Abstract
Background and Aims
Vegetative storage proteins (VSPs) are commonly bioactive in herbaceous plants but few VSPs with bioactivity have been identified in trees. In addition, information on the characterization of VSPs in evergreen trees is limited. The objective of this study was to characterize the VSPs with bioactivity in evergreen trees.
Methods
The VSP in lychee (Litchi chinensis), an evergreen fruit tree, was characterized by a combination of cytological, biochemical and molecular biological techniques.
Key Results
The VSP in lychee was a 22-kDa protein. It accumulated in the large central vacuoles of protein-storing cells (PSCs) in two distinguishable forms, granular and floccular. The PSCs were of a novel type. The 22-kDa protein is distributed in mature leaves, bark tissues of branches, trunk and large roots, paralleling the distribution of PSCs. Its homologues were present in mature seed. During young shoot development and fruiting, the 22-kDa protein decreased apparently, suggesting a nitrogen-storage function. The 22-kDa protein had several isoforms encoded by a small multigene family. One gene member, LcVSP1, was cloned. The LcVSP1 had no intron and contained a 675 bp open reading frame encoding a putative protein of 225 amino acids. LcVSP1 was homologous to Kunitz trypsin inhibitors. The 22-kDa protein inhibited trypsin and chymotrypsin, but had no inhibitory effect on subtilisin.
Conclusions
Lychee is rich in a 22-kDa VSP with trypsin inhibitor activity. The VSP plays an important role in nitrogen storage while its possible defensive function remains to be elucidated.
Key words: Defence, evergreen tree, Kunitz trypsin inhibitor, Litchi chinensis, nitrogen storage, vegetative storage protein
INTRODUCTION
Vegetative storage proteins (VSPs) occur in many herbaceous plants. The best characterized of these VSPs are the 27-, 29- and 94-kDa proteins in leaves of soybean where they specifically accumulate in large amounts in vacuoles of the paraveinal mesophyll cells (Franceschi et al., 1983; Wittenbach, 1983; Tranbarger et al., 1991) and act as temporary nitrogen reserves to support seed development (Staswick, 1994). By using transgenic soybean plants in which VSP synthesis is suppressed by a vspA antigene, however, Staswick et al. (2001) demonstrated that the VSPs in soybean play a very small, direct role in overall plant productivity. As the 27- and 29-kDa proteins are acid phosphatase active (De Wald et al., 1992) and the 94-kDa protein is a lipoxygenase (Tranbarger et al., 1991), VSPs could play some special roles under certain conditions (Tranbarger et al., 1991; De Wald et al., 1992). Proteins with biological activities are found in many other herbaceous plants, where they are present in flowers/flower buds (Berger et al., 1995; Liu et al., 2005), vegetative storage organs (Andrews et al., 1988; Volenec et al., 1996; Yeh et al., 1997; Flores et al., 2002; Meuriot et al., 2004; Dhont et al., 2006) and leaves (Graham et al., 1986; Ryan, 1990; Moura and Ryan, 2001; Horn et al., 2005). These proteins are generally regarded as VSPs in herbaceous plants and have been suggested to play a dual role in nitrogen storage and defence (Liu et al., 2005).
The best-characterized VSP in deciduous trees is the 32-kDa protein in poplar (for review, see Stepien et al., 1994). The protein consists of several isoforms encoded by a small multigene family (Langheinrich and Tischner, 1991; Stepien and Martin, 1992; Stepien et al., 1994). One gene member, bspA, has been cloned (Coleman and Chen, 1993). The 32-kDa protein acts as a seasonal nitrogen reserve on the basis of its seasonal fluctuation (for review, see Stepien et al., 1994). Unlike the VSPs in hebaceous plants, the 32-kDa protein in poplar has no enzymatic or other biological activities (Stepien and Matin, 1992; Stepien et al., 1994). Besides, WIN4 and PNI288 are also considered to be VSPs in poplar (Cooke and Weih, 2005). In addition to nitrogen storage, the two proteins as well as the 32-kDa protein are thought to play a role in defence against herbivores on the basis of the up-regulation of their genes by mechanical wounding (Davis et al., 1993; Cooke and Weih, 2005). VSPs are also found in many other temperate deciduous trees (for review, see Stepien et al. 1994) and tropical deciduous trees (Wu and Hao, 1986, 1991; Hao and Wu, 1993; Tian et al., 1998, 2003; Tian and Hu, 2004). The diversity in cytology and biochemistry of the VSPs suggests that VSPs with special bioactivities should be common in trees (Stepien et al., 1994; Tian et al., 1998). An open question is whether all of the VSPs in trees are generally biologically inactive.
Compared with deciduous trees, evergreen trees are poor in VSPs (Wetzel and Greenwood, 1989; Wetzel et al., 1991; Roberts et al., 1991; Arora et al., 1992) and none of the VSPs have been well characterized. The absence of VSPs from evergreen trees is consistent with their role as seasonal nitrogen storage in deciduous trees. However, by studying proteins in lychee (Litchi chinensis), a subtropical evergreen fruit tree, using a combination of cytological, biochemical and molecular biological techniques, it was found that lychee is rich in a VSP, a 22-kDa protein with trypsin inhibitor activity.
MATERIALS AND METHODS
Plant materials and treatments
Five-year-old ‘Feizixiao’ lychee (Litchi chinesis Sonn.) trees were grown in the Lychee Plantation of Chinese Academy of Tropical Agricultural Sciences on Hainan Island, China. Ten comparable trees, in terms of size, growth vigour and phenological phase (phenophase) were chosen from the plantation and controlled to sprout twice a year in late June (the shoots produced were called the first autumn shoots) and late September (the shoots produced were called the second autumn shoots). Four plant parts were harvested during the second autumn sprouting, blossom and fruit development: (1) mature leaves; (2) stems of the first autumn shoots (i.e. twigs) and the second autumn shoots (i.e. terminal branches); (3) trunk samples that were taken 50 cm up from the ground level; (4) large root samples that were taken at a distance of 50 cm from the trunk base from the larger lateral roots in the surface soil. Samples from different trees at each time point were analysed, and representative data are given in the Results.
Light- and electron-microscopy
For histochemical staining, samples were fixed in 4 % glutaraldehyde in 0·1 m phosphate buffer (pH 7·2) at 4 °C for 24 h. Paraffin blocks were prepared after dehydration through a graded series of ethanol. Microtome sections (10 µm thick) were stained with mercury–bromophenol blue, which is specific for protein (Wu and Hao, 1986).
