Abstract
Background and Aims
Hymenaea stigonocarpa (Fabaceae: Caesalpinioideae) is an endemic tree from the Brazilian cerrado (savanna vegetation), a biome classified as a hotspot for conservation priority. This study investigates the phylogeographic structure of H. stigonocarpa, in order to understand the processes that have led to its current spatial genetic pattern.
Methods
The polymorphism level and spatial distribution of variants of the plastid non-coding region between the genes psbC and trnS were investigated in 175 individuals from 17 populations, covering the greater part of the total distribution of the species. Molecular diversity indices were calculated and intra-specific relationships were inferred by the construction of haplotype networks using the median-joining method. Genetic differentiation among populations and main geographical groups was evaluated using spatial analysis of molecular variance (SAMOVA).
Key Results
Twenty-three different haplotypes were identified. The level of differentiation among the populations analysed was relatively high (FST = 0·692). Phylogeographic analyses showed a clear association between the haplotype network and geographic distribution of populations, revealing three main geographical groups: western, central and eastern. SAMOVA corroborated this finding, indicating that most of the variation can be attributed to differences among these three groups (58·8 %), with little difference among populations within groups (FSC = 0·252).
Conclusions
The subdivision of the geographic distribution of H. stigonocarpa populations into three genetically differentiated groups can be associated with Quaternary climatic changes. The data suggest that during glacial times H. stigonocarpa populations became extinct in most parts of the southern present-day cerrado area. Milder climatic conditions in the north and eastern portions of the cerrado resulted in maintenance of populations in these regions. Thus it is inferred that the most southern part of the present-day cerrado was re-colonized by different lineages from northern parts of this biome, after postglacial climate amelioration.
Key words: Biogeography, cerrado, genetic structure, Quaternary climate changes, Fabaceae, Leguminosae, Hymenaea stigonocarpa, neotropical savannas, Pleistocene, phylogeography, psbC-trnS
INTRODUCTION
Cerrado, the savannas of central Brazil, is the second most extensive biome in South America after the Amazon rain forest (Eiten, 1972). Recently, it was classified as a hotspot for conservation priority because of its rich biodiversity, with many endemic plants and animals. It is also extremely endangered by human action. Natural cerrado vegetation now covers only 20 % of its original area (about 1·7 million km2; Myers et al., 2000). The cerrado climate is characterized by conspicuous dry season during the southern winter (approx. April to September) with an average annual precipitation between 800 mm and 2000 mm and an average annual temperature between 18 °C and 28 °C (Ratter et al., 2006). The vegetation is composed of grasses with relatively shallow roots and deeply rooted evergreen and deciduous woody plants, growing in oligotrophic soils and subject to frequent fires (Bucci et al., 2005).
Environmental changes in neotropical savannas appear to have been spatially complex during glacial periods. The present-day areas of cerrado in south-eastern and mid-western Brazil are probably remnants of a large, continuous area that existed in the past (Behling and Hooghiemstra, 2001) because of markedly dry conditions during the last glaciation. Palaeopalynological studies have suggested that in the last glacial period, the vegetation of the southern cerrado was replaced by subtropical grassland (Behling and Lichte, 1997; Behling, 1998), which apparently expanded >750 km northwards, reflecting a drier and colder climate and the occurrence of heavy frosts (Behling and Hooghiemstra, 2001).
The changes in the vegetation coverage and in the distribution of plant species during the Pleistocene, associated with widespread climatic instability, have been considered to be important factors in the levels of genetic diversity and population differentiation within species (Caron et al., 2000; Dutech et al., 2000; Richardson et al., 2001; Collevatti et al., 2003; Hopper and Gioia 2004).
