Abstract
Background and Aims
The Alpine Meadow Grass Poa alpina is common in subalpine and alpine natural sites and agriculturally used land, where it is an important fodder grass. Natural factors and human land use are supposed to have been shaping its genetic diversity for hundreds of years. The species comprises sexually and vegetatively reproducing plants. The aim of this study was to investigate the effects of agricultural land use, environmental factors and the mode of reproduction on the distribution of its microsatellite diversity within and among populations and to analyse whether its genetic diversity is correlated with plant species diversity in grassland parcels.
Methods
Genetic diversity of P. alpina was assessed with five microsatellite markers for 569 plants originating from 20 natural sites and from 54 grassland parcels of different cultural tradition, land use and altitude in the Swiss Alps. Due to polyploidy and frequent aneuploidy of the species, data analyses were based on the presence of microsatellite bands.
Key Results
A low but significant differentiation was found in microsatellite bands among natural sites and agriculturally used parcels, while their microsatellite band diversity within populations did not differ. An increased differentiation was found in microsatellite bands with increasing geographic distance among parcels, and a differentiation among grazed and mown parcels, and among sexually and vegetatively reproducing populations. Band richness of sampled plants per village was higher for villages where parcels represented more different land-use types. Within populations, microsatellite band diversity was higher in grazed than in mown parcels.
Conclusions
The diversity of human land use in the Alps was associated with genetic diversity of P. alpina. Therefore, the ongoing socio-economically motivated land-use changes, which reduce the number of different land-use types, will affect the genetic diversity of P. alpina negatively.
Key words: Agriculture, cultural tradition, genetic diversity, grassland, land use, microsatellites, natural environment, Poa alpina, rarefaction, Swiss Alps
INTRODUCTION
Genetic diversity within and among populations is shaped by the balance between genetic drift, inbreeding, recombination, gene flow, mutation and selection (Loveless and Hamrick, 1984; Hartl and Clark, 1997). This balance depends on important life history traits of a plant species, such as the mode of reproduction or life form (Loveless and Hamrick, 1984; Hamrick and Godt, 1997; Godt and Hamrick, 1998). Moreover, both natural and anthropogenic factors are important for shaping genetic diversity. Potential natural determinants of genetic diversity include abiotic parameters, such as altitude or soil conditions. Furthermore, genetic diversity may be affected by the diversity of the surrounding community. Higher plant species richness was suggested to increase genetic diversity, if it increases the diversity of available niches (Odat et al., 2004; Vellend and Geber, 2005). A potential anthropogenic determinant of genetic diversity is land-use diversity, if different land management creates genetic differentiation among populations. Genetic diversity and its determinants were studied for a common and important fodder plant which occurs over a large altitudinal range at natural sites and in agriculturally used grassland, the Alpine Meadow grass Poa alpina, in the Swiss Alps.
The species can reproduce sexually via seeds and vegetatively by producing bulbils. In an accompanying common garden study, the proportion of genotypes reproducing vegetatively via bulbils was higher among samples from higher altitudes (Weyand, 2005), in line with the hypothesis of an adaptive advantage of vegetative reproduction in the harsher conditions at higher altitudes (Bauert, 1993; Pluess and Stöcklin, 2005; Weppler and Stöcklin, 2005). The presence of two different reproductive modes may affect population differentiation. Furthermore, as P. alpina occurs across a wide geographical range, isolation by distance is likely to have shaped the distribution of genetic diversity among populations of different regions (Wright, 1943). Genetic diversity within and differentiation among populations of P. alpina are probably also enhanced due to the highly variable polyploidy and frequent aneuploidy within the species (Duckert-Henriod and Favarger, 1987), which presumably restricts gene flow among individuals and populations and is likely to increase the ecological amplitude of the species (Briggs and Walters, 1997; Brochmann et al., 2004; Soltis et al., 2004).
For about 5000 years, the Alpine landscapes and in particular their grasslands have been shaped by human land use (Bätzing, 2003). In the European Alps, P. alpina is one of the most important fodder grasses for cattle (Conert, 1998). Therefore, P. alpina has been under agricultural selection pressure for hundreds of years. The species showed adaptation to anthropogenic land-use variation in a common garden experiment (Weyand, 2005), where plants from pastures allocated more biomass to reproduction than plants from natural sites, while plants from meadows allocated less biomass to reproduction than plants from natural sites. This suggests divergent selection between parcels of different land use. Higher allocation to reproduction in pastures may affect genetic diversity of P. alpina if it is related to higher seedling establishment. Genetic diversity in pastures could also be affected by the spatially more heterogeneous conditions created by grazing animals, offering more different niches. In the Alps, the relationship between land use and genetic diversity within a species is of particular interest, as due to land-use changes during the last decades many meadows have been converted to pastures (Bätzing, 2003) and the diversity of land-use types in the landscape has decreased.
In the Swiss Alps, the cultural traditions Romanic, Germanic and Walser contributed to a high landscape diversity through their different agricultural practices (Bätzing, 2003). If differences in land use lead to genetic divergence between populations, villages with higher land-use diversity may harbour higher genetic diversity of P. alpina than villages with lower land-use diversity.
The effects of natural factors and agricultural land use on genetic diversity of P. alpina within and among 12 villages in the Swiss Alps were studied. Each of the three cultural traditions, Romanic, Germanic and Walser, was represented by four villages. At the parcel level, genetic diversity was studied within and among populations from 20 natural sites and from 54 agriculturally used grasslands at different altitudes in these 12 villages. The agriculturally used parcels were either mown or grazed, and they were either additionally fertilized or unfertilized. Plant species diversity was known for all parcels from a previous study (Maurer et al., 2006).
As molecular markers, five polymorphic microsatellite loci were used (Maurer et al., 2005). Microsatellites offer high resolution of genetic diversity (Schlötterer, 1998). Therefore, they are ideal to investigate gene flow and genetic drift. Natural selection is unlikely to act on the investigated microsatellite loci themselves, but could affect their diversity if they were linked to loci under selection (Hartl and Clark, 1997; Merilä and Crnokrak, 2001).
The following questions were asked. (a) Are P. alpina populations from agriculturally used grasslands genetically differentiated from natural populations? (b) Is genetic differentiation among villages and among populations related to geographical distances, to differences in land use and to differences in reproductive modes? (c) Is genetic diversity within villages related to cultural traditions and to land-use diversity? (d) Is genetic diversity within grassland parcels related to altitude, land use and reproductive mode?