For indirect fluorescence localization, samples were cut to size 5 × 5 mm and fixed in glutaraldehyde (0·5 %) and paraformaldehyde (4 %) in 0·1 m phosphate buffer (pH 7·4) at 4 °C for 12 h. The samples were then dehydrated in ethanol and embedded in paraffin. Microtome sections (10 µm thick) were treated with the 22-kDa protein antiserum (1 : 1000) and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgGs (SABC, Shanghai, China), and then examined with a Leica DMLN microscope (Leica, Wetzlar, Germany).
Electron microscopy was accomplished according to Tian et al. (2003).
Protein extraction for sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and two-dimensional gel electrophoresis
Samples (2 g fresh weight) were mixed with 0·5 g of polyvinylpolypyrrolidone (PVPP; see Table 1 for abbreviations used in the text) and ground to a homogenate in 5 mL of extraction buffer [0·1 m Tris base, 0·05 m sodium borate, 0·005 m ascorbic acid, 1 % β-mercaptoethanol, 0·1 % (v/v) Triton X-100, 1 mm PMSF (phenylmethylsulphonyl fluoride), pH 8·4]. The homogenates were centrifuged at 17500 g for 30 min at 4 °C and the supernatants were collected. Protein in the supernatants was determined according to the method of Bradford (1976) using bovine serum albumin as standard. The supernatants were used for SDS–PAGE after boiling for 5 min with an equal volume of 2 × Laemmli sample buffer (Laemmli, 1970). Protein samples for two-dimensional gel electrophoresis were purified by Sephacryl S-100HR and DEAE-Sepharose in turn.
Table 1.
List of abbreviations used in the text
| CTAB | Cetyl trimethyl ammonium bromide |
| FITC | Fluorescein isothiocyanate |
| LDS | N-Lauroylsarcosine sodium salt |
| MALDI-TOF MS | Matrix-assisted laser desorption ionization–time of flight mass spectrometry |
| ORF | Open reading frame |
| PAGE | Polyacrylamide gel electrophoresis |
| PMF | Peptide mass fingerprinting |
| PSC | Protein-storing cell |
| PVPP | Polyvinylpolypyrrolidone |
| RACE | Rapid amplification of cDNA ends |
| SDS | Sodium dodecyl sulfate |
| UI | Inhibitor unit |
| UTR | Untranslated region |
| VSP | Vegetative storage protein |
SDS–PAGE
SDS–PAGE was performed using 14 % SDS–polyacrylamide gels (or alternatives as shown in the figures), 25 mm Tris base, 192 mm glycine and 0·1 % SDS running buffer, pH 8·3, at 25 mA gel−1. Twenty micrograms of protein were loaded per lane (unless as shown in the figures). Gels were routinely stained with Coomassie brilliant blue R-250.
Two-dimensional gel electrophoresis
Thirty micrograms of the purified 22-kDa protein were mixed with 125 µL of rehydration buffer containing 8 m urea, 2 % (v/v) Triton X-100, 65 mm DTT, 0·2 % (v/v) Bio-lyte (pH 3·5–10·0) and a trace of bromophenol blue. Isoelectric focusing was carried out automatically in IPGphor (Bio-Rad Laboratories, Inc., USA) using 7-cm IPG strips (linear pH 3·0–10·0) (Bio-Rad Laboratories, Inc.). The equilibration and application of the IPG strips to SDS–polyacrylamide gels were according to the manufacturer's instructions. The gels were stained with Coomassie brilliant blue R-250.
Production of antiserum
The 22-kDa protein was extracted from the bark tissues of branches, purified by preparative SDS–PAGE, and emulsified with Freund's adjuvant (Sigma). This emulsion was used to immunize two New Zealand male rabbits by hypodermic injection. The antiserum was purified by ammonium sulfate precipitation.
Western-blotting analysis
Electrophoretic transfer of proteins from SDS–polyacrylamide gels to unmodified nitrocellulose was based on Towbin et al. (1979). The other steps were carried out according to Tian et al. (2003).
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS)
The visualized protein spots were excised from gels by an Ettain Spot Cutter (Amersham Bioscience, Sweden). After being destained and dehydrated, the protein was digested in-gel with sequencing grade-modified trypsin (Roche) (10 ng ml−1 in 25 mm ammonium bicarbonate) for 16 h at 37 °C. The enzyme digest solution was spotted onto a sample plate with matrix (α-cyano-4-hydroxcinnamic acid, 8 mg mL−1 in 50 % trifluoroacetic acid) and allowed to air dry. Known trypsin autocleavable peptide masses (906·51 and 2273·16 Da) were used for a two-point internal calibration for each spectrum. Examples of peptide mass fingerprinting (PMF) were searched against NCBI protein databases with the search engine Matrix Science at http://www.matrixscience.com. All peptide masses were assumed to be monoisotopic and [M1H]1 (protonated molecular ions). Searches were conducted using a mass accuracy of 6100 ppm, and one missed cleavage site was allowed for each search.
Preparation of total RNA
Preparation of total RNA followed a routine protocol for RNA extraction (Sambrook et al., 1989) with small modifications. Plant materials were ground to a fine powder in liquid nitrogen using a mortar and pestle. Two grams of the powder were mixed with 15 mL of extraction buffer [2 % cetyl trimethyl ammonium bromide (CTAB), 25 mm EDTA, 2 m NaCl, 100 mm Tris–HCl, 1 % N-lauroylsarcosine sodium salt (LDS), 2 % PVPP, 1·25 m NaBO4, 2 % β-mercaptoethanol) in a 50-mL centrifuge tube for 3–5 min and incubated in water at 65 °C for 20 min. The RNA samples were treated with RNase-free DNase I extraction buffer (2 % CTAB, 25 mm EDTA, 2 m NaCl, 100 mm Tris–HCl, 1 % LDS, 2 % PVPP, 1·25 m NaBO4, 2 % mercaptoethanol) in a 50-mL centrifuge tube for 3–5 min and incubated in water at 65 °C for 20 min. The RNA samples were treated with RNase-free DNase I (Fermentas, Canada) before they were used for reverse transcription.
