Abstract
Background
Both sorghum (Sorghum bicolor) and sugarcane (Saccharum officinarum) are members of the Andropogoneae tribe in the Poaceae and are each other's closest relatives amongst cultivated plants. Both are relatively recent domesticates and comparatively little of the genetic potential of these taxa and their wild relatives has been captured by breeding programmes to date. This review assesses the genetic gains made by plant breeders since domestication and the progress in the characterization of genetic resources and their utilization in crop improvement for these two related species.
Genetic Resources
The genome of sorghum has recently been sequenced providing a great boost to our knowledge of the evolution of grass genomes and the wealth of diversity within S. bicolor taxa. Molecular analysis of the Sorghum genus has identified close relatives of S. bicolor with novel traits, endosperm structure and composition that may be used to expand the cultivated gene pool. Mutant populations (including TILLING populations) provide a useful addition to genetic resources for this species. Sugarcane is a complex polyploid with a large and variable number of copies of each gene. The wild relatives of sugarcane represent a reservoir of genetic diversity for use in sugarcane improvement. Techniques for quantitative molecular analysis of gene or allele copy number in this genetically complex crop have been developed. SNP discovery and mapping in sugarcane has been advanced by the development of high-throughput techniques for ecoTILLING in sugarcane. Genetic linkage maps of the sugarcane genome are being improved for use in breeding selection. The improvement of both sorghum and sugarcane will be accelerated by the incorporation of more diverse germplasm into the domesticated gene pools using molecular tools and the improved knowledge of these genomes.
Key words: Genomics, sorghum, Sorghum bicolor, sugarcane, Saccharum officinarum, crop improvement, domestication
INTRODUCTION
Crop plants were first cultivated around 10 000 years ago. However, crop domestication and development began much more recently (Doggett, 1970). Innumerable varieties, races and cultivars of agricultural plants have been developed to support human and animal demand for food, fibre and building materials. The Poaceae are an important global source of dietary protein, carbohydrates and other nutrients. Sorghum [Sorghum bicolor (L.) Moench] is the fourth most important cereal crop behind wheat, rice and maize, and is grown throughout the arid and semi-arid tropics (Smith and Frederiksen, 2000). Sugarcane (Saccharum officinarum L.) is the leading sugar-producing crop globally and is grown throughout tropical and subtropical parts of the world (Cordeiro et al., 2006b). Sorghum and sugarcane are each other's closest relatives among cultivated crops. Their evolutionary divergence is estimated as occurring as recently as 5 million years ago, with maize having separated 15–20 million years ago (Paterson et al., 2004). Intergeneric hybrids between the two groups have been reported, reinforcing their close relationship (Bowers et al., 2003). Both are more recent domesticates than the other major grass crops and despite ongoing breeding programmes using diverse germplasm, comparatively little of the genetic potential of these taxa and their wild crop relatives has been captured by breeding programmes to date.
SORGHUM AND ITS WILD RELATIVES
Sorghum domestication
Arthropological evidence suggests that hunter-gatherers consumed sorghum as early as 8000 bc (Smith and Frederiksen, 2000). The domestication of sorghum has its origins in Ethiopia and surrounding countries, commencing around 4000–3000 bc. Numerous varieties of sorghum were created through the practice of disruptive selection, whereby selection for more than one level of a particular character within a population occurs (Doggett, 1970). This results from a balance of farmer selection for cultivated traits and natural selection for wild characteristics, generating both improved sorghum types, wild types and intermediate types (Doggett, 1970). These improved sorghum types were spread via the movement of people and trade routes into other regions of Africa, India (approx. 1500–1000 bc), the Middle East (approx. 900–700 bc) and eventually into the Far East (approx. ad 400). By the time sorghum was transported to America during the late 1800s to early 1900s, the diversity of new sorghum types, varieties and races created through the movement of people, disruptive selection, geographic isolation and recombination of these types in different environments would have been large (Wright, 1931; Doggett, 1970).
Initial domestication of sorghum would have focused primarily on converting wild types with small, shattering (dehiscent) seed to improved types with larger, non-shattering seed. Disruptive selection resulted in sorghum types with vastly different characteristics in height, inflorescence type, and of course, end use (food, fodder, fibre, building materials, etc). Over time, sorghum has been described and redescribed by numerous taxonomists (Fig. 1), and is now described under the family Poaceae, tribe Andropogoneae, subtribe Sorghinae and genus Sorghum Moench (Clayton and Renvoize, 1986).
Fig. 1.
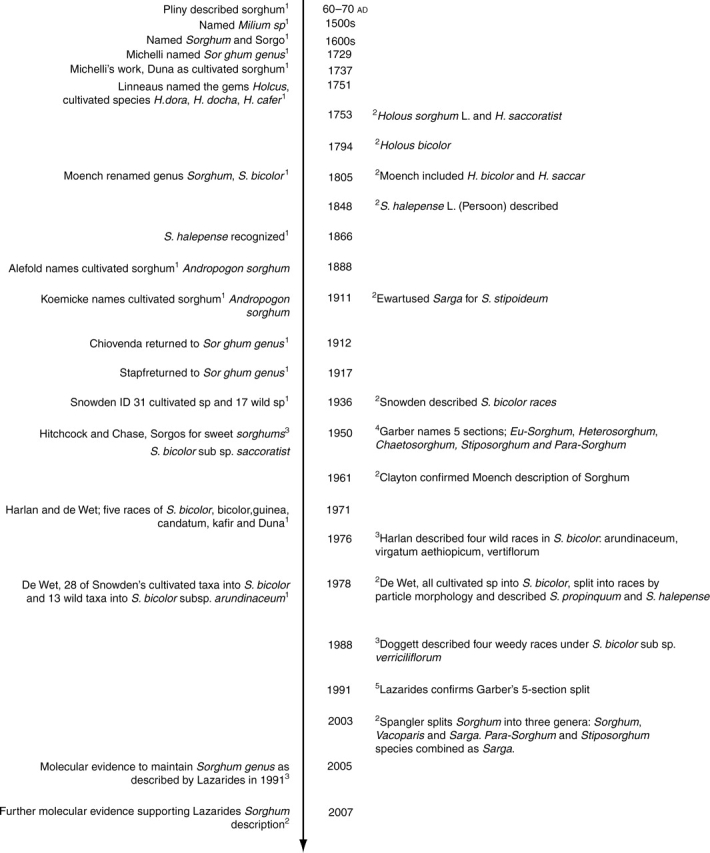
Time-line displaying the changes in Sorghum nomenclature over time. 1House et al. (1995); 2Spangler (2003); 3Smith and Frederiksen (2000); 4Garber (1950); 5Lazarides et al. (1991); 6Hodnett et al. (2005), Price et al. (2005a); 7Dillon et al. (2007).
The Sorghum genus as currently proscribed consists of 25 species (USDA ARS, 2007), although this varies in different scientific publications confirming the dynamic nature of the classification of cultivated sorghum and its wild relatives. The genus is separated into five taxonomic subgenera or sections: Eu-Sorghum, Chaetosorghum, Heterosorghum, Para-Sorghum and Stiposorghum (Garber, 1950). Section Eu-Sorghum contains all domesticated/cultivated sorghum races and varieties as Sorghum bicolor subsp. bicolor, as well as the wild and weed species S. halepense (L.) Pers. (Johnsons grass), S. propinquum (Kunth) Hitchc, S. × almum Parodi, S. × drummondii (Steud.) Millsp. & Chase, and S. arundinaceum (Desv.) Stapf. (the known progenitor of S. bicolor) (Harlan and de Wet, 1971; Doggett, 1988). All S. bicolor subsp. bicolor have 2n = 2x = 20 chromosomes, and are described as annual, with thick culms up to 5 m in height, often branched with many tillers. They have been classified into five basic races: bicolor, guinea, caudatum, kafir and durra, with ten intermediate races of these also recognized (Harlan and de Wet, 1972). These 15 races of cultivated sorghum are recognizable on spikelet/panicle morphology alone, and can be linked back to their specific environments and the nomadic peoples that first cultivated them (Smith and Frederiksen, 2000).
A comprehensive analysis of genetic diversity in sorghum landraces and core collections based on race, latitude of origin, photoperiod, seed quality, agronomic traits and DNA markers has demonstrated sorghum has considerable polymorphism that has been poorly exploited in terms of crop improvement (Wu et al., 2004; Abu Assar et al., 2005; Deu et al., 2006; Kayode et al., 2006). At the DNA level, two high-density maps have been completed, one intraspecific and another from an interspecific cross (between S. bicolor and S. propinquum). These maps showed a high colinearity from which the divergence between Sorghum species and the diversity within cultivated S. bicolor has been indicated (Feltus et al., 2006).
Changing characteristics/traits of domesticated sorghum and effects on yield
Early domestication of sorghum was associated with changing the small-seeded, shattering open panicles towards larger, non-shattering seeds and more compact panicles. This involved several factors: significantly increasing the number of branches within the inflorescence; decreasing the internode length of the rachis; and an increase in seed size so it protruded out of the glumes (House, 1985). These changes contributed to an increase in yield over the original sorghum landrace varieties.
Stable, high-yielding sorghum varieties have been recently developed through breeding/improvement programmes utilizing sorghum landrace varieties from Africa, India and China. This has involved selecting traits such as photoperiod insensitivity, reduced height (to reduce lodging), drought tolerance, and pest and disease resistance (Reddy et al., 2006).
Plant height and photoperiod insensitivity were the focus of conversion programmes that developed sorghum lines with desirable plant height and maturity that were usable in breeding programmes in both tropical, short-day environments and in long-day, temperate and subtropical environments. As sorghum originated in north-eastern Africa, the many landraces and early varieties were photoperiod sensitive, with a critical photoperiod of 12 h: once the day length is shorter than 12 h, the sorghum plant changes from vegetative to reproductive growth (Reddy et al., 2006). Growing these photoperiod-sensitive landraces/lines as a summer crop in temperate zones of America and Australia where the day length is longer than 13 h was difficult, especially as many growth-related characteristics are poorly expressed under these long-day conditions (Reddy et al., 2006). This made breeding improved varieties in semi-arid temperate and subtropical climates difficult. Cultivars and landraces were identified in India that had higher critical photoperiods, with no delay in flowering observed when grown in day lengths up to 17 h. These photoperiod-insensitive sorghum cultivars have since been widely adopted in breeding programmes throughout the world (Rai et al., 1999; Reddy et al., 2006).
Plant height and grain yield are highly correlated in some populations of sorghum, with maximum productivity achieved at heights of around 1·75–1·80 m and flowering at 68–70 d (Miller, 1982; Rao and Rana, 1982). However, plants of these heights easily lodge, and are not easily cultivated under modern farming practices. A selection of high-yielding, tall sorghum landraces/lines were crossed to shorter, photoperiod-insensitive sorghum lines to develop improved high-yielding cultivars with a shorter stature (Miller, 1980; Rosenow and Dahlberg, 2000).