MATERIALS AND METHODS
Study species
Poa alpina L. (Poaceae) is a common grass at subalpine and alpine levels in the northern hemisphere whose presence indicates high levels of nutrients and soil moisture (Conert, 1998). Accordingly, it occurs in pastures and nutrient-rich meadows, but also as a pioneer species in scree slopes and in snowbeds. In the European Alps, P. alpina is among the most important fodder grasses due to its high contents of fats and proteins (Bachmann, 1980; Conert, 1998). Similar to other species in the genus Poa and the Poideae (Brysting et al., 2004), P. alpina constitutes a polyploid complex. Because of frequent aneuploidy and the presence of multiple B-chromosomes, chromosome numbers are highly variable (Müntzing, 1980; Steiner and Heidenreich, 1997). In Switzerland, reported chromosome numbers range from 2n = 22 to 2n = 46 (Duckert-Henriod and Favarger, 1987), and > 60 chromosomes were found in Scotch plants (Müntzing, 1980). Chromosome numbers counted in the root tips of 25 plants of this study varied between 22 and 61 per plant. Presumably, variable polyploidy adds to genetic diversity in P. alpina, as heterozygosity increases strongly with ploidy level (Brochmann et al., 2004).
Some plants of P. alpina produce seeds, while others reproduce vegetatively by forming bulbils in the panicles instead of seeds (Müntzing, 1980). Such bulbils grow into little plantlets on the maternal plants, which therefore are called pseudoviviparous. Eventually, the plantlets may dehisce from the maternal plant and root (Pierce, 1998). Usually, pseudoviviparous plants also develop a sexual floret at the base of the plantlets (Philipson, 1934; Müntzing, 1980; Pierce et al., 2003). It is not known whether these sexual florets produce fertile pollen and viable seeds and whether there is gene flow between such florets and sexually reproducing plants. The mode of reproduction appears largely genetically determined, while phenotypic plasticity in the mode of reproduction plays a minor role (Schwarzenbach, 1953, 1956; Heide, 1989).
Study area
The study area comprises 12 villages in the Swiss Alps, four of each of the three cultural traditions Romanic, Germanic and Walser (Fig. 1), and covers an area of approx. 170 × 70 km. Each village belongs to a separate Alpine valley. To represent typical agricultural villages, the study villages were selected with the restriction that their agricultural character had only changed modestly during the last 50 years, and that they were not very touristy and did not have more than about 1500 inhabitants.
Fig. 1.
Map of Switzerland with the 12 study villages and their cultural traditions.
Study design and sampling
Poa alpina was searched after in parcels of land chosen for a vegetational survey of grasslands (Maurer et al., 2006) situated at three altitudinal levels, at the valley bottom [approx. 1000 metres above sea level (asl)], at intermediate altitudes (approx. 1500 m asl) and at the alp level (approx. 2000 m asl) in each village. The search for P. alpina L. was conducted in those of 216 grasslands parcels where, according to local farmers, the type of land use had never changed. These parcels were characterized by a combination of land use (mown or grazed) and fertilization (fertilized or unfertilized). In each parcel where P. alpina occurred, eight plants were sampled at interdistances of 5 m to minimize the probability of sampling the same genotype more than once. Altogether, plants from 54 agriculturally used grassland parcels, 13 meadows and 41 pastures were sampled, of which 19 were additionally fertilized and 35 were not. Because P. alpina did not occur in all parcels and not all land-use types were applied in each village, plants of all combinations of land use and fertilization could not be found for each village. However, the occurrence of certain land-use types revealed no geographical pattern that could have confounded differences in land use with differences due to geographic distance. In the same way as in the agricultural parcels, eight plants were sampled from each of 20 natural sites above tree line that had never been used agriculturally. The sampled plants were also used for a common garden experiment (Weyand, 2005). Single genotypes were obtained by separating collected plants into four single tillers which were planted in the corners of 7 cm × 7 cm pots. After 2 months of growth in a greenhouse, three of the four plants were discarded and one randomly selected plant per genotype was kept. After a further 2 months, leaf samples of each plant were collected. Because some of the plants died, 415 plants of P. alpina from 54 agriculturally used parcels and 154 plants from 20 natural sites were analysed, usually eight plants per parcel, and a few times only seven or six.
Microsatellite analysis
The collected leaf samples were dried immediately with silica gel. Then, about 30 mg of the material were ground in Eppendorf tubes with a glass bead in a shaking mill. DNA was extracted according to a Rogers and Bendich (1994) protocol modified by Steinger (1996), except that samples were incubated with CTAB (cetyltrimethyl ammonium bromide) buffer and mercaptoethanol at 65 °C.
All plants were screened for variation at five polymorphic microsatellite loci (Maurer et al., 2005). DNA was amplified with 10 µL reaction volumes containing 10 ng of genomic DNA, 0·5 µL each of the fluorescence labelled forward primer and of the reverse primer, 5 µL of Hotstar Taq Mastermix (Qiagen, Hombrechtikon, Switzerland) and 3 µL of sterilized H2O. After a preliminary denaturation step at 95 °C for 15 min, DNA was amplified by polymerase chain reaction (PCR) on a PTC-100 Programmable Thermo Controller (MJ Research Inc.) for 30 cycles of 30 s denaturing at 95 °C, 30 s of annealing at locus-specific temperatures (Maurer et al. 2005) and 30 s of extension at 72 °C, with a final 8 min extension step at 72 °C. A 1 µL aliquot of the PCR product was mixed with 10 µL of a 75 : 1 solution of formamide and GeneScan-500 (ROX) size standard (Applied Biosystems, Foster City, CA, USA). Fragment lengths were determined by capillary gel electrophoresis with an ABI PRISM 310 Genetic Analyser using GeneScan 2·1 (Applied Biosystems). Plants were combined in random order for PCR and the sequencer runs. Microsatellite bands were binned using Genotyper 2·1 (Applied Biosystems) by correcting peaks manually after automated scoring, and the assignment of each peak to the corresponding band was controlled. In each PCR run, at least one blank sample was added to control for a possible contamination of the samples. Preliminary tests of repeated PCR with a sample of eight plants showed a very high accuracy of the produced band pattern. Thus, band scoring appeared to be the most possible source of a potential genotyping error. Therefore, a sample of 50 plants (8·8 % of all analysed individuals) were scored independently twice for each of the five primers, and a genotyping error of 1·48 % was obtained.
Due to their polyploidy, plants could show more than two microsatellite bands per locus. In a subset of 25 analysed plants, the number of bands was positively correlated with the number of chromosomes, but variable chromosome numbers due to aneuploidy and frequent B-chromosomes do not allow assessment of ploidy levels (Maurer et al., 2005). Therefore, the data do not conform to standard statistics for co-dominant microsatellite markers of diploid organisms, such as observed and expected heterozygosities, and tests for deviation from Hardy–Weinberg equilibrium. Consequently, all analyses in this study are based on a presence–absence matrix of all bands across all plants.