Full-length cDNA cloning by rapid amplification of cDNA ends (RACE)
The N-terminal amino acid residues of the 22-kDa protein were sequenced by the Edman stepwise degradation method using an Applied Biosystem Procise 491 (ABI, USA) at Peking University (Beijing, China). The degenerate primers were designed as 5′-TA(CT) GA(CT) AT(ACT) AA(CT) GGH GA(CT) GA(AG) GTH AT-3′ (H = hypoxanthine). After reverse transcription (RT) with oligo dT-3site adaptor primer supplied by 3′-RACE kit (TaKaRa Biotech Co., Ltd, Dalian, China), the first-strand cDNA was used directly in a 3′-RACE PCR reaction. The amplified fragment was used to design one RT primer with its 5′-end phosphorylated (RTP: 5′-P-GCG GAC GAC AGC ACC TT-3′) and two pairs of PCR primers (P1: 5′-CCA ACA CAG GTC GGA TTT ACA-3′ and P2: 5′-GCA GCG TTC ATT TCT ACC AG-3′; P3: 5′-GTA GTC CAA TGA TGT TCT TTC CTG TCA A-3′ and P4: 5′-CGG ATG GCT GAG ACG ATG TAA TAT TC-3′) for the purpose of amplifying unknown 5′-end sequences of the cDNA by 5′-RACE according to the manufacturer's instructions (TaKaRa Biotech Co., Ltd, Dalian, China). To amplify the full-length cDNA, a pair of primers was designed (forward: 5′-ACC CAT CCC TCA AAA TAA GCG ACA TGA AGA G-3′; reverse: 5′-TTG GAT AAA CGA AAA GGA ACT CAT GTC CAT TAT TC-3′) according to the sequence information from the 3′- and 5′-RACE analyses.
Southern-blotting analysis
Genomic DNA was isolated from young leaves of seedlings using the CTAB method. The DNA was digested completely with EcoRI, HindIII or XhoI overnight, and then separated by electrophoresis in 1·0 % agarose gels, blotted onto Hybond-N membrane and probed with the LcVSP1 open reading frame (ORF) (32P labelled). The hybridization was performed according to the standard method of Sambrook et al. (1989).
Construction of expression vector systems in Escherichia coli and purification of fusion protein
Expression vector systems for LcVSP1 ORF were constructed using the Pgex6p1 vector (Invitrogen Biotech Co., USA). Restriction sites for EcoRI and XhoI were introduced by PCR at the 5′- and 3′-end of the LcVSP1 ORF, respectively. The amplified fragments were sequenced and digested with EcoRI and XhoI, and then ligated into Pgex6p1 vector. This expression plasmid was used to transform the E. coli BL21(DE3) strain and the fusion proteins were purified by GSTrap FF column (Invitrogen Biotech Co., USA) according to the manufacturer's instruction. The inhibitory activity of the purified fusion protein on trypsin was determined according to the method described below.
Assays for proteinase inhibitor activity
Assays for the inhibitory activity of the 22-kDa protein on trypsin and chymotrypsin, as well as subtilisin, were performed according to Erlanger et al. (1961). Inhibitory activity was measured against 16·2 µm trypsin (TPCK treated; Sigma, St Louis, MO, USA) or 333 ng of chymotrypsin and subtilisin (TLCK treated; Sigma,). Synthetic substrates N- ± benzoyl-dl-arginine-p-nitroanilide hydrochloride for trypsin and N-succinyl-l-alanyl-l-alanyl-l-prolyl-l-phenylalanine-p-nitroanilide for chymotrypsin and subtilisin were purchased from Sigma. Inhibitory activity was expressed as a percentage of the proteinase activity remaining after being incubated with inhibitor the 22 kDa protein for 30 min to the proteinase activity of control (without the 22 kDa protein). One trypsin inhibitor unit (UI) is defined as a decrease of 0·01 absorbance units at 410 nm 1 min−1 in 1 mL of reaction mixture under the assay conditions (Erlanger et al., 1961).
RESULTS
Protein-storing cells in perennial tissues
Paraffin sections stained with mercury–bromophenol blue revealed protein-storing cells (PSCs) with dense vacuolar inclusions which appeared clear blue under a light microscope (Fig. 1). The vacuolar inclusions were proteinaceous as evidenced by their staining property, and therefore served as a mark by which the PSCs could be recognized and their distribution in perennial tissues could be traced.
Fig. 1.
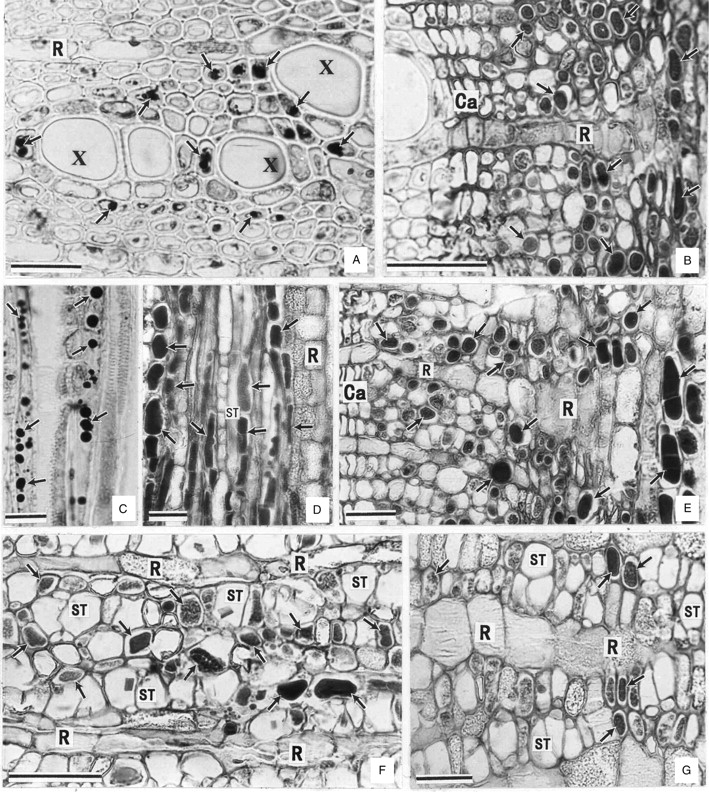
Distribution of protein-storing cells (arrows) in the perennial tissues. Note: no PSCs are distributed in the radial system (ray). (A, B) Cross-sections of xylem (A) and bark (B) of terminal branches. (C, D) Radial sections corresponding to (A) and (B), respectively. (E–G) Cross-sections of twigs (E), trunk (F) and large roots (G). Ca, Cambia; X, xylem; R, ray; ST, sieve tube. Scale bars = 20 µm.
In terminal branches (Fig. 1A–D) and twigs (Fig. 1E), PSCs were confined to the inner cortex and axial system of the secondary phloem and xylem. So, to a certain extent, the PSCs were not ordinary parenchyma cells. No PSCs were detected in the xylem of the trunk and large roots. In these organs, the PSCs were present in the secondary functional phloem or slightly beyond it and restricted to the axial system (Fig. 1F, G).