Phylogeographic studies have been used to investigate the effects of past climatic changes on the genetic structure of animal and plant species. These studies allow one to make inferences about species evolution within biomes, and these can be used to plan conservation strategies. Most phylogeographic studies of plants have been based on the variation found in organellar genomes, mainly the plastid DNA which is maternally inherited in most angiosperms. Gene flow of maternally inherited genes occurs via seed dispersal and is thus more restricted than that of nuclear genes, which are biparentally inherited and dispersed by pollen and seed (Birky et al., 1983; Ennos, 1994). The genetic structure of organellar genomes can be greatly influenced by their historical relationship of, and associated gene flow between, populations, as well as by climatic events such as glaciations that occur within a geological time frame (Avise et al., 1987; Avise, 1994; Schaal et al., 1998). Studies in which the genetic structure of angiosperm populations was characterized both by plastid DNA and nuclear DNA markers have shown that plastid DNA variation is more spatially structured than nuclear DNA variation (Ennos, 1994; El Mousadik and Petit, 1996; Petit et al., 2005), as expected due to the smaller effective size of plastid genes compared to nuclear genes.
Most phylogeographic studies in plants have been performed on holarctic species (e.g. Dumolin-Lapègue et al., 1997; Clark et al., 2000; Belahbib et al., 2001; Gugerli et al., 2001; Magni et al., 2005; Schierenbeck et al., 2005; Zhang et al., 2005). These studies have helped to reconstruct the history of the species distribution and to identify refugia and routes of postglacial colonization (Ferris et al., 1993; Petit et al., 1993; Dumolin-Lapègue et al., 1997; Petit et al., 2003). In the neotropical region, phylogeographic data are scarce (Caron et al., 2000; Dutech et al., 2000; Richardson et al., 2001; Cavers et al., 2003; Salgueiro et al., 2004; Lorenz-Lemke et al., 2005), especially for plants occurring in the Brazilian cerrado, for which only one study is known so far (Collevatti et al., 2003).
Hymenaea stigonocarpa, known as ‘jatobá-do-cerrado’, is an endemic species of the cerrado, occurring across almost the entire region occupied by this biome (approx. between 4–23°S and 41–55°W). It is among the dominant woody species in the cerrado flora, occurring in 236 of 316 sites analysed (Ratter et al., 2006). It is an economically valuable tree because its wood is long lasting and durable, and it is thus widely used in naval and civil construction (Rizzini, 1971). Its fruits have nutritional potential, both for wild animals and humans (Silva et al., 2001). Studies of the pollination biology and breeding system of H. stigonocarpa have shown that the species is an outcrosser, with pollination mainly by bats (Gibbs et al., 1999), as in H. courbaril (Crestana et al., 1985). There is no information in the literature about seed dispersal of H. stigonocarpa. However, it is widely accepted for H. courbaril that mammals are the principal seed dispersers (Asquith et al., 1999).
This study investigates the phylogeographic structure of H. stigonocarpa, in order to understand the processes that have resulted in its current spatial genetic pattern. The survey involved the analysis of populations of H. stigonocarpa sampled from the greater part of its range. The sequencing of the non-coding plastid DNA region, psbC-trnS, together with the palaeopalynological and palaeoclimatologic information available for south-eastern and central Brazil were used to infer the history of past changes leading to the present-day distribution of the species and also to identify possible colonization routes.
MATERIALS AND METHODS
Sampling populations and DNA extraction
Young leaves were collected from 175 adult individuals of H. stigonocarpa Mart. ex Hayne (Fabaceae: Caesalpinioideae) from 17 populations (Table 1), ranging from 10–23°S and 41–50°W and from 270–1080 m a.s.l., covering the greater part of its distribution (Table 1 and Fig. 1). Leaves were collected and stored in labelled plastic bags at −20 °C until DNA extraction. Voucher specimens from most of the populations collected were deposited in the Herbarium of the Departamento de Botânica da Universidade Federal de Minas Gerais (BHCB).
Table 1.