Analysis of differences between natural sites and agriculturally used grassland parcels
To test for differentiation between natural and agricultural sites, molecular variation among natural sites and agriculturally used parcels, among parcels and within parcels was partitioned with analysis of molecular variance (AMOVA; Excoffier et al. 1992) based on the pairwise Euclidean distance matrix of the presence of microsatellite bands in individuals of all 74 parcels. Then, to test whether regional differentiation was similar for natural and agricultural sites, molecular variation among villages, among parcels and within parcels was partitioned separately for the plants of the 54 agriculturally used parcels and for those of the 20 natural sites.
Analysis of differences among villages and among agriculturally used grassland parcels
To assess potential isolation by distance, a Mantel test (Mantel, 1967; Manly, 1997) was calculated using the matrix of Euclidean genetic distances among villages and among parcels, based on the relative abundance of each band per parcel, and the matrix of geographical distances among villages and among parcels. Furthermore, the Mantel test for the parcels with exclusively sexually and vegetatively reproducing P. alpina plants was calculated separately.
To examine potential differentiation among parcels of grassland of different land use (mown vs. grazed) and fertilization (fertilized vs. unfertilized), molecular variation among these groups, among parcels within these groups and within parcels was partitioned with AMOVA including plants of the 54 agriculturally used parcels. Furthermore, AMOVA was used to examine potential differentiation between ten parcels from which all sampled plants exclusively reproduced pseudoviviparously in the common garden (Weyand, 2005), and 22 parcels from which all sampled plants exclusively produced seeds.
In addition to AMOVA, among-parcel differentiation for microsatellite band richness rST(n) was measured among all 54 agriculturally used populations and also among meadows and pastures separately following El Mousadik and Petit (1996) and Petit et al. (1998). To obtain rST(n), first the expected band richness r′T(n) of a random sample of n = 6 plants out of all 415 plants and r′S(n) of a random sample of n = 6 plants for each parcel was calculated. Then, the differentiation for band richness was calculated as rST(n) = 1 – r′S(n)/r′T(n) (where S represents the single parcels and T the total population of all sampled plants) for each locus separately and the mean across the five loci.
Analysis of genetic diversity within villages and within parcels
Genetic diversity was measured within villages and within parcels as band richness r′(n) for each locus following El Mousadik and Petit (1996), except that plants were used as sample units instead of genes. The rarefaction procedure of Hurlbert (1971) was used to estimate band richness for a standardized sample size of n plants. As rarefaction sample size, n = 16 was used for villages and n = 6 for parcels. For each locus, the expected number of different bands r(n) in a sample of n plants was calculated according to the formula
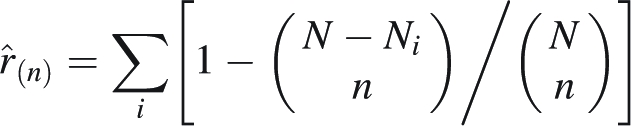 |
where Ni represents the number of occurrences of the ith band among the N sampled plants of a population and n the standardized sample size. One was subtracted from the band richness r(n) to obtain the corrected band richness r′(n), because a village or a parcel with only one single band is considered to be monomorphic. Then, mean band richness was calculated over all five loci, hereafter called band richness for simplicity.
Potential effects of the village characteristics latitude and longitude, ratio of the numbers of meadows and pastures in a village, altitude, number of land-use combinations (combination of land use and fertilization) investigated in a village and culture on band richness per village was tested with linear models and analysis of covariance (ANCOVA). For the two significant variables (number of land-use combinations and culture), ANCOVAs were calculated with sequential sums of squares including both variables. For these analyses only plants from agriculturally used parcels in each village were included. Furthermore, it was tested whether band richness per village was correlated with the accumulated number of plant species obtained from vegetation records in the corresponding parcels of each village.
To analyse within-parcel genetic diversity, the effects of cultural tradition, altitude, land use and fertilization on band richness per parcel and on the mean number of bands per plant for each parcel were investigated with hierarchical ANCOVA with sequential sums of squares. The effects of culture were tested against the remaining variation among villages and of all other factors against variation due to remaining differences among parcels. To account for differences among parcels because of different soil conditions or solar radiation, pH values and aspects of each parcel were used as covariates. However, as these covariates did not qualitatively change the results, the results are presented without covariates. Furthermore, a test was conducted to determine whether there was a difference in within-parcel diversity between populations from agriculturally used parcels and populations from natural sites. As there was no difference, the results are presented including only the agriculturally used parcels.
Further tests determined whether microsatellite diversity was affected by the abundance of P. alpina in the parcels. Spearman's rank correlations were calculated with the mean abundance of P. alpina of two plots (5 m × 5 m) per parcel and the measures of microsatellite diversity mean number of bands, band richness and mean Euclidean genetic distance of each parcel to all other parcels.
To test whether genetic diversity of P. alpina was correlated with plant species diversity, Spearman's rank correlation was calculated between mean plant species richness of two plots (5 m × 5 m) per parcel and mean number of bands per plant and parcel, and between total plant species richness of two plots per parcel and band richness per parcel.
All statistical analyses were performed using the software R (R Development Core Team 2004). For Mantel tests, the R-package vegan (Oksanen et al., 2006) was used and for AMOVAs the R-package ade4 (Chessel, 2004).
RESULTS
Overall microsatellite diversity
Among the 569 plants of P. alpina, altogether 209 bands were detected at the five microsatellite loci, between 25 and 61 per locus. Between one and eight bands were detected per plant and locus, with a mean of 3·35. In total, 531 multilocus microsatellite phenotypes were detected among all 569 plants and 386 multilocus microsatellite phenotypes among the 415 plants from agriculturally used parcels.
Differentiation between natural and agriculturally used grassland parcels
Natural and agriculturally used grassland parcels were genetically differentiated as low, but highly significant, 1·1 % of the variation in microsatellite bands resided between natural and agriculturally used grassland parcels (AMOVA, P < 0·004).
Genetic diversity among villages
Regional differentiation was slightly higher in natural sites, as 8·4 % of the variation in microsatellite bands in natural sites resided among villages, while the corresponding proportion was 6·8 % for agriculturally used sites (AMOVA, for both P < 0·001, Table 1). No statistically significant relationship of genetic distances with geographic distances was found (Mantel test with plants from agriculturally used parcels, rM = 0·22, P = 0·16) among pairs of the 12 villages.
Table 1.