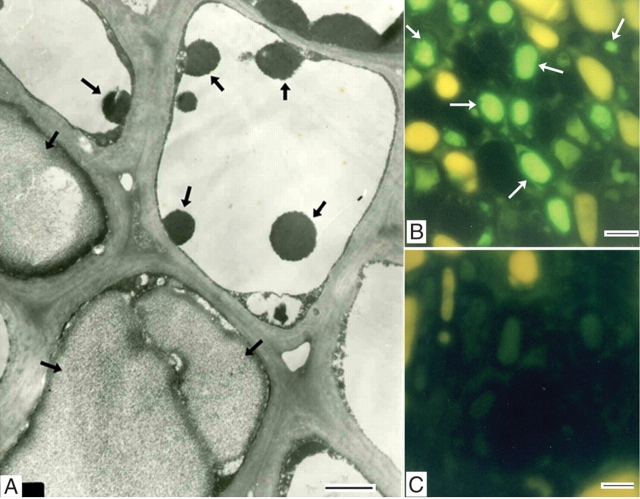
Electron microscopy revealed electron-dense masses in two different forms – granular and floccular – in the large central vacuoles of different phloem parenchyma cells (Fig. 2A). These masses should correspond to the proteinaceous inclusions in the PSCs observed under a light microscope (Fig. 1).
Fig. 2.
Identification of vacuolar protein inclusions. (A) Electron micrograph of the secondary phloem parenchyma cells in the stem of terminal branches, showing different forms of vacuolar inclusions (arrows). (B, C) Indirect immunofluorescence localization of the 22-kDa protein in terminal branches. Sections were treated with the polyclonal antibodies (B) and pre-immune-serum (C). Arrows indicate the materials labelled by FITC-specific fluorescence. Scale bars: A = 2 µm; B = 25 µm; C = 12·5 µm.
Isolation and identification of VSPs
A prominent protein band with molecular mass of about 22 kDa was detected in the SDS–PAGE profiles of soluble proteins from terminal branches (Fig. 3A, lane 2). By indirect immunofluorescence with polyclonal antibodies raised against the 22-kDa protein, materials labelled by FITC-specific blue-green fluorescence were observed in many of the axial parenchyma cells in the bark of terminal branches (Fig. 2B), corresponding to the proteinaceous inclusions in PSCs (Fig. 1B). The FITC-specific blue-green fluorescence could be easily distinguished from autofluorescence, such as the yellow autofluorescence emitted by tannin-like substances (Fig. 2B, C) and the weak autofluorescence by protein in the sections treated with pre-immune serum (Fig. 2C).
Fig. 3.
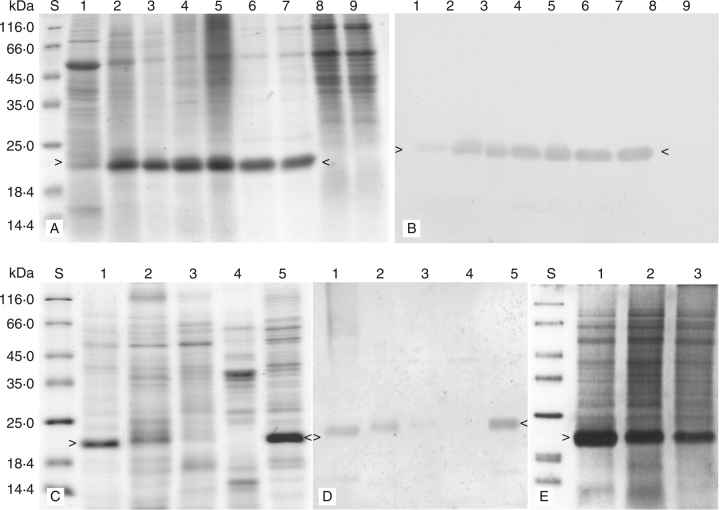
Distribution and seasonal fluctuation of the 22-kDa protein in lychee. (A, B) Polypeptide profiles from SDS–polyacrylamide gel electrophoresis (A) and western-immunoblots of their duplications (B), showing the distribution of a 22-kDa protein in leaves (1), bark tissues of terminal branches (2), twigs (3), trunk (4) and large roots (5), and xylem of terminal branches (6), twigs (7), trunk (8) and large roots (9). (C, D) Polypeptide profiles from 12 % SDS–polyacrylamide gel electrophoresis (C) and western immunoblots of their duplications (D), showing the distribution of the 22-kDa protein homologues in the peel of immature fruits (2) as well as the peel (3), pulp (4) and seeds (5) of mature fruits. Soluble proteins from the bark tissues of terminal branches (1) served as a positive control for the immuno-blot of the samples from fruits. (E) Polypeptide profiles from 12 % SDS–polyacrylamide gel electrophoresis, showing the 22-kDa protein mobilization. Samples were harvested from the bark tissues of terminal branches at bud sprouting stage (1), new shoot development (2) and fruiting (3).
Distribution of the 22-kDa protein in the vegetative organs was analysed by SDS–PAGE and western blot. The protein was largely absent from mature leaves (Fig. 3A, B, lane 1), but abundant in the bark tissues of terminal branches (Fig. 3A, B, lane 2), twigs (Fig. 3A, B, lane 3), trunk (Fig. 3A, B, lane 4) and large roots (Fig. 3A, B, lane 5). The protein was also present in considerable amounts in the xylem of terminal branches (Fig. 3A, B, lane 6) and twigs (Fig. 3A, B, lane 7), but could not be detected in the xylem of trunk (Fig. 3A, B, lane 8) and large roots (Fig. 3A, B, lane 9). Its homologues were present in the peel of immature (Fig. 3C, D, lane 2) and mature (Fig. 3C, D, lane 3) fruit as well as in the seed of mature fruit (Fig. 3C, D, lane 5), but absent from the pulp of mature fruit (Fig. 3C, D, lane 4).
The level of the 22-kDa protein in stems of the terminal branches was highest at the sprouting stage (Fig. 3E, lane 1) and then apparently decreased while the young shoot developed (Fig. 3E, lane 2) and during fruiting (Fig. 3E, lane 3).
Isolation and characterization of LcVSP1
The 22-kDa protein was transferred to a PVDF sheet and a partial amino acid sequence was determined by the automated Edman degradation. A sequence of the N-terminal 20 amino acid residues was: ASEPLYDINGDEVITGTEYY. Based on the genetic codes of the amino acid residues underlined, a degenerate primer was designed to amplify the 3′-end sequences of the protein cDNA by 3′-RACE. The nucleotide sequence of the amplified fragment was determined and used to design primers to amplify the unknown 5′-end sequences by 5′-RACE. In this way, a full-length cDNA from lychee was cloned. The cDNA, referred to as LcVSP1 (GenBank accession number: DQ659678) was 855 bp long containing a 675-bp ORF encoding a putative protein of 225 amino acids, flanked by a 23-bp 5′-UTR (untranslated region) and a 157-bp 3′-UTR including a poly(A) tail of 16 bp. The predicted molecular mass of the protein is 25 kDa, with a pI of 6·47. The amino acid sequence deduced from the residue alanine-25 to the residue tyrosine-44 was identical to that obtained by the automated Edman degradation. By the SignalP 3·0 Server (http://genome.cbs.dtu.dk/services/SignalP/), the sequence from residue methionine-1 to residue glycine-24 was predicted to be a signal peptide and the most likely cleavage site was between residues glycine-24 and alanine-25. Thus, the amino acid at the N-terminus of the mature protein is expected to be alanine, which is consistent with the result from the automated Edman degradation.