Sorghum is grown predominantly in low-rainfall, arid to semi-arid environments. The occurrence of drought stress is a major constraint to sorghum production globally. Two forms of drought stress have been identified in sorghum: ‘pre-anthesis’ where plants are stressed during panicle differentiation prior to flowering; and ‘post-anthesis’ when moisture stress occurs during the grain fill stage (Rosenow and Clark, 1995). The identification of varieties and lines with naturally high levels of pre-anthesis drought tolerance and the selection of these for higher yields has developed sorghum varieties with stable, high yields (Ellis et al., 1997). Post-anthesis drought stress can result in significant yield loss due to small grain size, premature plant death and susceptibility to diseases. Post-flowering drought tolerance is referred to as stay-green, with plants maintaining green leaf area and photosynthetic capability under severe moisture stress, which results in higher grain yields compared with senescent varieties (Borrell and Douglas, 1997; Borrell et al., 1999). The physiological components of stay-green (green leaf area at flowering; time of onset of senescence; rate of senescence) are independently inherited and easily combined through breeding, resulting in new sorghum varieties exhibiting high levels of stay-green with stable high yields and good levels of insect resistance (Borrell et al., 2000).
Sorghum production is affected by many pests and diseases globally. Some of the major pests include midge (Stenodiplosis sorghicola Coquillett), green bug (Schizaphis graminum Rondani), various aphids, shootfly (Atherigona soccata Rondani) and stem borer (Chilo partellus Swinhoe) (Sharma, 1993). Major diseases include downy mildew, anthracnose, sorghum rust, leaf blight, ergot and head and kernel smut (House, 1985). Success in breeding for insect resistance in sorghum varieties has been varied. Resistance to some pests is quantitatively inherited and therefore difficult to transfer into high-yielding cultivars (Tao et al., 2003). The exception to this is midge resistance, where high levels of midge immunity have been incorporated from Indian, American and Australian breeding lines into elite, high-yielding sorghum varieties in Australia, with greater than 80 % of the planted area utilizing these resistant varieties (Jordan et al., 1998; Tao et al., 2003).
Development of disease-resistant sorghum varieties has relied on identifying sorghum varieties/landraces with natural genetic resistance to the particular disease. To date, commercial sorghum varieties have been developed with resistance to grain moulds and anthracnose (Reddy et al., 2006).
The development of photoperiod-insensitive, dwarfed sorghum varieties with some levels of pest/disease resistance has improved the yields of cultivated sorghum varieties. However, the development of a hybrid cropping system is responsible for increases in yields of more than 300 % since the 1950s (Rooney and Smith, 2000). Hybrid cultivars make use of male sterility to enhance the combining abilities of the parental lines, resulting in heterosis and significant increases in phenotypic traits such as yield, plant height and days to flowering (Reddy et al., 2006).
Although the domestication and resulting super-domestication of sorghum has relied on principally S. bicolor subsp. bicolor varieties/landraces/lines for significant gains in agricultural production, the undomesticated Sorghum species offer an untapped wealth of novel traits for both biotic and abiotic stress resistance and yield.
Undomesticated Sorghum species as genetic resources for sorghum improvement
All cultivated sorghum varieties and landraces are S. bicolor subsp. bicolor of the Eu-Sorghum subgeneric section of the Sorghum genus. The other four sections, Chaetosorghum, Heterosorghum, Para-Sorghum and Stiposorghum contain 19, wild species native to Africa, Asia and Australia (Garber, 1950; Lazarides et al., 1991). These species are briefly outlined below, and contain new sources of genetic diversity for agronomic traits affecting yield, survivability and novel traits that may create new markets for sorghum products.
The monotypic sections Chaetosorghum and Heterosorghum contain the octaploid (2n = 40) Australian species S. macrospermum E.D. Garber and S. laxiflorum F.M. Bailey, respectively. Section Para-sorghum contains the five Australian species S. grande Lazarides, S. leiocladum (Hack.) C.E. Hubb., S. matarankense E.D. Garber & Snyder, S. nitidum (Vahl) Pers., S. timorense (Kunth) Buse, and the two African/Asian species S. purpureo-sericeum (Hochst. ex A. Rich.) Asch. & Schweinf. and S. versicolor Andersson. These species range in ploidy from 2n = 10 to 2n = 40, with S. grande, S. nitidum and S. timorense showing varying ploidy within species. Ten Australian endemic species form section Stiposorghum: Sorghum amplum Lazarides, S. angustum S.T. Blake, S. brachypodum Lazarides, S. bulbosum Lazarides, S. ecarinatum Lazarides, S. exstans Lazarides, S. interjectum Lazarides, S. intrans F. Muell. ex Benth., S. plumosum (R. Br.) P. Beauv., and S. stipoideum (Ewart & Jean White) C.A. Gardner & C.E. Hubb. (Garber, 1950; Lazarides et al., 1991). Most of these species are diploid with 2n = 10 chromosomes, while S. interjectum has 2n = 30, 40 and S. plumosum has 2n = 10, 20, 30 (Garber, 1950; Lazarides et al., 1991).
The adaptability of these undomesticated Sorghum species to colonize a wide range of soil and moisture conditions across a wide range of microenvironments is shown through their ability to survive very hot, dry, nutrient-limited environments. Due to their adaptability, many of the undomesticated Sorghum species have developed resistances to the many pests and diseases that affect sorghum grain production globally. Interestingly, many Australian undomesticated species contain resistances to the major pest/diseases of Africa and America, which are not yet present within Australia (Bapat and Mote, 1982; Karunakar et al., 1994; Franzmann and Hardy, 1996; Sharma and Franzmann, 2001; Kamala et al., 2002; Komolong et al., 2002).
Recent controlled-environment glasshouse trials have shown that the undomesticated Sorghum species, though adapted to specific abiotic conditions in the wild, showed prolific growth under moderate temperature in a standard potting mix and watered regularly (Table 1). These data show useful variations to germination times and time to flowering. Representatives of the undomesticated Heterosorghum, Para-Sorghum, Stiposorghum and a Eu-Sorghum were grown concurrently to compare their development under controlled conditions (Fig. 2). Cultivated S. bicolor takes 3–10 d to germinate depending on soil temperatures, with the first 30–35 d post-germination undergoing lower leaf growth followed by a rapid elongation in non-dwarf varieties. Flowering in S. bicolor occurs 55–70 d post-germination and seeds reach physiological maturity 30–40 d post-anthesis. It then takes 20–25 d to reduce the moisture content to the 12 % required for post-harvest storage (House et al., 1995). There appears to be limited differences between undomesticated species and S. bicolor for these traits (Table 1).
Table 1.
Germination, growth rates and flowering times for undomesticated Sorghum species
| Species | DTT* | Height (cm) |
Days till flower | Number of panicles |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 30 d | 60 d | 90 d | 130 d | 10 dpa | 20 dpa | 30 dpa | 40 dpa | Final max count | |||
| S. amplum | 4·5 | 25 | 40 | 40 | 125 fl | 136 | md | 16 | 29 | – | – |
| S. angustrum | 5·5 | 18 | 40 | 62 fl | 95 fl | 86 | 9 | 20·5 | 58 | 146 | >300 |
| S. brachypodum | 3·5 | 35 | 70 | 150 fl | 210 fl | 86 | 4 | 26·5 | 45 | 62·5 | >300 |
| S. bulbosum | 3·0 | 30 | 55 | 120 fl | 170 fl | 95 | 22 | 67 | 89 | – | – |
| S. exstans (p) | 3·5 | 12 | 45 fl | 70 fl | 95 fl | 65 | 10 | 17 | 47 | 113 | 235 |
| S. intrans (p) | 3·5 | 5 | 20 p | 20 p | 85·0 | >160 | – | – | – | – | – |
| S. laxiflorum | 5·5 | 60 | 60 fl | 90 fl | 95 fl | 61 | md | 16·5 | 44 | 94 | 214 |
| S. leiocladum | 5·5 | 27 | 30 | 35 | 85·0 | 141 | 1 | – | – | – | – |
| S. matarankense | 4·5 | 58 | 88 fl | 130 fl | 150 fl | 81 | 2 | 5 | 23 | 53 | 69 |
| S. nitidum | 4·0 | 25 | 48 | 60 | 90 | >160 | – | – | – | – | – |
| S. plumosum | 8·0 | 8 | 15 | 15 | 60 | >160 | – | – | – | – | – |
| S. propinqeum | 3·0 | 45 | 50 | 100 | 240 fl | 106 | 19 | 35 | – | – | >300 |
| S. stipoideum | 4·0 | 45 | 70 | 60 fl | 130 fl | 91 | 18 | 60 | 112 | – | – |
| S. timorense | 3·0 | 45 | 100 fl | 100 fl | 145 fl | 69 | md | 3 | 14 | 18 | 63 |
* DTT, Days till transplant: seeds were germinated on damp filter paper; once a strong radicle and the first coleoptile had emerged they were transplanted to potting mix.
md, Missing data, (p), prostrate growth habit; fl, height was measured to the top of flag leaf or seed head.
A dash indicates that delayed onset of flowering caused the trial to be terminated before counts could be made.
Fig. 2.
Growth trial of Sorghum species at seedling stage. Note the broader leaf (far left) of the Eu-sorghum, S. propinquum compared with the Para-Sorghum, Stiposorghum and Heterosorghum species.
Undomesticated Sorghum species: grain attributes
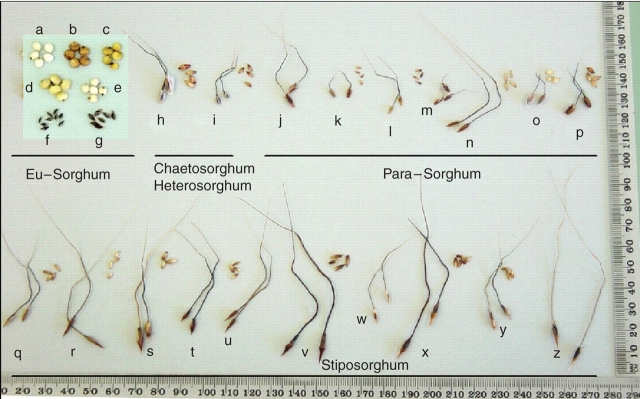
The morphology of seed size and shape within the Sorghum genus varies greatly. Figure 3 shows the morphology of the domesticated S. bicolor subsp. bicolor and undomesticated Eu-Sorghum, Chaetosorghum, Heterosorghum, Para-Sorghum and Stiposorghum species. Variation in the grain morphology of representatives of the undomesticated Heterosorghum, Para-Sorghum and Stiposorghum species have also been evaluated at the microscopic level. Mature caryopses of 13 species were critically point dried, snap fractured and examined using a Leostereoscan 440 scanning electron microscope to determine if novel variations existed in the undomesticated species (Shapter et al., 2007).
Fig. 3.
Variation in Sorghum species seed and caryopsis morphology and size. Letters on the figure denote different species: a–e, S. bicolor caryopsis AusTRCF 322649, 322618, 322620, 322666 and 322611, respectively; f, S. propinquum; g, S. halepense; h, S. macrospermum 322277 seed and caryopsis; i, S. laxiflorum 302503 seed and caryopsis; j, S. grande 302580 seed; k, S. leiocladum 300170 seed and caryopsis; l, S. matarankense 302521 seed and caryopsis; m, S. nitidum 302539 seed; n, S. timorense 302660 seed and caryopsis; o, S. purpureo-sericeum 321134 seed and caryopsis; p, S. versicolor 321126 seed and caryopsis; q, S. amplum 302623 seed and caryopsis; r, S. angustum 302604 seed and caryopsis; s, S. brachypodium 302480 seed and caryopsis; t, S. bulbosum 302646 seed and caryopsis; u, S. ecarinatum 302661 seed; v, S. exstans 302577 seed and caryopsis; w, S. interjectum 302563 seed; x, S. intrans 302390 seed and caryopsis; y, S. plumosum 302489 seed and caryopsis; z, S. stipoideum 302644 seed.