Geographical location of Hymenaea stigonocarpa populations, altitude, number of individuals sampled per population, number of haplotypes per population and diversity indices based on the psbC/trnS3 region of plastid DNA
| Populations/state | Abbr. | Latitude/longitude | Altitude (m) | n | nh | A | k | h | π |
|---|---|---|---|---|---|---|---|---|---|
| São Manuel/SP | SMC | 22°43′18″S/48°25′49″W | 530 | 6 | 1 | 0·000 | 0 | 0 | 0 |
| Itu/SP | ITC | 23°23′44″S/47°20′00″W | 634 | 3 | 2 | 0·667 | 0·667 | 0·00127 | |
| Pirenópolis/GO | PIC | 15°30′58″S/49°08′11″W | 730 | 11 | 2 | 0·818 | 0·327 | 0·327 | 0·00063 |
| Faina/GO | FAC | 15°32′53″S/50°17′45″W | 400 | 9 | 2 | 0·988 | 0·500 | 0·500 | 0·00096 |
| Chapada dos Veadeiros/GO | CVC | 14°10′39″S/47°49′04″W | 800 | 15 | 2 | 0·400 | 0·133 | 0·133 | 0·00026 |
| Tamanduá/DF | JTC | 15°45′23″S/47°49′36″W | 1000 | 6 | 2 | 1·000 | 0·533 | 0·533 | 0·00102 |
| Palmas/TO | TOC | 10°12′47″S/48°21′38″W | 270 | 7 | 2 | 1·000 | 0·571 | 0·571 | 0·00109 |
| Tupaciguara/MG | TUC | 18°31′32″S/48°59′29″W | 650 | 10 | 2 | 0·600 | 0·200 | 0·200 | 0·00058 |
| Furnas/MG | FUC | 20°51′49″S/46°23′16″W | 880 | 16 | 3 | 1·858 | 0·958 | 0·708 | 0·00115 |
| Abadia dos Dourados/MG | ADC | 18°29′03″S/47°22′35″W | 800 | 9 | 1 | 0·000 | 0 | 0 | 0 |
| Dores do Indaia/MG | DIC | 19°26′48″S/45°35′35″W | 730 | 7 | 1 | 0·000 | 0 | 0 | 0 |
| Fazenda Brejão/MG | FBC | 17°00′00″S/45°54′00″W | 550 | 10 | 2 | 0·967 | 0·467 | 0·467 | 0·00089 |
| Vale do Peruaçu National Park/MG | PEC | 15°07′20″S/44°14′53″W | 700 | 14 | 2 | 0·692 | 0·264 | 0·264 | 0·00051 |
| Cachoeira do Pajeú/MG | CHC | 15°58′00″S/41°30′00″W | 750 | 10 | 2 | 0·600 | 0·400 | 0·200 | 0·00077 |
| Montes Claros/MG | MCC | 16°18′41″S/42°53′26″W | 800 | 15 | 6 | 2·766 | 1·391 | 0·771 | 0·00267 |
| Rio Preto State Park/MG | RPC | 18°00′00″S/43°23′00″W | 900 | 8 | 3 | 1·750 | 1·393 | 0·679 | 0·00267 |
| Chapada da Diamantina/BA | MUC | 13°00′00″S/41°29′24″W | 1080 | 19 | 6 | 2·644 | 1·392 | 0·754 | 0·00267 |
n, Sample size; nh, number of haplotypes; A, haplotypic richness; k, average number of nucleotide differences; h, haplotype diversity; π, nucleotide diversity.
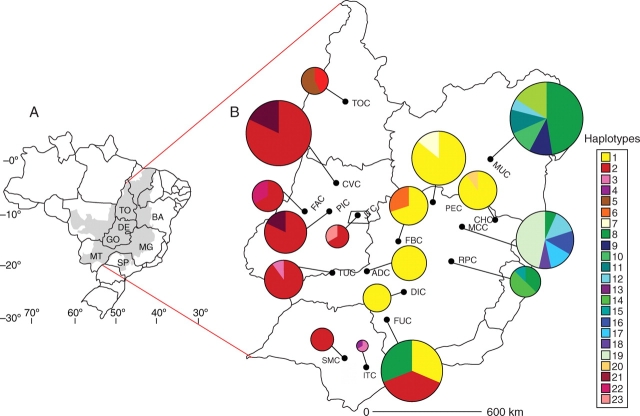
Fig. 1.