Summary of analysis of molecular variance (AMOVA) of microsatellite phenotypes of plants of Poa alpina from 54 agriculturally used parcels and from 20 natural sites grouped in 12 villages
| Source of variation | Variance component | |||
|---|---|---|---|---|
| d.f. | Absolute | % | P | |
| 54 populations from agriculturally used parcels | ||||
| Among villages | 11 | 0·7981 | 6·76 | <0·001 |
| Among parcels within villages | 42 | 2·1790 | 18·39 | <0·001 |
| Within parcels | 361 | 8·8422 | 74·86 | <0·001 |
| Total | 414 | 11·8122 | 100·00 | |
| 20 populations from natural sites | ||||
| Among villages | 10 | 1·0184 | 8·42 | <0·001 |
| Among parcels within villages | 9 | 2·2502 | 18·61 | <0·001 |
| Within parcels | 134 | 8·8203 | 72·96 | <0·001 |
| Total | 153 | 12·0889 | 100·00 | |
AMOVA was based on the matrix of pairwise Euclidean distance between individuals in the presence of microsatellite bands. % = percentage of total variation.
Genetic diversity within villages
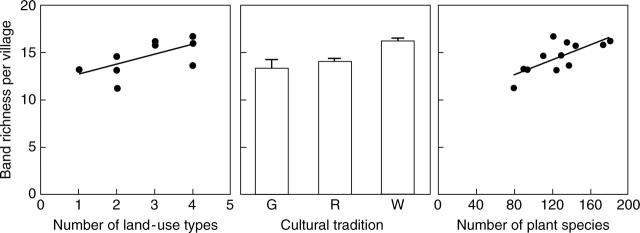
Band richness per village, based on a standardized sample size of 16 plants, was between 11·2 and 16·6 bands, with a mean of 14·5, and it was higher in villages with higher numbers of land-use combinations among the sampled parcels (P < 0·05, Fig. 2A). Furthermore, cultural traditions affected genetic diversity within villages because in villages with Walser tradition, band richness was higher than in Germanic villages (Tukey HSD, P < 0·05, Fig. 2B). When both these significant variables were fitted in a model with sequential sums of squares, the effect of cultural tradition only was significant when introduced into the model before the number of land-use combinations (introduced first: P < 0·05, second: P = 0·15), while the significant effect of the number of land-use combinations was independent of the fitting sequence. Moreover, band richness per village was positively correlated with the accumulated number of plant species occurring in the parcels with P. alpina per village (r = 0·73, P < 0·01, Fig. 2C).
Fig. 2.
Relationship between microsatellite band richness of Poa alpina per village sample and the number of land-use types investigated per village (ANOVA model with sequential sums of squares, P < 0·05), cultural tradition of 12 villages in the Swiss Alps (Tukey HSD, P < 0·05) and number of plant species recorded in the same parcels per village (P < 0·01). Band richness is based on a standardized sample size of 16 plants. G, Germanic tradition; R, Romanic; W, Walser. In the figure with the number of land-use types, three data points are hidden by others.
Genetic diversity among agriculturally used parcels
Of the variation in bands, 25·1 % resided among parcels (AMOVA, P < 0·001) and 18·4 % among parcels within villages (AMOVA; P < 0·001, Table 1). Parcel differentiation for band richness rST(n) was 0·25 across all five loci (Table 2), in line with the AMOVA result. Differentiation for band richness was higher among the 13 meadows than among the 41 pastures (Table 2).
Table 2.
Population genetic measures for 54 populations of Poa alpina from agriculturally used parcels of grassland in the Swiss Alps
| Microsatellite locus | Mean no. of bands per plant and population | r′S(6) | r′T(6) | rST(6) | rST(6) among meadows | rST(6) among pastures |
|---|---|---|---|---|---|---|
| Poa CA1D4 | 2·88 | 7·19 | 9·03 | 0·20 | 0·29 | 0·18 |
| Poa GAC1 | 4·44 | 11·62 | 16·35 | 0·29 | 0·36 | 0·27 |
| Poa GA1C3 | 2·23 | 5·12 | 6·75 | 0·24 | 0·34 | 0·21 |
| Poa CA1F4 | 3·87 | 8·39 | 10·57 | 0·21 | 0·25 | 0·19 |
| Poa CAB12 | 3·37 | 7·29 | 10·33 | 0·29 | 0·34 | 0·28 |
| Mean | 3·36 | 7·92 | 10·61 | 0·25 | 0·32 | 0·23 |
r′S(6), band richness per population with a standardized sample size of six plants; r′T(6), band richness of the hypothetical total population with a sample size of six plants; rST(6), differentiation for band richness among populations; rST(6) among meadows, differentiation for band richness among 13 meadows; rST(6) among pastures, differentiation for band richness among 41 pastures.
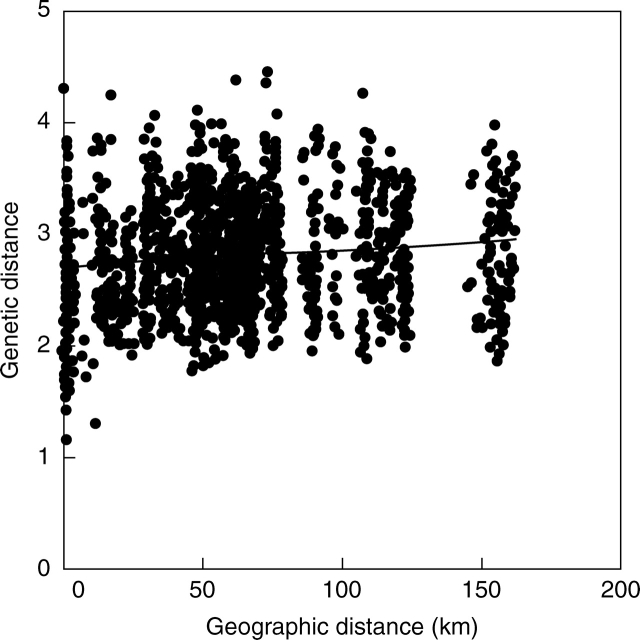
A tendency to isolation by distance was found among agriculturally used parcels, as geographically more distant parcels tended to be genetically more distant (Mantel test, rM = 0·12, P = 0·057, Fig. 3A). The pattern was significant among parcels with exclusively sexually reproducing plants (rM = 0·22, P < 0·05), but not among parcels with exclusively vegetatively reproducing plants (rM = 0·19, P = 0·213). When variation in microsatellite bands among parcels was partitioned according to land use, 1·18 % was found to reside between mown and grazed grassland parcels (AMOVA, P < 0·02), and only 0·02 % between fertilized and unfertilized grassland parcels (AMOVA, P > 0·42).
