
- Vaughan DA, Tomooka N, Kaga A. Azuki bean. In: Singh RJ, Jauhar PP, editors. Genetic resources, chromosome engineering, and crop improvement: grain legumes. Boca Raton, FL: CRC Press; 2004. pp. 341–353. [Google Scholar]
- Vallejos CE, Sakiyama NS, Chase CD. A molecular marker-based linkage map of Phaseolus vulgaris L. Genetics. 1992;131:733–740. doi: 10.1093/genetics/131.3.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdcourt B. Studies of the Leguminosae-Papilionoideae for the Flora of Tropical East Africa. IV. Kew Bulletin. 1970;24:507–569. [Google Scholar]
- Wang XW, Kaga A, Tomooka N, Vaughan DA. The development of SSR markers by a new method in plants and their application to gene flow studies in azuki bean [Vigna angularis (Willd.) Ohwi & Ohashi] Theoretical and Applied Genetics. 2004;109:352–360. doi: 10.1007/s00122-004-1634-8. [DOI] [PubMed] [Google Scholar]
- Wojciechowski MF, Lavin M, Sanderson MJ. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. American Journal of Botany. 2004;91:1846–1862. doi: 10.3732/ajb.91.11.1846. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H. Wild and weedy azuki beans in Japan. Economic Botany. 1992;46:384–394. [Google Scholar]
- Yu K, Park SJ, Poysa V, Gepts P. Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.) Journal of Heredity. 2000;91:429–434. doi: 10.1093/jhered/91.6.429. [DOI] [PubMed] [Google Scholar]
- Zong XX, Kaga A, Tomooka N, Wang XW, Han OK, Vaughan DA. The genetic diversity of the Vigna angularis complex in Asia. Genome. 2003;46:647–658. doi: 10.1139/g03-041. [DOI] [PubMed] [Google Scholar]