(A) Map of Brazil and distribution of cerrado vegetation in grey. (B) Approximate geographic location and plastid DNA haplotype frequencies of the populations of H. stigonocarpa studied. Circle size is proportional to sample size and colours represent the different haplotypes as shown in the key.
Total DNA was extracted by the protocol originally described by Doyle and Doyle (1987) with the modifications suggested by Ferreira and Grattapaglia (1995). Quantity and quality of DNA were assessed by visualization on a 0·8 % agarose gel.
Plastid DNA sequencing
To screen for variation in plastid DNA, nine regions were investigated using the nine ‘universal’ primer combinations: trnK1/trnK2, trnH/trnK, psbC/trnS3 (Demesure et al., 1995), trnQ/trnS2 (Dumolin-Lapègue et al., 1997), ccmp4-L/atpH (Weising and Gardner, 1999), psbB/psbF, rpl20/rps12 (Hamilton, 1999), trnL-c/trnL-d and trnL-e/trnF-f (Taberlet et al., 1991). Of these, trnH/trnK, psbC/trnS3, trnL-c/trnL-d and trnL-e/trnF-f produced clear single products, but only the first two regions showed variation in the samples analysed. Sequences for trnH/trnK were of low quality. The psbC/trnS3 region was approx. 1600 base pairs (bp) long and was sequenced for all individuals of H. stigonocarpa.
Polymerase chain reactions (PCR) were carried out in 25 µL final volume, containing 10 ng template DNA, 1 × PCR buffer (IC; Phoneutria), 200 µm dNTPs; 0·5 µm each primer, 5 µg of bovine serum albumin (BSA) and 1 U Taq polymerase (Phoneutria). After amplification, PCR products were visualized on 1 % agarose gels stained with ethidium bromide, and were purified using polyethylene glycol (PEG) 20 %/2·5 m NaCl precipitation. To sequence the psbC/trnS3 region, psbC (Demesure et al., 1995) and RCS 5′-AAGATATGCCAGATTCCACC-3′ (designed using a sequence alignment for six species of Fabaceae: H. courbaril, H. stigonocarpa, H. reticulata, H. aurea, Dalbergia nigra, Plathymenia reticulata) primers were used.
Sequencing was conducted in 10-μL reactions with 3 µL of purified PCR product, 2 µL of milliQ water, 1 µL of primer (5 µm) and 4 µL of ET-DYE Terminator Kit (Amersham Biosciences). The thermocycling programme was as follows: 35 cycles of 25 s at 95 °C, 15 s at 54 °C and 3 min at 60 °C. Sequencing products were precipitated and cleaned with ammonium acetate and ethanol, and then dried at room temperature, dissolved in loading buffer (formamide 70 % and 1 mm EDTA) and run on a MegaBACE sequencer (80 s injection time, 240 min run length).
Data analysis
Consensus sequences were assembled for each individual using at least two forward and two reverse sequences made from independent PCR products, using the software Phred v. 0·20425 (Ewing and Green, 1998; Ewing et al., 1998), Phrap v. 0·990319 (http://www.phrap.org/) and Consed 12·0 (Gordon et al., 1998). Multiple sequence alignments were made using Clustal X (Thompson et al., 1997) implemented in MEGA 3·0 (Kumar et al., 2004). Clustal alignments were also checked and edited by hand to minimize software artefacts.