Fig. 3.
Relationship of pairwise Euclidean genetic distance between populations of Poa alpina from 54 agriculturally used parcels of grassland in the Swiss Alps with pairwise geographic distances (Mantel test, rM=0·12, P = 0·057).
Of the variation in microsatellite bands, 4·2 % resided between the 22 parcels with exclusively seed-producing samples of P. alpina and the ten parcels with exclusively pseudoviviparous samples (AMOVA, P < 0·001).
Overall, the results of this section indicate substantial regional genetic differentiation of P. alpina among different villages and a genetic differentation among parcels with different reproductive modes of P. alpina. Genetic differentiation among mown and grazed parcels was less pronounced.
Genetic diversity within agriculturally used parcels
Of the variation in microsatellite bands, 74·9 % resided within parcels (AMOVA, P < 0·001; Table 1). Band richness, based on a rarefaction sample size of six plants, varied between 5·12 and 11·62 per parcel and locus (Table 2), and the mean number of bands per plant was between 2·88 and 4·44 per locus (Table 2) and 16·8 across all loci.
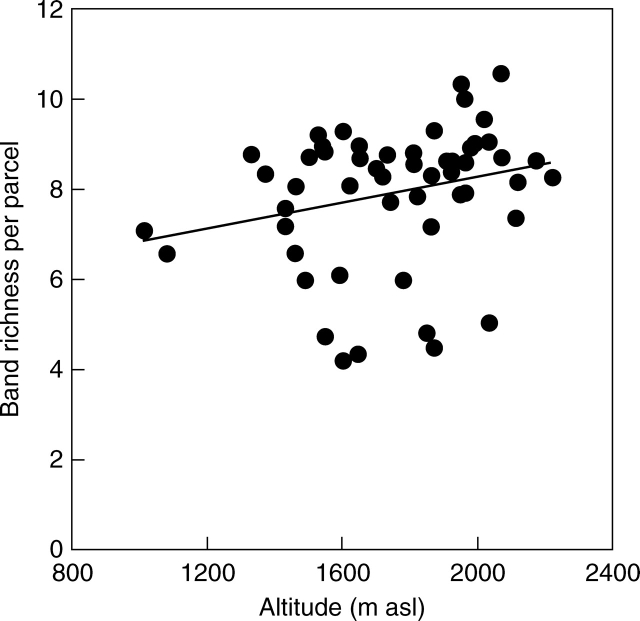
Band richness increased with increasing parcel altitude (Table 3, Fig. 4), while the mean number of bands per plant was independent of altitude (Table 3). Cultural traditions did not affect genetic diversity within parcels (Table 3).
Table 3.
Summary of analyses of the mean number of microsatellite bands per plant (MB) and band richness per parcel (BR) in 54 populations of Poa alpina from agriculturally used parcels of land in the Swiss Alps
| Source of variation | d.f. | SSMB | FMB | PMB | SSBR | FBR | PBR |
|---|---|---|---|---|---|---|---|
| Culture | 2 | 4·34 | 0·24 | NS | 7·72 | 2·04 | NS |
| Village[culture] | 9 | 81·38 | 7·54 | P < 0·01 | 17·01 | 1·12 | NS |
| Altitude | 1 | 3·94 | 3·28 | P < 0·1 | 9·60 | 5·67 | P < 0·05 |
| Fertilization | 1 | 0·25 | 0·21 | NS | 0·05 | 0·03 | NS |
| Land use | 1 | 8·78 | 7·32 | P < 0·05 | 6·83 | 4·04 | P < 0·1 |
| Fertilization × land use | 1 | 0·60 | 0·50 | NS | 0·55 | 0·33 | NS |
| Culture × altitude | 2 | 4·00 | 1·00 | NS | 1·45 | 0·31 | NS |
| Village[culture] × altitude | 9 | 18·05 | 1·67 | NS | 20·80 | 1·37 | NS |
| Culture × fertilization | 2 | 1·34 | 0·39 | NS | 3·90 | 1·16 | NS |
| Culture × land use | 2 | 5·47 | 1·16 | NS | 0·98 | 0·08 | NS |
| Village[culture] × fertilization | 7 | 12·12 | 1·44 | NS | 11·78 | 0·99 | NS |
| Village[culture] × land use | 3 | 7·09 | 1·97 | NS | 18·10 | 3·57 | P < 0·1 |
| Altitude × fertilization | 1 | 0·11 | 0·09 | NS | 0·00 | 0·00 | NS. |
| Altitude × land use | 1 | 0·40 | 0·33 | NS | 6·26 | 3·70 | P < 0·1 |
| Residuals | 11 | 13·19 | 18·61 |
Culture denotes the Romanic, Germanic and Walser cultural traditions. Village denotes the 12 study villages. Fertilization denotes the difference between unfertilized and fertilized parcels. Land use denotes differences between mown and grazed grasslands. In the sequential sums of squares ANCOVA, effects of culture were tested against the remaining variation among villages. NS denotes values of P > 0·1.
Fig. 4.
Relationship between microsatellite band richness, based on a rarefaction sample size of six plants, and altitude of 54 populations of Poa alpina from agriculturally used parcels of land in the Swiss Alps (ANCOVA model with sequential sums of squares, P < 0·05).
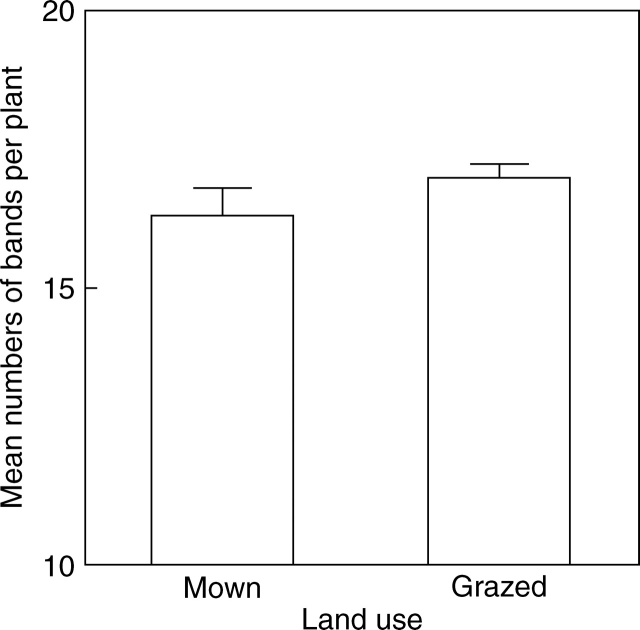
Land use affected within-parcel microsatellite diversity, as the mean number of bands per plant was 3·0 % higher in pastures than in meadows (F1,53 = 7·32; P < 0·05, Table 3, Fig. 5). Moreover, band richness tended to be higher in pastures than in meadows (F1,53 = 4·04; P = 0·07, Table 3).