The endosperm of cultivated S. bicolor is described as having two distinct regions or layers. The floury central endosperm (Fig. 4A) contains simple round or lenticellar starch granules in a discontinuous protein matrix with few if any protein bodies present. The vitreous or corneous outer endosperm (Fig. 4B) is characterized by polygonal starch granules, 4–25 µm in diameter, the surface of which is typically indented from the protein bodies that are part of the continuous protein matrix surrounding the granules. Variations to the distribution and configuration of these two regions have been shown to alter the functional and putatively the nutritional value of sorghum flours and other foods (Serna-Saldivar and Rooney, 1995; Lindeboom et al., 2004; Tesso et al., 2006).
Fig. 4.
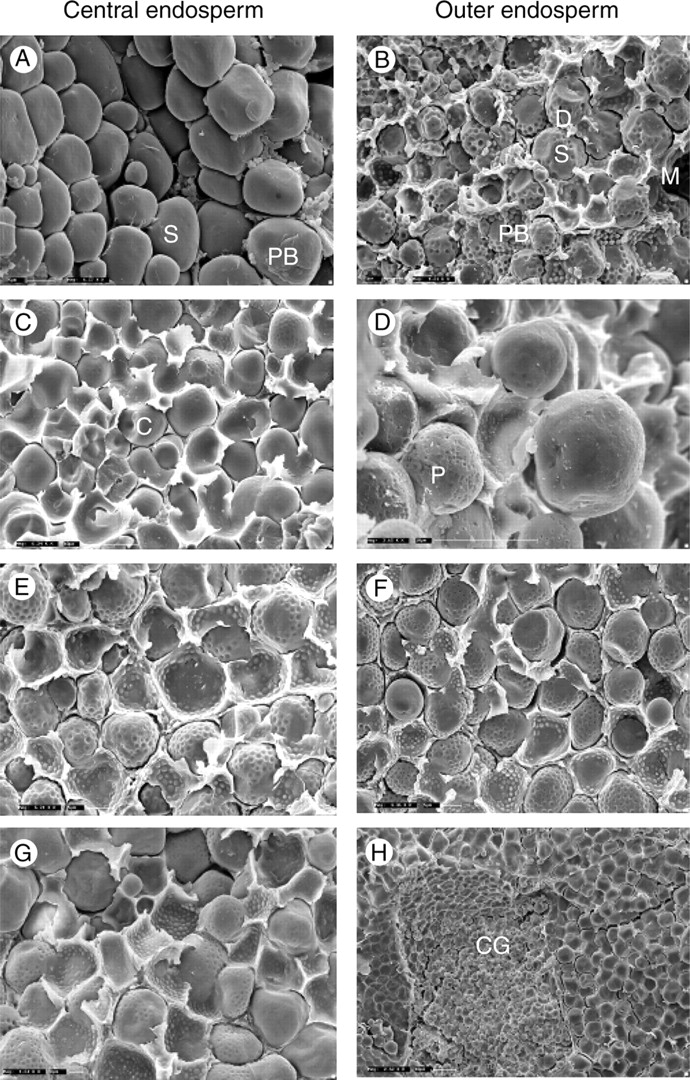
The left-hand column shows the variation in the central endosperm and the right-hand column compares the outer layers. (A, B) Representative images of S. bicolor, showing the standard floury and vitreous endosperm, respectively. (C–H) Images from outside the Eu-Sorghums are representative of the variations observed across the species. PB, Protein bodies; M, matrix; S, starch granule; D, indentations left by protein bodies; C, channels; P, pores; CG, small polygonal starch granules forming compound granules.
The undomesticated Sorghum species showed varied distribution of protein bodies throughout the endosperm (Fig. 4C–H). Similarly, variation in the starch granule size and shape was also noted (Shapter et al., 2007). Some of the undomesticated species had distinctly smaller, more spherical granules throughout the endosperm (Fig. 4D). Importantly, several species showed native channelling of the starch granules and pores on their surface (Fig. 4C) which have been shown to improve the digestion of sorghum starches (Fannon et al., 2003, 2004; Benmoussa et al., 2006). One species appeared to have sections of the endosperm with small rice-like starch granules, usually only seen in the sub-aleurone layer in S. bicolor (Shapter et al., 2007). Several wild species also maintained a single morphology across the entire endosperm, rather than the two layers seen in S. bicolor. Amongst these differences some species retained the characteristic morphology of the S. bicolor vitreous layer (Fig. 4F).
The sub-aleurone of S. bicolor is described as being 15–30 µm wide and is an area of very small starch granules and denser protein matrix, the endosperm proper (Fig. 5A and B).
Fig. 5.
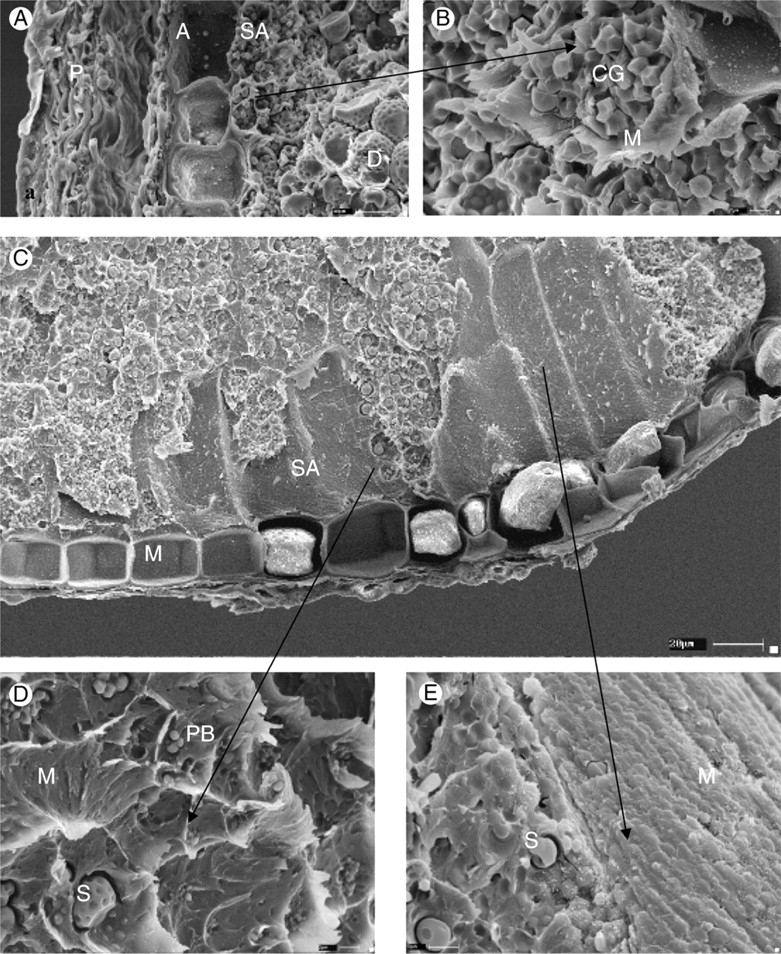
Novel sub-aleurone morphology of the Para-Sorghums and Stiposorghums: (A) the characteristic S. bicolor outer endosperm and pericarp; (B) an increased magnification of the sub-aleurone layer itself; (C) a representative image of the novel morphology found in the undomesticated sorghums; (D) and (E) magnified features of the morphology shown in (C). SA, Sub-aleurone layer; A, aleurone; PB, protein bodies; M, matrix,; S, starch granule; D, indentations left by protein bodies; P, pericarp; CG, small polygonal starch granules forming compound granules.
In the Para-Sorghum and Stiposorghum species examined, areas of the sub-aleurone have a striated appearance (Fig. 5C) not previously reported in microscopy studies (F. M. Shapter et al., unpubl. res.). Investigation of these areas under high magnification showed what appeared to be a much denser protein matrix, embedded with spherical-shaped bodies reminiscent of protein bodies. Within this layer, small starch granules typical of sub-aleurone starch granules are interspersed (Fig. 5D and E). More investigation is needed to confirm if this layer is proteinaceous. From an adaptive point of view, the development of a highly proteinaceous layer directly below the aleurone would provide a rich nitrogen source for the germinating seedling when establishing itself in low nitrogen soils, typical of northern Australia where many of these undomesticated species are endemic. Protein/starch interactions in sorghum have been shown to decrease starch digestibility, especially after cooking (Duodo et al., 2003). The occurrence of increased protein content in the endosperm may therefore result in a further decrease in starch digestibility which has utility for raising the glycaemic index of foods for Western diets.
Hybridizing potential of undomesticated sorghum species
Modern sorghum breeding programmes have not used species outside of section Eu-Sorghum as sources of genetic diversity due to a lack of information regarding the genetic relationships between the species. Recent phylogenetic analysis of all 25 Sorghum species based on the three gene sequences ITS1, ndhF and Adh1 has identified S. macrospermum and S. laxiflorum as the undomesticated species outside of Eu-Sorghum most closely related to cultivated sorghum varieties (Dillon et al., 2007). The relationships identified can now act as a guide for plant breeders.
Most of the undomesticated Sorghum species fall within the tertiary genepool, making gene transfer to domesticated species very difficult due to strong sterility barriers (Harlan and de Wet, 1971). The nature of the sterility barriers in Sorghum have recently been identified as pollen–pistil incompatibilities whereby the pollen of undomesticated species behaves abnormally in the pistils of S. bicolor, resulting in no hybrid embryo formation (Hodnett et al., 2005). As a result, pollen rarely grew beyond the stigma of S. bicolor; however, a single embryo was formed using S. macrospermum pollen. The embryo of this S. bicolor × S. macrospermum cross was rescued and raised through tissue culture, with the seedling verified as a hybrid based upon cytological and morphological characteristics (Price et al., 2005b).
Although a hybrid embryo was formed and able to be rescued via tissue culture, pollen–pistil incompatibilities make this an extremely rare occurrence. Methods of increasing the frequency of hybridization are required to successfully utilize the undomesticated Sorghum species. An S. bicolor accession was discovered containing a recessive gene (inhibition of alien pollen = iap) that allowed maize (Zea mays L.) pollen tubes to grow through S. bicolor pistils (Laurie and Bennett, 1989). This S. bicolor accession can successfully override the pollen–pistil incompatibilities between S. bicolor and undomesticated Sorghum species and lead to the production of hybrid embryos and plants (Price et al., 2006). Hybrids between S. bicolor × S. macrospermum were obtained from germinated seeds, while the hybrids between S. bicolor × S. angustum and S. bicolor × S. nitidum were recovered through embryo rescue and tissue culture. The hybrid nature of these seedlings was again confirmed by the presence of genomes from both parental species that could be readily identified based upon chromosome size and number (Price et al., 2006).
Introgression of the undomesticated S. macrospermum genome with cultivated S. bicolor has been tracked using FISH (fluorescent in situ hybridization) (Kuhlman et al., 2006). FISH discriminated between the chromosomes of the two parent species, and confirmed through bivalent formation and allosyndetic pairing that recombination was occurring. Progeny of this novel hybrid when backcrossed to S. bicolor expressed altered fertility, again confirming that introgression from the undomesticated parent has occurred (Kuhlman et al., 2006). The analysis of the amount of DNA introgressed from the undomesticated S. macrospermum is currently being undertaken using AFLPs (L. C. Kuhlman et al., unpubl. res.).