Molecular diversity indices (π, nucleotide diversity; h, haplotype diversity; k, mean number of nucleotide substitutions) were calculated using MEGA 3·0 and DNAsp 3·99 (Rozas et al., 2003). The haplotypic richness was estimated by RAREFAC that uses the technique of rarefaction to correct for sample size (Petit et al., 1998). Typically, rarefaction is used to standardize allelic richness to the smallest n in a comparison (Petit et al., 1998). However, the ITC population was not included in this analysis due to its small sample size (n = 3) and a rarefaction size of n = 6 was used. Intra-specific relationships were inferred by the construction of haplotype networks using the median-joining algorithm (Bandelt et al., 1999) implemented in NETWORK 4·1 (Forster et al., 2000). Hymenaea reticulata and H. aurea were designated as outgroups. To test the influence of geography in population genetic structure, simple linear regressions were made to correlate geographical distances with genetic distance index (FST values) using the Barrier 2·2 software (Manni et al., 2004). Estimates of differentiation and F statistics were calculated taking into account the pairwise distance between plastid DNA haplotypes. The program SAMOVA (spatial analysis of molecular variation; Dupanloup et al., 2002) was used in order to explore the population structure without a priori hypotheses of the expected structure. This method uses a simulated annealing procedure to define K groups of populations that are geographically homogenous and maximally differentiated from each other. The method requires the a priori definition of the number of groups (K) of populations that exist, and generates F statistics (FSC, FST and FCT) using an AMOVA approach (Excoffier et al. 1992). By exploring the behaviour of the indices FCT and FSC for different values of K, it is possible (Dupanloup et al., 2002) to identify the optimum number of population groups for a set of sample populations. One hundred simulated annealing processes were used for each value of K from K = 2 to K = 8. Pairwise comparisons of FST between populations were analysed using an AMOVA implemented in ARLEQUIN ver. 3·01 (Excoffier et al., 2005).
RESULTS
Genetic diversity
The amplification of the non-coding plastid DNA region psbC/trnS3 produced a fragment of approx. 1600 bp, of which 524 bp were sequenced for all individuals. The aligned region included four indels at positions 14, 30, 402 and 509 (Table 2). There were 507 conserved positions and 13 variable (excluding the four indels) sites (total number of mutations: 15), 11 potentially parsimony informative sites with two variants and two with three variants each (Table 2). This region had a high AT content (57·8 %), with the presence of several mononucleotide repeats.
Table 2.
Distribution and frequency of plastid DNA haplotypes based on the psbC/trnS3 region in each population of Hymenaea stigonocarpa
| Polymorphic sites |
|||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 1 | 1 | 2 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 4 | 4 | 5 | 5 | |||||||||||||||||||
| 1 | 3 | 7 | 8 | 9 | 5 | 2 | 3 | 7 | 8 | 0 | 4 | 5 | 6 | 8 | 0 | 1 | |||||||||||||||||||