Fig. 5.
Relationship between number of bands per plant and type of land use among 54 populations of Poa alpina from agriculturally used parcels of land in the Swiss Alps (ANCOVA model with sequential sums of squares, P < 0·05).
Within-parcel microsatellite band diversity did not appear to be affected by more pronounced genetic drift in smaller populations of P. alpina, because the mean number of bands per plant (P = 0·3714), band richness per parcel (P = 0·6486) and mean Euclidean genetic distance of each parcel to all other parcels (P = 0·6599) were independent of mean abundance of P. alpina.
Correlations between mean plant species richness in two plots per parcel and the mean number of bands or total plant species richness in two plots per parcel and band richness were not statistically significant (P = 0·1875 and P = 0·9031, respectively), indicating that genetic diversity of P. alpina was not affected by plant species diversity of the 54 agriculturally used grassland parcels.
DISCUSSION
Genetic differentiation among natural and agriculturally used parcels
The small but significant microsatellite band differentiation among natural sites and agriculturally used parcels suggests that human land use caused not only a divergence in genetic diversity among populations of P. alpina from mown and grazed grasslands, but also differentiation from natural habitats. This is in line with an accompanying quantitative genetic experiment, where plants from pastures and from meadows both differed from plants from natural sites in biomass allocation to reproduction (Weyand, 2005). To our knowledge, only one study considering populations from natural sites and different agriculturally used parcels of land has been carried out, which investigated Sesleria albicans (Reisch et al., 2003). Unfortunately, the authors did not separate natural from agricultural populations but rather analysed whether there was general variation in genetic diversity among habitat types, which prevents a comparison with their results.
Genetic differentiation among villages and among populations
Probably because of the large 170 km east–west range comprising different Alpine valleys, genetic differentiation among grasslands was pronounced and explained 25 % of the variation, and pairs of parcels tended to be genetically isolated by distance. The tests using parcels with exclusively sexually and exclusively vegetatively reproducing plants show that the pattern including all parcels is weakened by the lack of isolation by distance in plants reproducing vegetatively by bulbils. Previously in P. alpina, isolation by distance had been studied and observed only within a population in Norway (Bjørnstad et al., 1995). In addition to geographic distance, variable polyploidy, which can restrict gene flow (Briggs and Walters, 1997), may have contributed to population differentiation in P. alpina. Plants of P. alpina were highly variable in chromosome number (between 22 and 61 in this study) due to frequent aneuploidy and multiple B-chromosomes (Müntzing, 1980; Duckert-Henriod and Favarger, 1987; Maurer et al., 2005).
Differentiation due to land use
Different habitat conditions can lead to genetically based ecotypic differentiation in grass species (Stapledon, 1928). In the present study, mowing and grazing over hundreds of years apparently led to genetic differentiation between mown and grazed populations of P. alpina, while fertilization had no effect. This corresponds to the results of a common garden experiment using the same genotypes, which showed a divergence in biomass allocation between plants from mown and grazed parcels, but no difference between plants from fertilized and unfertilized parcels (Weyand, 2005). Although the effect of mowing and grazing on genetic divergence between parcels was smaller than the effects of isolation by distance or reproductive mode, it adds to the evidence that land use affects biodiversity not only at the plant community level, but also at the level of genetic diversity within species (Odat et al., 2004). The result is especially remarkable, as it suggests that land use affects biodiversity independently of regional differences. Land use-induced ecotypes have also been reported for the grassland forbs Rhinanthus alectorolophus (Zopfi, 1993) and Euphrasia rostkoviana (Zopfi, 1998). As in the common garden, there was no difference in reproductive modes among plants from meadows and pastures, and as the reproductive mode appeared to be largely genetically determined (Weyand, 2005), mowing and grazing are likely to act on genetic diversity of P. alpina directly rather than via its reproductive mode.
For the observed higher population differentiation among populations of meadows than among populations of pastures there are two mutually non-exclusive explanations. First, endozoochorous or exozoochorous seed transport by cattle could increase gene flow among pastures. Secondly, the result could indicate more diverse habitat conditions among meadows than among pastures. Land-use intensity of the investigated meadows with P. alpina varies between mowing every second year to twice per season and may thus be responsible for differentiation among mown parcels as different mowing intensity was reported to exert differential selection in Festuca pratensis (Kölliker et al., 1998). In contrast, among pastures, land-use intensity probably varies more between seasons than it does among parcels, which results in more uniform selection, and may thus contribute to the weaker genetic differentiation among pastures than among meadows. A third alternative explanation of higher genetic drift among meadows than among pastures can be ruled out, because the abundance of P. alpina, which determines the importance of genetic drift, was correlated neither with within-parcel genetic diversity nor with genetic distance of a parcel to all other parcels.
Differentiation due to reproductive mode
The observed substantial genetic differentiation among parcels with exclusively seed-producing samples of P. alpina and those with exclusively pseudoviviparous samples suggests that gene flow between seed-producing and pseudoviviparous plants of P. alpina, the latter of which produce a sexual floret at the basis of their inflorescence (Philipson, 1934; Müntzing, 1980), is rather low. Furthermore, the lack of isolation by distance among parcels with exclusively pseudoviviparous plants is an indication that gene flow is largely restricted to sexually reproducing plants.
Genetic diversity within villages
In Walser villages, more different land-use combinations with P. alpina tended to be present than in Romanic and Germanic villages (result not shown). This is most probably due to the combination of the alpine to subalpine altitudinal distribution of P. alpina with the higher altitudes of Walser villages than villages of the other traditions. Due to settlement history, the later arriving Walser people had to settle at higher altitudes than Romanic and Germanic people (Bätzing, 2003). At the valley bottom of villages at lower altitudes, P. alpina was not present in all types of parcels. Accordingly, this reduced the number of investigated land-use combinations in Romanic and Germanic villages and probably caused the observed dependence of the statistical significance of cultural tradition on band richness of P. alpina within villages on the fitting sequence. The higher genetic diversity of P. alpina in villages where P. alpina occurred in a larger number of different land-use combinations is in line with the observed microsatellite differentiation among parcels of different land use. Moreover, it corresponds to the result of a study of plant species diversity of the same 12 villages, which revealed a significantly positive relationship between plant species richness per village and the number of different land-use combinations present in the villages (Maurer et al., 2006). Accordingly, genetic diversity of P. alpina was also positively correlated with the total number of plant species recorded in all the parcels per village where the Poa plants had been sampled.