The identification and use of the iap S. bicolor accession has enabled the successful introgression of genes from undomesticated Sorghum species into cultivated sorghum, and is the first step towards accessing these unique unexploited genes for both biotic and abiotic stresses and agronomic traits. The potential for improving the yield productivity through these traits in commercial sorghum varieties is now a reality.
The role of genomics in improving domesticated S. bicolor
Sorghum bicolor, a diploid, has a relatively small genome (735 Mbp), which although larger than rice (389 Mbp) is smaller than the other important cereals (wheat 16 900 Mbp, maize 2600 Mbp). The last genome duplication event for the S. bicolor genome seems to have occurred much earlier than the divergence of the major cereal crops from a common ancestor (Paterson et al., 2004). Completion of the whole genome sequencing project in 2007 will exponentially increase the sequence data available for Sorghum and will provide valuable information on cereal domestication in the African continent, an event that appears to have occurred independently of other continents though by similar reinforced selective pressures (Paterson et al., 2004). In a way, the sorghum genome sequencing will close a biographic triangle into the knowledge of the polymorphism shared before the divergence of these important grasses and ultimately in the understanding of the evolution in cereals crops between Africa, America and Asia (Kresovich et al., 2005). The tenets of colinearity and microlinearity of grass genomes mean that our knowledge of other cereals and their evolutionary ties will also greatly improve. Due to their economic and scientific value, cereal genomes have been studied over the last 15 years using highly advanced technologies. The similarity at the DNA level makes it possible to use comparative genetics to look for particular genes of unknown sequence between the genomes with the aim of using that information to develop new varieties or discovering new genes that could have a potential impact on traits that are of global importance (e.g. food quality, drought resistance).
The genetic diversity existing within and between Australian Sorghum species was recently evaluated using simple sequence repeats (SSRs) (Dillon et al., 2005). SSRs were sourced from the cultivated S. bicolor (Brown et al., 1996; Taramino et al., 1997; Kong et al., 2000) to determine diversity in these closely related taxa. This method has successfully evaluated diversity in the related species of many crop groups (e.g. Peakall et al., 1998; Hernández et al., 2001; Chen et al., 2002; Scott et al., 2003; González-Martínez et al., 2004; Sudupak, 2004). This evaluation of the Australian species has shown significantly higher levels of genetic diversity both between (inter-) and within (intra-) species compared with the intra-specific diversity of S. bicolor varieties. The relatively high transfer rate of S. bicolor-derived SSRs to the wild species and their high level of diversity suggests that these SSRs are an efficient, highly informative source of molecular markers for the undomesticated Sorghum species.
Screening for novel genetic variation in S. bicolor
Mutations, both natural and artificially induced, provide an alternate source of genetic diversity. Mutants have long been a valuable resource in plant breeding (van Harten, 1998) and, in recent times, in plant genomics research (Henikoff and Comai, 2003; Till et al., 2003; Henikoff et al., 2004). However, the method employed (irradiation or chemical) to induce a mutated population will affect its usefulness and application for genomics research. A review of the comprehensive International Atomic Energy Agency's Mutant Varieties Database (http://www-mvd.iaea.org/MVD/default.htm) shows only 15 induced sorghum mutant accessions amongst more than 2500 registered mutants.
As a result of the random nature of mutation induction, by physical and chemical means, each individual in a population will contain a unique range of gene mutations. This provides a powerful resource for genome analysis employing recent molecular technologies. It is well established that the ultimate goal in DNA research is to ascertain the DNA sequence of a gene. However, the existing technology for genotyping has become a powerful way to avoid the sequencing step or at least for reducing dramatically the number of samples needed to be sequenced. Analysis of DNA polymorphism in natural and mutated populations is more efficient with the use of capillary electrophoresis (Szantai et al., 2005; Davies et al., 2006) which has the advantages of improved efficiency, sensitivity and throughput (Tang et al., 2004) when compared with gel electrophoresis (Vouk et al., 2000; Cordeiro et al., 2006b). Additionally, the use of capillary electrophoresis has the advantage of reducing costs and time through multiplexing (Kan et al., 2004).
Gamma irradiation and EMS (ethyl-methane-sulfonate) mutation protocols have been optimized for selected S. bicolor populations to generate random changes in the sorghum genome. The second generation of plants was screened to assess the amount of polymorphism that has been generated and now mutations can be identified in candidate genes by utilizing an approach to genetic analysis called TILLING (Targeting Induced Local Lesions IN Genomes), which was first applied in plants by McCallum et al. (2000). A significant body of scientific literature is now available on this technique (Comai and Henikoff, 2006).
TILLING allows for genotypic screening for allelic variations prior to commencing with the more costly and labour-intensive phenotyping (Henikoff et al., 2004). EMS-induced TILLING populations have been produced for the major cereal crops: wheat (Slade et al., 2005), rice (Wu et al., 2005), barley (Caldwell et al., 2004), maize (Till et al., 2004) and sorghum (in the authors' laboratory). TILLING is fast becoming a mainstream technology for mutation characterization (Comai and Henikoff, 2006) and for analysing single nucleotide polymorphisms (SNP) (Cordeiro et al., 2006a). A very sensitive high-throughput screening method based on capillary electrophoresis has been developed (Cross et al., 2007) using Endonucleolytic Mutation Analysis by Internal Labelling (EMAIL) to greatly improve the effectiveness of this new reverse genetics approach to crop improvement.
THE GENOME OF SUGARCANE AND ITS WILD RELATIVES
Sugarcane is an important vegetatively propagated crop which is cultivated for its sugar-rich stalks. It contributes an estimated 75 % of the world's sucrose supply with its mature stem capable of accumulating 12–16 % of its fresh weight and approx. 50 % of its dry weight as sucrose (Bull and Glasziou, 1963). Sugarcane originated in South-east Asia and New Guinea (Lebot, 1999). Modern cultivated sugarcane (Saccharum spp.) is a hybrid complex originating from crosses between S. officinarum L. and S. spontaneum L., and in some lineages S. sinense Roxb., or S. barberi Jesw (Edme et al., 2005). Limited numbers of clones, and hence genetic variation, of the two major progenitors have been captured by commercial breeding programmes. Sugarcane, like sorghum, is a relatively recently domesticated species with little of the available genetic diversity having been incorporated or actively analysed for introgression into domesticated varieties. Breeding programmes in the early 1900s focused on hybridization of S. officinarum clones but soon progressed to interspecific crosses incorporating S. spontaneum. This resulted in improved agronomic traits, such as ratooning and disease resisitance, but required a backcrossing programme to S. officinarum, called ‘nobilization’, to elevate the sucrose content (Roach, 1989; Edme et al., 2005). Since then the majority of breeding programmes have focused on intercrossing between the hybrids, though in recent decades the larger increases in genetic gain have been made by incorporating more diverse germplasm into the cultivated backgrounds (Edme et al., 2005).
Taxonomy of sugarcane
Sugarcane belongs to the genus Saccharum, first established by Linnaeus in Species Plantarum in 1753 with two species: S. officinarum and S. spicalum L. The original classification of Linnaeus' has since been revised to contain six species: S. officinarum, known as the noble cane; S. spontaneum L., S. robustum E.W. Brandes & Jeswiet ex Grassl, and S. edule Hassk., classified as wild species; and S. sinense Roxb. and S. barberi Jeswiet, classified as ancient hybrids (Buzacott, 1965; Daniels and Roach, 1987; D'Hont and Layssac, 1998). The genus falls in the tribe Andropogoneae in the grass family, Poaceae, that includes other tropical grasses such as Sorghum and Zea (maize). Closely related to Saccharum are another four genera (Erianthus section Ripidum, Miscanthus section Diandra, Narenga and Sclerostachya) that purportedly readily interbreed, forming the ‘Saccharum complex’ (Daniels and Roach, 1987). They have in common a high level of polyploidy and aneuploidy (unbalanced number of chromosomes) that creates a challenge for both the taxonomist and molecular biologist (Daniels and Roach, 1987; Sreenivasan et al., 1987).
The sugarcane genome
The complexity and size of the sugarcane genome is a major limitation in genetic improvement. Whilst continued selective breeding for enhanced sucrose accumulation has been able to achieve over half of the yield increase in the past 50 years, it has been reported as having reached a plateau due to limits to the gene pool exploited in traditional breeding programmes (Mariotti, 2002). Individual research programmes, however, have been shown to still be making significant annual genetic gains by maintaining a diverse gene pool (Edme et al., 2005). The employment of new technologies to assist in the association of traits with genetic markers and genetic maps can aid in achieving further yield increases in breeding programmes.
Most sugarcane cultivars contain more than 100 chromosomes which can be assigned to eight homology groups (Rossi et al., 2003; Aitken et al., 2005). Over the past two decades, studies utilizing various molecular techniques to unravel the complexity of this important crop species have provided a greater understanding of its complex genetic make-up (Bonierbale et al., 1988; Wu et al., 1992; D'Hont, 1994; Sills et al., 1995; Grivet et al., 1996; Ming et al., 2001; Rossi et al., 2003). Significant achievements include milestones that demonstrate the use of single (markers present on one chromosome only) and double dose (marker present on two chromosomes) markers for mapping and QTL analysis (Ming et al., 2001, 2002; Hoarau et al., 2002; Aitken et al., 2004), and large-scale EST sequencing projects by SUCEST-Sugar Cane EST Genome Project (Vettore et al., 2001), SASRI-South African Sugar Research Institute (Carson and Botha, 2000), UGA-University of Georgia, USA (Ma et al., 2004), and CSIRO- Australia's Commonwealth Scientific and Industrial Research Organization (Casu et al., 2004). Unfortunately, despite these achievements, the pace of progress with sugarcane genomics has lagged behind that achieved with other agricultural crops (Ramsay et al., 2000; Delseny et al., 2001; Mullet et al., 2002).
Analysis of variation in the sugarcane genome
In 1997, an effort was made by the International Consortium for Sugarcane Biotechnology to develop and evaluate simple sequence repeats (SSRs) or microsatellite sequences as a marker system for sugarcane. Markers were developed from an enriched microsatellite library and were shown to have the capacity to distinguish between sugarcane genotypes due to their ability to detect large numbers of alleles (Cordeiro et al., 2000). To date, this marker system has delivered a number of applications that have advanced both sugarcane research and breeding. Published applications include the mapping of alleles generated from 72 SSR primer pairs onto a genetic map constructed on the Australian hybrid cultivar, Q165A (Aitken et al., 2005); validation of the introgression of genes into F1 hybrids of crosses made between S. spontaneum and elite commercial clones (Pan et al., 2004); the confirmation of fertile intergeneric F1 hybrids of S. officinarum and E. arundinaceus as well as backcross (BC1) progeny from the F1 to hybrid sugarcane (Cai et al., 2005); and the use of the markers to register and confirm sugarcane varieties by the United States Department of Agriculture (USDA) (Tew et al., 2003). SSR markers have also been used to draw useful information on the relationships between various members of the ‘Saccharum complex’ (Cordeiro et al., 2003; Cai et al., 2005) as well as relationships between clonal cultivars of hybrid canes (Pan et al., 2003a). A fingerprint database of major Australian sugarcane cultivars has been developed using these markers (Piperidis et al., 2001) as has molecular genotyping of elite clones produced by the USDA (Pan et al., 2003a, b).