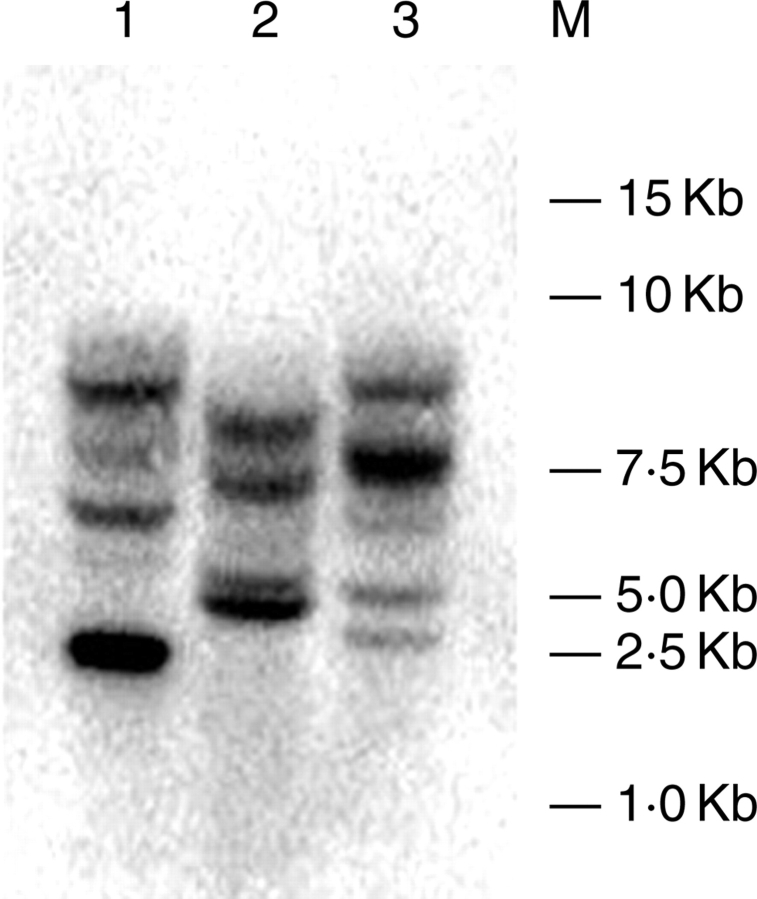
To test whether LcVSP1 contained intron(s), the primers used to amplify the full-length cDNA were used to amplify the gene from genomic DNA. A specific DNA fragment was obtained and its nucleotide sequence was identical to that of the full-length cDNA, suggesting that there was no intron in LcVSP1. Southern-blotting analysis confirmed that the LcVSP1 identified belongs to a small multigene family (Fig. 4).
Fig. 4.
DNA-blot profiles of the genomic DNA digested with EcoRI (1), HindIII (2) and XhoI (3), hybridized with the LcVSP1 ORF probe. Five hybridizing regions could be detected in the three samples digested by different endonucleases, respectively, under highly stringent washing conditions.
Blast analysis showed that LcVSP1 was homologous to Kunitz trypsin inhibitors. The deduced amino acid sequence shares 42·4 %, 42·4 %, 39·0 % identity with the trypsin inhibitors from Glycine max, Theobroma microcarpum and Populus balsamifera subsp. trichocarpa, respectively. It also shares 44·3 % identity with the miraculin-like protein 2 from Citrus jambhiri, 41·5 % identity with the tumour-related protein from Nicotiana tabacum, 36·8 % identity with lemir from Lycopersicon esculentum, and 27·2 % indentity with the endopeptidase inhibitor from Arabidopsis thaliana (Fig. 5).
Fig. 5.

Amino acid alignment of LcVSP1 and other proteins. The lemir from Lycopersicon esculentum (AAC63057), tumour-related protein from Nicotiana tabacum (AAC49969), endopeptidase inhibitor from Arabidopsis thaliana (NP_173228), trypsin inhibitor homologue from Glycine max (S16252), putative 21-kDa trypsin inhibitor from Theobroma microcarpum (AAV41234), miraculin-like protein2 from Citrus jambhiri (BAE79511) and Kunitz trypsin inhibitor 4 from Populus balsamifera subsp. trichocarpa (AAQ84217). Residues that are identical throughout the proteins are shown in black. Residues conserved in > 50 % of the proteins are shown in pink. Similar amino acid residues are shown in blue. Except for LcVSP1, all of the sequences were named conferring to their GenBank accession numbers.
Biochemical properties of the 22-kDa protein
To demonstrate that the LcVSP1 possessed trypsin inhibitor activity, the ORF was expressed heterologously as a fusion protein with the glutathione S-transferase fusion vector in E. coli strain BL21(DE3). The fusion protein with the prospective size of about 50 kDa was produced (Fig. 6A, lanes 2, 3). It was recognized by the polycolonal antibodies raised against the 22-kDa protein (Fig. 6B, lanes 2, 3) and had an inhibitory effect on trypsin. The average inhibitory activity of the fusion protein was 973 UI mg−1 protein at half maximum velocity of inhibition against 16·2 µg of trypsin.
Fig. 6.