Genetic diversity within populations
In contrast to findings with F. pratensis populations (Kölliker et al., 1998), genetic diversity of P. alpina was not affected by fertilization. Nevertheless, agricultural land use significantly affected within-population genetic diversity of P. alpina. Populations originating from pastures were genetically more diverse, as they showed more bands per plant and tended to have higher band richness than meadow populations. Because samples of P. alpina from meadows and those from pastures did not differ in their reproductive modes (Weyand, 2005), variation in reproductive mode cannot be responsible for these effects of land use on genetic diversity. Rather, this may be due to higher recruitment in grazed sites, either because of the higher biomass allocation of plants from pastures to reproduction (Weyand, 2005), or because of the higher probability of establishment of seedlings and pseudoviviparous plantlets in pastures, which offer more vegetation gaps as safe sites for establishment (Grubb, 1977). Moreover, selection in mown sites may be more uniform than in spatially more heterogeneous grazed sites, which may reduce genetic diversity more strongly in meadows than in pastures. Accordingly, in F. pratensis, molecular genetic diversity was lower the more intense the cutting regime was (Kölliker et al., 1998). Furthermore, genetic diversity could have been enhanced in pastures because of higher gene flow due to seed transport by cattle.
Parcels from higher altitudes had higher band richness. In contrast, the number of bands per plant, which did not differ significantly between exclusively pseudoviviparous and exclusively seed-producing samples, did not increase with altitude and neither did the number of chromosomes per plant (data not shown). A potential biological explanation for this set of results could be the differentiation between pseudoviviparous and seed-producing samples of P. alpina in combination with an increase in pseudoviviparous reproduction from 20 % in valley genotypes to 50 % in alpine genotypes observed in the common garden (Weyand, 2005) and a resulting accumulation of bands typical for each of the two reproductive modes with increasing altitude.
Correlations between genetic diversity of Poa alpina and plant species richness
A high niche diversity has been suggested to maintain higher genetic diversity in plant species (Odat et al., 2004; Vellend and Geber, 2005) and can also increase species richness, at least in nutrient-rich habitats (Gigon and Leutert, 1996; Proulx and Mazumder, 1998; Austrheim and Eriksson, 2001; Maurer et al., 2006). Therefore, higher genetic diversity of P. alpina in parcels with higher plant species richness might have been expected. However, in the present study, genetic diversity of P. alpina was not correlated with plant species richness.
CONCLUSIONS
The genetic complexity of the polyploid and aneuploid P. alpina constrained the present study of molecular diversity to an analysis of the presence of microsatellite bands. Nevertheless, the distribution of genetic diversity within and among populations of P. alpina turned out to be affected by land-use diversity in villages and by the specific land use within parcels. Higher genetic diversity within pastures than within meadows, genetic differentiation among populations from meadows and pastures, and higher genetic diversity within villages with more diverse land use lead to two important conclusions. First, the findings demonstrate that the ongoing socio-economically motivated land-use changes in the Swiss Alps not only affect diversity at the landscape and community levels, but they also change genetic diversity within species. Moreover, promoting genetic diversity cannot be achieved by just maintaining the single type of land use associated with the highest within-population diversity, but requires the maintenance of a high diversity of land-use types.
ACKNOWLEDGEMENTS
We thank R. Husi and student workers for support in the laboratory. We are grateful to B. Hefti-Gautschi (ecogenics, Zürich, Switzerland) for sharing her knowledge about microsatellite analysis. We thank all involved municipalities and authorities for their support, and especially the local contact persons and all farmers who allowed us to collect plants on their land. F. Spinnler collected plants from natural sites. We acknowledge support by the Swiss National Research Programme 48 ‘Landscapes and Habitats of the Alps’ (Grant 4048-064494/1) and by the Institute for Environmental Sciences of the University of Zürich.
LITERATURE CITED
- Austrheim G, Eriksson O. Plant species diversity and grazing in the Scandinavian mountains – patterns and processes at different spatial scales. Ecography. 2001;24:683–695. [Google Scholar]
- Bachmann MA. Oekologie und Breeding System bei Poa alpina L. University of Zurich; 1980. PhD thesis. [Google Scholar]
- Bauert MR. Vivipary in Polygonum viviparum – an adaptation to cold climate? Nordic Journal of Botany. 1993;13:473–480. [Google Scholar]
- Bjørnstad ON, Iversen A, Hansen M. The spatial structure of the gene pool of a viviparous population of Poa alpina – environmental controls and spatial constraints. Nordic Journal of Botany. 1995;15:347–354. [Google Scholar]
- Briggs D, Walters SM. Plant variation and evolution. 3rd edn. Cambridge: Cambridge University Press; 1997. [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG, Borgen L, Gundt HH, Scheen AC, Eleven R. Polyploidy in arctic plants. Biological Journal of the Linnean Society. 2004;82::521–536. [Google Scholar]
- Brysting AK, Fay MF, Leitch IJ, Aiken SG. One or more species in the arctic grass genus Dupontia? – A contribution to the Panarctic Flora project. Taxon. 2004;53:365–382. [Google Scholar]
- Bätzing W. Die Alpen – Geschichte und Zukunft einer europäischen Kulturlandschaft. 2nd edn. München: C.H. Beck; 2003. [Google Scholar]
- Chessel DD, Dufour A-B, Thioulouse J. The ade4 package-I-One-table methods. R News. 2004;4:5–10. [Google Scholar]
- Conert HJ, Jäger EJ, Kadereit JW, Schulze-Motel W, Wagenitz G, Weber HE. 14. Poa alpina. In: Conert HJ, Jäger EJ, Kadereit JW, et al., editors. Gustav Hegi Illustrierte Flora von Mitteleuropa. Berlin: Parey Buchverlag; 1998. pp. 690–693. [Google Scholar]
- Duckert-Henriod MM, Favarger C. Contribution à la cytotaxonomie et à la cytogéographie des Poa de la Suisse. Basel: Birkhäuser Verlag; 1987. [Google Scholar]
- El Mousadik A, Petit RJ. High level of genetic differentiation for allelic richness among populations of the argan tree Argania spinosa (L) Skeels endemic to Morocco. Theoretical and Applied Genetics. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes – application to human mitochondrial-DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigon A, Leutert AG. The dynamic keyhole key model of coexistence to explain diversity of plants in limestone and other grasslands. Journal of Vegetation Science. 1996;7:29–40. [Google Scholar]