High-throughput SNP genotyping
High-throughput genotyping technologies based on single nucleotide polymorphisms (SNPs) or small-scale insertion/deletions (indel) could become efficient alternative tools for traditional markers because of their greater abundance in the genome and ease of measurement. SNPs are being identified and rapidly mapped to provide a rich source of genetic information with the potential for allowing a greater insight into understanding the genetic complexity of many organisms. SNPs are present in high frequency in any genome, amenable to high throughput analysis and have the ability to reveal hidden polymorphisms where other methods fail (Bhattramakki and Rafalski, 2001). In plants, a number of studies have been able to link SNPs with phenotypic traits of agronomic interest, such as the putative betaine aldehyde dehydrogenase gene responsible for the fragrance trait in rice (Bradbury et al., 2005) and SNPs found in the starch synthase IIa gene associated with starch gelatinization temperature in rice (Waters et al., 2005). These studies highlight the usefulness of SNP markers, demonstrating both the abundance of this marker type and the potential causal association between a single nucleotide alteration and organism phenotype. A further major advantage of SNP markers is that they allow easy and unambiguous identification of alleles or haplotypes.
Whilst numerous technical methods have been developed for their detection (Gut, 2001), the majority are applicable mainly to diploid genomes where a simple presence/absence of either one or both of the alternative bases would indicate homozygosity or heterozygosity. Sugarcane, with its complex genome comprising an estimated 8–14 copies of every chromosome (Rossi et al., 2003; Aitken et al., 2004), can have up to 14 different alleles present, with individual alleles in varying numbers. Thus, the frequency of an SNP base at a gene locus will be determined by both the number of chromosomes carrying the gene, and the number of different alleles (or haplotypes) and frequency of each allele possessing each SNP base. Hence, any method used to detect SNPs at a particular locus in sugarcane must be able to determine the frequency of each SNP base in different genotypes, rather than simply detecting the presence and absence of SNPs. Such detection systems are generally more complex and expensive than simpler and more common methods used for detecting less complex genomes (Ross et al., 1998; Ahmadian et al., 2000; Alderborn et al., 2000; Nurmi et al., 2001; Storm et al., 2003).
Use of SNPs in sugarcane
Currently, whilst there are only a limited number of papers describing the use of SNPs to understand the sugarcane genome, they point to this marker system as a valuable means of mapping candidate genes and for identifying the genetic basis of QTLs of agronomically important traits. These studies include a discussion on the ability of SNPs to: delineate a set of 64 ESTs into two groups that are likely to represent two gene family members of 6-phosphogluconate dehydrogenase (Grivet et al., 2001); delineation of 178 ESTs into three paralogous genes to reveal the expression of an Adh2 and two Adh1 genes in sugarcane (Grivet et al., 2003); the development of co-dominant cleaved amplified polymorphic sequence (CAPS) markers (Quint et al., 2002); and to map several candidate genes and ESTs (McIntyre et al., 2005).
In sugarcane, the proportional frequencies of each SNP base will vary depending on the number of alleles of the gene containing the SNP locus. The ability to capture this information accurately across several SNPs within a set of homo(eo)logous alleles can give an indication of the number of allele haplotypes present for a gene and potentially provide the haplotype sequences. This information could have implications for sugarcane breeding. High yield potential may be due to the presence of, or different number of copies of a specific allele(s) present at a gene locus, or possibly a combination of both. Knowledge of the sequence underlying each allele haplotype has the potential to allow allele-specific markers to be designed.
Quantitative methods to detect allele dosage in sugarcane are now possible with such techniques as pyrophosphate sequencing using the Pyrosequencer™ platform (Cordeiro et al., 2006b) and mass-spectrometry using the Sequenom™ platform (Cordeiro et al., 2006b). These methods have allowed the quantitative detection of frequencies of consensus to alternate SNP bases at any particular SNP locus. Utilizing a group of SNP markers developed to the same EST or gene, it becomes possible to infer the likely copy number of the EST or gene. This information then allows for possible haplotypes of a gene present in hybrid cane to be determined through statistical approaches (Cordeiro et al., 2006b).
In theory, the association of SNP variations with either the presence or absence of different phenotypes among individuals or among individuals from different populations appears straightforward. This simplistic view does not account for the majority of base polymorphisms that do not result in any amino acid change. Determining the haplotypes is more important for predicting individual phenotypes than are the underlying SNPs. Determining haplotypes also allows the ability to infer the evolutionary history of a DNA region (Templeton et al., 1988; Tishkoff et al., 1998). However, difficulties are encountered in determining SNP haplotypes when inbred or homozygous individuals are not available (Rafalski, 2002) as is usually the case with sugarcane.
The ability to determine SNP base frequencies provides the means to determine the likely copy number of homo(eo)logous loci in sugarcane. Where chromosome counts have been performed for a genotype, this information can be used to support the inference of the most likely copy number of homo(eo)logous loci. Knowledge of the number of homo(eo)logous loci will assist in the deduction of the allelic composition of the locus in any particular sugarcane genotype. The ability to determine haplotypes also opens possibilities in unraveling the complexities of the sugarcane genome. By defining haplotypes in parents of crosses, it may be possible to deduce their segregation in progeny; or to determine allele dosage and composition in any particular genotype in relation to phenotypic performance. A further level of analysis is required to determine the level of expression of each of the haplotypes in this complex genome.
Genetic mapping
Genetic maps are widely used in plant breeding to identify genomic regions controlling traits of interest. Such information assists in understanding the genetic basis of the target trait, as well as providing DNA markers for use in marker-assisted breeding. In sugarcane, only markers that are present as a single copy in one parent and absent in the second [i.e. single-dose (SD) marker] can be incorporated into maps using populations of conventional size (approx. 250 progeny) (Wu et al., 1992). In these populations, SD markers segregate in a 1 : 1 ratio.
The first maps of a cultivar were initiated on the selfed progeny of SP70-1006 (D'Hont, 1994). This map was later transferred and further developed on the cultivar R570 (Grivet et al., 1996) using RFLP probes from maize and sugarcane. By 2001, the R570 map, as it had become commonly known, contained some 600 RFLP markers derived from a number of grass (Poaceae) species (D'Hont and Glaszmann, 2001). The markers on this map distribute over 98 cosegregation groups covering a total length of 2008 cM. A parallel mapping effort was also carried out to place 939 single-dose AFLP markers on R570, of which 887 were distributed into 120 cosegregation or linkage groups (Hoarau et al., 2001). A more recent map has been developed on a cross between the Australian commercial variety Q165A (2n = 115) with the S. officinarum clone IJ76-514 (2n = 80) using a combination of AFLP and SSR markers. A total of 967 single dose markers were generated from the two marker systems, and 910 were distributed across 116 linkage groups covering a total map length of 9058·3 cM (Aitken et al., 2005). Markers on these maps have all been generated through anonymous marker systems. However, the use of SNP markers are resulting in ESTs mapped onto the Q165A map.
EcoTILLING for mapping ESTs
Parallel to the development of quantitative SNP frequency scoring methods has been the adaptation of the EcoTILLING method for detecting and mapping sugarcane ESTs. TILLING utilizes the CelI mismatch-cleavage enzyme on heteroduplexed DNA strands with detection of end-labelled cleavage product (McCallum et al., 2000). A variant of this method utilizes natural populations for the discovery of polymorphisms (SNPs, SSRs and indels), and is referred to as EcoTILLING (Comai et al., 2004). Both methods as published rely largely on electrophoretic gels to separate and visualize the products. In sugarcane, this does not allow SNPs that occur on a single allele to be clearly detected. Modifying the protocol and moving the detection system to capillary electrophoresis has allowed the detection of single-dose SNPs in sugarcane to be identified (Cordeiro et al., 2006b) and mapped (McIntyre et al., 2006). Our early experience with family members of the sucrose phosphate synthase gene indicate straightforward detection of the presence of 5–11 SNPs in fragment lengths of genomic DNA between 300 bp and 400 bp in length. Neither prior knowledge of any SNP in the fragment nor the alignment of multiple ESTs are required to identify putative SNPs and their location. Whilst the method is as yet unable to indicate the frequency at which an individual SNP base is present, it has been demonstrated that the detected variation in base composition segregates as expected in progeny of mapping populations. Using the SPS gene family members as an example, the mapping of the gene family members through the EcoTILLING approach supports sequence information that three of the five gene family members may contain more than one gene, with each gene possessing from one to five alleles (McIntyre et al., 2006). This observation will in time allow further unravelling of the complexities of the sugarcane genome.
Sorghum genome information as a resource for sugarcane
Sorghum is the closest cultivated relative of sugarcane. Sugarcane has a large genome that has duplicated at least twice since it diverged from sorghum, around 5 million years ago (Al-Janbi et al., 1997). The extensive similarity in the gene order between these two genomes, where intercrosses are still possible (Ming et al., 1998), makes sorghum the best model crop for the Androponeae tribe (Price et al., 2005a) with the aim of understanding the extensive gene rearrangements and assisting the development of genetic maps in sugarcane.
Sequencing of Sorghum provides another model genome within the grasses, which particularly when utilized in conjunction with rice, will stimulate evolutionary understanding of the entire Poaceae. Sequencing will stimulate gene and allele discovery and crop improvement in Sorghum as it did in rice. Sugarcane genomics will be supported by the Sorghum sequence data. The sequences of Sorghum genes and to a lesser extent the location of genes in the genome should be useful in sugarcane.
Genetic resources for sorghum and sugarcane improvement have been enhanced by the application of genomic tools to analysis of wild relatives in the Sorghum and Saccharum genera. Mutant populations (including TILLING populations) of Sorghum expand the options for gene discovery and genetic manipulation. Protocols for EcoTILLING (Cordeiro et al., 2006a) and quantitative SNP analysis in the complex sugarcane genome should be valuable tools for gene mapping, gene discovery and association genetics in sugarcane. The availability of a Sorghum genome sequence will further accelerate the potential to apply these techniques in both Sorghum and sugarcane. Gene discovery in this germplasm will also be supported by application of advances in expression profiling tools as has been applied to other crop species in the Poaceae (McIntosh et al., 2007).
ACKNOWLEDGEMENTS
We thank the Grain Foods CRC, CRC for Sugarcane Industry Innovation through Biotechnology and the Australian Research Council for their support of this research. Funding to pay the Open Access publication charges for this article was provided by the OECD.