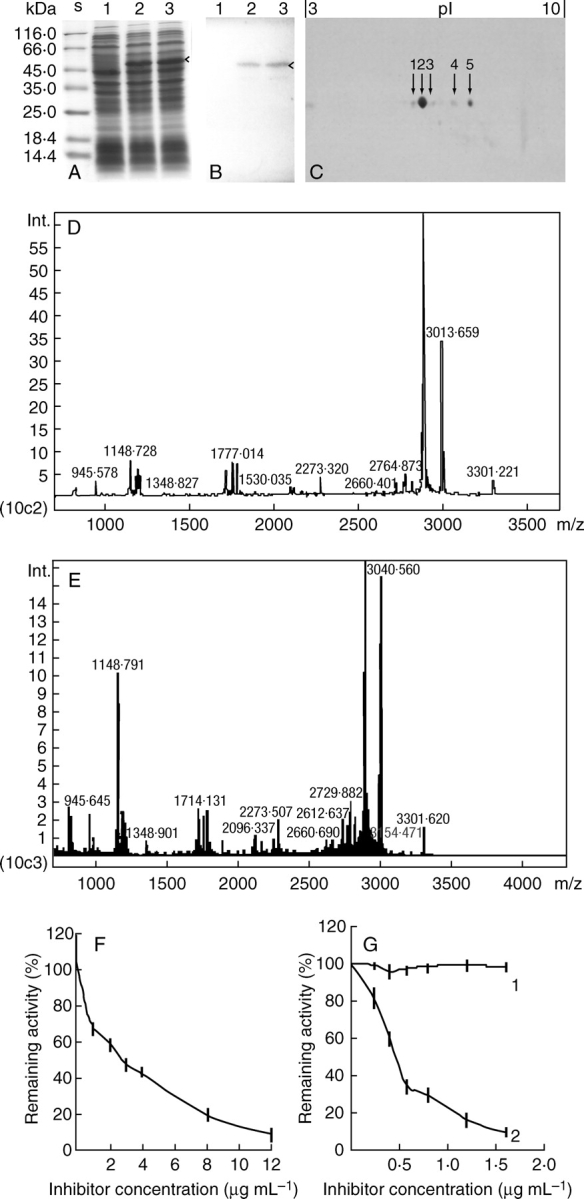
Biochemical analysis of the VSP in lychee. (A, B) Polypeptide profiles from 10 % SDS–polyacrylamide gel electrophoresis (A) and western immunoblots of their duplications (B), showing LcVSP1 heterologously expresessed in E. coli strain BL21(DE3). Samples from the IPTG-untreated control (1) and those treated with 3 mm IPTG for 2 h (2) and 4 h (3). (C) Polypeptide profiles of the purified 22-kDa protein from two-dimensional gel electrophoresis. The pH range used for isoelectric focusing is 3–10 as indicated on top of the gel. Of the five spots detected, spots 2 and 5 were identified by MALDI-TOF MS. (D, E) PMF pattern of spot 2 (D) and spot 5 (E) generated by MALDI-TOF MS. (F) Inhibitory effect of the 22-kDa protein on trypsin. (G) Inhibitory effect of the 22-kDa protein on subtilisin (1) and chymotypsin (2).
The purified 22-kDa protein from lychee was separated by two-dimensional polyacrylamide gel into five detectable spots with a pI of about 6·13, 6·47, 6·75, 7·42 and 7·94, respectively (Fig. 6C). Spot 2 was the most prominent and its pI (6·47) was the same as the predicted pI of the putative protein LcVSP1.
MALDI-TOF MS showed that the PMF pattern of spot 2 (Fig. 6D) was very similar to that of spot 5 (Fig. 6E). Based on the criteria that the proteins identified must rank at the top hit with more than four matched peptides and a sequence coverage of more than 10 % (Dai et al., 2006), both of the PMF patterns of spot 2 and spot 5 were matched to the putative protein LcVSP1 (DQ659678). By searching against the NCBI protein database with the search engine Matrix science, five mass values matched among the six mass values searched with a sequence coverage of 20 %. Thus, spots 2 and 5 and, probably, spots 1, 3 and 4 were isoforms of the 22-kDa protein in lychee.
The proteinase inhibitor activity of the purified 22-kDa protein was further tested in vitro. A typical purification is summarized in Table 2. The protein inhibited trypsin (Fig. 6F) and chymotrypsin (Fig. 6G, line 2) with a concentration of 2·5 µg mL−1 and 0·45 µg mL−1 at half maximum velocity of inhibition against 16·2 µg of trypsin and 333 ng of chymotrypsin, respectively. The inhibitory effect of the 22-kDa protein on subtilisin could not be detected (Fig. 6G, line 1).
Table 2.
Purification of the 22-kDa protein from the soluble proteins in bark
| Fraction | Total activity (UI) | Protein (mg) | Specific activity (UI mg−1) | Yield ( %) | Purification (-fold) |
|---|---|---|---|---|---|
| Bark protein supernant | 16575·30 | 78·93 | 210 | 100 | 1 |
| Sephacryl S-100HR | 6415·97 | 13·65 | 470 | 38·71 | 2·24 |
| DEAE-Sepharose | 6057·8 | 6·06 | 1000 | 36·55 | 4·76 |
DISCUSSION
The 22-kDa protein in Litchi chinensis fits the criteria applied to the typical VSPs in plants (Clausen and Apel, 1991; Staswick, 1994), i.e. it is sequestered in the vacuoles of PSCs to a relatively high content and with an evident seasonal fluctuation.
The PSCs in the vegetative organs of trees have been divided into two types: Hevea-type and Populus-type (Tian et al., 1998). PSCs in lychee (Litchi chinensis), however, are different from either the Hevea-type or the Populus-type of PSC. The PSCs in lychee are similar to the Populus-type of PSCs in that they occur in the xylem as well as in the bark tissues (phloem and cortex) of branches and the molecular mass of the VSP, 22 kDa, is within the range of 15- to 45-kDa, which is considered to be a feature of the VSPs in the Populus-type of PSC (Stepien et al., 1994; Tian et al., 2003). On the other hand, the PSCs in lychee differ from the Populus-type of PSC in that their distribution is confined to the axial system of the secondary phloem, which is the characteristic of the Hevea-type of PSC (Tian et al., 1998). Thus, the PSCs in lychee can be considered as a novel type of PSC.
Previous data have shown that evergreen trees are poor in VSPs and the VSPs in the perennial tissues disappear completely in summer (Wetzel et al., 1991; Arona et al., 1992). Here, it is demonstrated that lychee, an evergreen subtropical fruit tree, is rich in a 22-kDa VSP. The 22-kDa protein is mainly distributed in the bark tissues of branches, trunk and large roots, and occurs in large amounts in the stems of branches all the year round. The protein decreased during young shoot and fruit development, suggesting a role in nitrogen storage. The seasonal fluctuation of the 22-kDa protein in lychee (Fig. 3E) is similar to that of general stored nitrogen compounds in other evergreen trees (Kato, 1981, 1986), which is much less dramatic than that of the VSPs in deciduous trees where the abundant VSPs accumulated in the stem of branches are mobilized almost completely during young shoot development (Wetzel et al., 1989; Tian et al., 2003). Nevertheless, based on the fact that nitrogen application has little effect on fruit production of lychee (Menzel et al., 1994), the stored nitrogen compounds in the vegetative organs, one of the main components of which should be the 22-kDa protein, may therefore play a pivotal role in supporting fruit growth and development.