- Godt MJW, Hamrick JL. Allozyme diversity in the grasses. In: Cheplick GP, editor. Population biology of grasses. Cambridge: Cambridge University Press; 1998. pp. 11–29. [Google Scholar]
- Grubb PJ. The maintenance of species-richness in plant communities – the importance of the regeneration niche. Biological Reviews of the Cambridge Philosophical Society. 1977;52:107–145. [Google Scholar]
- Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. In: Silvertown J, Franco M, Harper JL, editors. Plant life histories. Cambridge: Cambridge University Press; 1997. pp. 102–117. [Google Scholar]
- Hartl DL, Clark AG. Principles of population genetics. 3rd edn. Sunderland, MA: Sinauer Associates, Inc; 1997. [Google Scholar]
- Heide OM. Environmental control of flowering and viviparous proliferation in seminiferous and viviparous arctic populations of two Poa species. Arctic and Alpine Research. 1989;21:305–315. [Google Scholar]
- Hurlbert SH. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- Kölliker R, Stadelmann FJ, Reidy B, Nösberger J. Fertilization and defoliation frequency affect genetic diversity of Festuca pratensis Huds. in permanent grasslands. Molecular Ecology. 1998;7:1557–1567. [Google Scholar]
- Loveless MD, Hamrick JL. Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics. 1984;15:65–95. [Google Scholar]
- Manly BJF. Randomization, bootstrap and Monte Carlo methods in biology. 2nd edn. London: Chapman & Hall; 1997. [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- Maurer K, Gautschi B, Weyand A, Stöcklin J, Fischer M. Isolation and characterization of microsatellite DNA markers in the grass Poa alpina L. Molecular Ecology Notes. 2005;5:719–720. [Google Scholar]
- Maurer K, Weyand A, Fischer M, Stöcklin J. Old cultural traditions, in addition to land use and topography, are shaping plant diversity of grasslands in the Alps. Biological Conservation. 2006;130:438–446. [Google Scholar]
- Merilä J, Crnokrak P. Comparison of genetic differentiation at marker loci and quantitative traits. Journal of Evolutionary Biology. 2001;14:892–903. [Google Scholar]
- Müntzing A. Mode of propagation and chromosomal peculiarities in Scotch material of Poa alpina. Hereditas. 1980;92:291–296. [Google Scholar]
- Odat N, Jetschke G, Hellwig FH. Genetic diversity of Ranunculus acris L. (Ranunculaceae) populations in relation to species diversity and habitat type in grassland communities. Molecular Ecology. 2004;13:1251–1257. doi: 10.1111/j.1365-294X.2004.02115.x. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O'Hara RB. vegan: community ecology package version 1·8–2. 2006. Oulu.
- Petit RJ, El Mousadik A, Pons O. Identifying populations for conservation on the basis of genetic markers. Conservation Biology. 1998;12:844–855. [Google Scholar]
- Philipson WR. The morphology of the lemma in grasses. New Phytologist. 1934;33:359–371. [Google Scholar]
- Pierce S. UK: University of Durham; 1998. Resource allocation in the pseudoviviparous Alpine Meadow Grass (Poa alpina L.) PhD Thesis. [Google Scholar]
- Pierce S, Stirling CM, Baxter R. Pseudoviviparous reproduction of Poa alpina var. vivipara L. (Poaceae) during long-term exposure to elevated atmospheric CO2. Annals of Botany. 2003;91:613–622. doi: 10.1093/aob/mcg067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluess AR, Stöcklin J. The importance of population origin and environment on clonal and sexual reproduction in the alpine plant Geum reptans. Functional Ecology. 2005;19:228–237. [Google Scholar]
- Proulx M, Mazumder A. Reversal of grazing impact on plant species richness in nutrient-poor vs. nutrient-rich ecosystems. Ecology. 1998;79:2581–2592. [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna: 2004. http://www.r-project.org/ [Google Scholar]
- Reisch C, Poschlod P, Wingender R. Genetic differentiation among populations of Sesleria albicans Kit. ex Schultes (Poaceae) from ecologically different habitats in central Europe. Heredity. 2003;91:519–527. doi: 10.1038/sj.hdy.6800350. [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin SB, Schilperoort RA, editors. Plant molecular biology manual. Dordrecht: Kluwer Academic Publishers; 1994. pp. 1–8. [Google Scholar]
- Schlötterer C. Microsatellites. In: Hoelzel A, editor. Molecular genetic analysis of populations – a practical approach. Oxford: IRL Press at Oxford University Press; 1998. pp. 237–261. [Google Scholar]
- Schwarzenbach FH. Die Abhängigkeit der Bulbillenbildung bei Poa alpina vivipara von Photoperiodismus und Frost. Experientia. 1953;9:96–96. doi: 10.1007/BF02178333. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach FH. Die Beeinflussung der Viviparie bei einer grönländischen Rasse von Poa alpina L. durch den jahreszeitlichen Licht- und Temperaturwechsel. Berichte der Schweizerischen Botanischen Gesellschaft. 1956;66:204–223. [Google Scholar]
- Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since plant speciation. New Phytologist. 2004;161:173–191. [Google Scholar]
- Stapledon RG. Cocksfoot grass (Dactylis glomerata L.): ecotypes on relation to the biotic factor. Journal of Ecology. 1928;16:72–104. [Google Scholar]
- Steiner AM, Heidenreich SC. Verification of varieties of alpine meadow-grass (Poa alpina L.) floret morphology, chromosome number and single seed storage protein electrophoresis. Plant Varieties and Seeds. 1997;10:129–134. [Google Scholar]
- Steinger T, Körner C, Schmid B. Long-term persistence in a changing climate: DNA analysis suggests very old ages of clones of alpine Carex curvula. Oecologia. 1996;105:94–99. doi: 10.1007/BF00328796. [DOI] [PubMed] [Google Scholar]
- Vellend M, Geber MA. Connections between species diversity and genetic diversity. Ecology Letters. 2005;8:767–781. [Google Scholar]
- Weppler T, Stöcklin J. Variation of sexual and clonal reproduction in the alpine Geum reptans in contrasting altitudes and successional stages. Basic and Applied Ecology. 2005;6:305–316. [Google Scholar]
- Weyand A. Drivers of grassland biodiversity in the Swiss Alps. University of Zurich; 2005. PhD Thesis. [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopfi HJ. Ecotypic variation in Rhinanthus alectorolophus (Scopoli) Pollich (Scrophulariaceae) in relation to grassland management. II. The genotypic basis of seasonal ecotypes. Flora. 1993;188:153–173. [Google Scholar]
- Zopfi HJ. The genetic basis of ecotypic variants of Euphrasia rostkoviana Hayne (Scrophulariaceae) in relation to grassland management. Flora. 1998;193:41–58. [Google Scholar]