LITERATURE CITED
- Abu Assar AH, Uptmoor R, Abdelmula AA, Salih M, Ordon F, Friedt W. Genetic variation in sorghum germplasm from Sudan, ICRISAT, and USA assessed by simple sequence repeats (SSRs) Crop Science. 2005;45:1636–1644. [Google Scholar]
- Ahmadian A, Gharizadeh B, Gustafsson AC, Sterky F, Nyrén P, Uhlén M, et al. Single-nucleotide polymorphism analysis by pyrosequencing. Analytical Biochemistry. 2000;280:103–110. doi: 10.1006/abio.2000.4493. [DOI] [PubMed] [Google Scholar]
- Aitken K, Jackson P, Piperidis G, McIntyre L. QTL identified for yield components in a cross between a sugarcane (Saccharum spp.) cultivar Q165A and a S. officinarum clone IJ76-514. In: Fischer T, Turner N, Angus J, McIntyre L, Robertson M, Borrell A, et al., editors. Proceedings for the 4th International Crop Science Congress; 26 September to 1 October 2004; Brisbane, Australia. 2004. New directions for a diverse planet. [Google Scholar]
- Aitken KS, Jackson PA, McIntyre CL. A combination of AFLP and SSR markers provides extensive map coverage and identification of homo(eo)logous linkage groups in a sugarcane cultivar. Theoretical and Applied Genetics. 2005;110:789–801. doi: 10.1007/s00122-004-1813-7. [DOI] [PubMed] [Google Scholar]
- Alderborn A, Kristofferson A, Hammerling U. Determination of single-nucleotide polymorphisms by real-time pyrophosphate DNA sequencing. Genome Research. 2000;10:1249–1258. doi: 10.1101/gr.10.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Janbi SM, Honeycutt RJ, Peterson C, Sobral BWS. Phylogenetics analysis of organellar DNA sequences in the Andropogoneae. Saccharum. Theoretical and Applied Genetics. 1997;88:933–944. doi: 10.1007/BF00220799. [DOI] [PubMed] [Google Scholar]
- Bapat DR, Mote UN. Sources of shootfly resistance in. Sorghum. Journal of the Maharashtra Agricultural University. 1982;7:238–240. [Google Scholar]
- Benmoussa M, Suhendra B, Aboubacar A, Hamaker BR. Distinctive sorghum starch granule morphologies appear to improve raw starch digestibility. Starch – Starke. 2006;58:92–99. [Google Scholar]
- Bhattramakki D, Rafalski A. Discovery and application of single nucleotide polymorphism markers in plants. In: Henry RJ, editor. Plant genotyping: the DNA fingerprinting of plants. Wallingford: CAB International; 2001. pp. 179–191. [Google Scholar]
- Bonierbale MW, Plaisted RL, Tanksely SD. RFLP maps based on common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics. 1988;120:1095–1103. doi: 10.1093/genetics/120.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell AK, Douglas ACL. Maintaining green leaf area in grain sorghum increased nitrogen uptake under post-anthesis drought. International Sorghum and Millets Newsletter. 1997;38:89–91. [Google Scholar]
- Borrell AK, Bidinger FR, Sunitha K. Stay-green associated with yield in recombinant inbred sorghum lines varying in rate of leaf senescence. International Sorghum and Millets Newsletter. 1999;40:31–33. [Google Scholar]
- Borrell AK, Hammer GL, Douglas ACL. Does maintaining green leaf area in sorghum improve yield under drought? I. Leaf growth and senescence. Crop Science. 2000;40:1026–1037. [Google Scholar]
- Bowers JE, Abbey C, Anderson S, Chang C, Draye X, Hoppe AH, et al. A high density genetic recombination map of sequence-tagged sites for Sorghum, as a framework for comparative structural and evolutionary genomics of tropical grasses. Genetics. 2003;165:367–386. doi: 10.1093/genetics/165.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury LMT, Fitzgerald TL, Henry RJ, Jin QS, Waters DLE. The gene for fragrance in rice. Plant Biotechnology Journal. 2005;3:363–370. doi: 10.1111/j.1467-7652.2005.00131.x. [DOI] [PubMed] [Google Scholar]
- Brown SM, Hopkins MS, Mitchell SE, Senior ML, Wang TY, Duncan RR, et al. Multiple methods for the identification of polymorphic simple sequence repeats (SSRs) in sorghum [Sorghum bicolor (L.) Moench] Theoretical and Applied Genetics. 1996;93:190–198. doi: 10.1007/BF00225745. [DOI] [PubMed] [Google Scholar]
- Bull TA, Glasziou KT. The evolutionary significance of sugar accumulation in. Saccharum. Australian Journal of Biological Science. 1963;16:737–742. [Google Scholar]
- Buzacott JH. Cane varieties and breeding. In: Kim NJ, Mungomery RW, Hughes CG, editors. Manual of cane growing. Sydney: Angus and Robertson; 1965. pp. 220–253. [Google Scholar]
- Cai Q, Aitken K, Deng HH, Chen XW, Fu C, Jackson PA, et al. Verification of the introgression of Erianthus arundinaceus germplasm into sugarcane using molecular markers. Plant Breeding. 2005;124:322–328. [Google Scholar]
- Caldwell DG, McCallum N, Shaw P, Muehlbauer GJ, Marshall DF, Waugh R. A structured mutant population for forward and reverse genetics in barley (Hordeum vulgare L.) The Plant Journal. 2004;4:143–150. doi: 10.1111/j.1365-313X.2004.02190.x. [DOI] [PubMed] [Google Scholar]
- Carson DL, Botha FC. Preliminary analysis of expressed sequence tags for sugarcane. Crop Science. 2000;40:1769–1779. [Google Scholar]
- Casu RE, Dimmock CM, Chapman SC, Grof CPL, McIntyre CL, Bonnett GD, et al. Identification of differentially expressed transcripts from maturing stem of sugarcane by in silico analysis of stem expressed sequence tags and gene expression profiling. Plant Molecular Biology. 2004;54:503–517. doi: 10.1023/B:PLAN.0000038255.96128.41. [DOI] [PubMed] [Google Scholar]
- Chen X, Cho YG, McCouch SR. Sequence divergence of rice microsatellites in Oryza and other plant species. Molecular Genetics and Genomics. 2002;268:331–343. doi: 10.1007/s00438-002-0739-5. [DOI] [PubMed] [Google Scholar]
- Clayton WD, Renvoize SA. Kew Bulletin Addition Series. XIII. London: Royal Botanic Gardens, Kew; 1986. Genera Graminum grasses of the world; pp. 338–345. [Google Scholar]
- Comai L, Henikoff S. TILLING: practical single-nucleotide mutation discovery. The Plant Journal. 2006;45:684–694. doi: 10.1111/j.1365-313X.2006.02670.x. [DOI] [PubMed] [Google Scholar]
- Comai L, Young K, Till BJ, Reynolds SH, Greene EA, Codomo CA, et al. Efficient discovery of DNA polymorphisms in natural populations by EcoTILLING. The Plant Journal. 2004;37:778–786. doi: 10.1111/j.0960-7412.2003.01999.x. [DOI] [PubMed] [Google Scholar]
- Cordeiro GM, Taylor GO, Henry RJ. Characterisation of microsatellite markers from sugarcane (Saccharum sp.), a highly polyploid species. Plant Science. 2000;155:161–168. doi: 10.1016/s0168-9452(00)00208-9. [DOI] [PubMed] [Google Scholar]
- Cordeiro GM, Pan YB, Henry RJ. Sugarcane microsatellites for the assessment of genetic diversity in sugarcane germplasm. Plant Science. 2003;165:181–189. [Google Scholar]
- Cordeiro GM, Eliott F, McIntyre CL, Casu RE, Henry RJ. Characterisation of single nucleotide polymorphisms in sugarcane ESTs. Theoretical and Applied Genetics. (a) 2006;113:331–343. doi: 10.1007/s00122-006-0300-8. [DOI] [PubMed] [Google Scholar]
- Cordeiro G, Eliott F, Henry R. An Optimised Ecotilling protocol for polyploids or pooled samples using a capillary electrophoresis system. Analytical Biochemistry. (b) 2006;355:145–147. doi: 10.1016/j.ab.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Cross M, Lee SL, Henry RJ. A novel detection strategy for scanning multiple mutations using CEL I; Paper presented at the Plant and Animal Genomes Conference XV, Mutation Screening Workshop; 13–17; San Diego, USA: 2007. Jan, 2007. [Google Scholar]
- Daniels J, Roach BT. Taxonomy and evolution in sugarcane. In: Heinz DJ, editor. Sugarcane improvement through breeding. Amsterdam: Elsevier Press; 1987. pp. 7–84. [Google Scholar]
- Davies H, Dicks E, Stephens P, Cox C, Teague J, Greenman C, et al. High throughput DNA sequence variant detection by conformation sensitive capillary electrophoresis and automated peak comparison. Genomics. 2006;87:427–432. doi: 10.1016/j.ygeno.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Delseny M, Salses J, Cooke R, Sallaud C, Regad F, Lagoda P, et al. Rice genomics: present and future. Plant Physiology and Biochemistry. 2001;39:323–334. [Google Scholar]
- Deu M, Rattunde F, Chantereau J. A global view of genetic diversity in cultivated sorghums using a core collection. Genome. 2006;49:168–180. doi: 10.1139/g05-092. [DOI] [PubMed] [Google Scholar]
- D'Hont A. A molecular approach to unravelling the genetics of sugarcane, a complex polyploid of the Andropogoneae tribe. Genome. 1994;37:222–230. doi: 10.1139/g94-031. [DOI] [PubMed] [Google Scholar]
- D'Hont A, Glaszmann JC. Sugarcane genome analysis with molecular markers: a first decade of research. In: Hogarth DM, editor. Proceedings of the International Society of Sugarcane Technologists XXVI Congress; Brisbane: The Australian Society of Sugar Cane Technologists; 2001. pp. 556–559. [Google Scholar]
- D'Hont A, Layssac M. 1998. Analysis of cultivars genome structure by molecular cytogenetics and the study of introgression mechanisms. France: Centre de coopération internationale en recherche agronomique pour le développement, Annual Crops Department (CIRAD-CA) pp. 9–10. [Google Scholar]
- Dillon SL, Lawrence PK, Henry RJ. The new use of Sorghum bicolor derived SSR markers to evaluate genetic diversity in seventeen Australian Sorghum species. Plant Genetic Resources. 2005;3:19–28. [Google Scholar]
- Dillon SL, Lawrence PK, Henry RJ, Price HJ. Sorghum resolved as a distinct genus based on combined ITS1, ndhF and Adh1 analyses. Plant Systematics and Evolution. 2007 (in press) [Google Scholar]
- Doggett H. Sorghum. London, New York: Longman; published by Wiley; 1970. [Google Scholar]
- Doggett H. Sorghum. 2nd edn. London, New York: Longman; published by Wiley; 1988. [Google Scholar]
- Duodo K, Taylor J, Belton P, Hamaker B. Factors affecting sorghum protein digestibility. Journal of Cereal Science. 2003;38:117–131. [Google Scholar]
- Edme SJ, Miller JD, Glaz B, Tai PY-P, Comstock JC. Genetic contribution to yield in the Florida sugarcane industry accross 33 years. Crop Science. 2005;45:92–97. [Google Scholar]
- Ellis RH, Qi A, Craufurd PQ, Summerfield RJ, Roberts EH. Effects of photoperiod, temperature and asynchrony between thermoperiod and photoperiod on development to panicle initiation in. Sorghum. Annals of Botany. 1997;79:169–178. [Google Scholar]
- Fannon J, Gray J, Gunawan N, Huber K, BeMiller J. The channels of starch granules. Food Science and Biotechnology. 2003;12:700–704. [Google Scholar]
- Fannon J, Gray J, Gunawan N, Huber K, BeMiller J. Heterogeneity of starch granules and the effect of granule channelization on starch modification. Cellulose. 2004;11:247–254. [Google Scholar]