Although the VSPs in many herbaceous plants are generally bioactive, none of the VSPs identified in trees, as far as is known, have been reported as having special bioactivities, except for the lectins, which behave as storage proteins (Nsimba-Lubaki et al., 1986; Herman et al., 1988; Baba et al., 1991; Hankins et al., 1998) and have a role in plant defence (Peumans and van Damme, 1995). The proteinase inhibitors that are considered to be VSPs in the leaves of some herbaceous plants belong to the Bowman–Birk trypsin inhibitor family. They accumulate in the large central vacuoles under biotic and/or abiotic stress conditions and their defence function has been well demonstrated (Graham et al., 1986; Ryan et al., 1990; Moura and Ryan 2001; Horn et al., 2005). Here evidence is provided that a typical VSP, the 22-kDa protein in lychee, possesses trypsin inhibitor activity. The protein inhibits trypsin and chymotrypsin as is the case of the Kunitz trypsin inhibitor in Peltophorum dubium (Troncoso et al., 2003), but has no inhibitory effect on subtilisin. Meanwhile, the putative LcVSP1 protein is homologous to Kunitz trypsin inhibitors. Taken together, the 22-kDa protein in lychee belongs to Kunitz trypsin inhibitor family. In poplar, a number of Kunitz trypsin inhibitor genes are up-regulated shortly after leaf damage by wounding (Christopher et al., 2004). Considering the fact that all of the trypsin inhibitors that behave as VSPs in herbaceous plants are Bowman–Birk trypsin inhibitors, the possible defence function of the 22 kDa VSP, a Kunitz trypsin inhibitor in lychee, remains to be elucidated.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (No. 30460107).
LITERATURE CITED
- Andrews DL, Beames B, Summers MD, Park WD. Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochemical Journal. 1988;252:199–206. doi: 10.1042/bj2520199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Wisniewski ME, Scorza R. Cold acclimation in genetically related (sibing) deciduous and evergreen peach [Prunus persica (L.) Batsch]. I. Seasonal changes in cold hardiness and polypeptides of bark and xylem tissues. Plant Physiology. 1992;99:1562–1568. doi: 10.1104/pp.99.4.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K, Ogawa M, Nagano A, Kuroda H, Sumiya K. Developmental changes in the bark lectin of Sophora japonica L. Planta. 1991;183:462–470. doi: 10.1007/BF00197746. [DOI] [PubMed] [Google Scholar]
- Berger S, Bell E, Sadka A, Mullet JE. Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Molecular Biology. 1995;27:933–942. doi: 10.1007/BF00037021. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Christopher ME, Miranda M, Major IT, Constabel CP. Gene expression profiling of systemically wound-induced defenses in hybrid poplar. Planta. 2004;219:936–947. doi: 10.1007/s00425-004-1297-3. [DOI] [PubMed] [Google Scholar]
- Clausen S, Apel K. Seasonal changes in the concentration of the major storage protein and its mRNA in xylem ray cells of poplar trees. Plant Molecular Biology. 1991;17:669–678. doi: 10.1007/BF00037052. [DOI] [PubMed] [Google Scholar]
- Coleman GD, Chen THH. Sequence of a poplar bark storage protein gene. Plant Physiology. 1993;102:1347–1348. doi: 10.1104/pp.102.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke JEK, Weih M. Nitrogen storage and seasonal nitrogen cycling in Populus: bridging molecular physiology and ecophysiology. New Phytologist. 2005;167:19–30. doi: 10.1111/j.1469-8137.2005.01451.x. [DOI] [PubMed] [Google Scholar]
- Dai S-J, Li L, Chen T-T, Chong K, Xue Y-B, Wang T. Proteomic analyses of Oryza sativa mature pollen reveal novel proteins associated with pollen germination and tube growth. Proteomics. 2006;6:2504–2529. doi: 10.1002/pmic.200401351. [DOI] [PubMed] [Google Scholar]
- Davis JM, Egelkrout EE, Coleman GD, Chen THH, Haissig BE, Riemenschneider DE, et al. A family of wound-induced genes in Pupulus shares common features with genes encoding vegetative storage proteins. Plant Molecular Biology. 1993;23:135–143. doi: 10.1007/BF00021426. [DOI] [PubMed] [Google Scholar]
- De Wald DB, Mason HS, Mullet JE. The soybean vegetative storage proteins VSP-alpha and VSP-beta are acid-phosphatases active on polyphosphates. Journal of Biological Chemistry. 1992;267:15958–15964. [PubMed] [Google Scholar]
- Dhont C, Castonguay Y, Avice J-C, Chalifour F-P. VSP accumulation and cold-inducible gene expression during autumn hardening and overwintering of alfalfa. Journal of Experimental Botany. 2006;57:2325–2337. doi: 10.1093/jxb/erj204. [DOI] [PubMed] [Google Scholar]
- Erlanger BF, Kokowski N, Cohen W. The preparation and properties of two new chromogenic substrates of trypsin. Archives of biochemistry and biophysics. 1961;95:271–278. doi: 10.1016/0003-9861(61)90145-x. [DOI] [PubMed] [Google Scholar]
- Flores T, Alape-Giron A, Flores-Diaz M, Flores HE. Ocatin: a novel tuber storage protein from the Andean tuber crop oca with antibacterial and antifungal activities. Plant Physiology. 2002;128:1291–1302. doi: 10.1104/pp.010541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi VR, Vittennbach VA, Giaquinta RT. Paraveinal mesophyll of soybean leaves in relation to assimilate transfer and compartmentation. III. Immunohistochemical localization of specific glycopeptides in the vacuole after depodding. Plant Physiology. 1983;72:586–589. doi: 10.1104/pp.72.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JS, Hall G, Pearce G, Ryan CA. Regulation of synthesis of proteinase inhibitor I and II mRNAs in leaves of wounded tomato plants. Planta. 1986;169:399–405. doi: 10.1007/BF00392137. [DOI] [PubMed] [Google Scholar]
- Hankins CN, Kindinger JI, Shannon LM. The lectins of Sophora japonica: purification, properties, and N terminal amino acid sequences of five lectins from bark. Plant Physiology. 1998;86:67–70. doi: 10.1104/pp.86.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B-Z, Wu J-L. Vacuole proteins in parenchyma cells of secondary phloem and xylem of Dalbergia odorifera. Trees. 1993;8:104–109. [Google Scholar]