| Haplotype | 4 | 0 | 7 | 8 | 1 | 7 | 9 | 4 | 1 | 8 | 2 | 2 | 7 | 2 | 5 | 9 | 6 | SMC | ITC | PIC | FAC | CVC | JTC | TOC | TUC | FUC | ADC | DIC | FBC | PEC | CHC | MCC | RPC | MUC | Frequency |
| H1 | – | A | T | T | G | T | A | T | T | C | – | A | C | C | G | T | G | 5 | 9 | 7 | 7 | 12 | 9 | 49 | |||||||||||
| H2 | – | . | C | . | . | . | . | . | . | . | – | . | . | . | . | . | . | 6 | 9 | 6 | 14 | 4 | 3 | 9 | 6 | 57 | |||||||||
| H3 | – | . | C | . | . | . | . | . | . | . | A | . | . | . | . | . | . | 2 | 1 | 3 | |||||||||||||||
| H4 | – | . | C | . | . | . | . | . | . | . | A | . | G | . | . | . | . | 1 | 1 | ||||||||||||||||
| H5 | – | . | C | . | . | . | . | . | . | . | – | T | . | . | . | . | . | 4 | 4 | ||||||||||||||||
| H6 | – | . | . | A | . | . | . | . | . | . | – | . | . | . | . | . | . | 3 | 3 | ||||||||||||||||
| H7 | – | . | . | . | . | . | . | G | . | . | – | . | . | . | . | . | . | 2 | 2 | ||||||||||||||||
| H8 | – | . | . | . | . | . | . | . | . | . | – | . | . | . | . | . | A | 5 | 1 | 3 | 9 | 18 | |||||||||||||
| H9 | – | – | . | . | . | . | . | . | . | . | – | . | . | . | C | . | A | 2 | 2 | ||||||||||||||||
| H10 | – | . | . | . | . | A | . | . | . | . | – | . | . | . | . | . | A | 2 | 2 | ||||||||||||||||
| H11 | – | – | . | . | . | A | . | . | . | . | – | . | . | . | . | . | A | 2 | 2 | ||||||||||||||||
| H12 | – | . | . | . | . | . | . | . | . | . | – | . | . | A | . | – | A | 2 | 1 | 3 | |||||||||||||||
| H13 | – | . | . | . | . | . | . | . | . | . | – | C | . | . | . | . | A | 3 | 3 | ||||||||||||||||
| H14 | – | . | . | . | . | . | G | . | A | . | – | . | . | . | . | . | A | 4 | 4 | ||||||||||||||||
| H15 | – | . | . | . | . | . | . | . | . | . | – | . | . | . | C | . | A | 1 | 1 | ||||||||||||||||
| H16 | – | . | . | . | . | . | . | . | . | G | – | . | . | . | . | . | A | 2 | 2 | ||||||||||||||||
| H17 | – | . | . | . | . | . | . | . | A | . | – | . | . | . | . | . | A | 2 | 2 | ||||||||||||||||
| H18 | – | . | . | . | . | . | . | . | . | . | – | . | G | . | . | . | A | 1 | 1 | ||||||||||||||||
| H19 | – | . | . | . | . | . | . | . | . | . | – | . | . | . | . | – | A | 7 | 7 | ||||||||||||||||
| H20 | – | . | . | . | . | . | G | . | . | . | – | C | . | . | . | . | . | 1 | 1 | ||||||||||||||||
| H21 | T | . | C | . | . | . | . | . | . | . | – | . | . | . | . | . | . | 2 | 1 | 3 | |||||||||||||||
| H22 | – | . | C | . | A | . | . | . | . | . | – | . | . | . | . | . | . | 3 | 3 | ||||||||||||||||
| H23 | – | . | C | . | . | . | C | . | . | . | – | . | . | . | . | . | . | 2 | 2 | ||||||||||||||||
| Total | 175 | ||||||||||||||||||||||||||||||||||
Twenty-three haplotypes were found (Fig. 2) defined by the 13 sites and four indels. Total haplotype diversity (h), nucleotide diversity (π) and the mean number of nucleotide differences (k) were 0·804, 0·003 and 1·598, respectively. Haplotype diversity for each population (h) ranged from 0 to 0·771, haplotypic richness (A) from 0 to 2·766, nucleotide diversity from 0 to 0·00267 and the mean number of nucleotide differences from 0 to 1·393 (Table 1).
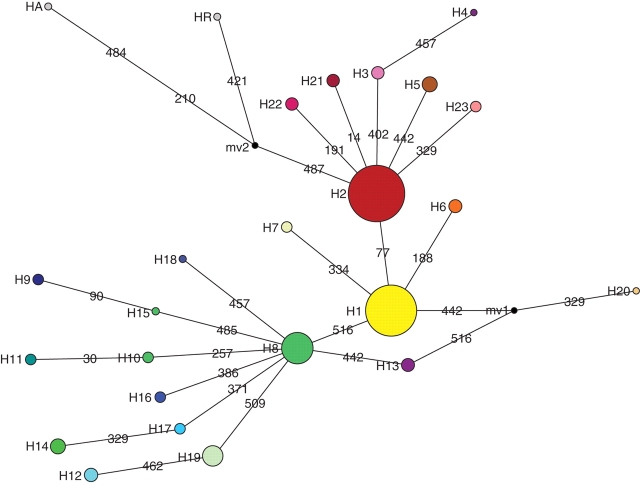
Fig. 2.