- Feltus FA, Hart GE, Schertz KF, Casa AM, Kresovich S, Abraham S, et al. Alignment of genetic maps and QTLs between inter- and intra-specific sorghum populations. Theoretical and Applied Genetics. 2006;112:1295–1305. doi: 10.1007/s00122-006-0232-3. [DOI] [PubMed] [Google Scholar]
- Franzmann BA, Hardy AT. Testing the host status of Australian indigenous sorghums for the sorghum midge. In: Foale MA, Henzell RG, Kneip JF, editors. Proceedings of the Third Australian Sorghum Conference; Tamworth, NSW, Australia. 1996. pp. 365–367. [Google Scholar]
- Garber ED. Cytotaxonomic studies in the genus Sorghum. University of California Publications in Botany. 1950;23:283–361. [Google Scholar]
- González-Martínez SC, Robledo-Arnuncio JJ, Collada C, Díaz A, Williams CG, Alía R, et al. Cross-amplification and sequence variation of microsatellite loci in Eurasian hard pines. Theoretical and Applied Genetics. 2004;109:103–111. doi: 10.1007/s00122-004-1596-x. [DOI] [PubMed] [Google Scholar]
- Grivet L, D'Hont A, Roques D, Feldmann P, Lanaud C, Glaszmann J-C. RFLP mapping in a highly polyploid and aneuploid interspecific hybrid. Genetics. 1996;142:987–1000. doi: 10.1093/genetics/142.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivet L, Glaszmann JC, Arruda P. Sequence polymorphism from EST data in sugarcane: a fine analysis of 6-phosphogluconate dehydrogenase genes. Genetics and Molecular Biology. 2001;24:161–167. [Google Scholar]
- Grivet L, Glaszmann JC, Vincentz M, da Silva F, Arruda P. ESTs as a source for sequence polymorphism discovery in sugarcane: example of the Adh genes. Theoretical and Applied Genetics. 2003;106:190–197. doi: 10.1007/s00122-002-1075-1. [DOI] [PubMed] [Google Scholar]
- Gut IG. Automation in genotyping of single nucleotide polymorphisms. Human Mutation. 2001;17:475–492. doi: 10.1002/humu.1131. [DOI] [PubMed] [Google Scholar]
- Harlan JR, de Wet JMJ. Toward a rational classification of cultivated sorghums. Crop Science. 1971;12:172–176. [Google Scholar]
- Harlan JR, de Wet JMJ. A simplified classification of cultivated plants. Taxon. 1972;20:509–517. [Google Scholar]
- van Harten AM. Mutation breeding theory and applications. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- Henikoff S, Comai L. Single-nucleotide mutations for plant functional genomics. Annual Review of Plant Biology. 2003;54:375–401. doi: 10.1146/annurev.arplant.54.031902.135009. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Till BJ, Comai L. TILLING: traditional mutagenesis meets functional genomics. Plant Physiology. 2004;135:630–636. doi: 10.1104/pp.104.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández P, Dorado G, Laurie DA, Martin A, Snape JW. Microsatellites and RFLP probes from maize are efficient sources of molecular markers for the biomass energy crop. Miscanthus. Theoretical and Applied Genetics. 2001;102:616–622. [Google Scholar]
- Hoarau JY, Offmann B, D'Hont A, Risterucci AM, Roques D, Glaszmann JC, et al. Genetic dissection of a modern sugarcane cultivar (Saccharum spp.). I. Genome mapping with AFLP markers. Theoretical and Applied Genetics. 2001;103:84–97. [Google Scholar]
- Hoarau JY, Grivet L, Offmann B, Raboin LM, Diorflar JP, Payet J, et al. Genetic dissection of a modern sugarcane cultivar (Saccharum spp.). II. Detection of QTLs for yield components. Theoretical and Applied Genetics. 2002;105:1027–1037. doi: 10.1007/s00122-002-1047-5. [DOI] [PubMed] [Google Scholar]
- Hodnett GL, Burson BL, Rooney WL, Dillon SL, Price HJ. Pollen–pistil interactions result in reproductive isolation between Sorghum bicolor and divergent Sorghum species. Crop Science. 2005;45:1403–1409. [Google Scholar]
- House LR. A guide to sorghum breeding. 2nd edn. Patancheru, India: International Crops Research Institute for the Semi-Arid tropics; 1985. p. 206. [Google Scholar]
- House LR, Osmanzai M, Gomez MI, Monyo ES, Gupta SC. Agronomic principles. In: Dendy DAV, editor. Sorghum and millets, chemistry and technology. St Paul, MN: American Association of Cereal Chemists; 1995. pp. 27–67. [Google Scholar]
- Jordan DR, Tao YZ, Godwin ID, Henzell RG, Cooper M, McIntyre CL. Loss of genetic diversity associated with selection for resistance to sorghum midge in Australian sorghum. Euphytica. 1998;102:1–7. [Google Scholar]
- Kamala V, Singh SD, Bramel PJ, Rao DM. Sources of resistance to downy mildew in wild and weedy sorghums. Crop Science. 2002;42:1357–1360. [Google Scholar]
- Kan CCPF, Doherty EAS, Barron AE. DNA sequencing and genotyping in miniaturized electrophoresis systems. Electrophoresis. 2004;25:3564–3588. doi: 10.1002/elps.200406161. [DOI] [PubMed] [Google Scholar]
- Karunakar RI, Narayana YD, Pande S, Mughogho LK, Singh SD. Evaluation of wild and weedy sorghums for downy mildew resistance. International Sorghum and Millets Newsletter. 1994;35:104–106. [Google Scholar]
- Kayode APP, Linnemann AR, Nout MJR, Hounhouigan JD, Stomph TJ, Smulders MJM. Diversity and food quality properties of farmers' varieties of sorghum from Benin. Journal of the Science of Food and Agriculture. 2006;86:1032–1039. [Google Scholar]
- Komolong B, Chakraborty S, Ryley M, Yates D. Identity and genetic diversity of the sorghum ergot pathogen in Australia. Australian Journal of Agricultural Research. 2002;53:621–628. [Google Scholar]
- Kong L, Dong J, Hart GE. Characteristics, linkage-map positions, and allelic differentiation of Sorghum bicolor (L.) Moench DNA simple-sequence repeats (SSRs) Theoretical and Applied Genetics. 2000;101:438–448. [Google Scholar]
- Kresovich S, Barbazuk B, Bedell JA, Borrell A, Buell CR, Burke J, et al. Toward sequencing the sorghum genome. A US National Science Foundation-sponsored Workshop Report. Plant Physiology. 2005;138:1898–1902. doi: 10.1104/pp.105.065136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman LC, Burson BL, Klein PE, Stelly DM, Rooney WL. Interspecific sorghum breeding using S. macrospermum. Proceedings of the ASA-CSSA-SSA 2006 International Meetings; Indianapolis. 2006. Nov, pp. 12–16. 2006. [Google Scholar]
- Laurie D, Bennett MD. Genetic variation in Sorghum for the inhibition of maize pollen tube growth. Annals of Botany. 1989;64:675–681. [Google Scholar]
- Lazarides M, Hacker JB, Andrew MH. Taxonomy, cytology and ecology of indigenous Australian sorghums (Sorghum Moench: Andropogoneae: Poaceae) Australian Systematic Botany. 1991;4:591–635. [Google Scholar]
- Lebot V. Biomolecular evidence for plant domestication in Sahul. Genetic Resources and Crop Evolution. 1999;46:619–628. [Google Scholar]
- Lindeboom N, Chang PR, Tyler RT. Analytical, biochemical and physicochemical aspects of starch granule size, with emphasis on small granule starches: a review. Starch – Starke. 2004;56:89–99. [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S. Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiology. 2000;123:439–442. doi: 10.1104/pp.123.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh SR, Watson L, Bundock PC, Crawford AC, White J, Cordeiro GM, et al. SAGE of the most abundant transcripts in the developing wheat Caryopsis. Plant Biotechnology Journal. 2007;5:69–83. doi: 10.1111/j.1467-7652.2006.00218.x. [DOI] [PubMed] [Google Scholar]
- McIntyre CL, Casu RE, Drenth J, Knight D, Whan VA, Croft BJ, et al. Resistance gene analogues in sugarcane and sorghum and their association with quantitative trait loci for rust resistance. Genome. 2005;48:391–400. doi: 10.1139/g05-006. [DOI] [PubMed] [Google Scholar]
- McIntyre CL, Jackson M, Cordeiro G, Amouyal O, Eliott F, Henry RJ, et al. The identification and characterisation of alleles of sucrose phosphate synthase gene family III in sugarcane. Molecular Breeding. 2006;18:39–50. [Google Scholar]
- Ma HM, Schulze S, Lee S, Yang M, Mirkov E, Irvine J, et al. An EST survey of the sugarcane transcriptome. Theoretical and Applied Genetics. 2004;108:851–863. doi: 10.1007/s00122-003-1510-y. [DOI] [PubMed] [Google Scholar]
- Mariotti JA. 2002. Mar–Apr. Selection for sugar cane yield and quality components in subtropical climates. Sugar Cane International; pp. 22–26. [Google Scholar]
- Miller FR. The breeding of sorghum. Texas Agricultural Experiment Station. 1980;1451:128–136. [Google Scholar]
- Miller FR. Genetic and environmental response characteristics of sorghum. Sorghum in the Eighties, Proceedings of International Symposium on Sorghum; ; 2–7 November 1981. Patancheru, India. ICRISAT; 1982. pp. 393–402. [Google Scholar]
- Ming R, Liu SC, Lin YR, da Silva J, Wilson W, Braga D, et al. Detailed alignment of Saccharum and Sorghum chromosomes: comparative organization of closely related diploid and polyploid genomes. Genetics. 1998;150:1663–1682. doi: 10.1093/genetics/150.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Liu S-C, Moore PH, Irvine JE, Paterson AH. QTL analysis in a complex autopolyploid: genetic control of sugar content in sugarcane. Genome Research. 2001;11:2075–2084. doi: 10.1101/gr.198801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Del Monte TA, Hernandez E, Moore PH, Irvine JE, Paterson AH. Comparative analysis of QTLs affecting plant height and flowering among closely-related diploid and polyploid genomes. Genome. 2002;45:794–803. doi: 10.1139/g02-042. [DOI] [PubMed] [Google Scholar]
- Mullet JE, Klein RR, Klein PE. Sorghum bicolor — an important species for comparative grass genomics and a source of beneficial genes for agriculture. Current Opinion in Plant Biology. 2002;5:118–121. doi: 10.1016/s1369-5266(02)00232-7. [DOI] [PubMed] [Google Scholar]
- Nurmi J, Kiviniemi M, Kujanpaa M, Sjoroos M, Ilonen J, Lovgren T. High-throughput genetic analysis using time-resolved fluorometry and closed-tube detection. Analytical Biochemistry. 2001;299:211–217. doi: 10.1006/abio.2001.5434. [DOI] [PubMed] [Google Scholar]
- Pan Y-B, Cordeiro GM, Richard EP, Henry RJ. Molecular genotyping of sugarcane clones with microsatellite DNA markers. Maydica. (a) 2003;48:319–329. [Google Scholar]
- Pan Y-B, Miller JD, Schnell RJ, Richard J,RP, Wei Q. Application of microsatellite and RAPD fingerprints in the Florida sugarcane variety program. Sugar Cane International. (b) 2003;2003:19–28. [Google Scholar]