- Herman EM, Charles HN, Leland SM. Bark and leaf lectins of Sophora japonica are sequestered in protein-storage vacuoles. Plant Physiology. 1988;86:1027–1031. doi: 10.1104/pp.86.4.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M, Patankar AG, Zavala JA, Wu J-Q, Dolecková-Marešová L, Vujtchová M, et al. Differential elicitation of two processing proteases controls the processing pattern of the trypsin proteinase inhibitor precursor in Nicotiana attenuata. Plant Physiology. 2005;135:375–388. doi: 10.1104/pp.105.064006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T. Major nitrogen compounds transported in xylem vessels from roots to top in citrus trees. Physiologia Plantarum. 1981;52:275–279. [Google Scholar]
- Kato T. Nitrogen metabolism and utilization in citrus. Horticultural Reviews. 1986;8:181–216. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langheinrich U, Tischner R. Vegetative storage proteins in poplar: induction and characterization of a 32- and a 36-kilodalton polypeptide. Plant Physiology. 1991;97:1017–1025. doi: 10.1104/pp.97.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-L, Ahn J-E, Datta S, Salzman RA, Moon J, Huyghues-Despointes B, et al. Arabidopsis vegetative storage protein is an anti-insect acid phosphatase. Plant Physiology. 2005;139:1545–1556. doi: 10.1104/pp.105.066837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel CM, Haydon GF, Doogan VJ, Simpson DR. Time of nitrogen application and yield of Bengal lychee on a sandy loam soil in subtropical Queensland. Australian Journal of Experimental Agriculture. 1994;34:803–811. [Google Scholar]
- Meuriot F, Noquet C, Avice JC, Volenec JJ, Cunningham SM, Sors TG, et al. Methyl jasmonate alters N partitioning, N reserves accumulation and induces gene expression of a 32-kDa vegetative storage protein that possesses chitinase activity in Medicago sativa taproots. Physiologia Plantarum. 2004;120:113–123. doi: 10.1111/j.0031-9317.2004.0210.x. [DOI] [PubMed] [Google Scholar]
- Moura DS, Ryan CA. Wound-inducible proteinase inhibitors in pepper: differential regulation upon wounding, systemin, and methyl jasmonate. Plant Physiology. 2001;126:289–298. doi: 10.1104/pp.126.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsimba-Lubaki M, Allen AK, Permans WJ. Isolation and characterization of glycoprotein lectins from the bark of three species of elder. Sambucus ebulus, Sambucus nigra and Sambucus racemosa. Planta. 1986;168:113–118. doi: 10.1007/BF00407017. [DOI] [PubMed] [Google Scholar]
- Peumans WJ, van Damme EJM. The role of lectins in plant defence. Histochemical Journal. 1995;27:253–271. doi: 10.1007/BF00398968. [DOI] [PubMed] [Google Scholar]
- Roberts DR, Toivonen P, McInnis SM. Discrete proteins associated with overwintering of spruce and Douglas-fir seedlings. Canadian Journal of Botany. 1991;69:437–441. [Google Scholar]
- Ryan CA. Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annual Review of Phytopathology. 1990;28:425–449. [Google Scholar]
- Sambrook J, Fritsh EF, Manniatis T. Molecular cloning, a laboratory manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Staswick PE. Storage proteins of vegetative plant tissues. Annual Review of Plant Physiology and Plant Molecular Biology. 1994;45:303–322. [Google Scholar]
- Staswick PE, Zhang Z, Clemente TE, Specht JE. Efficient down-regulation of the major vegetative storage protein genes in transgenic soybean does not compromise plant productivity. Plant Physiology. 2001;127:1819–1826. [PMC free article] [PubMed] [Google Scholar]
- Stepien V, Martin F. Purification, characterization and localization of the bark storage proteins of poplar. Plant Physiology and Biochemistry. 1992;30:399–407. [Google Scholar]
- Stepien V, Sauter JJ, Martin F. Vegetative storage proteins in woody plants. Plant Physiology and Biochemistry. 1994;32:185–192. [Google Scholar]
- Tian W-M, Hu Z-H. Distribution and ultrastructure of vegetative storage proteins in Leguminosae. IAWA Journal. 2004;25:459–469. [Google Scholar]
- Tian W-M, Han Y-Q, Wu J-L, Hao B-Z. Characteristics of protein-storing cells associated with a 67 kDa protein in Hevea brasiliensis. Trees. 1998;12:153–159. [Google Scholar]
- Tian W-M, Wu J-L, Hao B-Z, Hu Z-H. Vegetative storage proteins in the tropical tree Swietenia macrophylla: seasonal fluctuation in relation to a fundamental role in the regulation of tree growth. Canadian Journal of Botany. 2003;81:492–500. [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of protein from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proceedings of the National Academy of Sciences of the USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranbarger TJ, Franceschi VR, Hildebrand DF, Grimes HD. The soybean 94-kilodalton vegetative storage protein is a lipoxygenase that is localized in paraveinal mesophyll cell vacuoles. The Plant Cell. 1991;3:973–987. doi: 10.1105/tpc.3.9.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso MF, PC Zolezzi, U Hellman, C Wolfenstein-Todel. A novel trypsin inhibitor from Peltophorum dubium seeds, with lectin-like properties, triggers rat lymphoma cell apoptosis. Archives of Biochemistry and Biophysics. 2003;411:93–104. doi: 10.1016/s0003-9861(02)00726-9. [DOI] [PubMed] [Google Scholar]
- Volenec JJ, Ourry A, Joern BC. A role for nitrogen reserves in forage regrowth and stress tolerance. Physiologia Plantarum. 1996;97:185–193. [Google Scholar]
- Wetzel S, Greenwood JS. Proteins as a potential storage compound in bark and leaves of several softwoods. Trees. 1989;3:149–153. [Google Scholar]
- Wetzel S, Demmers C, Greenwood JS. Seasonally fluctuating bark proteins are a potential form of nitrogen storage in three temperate hardwoods. Planta. 1989;178:275–281. doi: 10.1007/BF00391854. [DOI] [PubMed] [Google Scholar]
- Wetzel S, Demmers C, Greenwood JS. Protein-storing vacuoles in inner bark and leaves of softwoods. Trees. 1991;5:196–202. [Google Scholar]
- Wittenbach VA. Purification and characterization of a soybean leaf storage glycoprotein. Plant Physiology. 1983;73:125–129. doi: 10.1104/pp.73.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-L, Hao B-Z. Protein-storing cells in the secondary phloem of Hevea brasiliensis. Chinese Science Bulletin. 1986;3:221–223. [Google Scholar]
- Wu J-L, Hao B-Z. Vacuole protein in secondary phloem parenchyma cells of the Meliaceae species. IAWA Bulletin. 1991;12:51–56. [Google Scholar]
- Yeh KW, Chen JC, Lin MI, Chen YM, Lin CY. Functional activity of sporamin from sweet potato (Ipomoea batatas Lam): a tuber storage protein with trypsin inhibitory activity. Plant Molecular Biology. 1997;33:565–570. doi: 10.1023/a:1005764702510. [DOI] [PubMed] [Google Scholar]