- Pan YB, Burner DM, Wei Q, Cordeiro GM, Legendre BL, Henry RJ. New Saccharum hybrids in S. spontaneum cytoplasm developed through a combination of conventional and molecular breeding approaches. Plant Genetic Resources. 2004;2:131–139. [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proceedings of the National Academy of Sciences of the USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Gilmore S, Keys W, Morgante M, Rafalski A. Cross-species amplification of soybean (Glycine max) simple sequence repeats (SSRs) within the genus and other legume genera: implications for the transferability of SSRs in plants. Molecular Biology and Evolution. 1998;15:1275–1287. doi: 10.1093/oxfordjournals.molbev.a025856. [DOI] [PubMed] [Google Scholar]
- Piperidis G, Taylor GO, Smith GR. A microsatellite marker database for fingerprinting sugarcane clones. XXIV Proceedings of the International Society of Sugar Cane Technologists; Mackay: Australian Society of Sugar Cane Technologists; 2001. pp. 632–633. [Google Scholar]
- Price HJ, Dillon SL, Hadnett G, Rooney WL, Ross L, Johnston JS. Genome evolution in the genus Sorghum (Poaceae) Annals of Botany. (a) 2005;95:219–227. doi: 10.1093/aob/mci015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price HJ, Hodnett GL, Burson BL, Dillon SL, Rooney WL. A Sorghum bicolor × S. macrospermum hybrid recovered by embryo rescue and culture. Australian Journal of Botany. (b) 2005;53:579–582. [Google Scholar]
- Price HJ, Hodnett GL, Burson BL, Dillon SL, Stelly DM, Rooney WL. Genome dependent interspecific hybridisation of Sorghum bicolor (Poaceae) Crop Science. 2006;46:2617–2622. [Google Scholar]
- Quint M, Mihaljevic R, Dussle CM, Xu ML, Melchinger AE, Lubberstedt T. Development of RGA-CAPS markers and genetic mapping of candidate genes for sugarcane mosaic virus resistance in maize. Theoretical and Applied Genetics. 2002;105:355–363. doi: 10.1007/s00122-002-0953-x. [DOI] [PubMed] [Google Scholar]
- Rai KN, Murty DS, Andrews DJ, Bramel-Cox PJ. Genetic enhancement of pearl millet and sorghum for the semi-arid tropics of Asia and Africa. Genome. 1999;42:617–628. [Google Scholar]
- Rafalski A. Applications of single nucleotide polymorphisms in crop genetics. Current Opinion in Plant Biology. 2002;5:94–100. doi: 10.1016/s1369-5266(02)00240-6. [DOI] [PubMed] [Google Scholar]
- Ramsay L, Macaulay M, degli Ivanissevich S, MacLean K, Cardle L, Fuller J, et al. A simple sequence repeat-based linkage map of barley. Genetics. 2000;156:1997–2005. doi: 10.1093/genetics/156.4.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao NPG, Rana BS. Selection in temperate and tropical crosses of sorghum. Sorghum in the Eighties, Proceedings of International Symposium on Sorghum; 2–7; Patancheru, India. ICRISAT; 1982. Nov, pp. 403–420. 1981. [Google Scholar]
- Reddy BBS, Ramesh S, Reddy PS. Sorghum genetic resources, cytogenetics and improvement. In: Singh RJ, Jauhar PP, editors. Cereals. Vol. 2. Boca Raton, FL: CRC Taylor and Francis; 2006. pp. 309–363. Genetic resources, chromosome engineering, and crop improvement. [Google Scholar]
- Roach BT. Origin and improvement of the genetic base of sugarcane. Proceedings of the Australian Society of Sugar Cane Technology, 34–47. 1989 [Google Scholar]
- Rooney WL, Smith CW. Techniques for developing new cultivars. In: Smith CW, Frederiksen RA, editors. New York, NY: John Wiley & Sons; 2000. pp. 329–347. Sorghum, origin, history, technology and production. [Google Scholar]
- Rosenow DT, Clark LE. Drought and lodging resistance for a quality sorghum crop. Proceedings of the Fiftieth Annual Corn and Sorghum Industry Research Conference; 6–7 December; Chicago, IL. 1995. pp. 82–97. [Google Scholar]
- Rosenow DT, Dahlberg JA. Collection, conversion and utilisation of sorghum. In: Smith CW, Frederiksen RA, editors. New York, NY: John Wiley, Sons; 2000. pp. 309–328. Sorghum, origin, history, technology and production. [Google Scholar]
- Ross P, Hall L, Smirnow I, Haff L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nature Biotechnology. 1998;16:1347–1351. doi: 10.1038/4328. [DOI] [PubMed] [Google Scholar]
- Rossi M, Araujo PG, Paulet F, Garsmeur O, Dias VM, Chen H, et al. Genomic distribution and characterization of EST-derived resistance gene analogs (RGAs) in sugarcane. Molecular Genetics and Genomics. 2003;269:406–419. doi: 10.1007/s00438-003-0849-8. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Shepherd M, Henry RJ. Characterization of highly conserved microsatellite loci in Araucaria cunninghamii and related species. Plant Systematics and Evolution. 2003;236:115–123. [Google Scholar]
- Serna-Saldivar S, Rooney L. Structure and chemistry of sorghum and millets. In: Dendy DAV, editor. St Paul, MN: American Association of Cereal Chemists; 1995. pp. 69–124. Sorghum and millets, chemistry and technology. [Google Scholar]
- Shapter FM, Lee LS, Henry R. Endosperm and starch granule morphology in wild cereal relatives. Plant Genetic Resources. 2007 (in press) [Google Scholar]
- Sharma HC. Host-plant resistance to insects in sorghum and its role in integrated pest management. Crop Protection. 1993;12:11–34. [Google Scholar]
- Sharma HC, Franzmann BA. Host-plant preference and oviposition responses of the sorghum midge, Stenodiplosis sorghicola (Coquillett) (Dipt., Cecidomyiidae) towards wild relatives of sorghum. Journal of Applied Entomology. 2001;125:109–114. [Google Scholar]
- Sills G, Bridges W, Al-Janabi S, Sobral BWS. Genetic analysis of agronomic traits in a cross between sugarcane (Saccharum officinarum L.) and its presumed progenitor (S. robustum Brandes, Jesw. Ex. Grassl) Molecular Breeding. 1995;1:355–363. [Google Scholar]
- Slade AJ, Fuerstenberg SI, Loeffler D, Steine MN, Facciotti D. A reverse genetic, non-transgenic approach to wheat crop improvement by TILLING. Nature Biotechnology. 2005;23:75–81. doi: 10.1038/nbt1043. [DOI] [PubMed] [Google Scholar]
- Smith CW, Frederiksen RA. Sorghum: origin, history, technology, and production. New York, NY: John Wiley and Sons; 2000. p. 824. [Google Scholar]
- Spangler RE. Taxonomy of Sarga, Sorghumand Vacoparis (Poaceae: Andropogoneae) Australian Systematic Botany. 2003;16:279–299. [Google Scholar]
- Sreenivasan TV, Ahloowalia BS, Heinz DJ. Cytogenetics. In: Heinz DJ, editor. Amsterdam: Elsevier; 1987. pp. 211–253. Sugarcane improvement through breeding. [Google Scholar]
- Storm N, Darnhofer-Patel B, van den Boom D, Rodi CP. MALDI-TOF mass spectrometry-based SNP genotyping. Methods in Molecular Biology. 2003;212:241–262. doi: 10.1385/1-59259-327-5:241. [DOI] [PubMed] [Google Scholar]
- Sudupak MA. Inter- and intra-species inter simple sequence repeat (ISSR) variations in the genus. Cicer. Euphytica. 2004;135:229–238. [Google Scholar]
- Szantai E, Ronai Z, Szilagyi A, Sasvari-Szekely M, Guttman A. Haplotyping by capillary electrophoresis. Journal of Chromatography. 2005;1079:41–49. doi: 10.1016/j.chroma.2005.03.078. [DOI] [PubMed] [Google Scholar]
- Tang T, Huang JZ, Zhong Y, Shi SH. High-throughput S-SAP by fluorescent multiplex PCR and capillary electrophoresis in plants. Journal of Biotechnology. 2004;114:59–68. doi: 10.1016/j.jbiotec.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Tao YZ, Hardy A, Drenth J, Henzell RG, Franzmann BA, Jordan DR, et al. Identifications of two different mechanisms for sorghum midge resistance through QTL mapping. Theoretical and Applied Genetics. 2003;107:116–122. doi: 10.1007/s00122-003-1217-0. [DOI] [PubMed] [Google Scholar]
- Taramino G, Tarchini R, Ferrario S, Lee M, Pé ME. Characterisation and mapping of simple sequence repeats (SSRs) in. Sorghum bicolor. Theoretical and Applied Genetics. 1997;95:66–72. [Google Scholar]
- Templeton AR, Sing CF, Kessling A, Humphries S. A cladistic analysis of phenotype associations with haplotypes inferred from restriction endonuclease mapping. II. The analysis of natural populations. Genetics. 1988;120:1145–1154. doi: 10.1093/genetics/120.4.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesso T, Ejeta G, Chandrashekar A, Huang CP, Tandjung A, Lewamy M, et al. A novel modified endosperm texture in a mutant high-protein digestibility/high-lysine grain sorghum (Sorghum bicolor (L.) Moench) Cereal Chemistry. 2006;83:194–201. [Google Scholar]
- Tew TL, White WH, Grisham MP, Dufrene EO, Jr, Garrison DD, Veremis JC, et al. Registration of ‘HoCP 13 96–540’ Sugarcane. Crop Science. 2003;45:785–786. [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Research. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till BJ, Reynolds SH, Weil C, Springer N, Burtner C, Young K, et al. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biology. 2004;4:12. doi: 10.1186/1471-2229-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Goldman A, Calafell F, Speed WC, Deinard AS, Bonne-Tamir B, et al. A global haplotype analysis of the mytonic dystrophy locus: implications for the evolution of modern humans and for the origin of myotonic dystrophy mutations. American Journal of Human Genetics. 1998;62:1389–1402. doi: 10.1086/301861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA ARS. Germplasm Resources Information Network (GRIN) Beltsville, MD: National Germplasm Resources Laboratory. URL; 2007. National Genetic Resources Program. [Online Database] http://www.ars-grin.gov/cgi-bin/npgs/html/tax_search.pl. (17 May 2007) [Google Scholar]
- Vettore AL, da Silva FR, Kemper EL, Arruda P. The libraries that made SUCEST. Genetic Molecular Biology. 2001;24:1–7. [Google Scholar]
- Vouk K, Gazvoda B, Komel R. Fluorescent multiplex PCR and capillary electrophoresis for analyisis of PKD1 and PKD2 associated microsatellite markers. BioTechniques. 2000;29:1187–1190. doi: 10.2144/00296bm06. [DOI] [PubMed] [Google Scholar]
- Waters DLE, Henry RJ, Reinke RF, Fitzgerald MA. Gelatinisation temperature of rice explained by polymorphisms in starch synthase. Plant Biotechnology Journal. 2005;4:115–122. doi: 10.1111/j.1467-7652.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JL, Wu CJ, Lei CL, Baraoidan M, Bordeos A, Madamba MRS, et al. Sorghum diversity evaluated by simple sequence repeat (SSR) markers and phenotypic performance. Plant Production Science. 2004;7:301–308. [Google Scholar]
- Wu JL, Wu CJ, Lei CL, Baraoidan M, Bordeos A, Madamba MRS, et al. Chemical- and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Molecular Biology. 2005;59:85–97. doi: 10.1007/s11103-004-5112-0. [DOI] [PubMed] [Google Scholar]
- Wu K, Burnquist W, Sorrels M, Tew TL, Moore P, Tanksley S. The detection and estimation of linkage in polyploids using single-dose restriction fragments. Theoretical and Applied Genetics. 1992;83:294–300. doi: 10.1007/BF00224274. [DOI] [PubMed] [Google Scholar]