MJ network analysis of the relationships between haplotypes of the psbC/trnS3 (524 pb) plastid DNA region from 175 H. stigonocarpa individuals and two outgroups (HA and HR). Circle area is proportional to haplotype frequency and colours are as in Fig. 1. Lines drawn between haplotypes represent mutation events identified by the numbers corresponding to the positions at which the mutations were observed. Black points represent hypothetical haplotypes (median vector).
The two most diverse populations in terms of haplotype number were MUC and MCC with six haplotypes (Fig. 1 and Table 2). Populations FUC and RPC each only had three haplotypes, although found in similar frequencies, resulting in h values close to MUC and MCC (Table 1). The populations MUC, MCC, RPC and FUC also exhibited the highest indices of haplotypic richness after rarefaction to correct for sample size. Populations SMC, ADC and DIC only had one haplotype each (diversity indices = 0; Fig. 1 and Table 2).
Phylogeographic structure
The relationships among the 23 haplotypes observed and the outgroups H. aurea (HA) and H. reticulata (HR) are shown in the network in Fig. 2, analysed using the median-joining method. The most frequent haplotypes were H1, H2 and H8, occurring in 28, 33 and 11 % of individuals sampled, respectively. Haplotypes H2 and H8 were each linked to H1 by a single nucleotide substitution at positions 77 and 516, respectively (Fig. 2). Most haplotypes (17) were only found in one population (Table 2). Haplotypes H16, H17, H18 and H19 were only found in the MCC population, H9, H10, H11 and H13 in MUC and haplotypes H14 and H15 in RPC (Table 2). Other exclusive haplotypes were found in populations ITC, FAC, JTC, TOC, FBC, PEC and CHC (Table 2).
The SAMOVA analyses clearly indicated that there were distinct groups of genetically defined sampling areas. In analyses where K = 2, partitions of the sampling areas were identified that suggested two groups (groups: FUC, ADC, DIC, FBC, PEC, CHC, MCC, RPC, MUC vs. SMC, ITC, PIC, FAC, CVC, JTC, TOC, TUC; FCT = 0·476). In analyses where K = 3, an additional partition was identified that subdivided the first group into two areas, with an FCT value of 0·588. With K = 4 the FCT decreased to 0·473 and after K = 5 to K = 8 the FCT values became stable, ranging from 0·579 to 0·617. Thus, the present analysis suggested the presence of three FST geographical groups: a western group comprising SMC, ITC, PIC, FAC, CVC, JTC, TOC and TUC, a central group comprising FUC, ADC, DIC, FBC, CHC and PEC and an eastern group comprising MCC, RPC and MUC. The SAMOVA performed with eastern, central and western clusters resulted in an FST value of 0·692, indicating that 69·2 % of the variation was due to differences among the populations, and an FSC of 0·252, indicating that 25·2 % of the genetic variation was due to differences among populations within these groups (Table 3). The analysis using the pairwise FST distances in Barrier 2·2 (Manni et al., 2004) corroborated the SAMOVA analysis, showing the existence of three main geographical population clusters. The FST values calculated for each pair of populations ranged from 0 to 1·00 and most values observed were significant (P < 0·05; Table 4). The majority of non-significant pairwise FST values were observed among population pairs within groups (Table 4). The mean FST within groups (0·258) was much lower than the mean FST among groups (0·701), agreeing with the division into three groups.
Table 3.
Analysis of molecular variance based on the psbC/trnS3 region of plastid DNA for 17 populations of Hymenaea stigonocarpa
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation | Fixation indices |
|---|---|---|---|---|---|
| Among groups | 2 | 71·17 | 0·594 Va* | 58·84 | FCT: 0·588 |
| Among populations within groups | 14 | 18·76 | 0·104 Vb* | 10·36 | FST: 0·692 |
| Within populations | 158 | 49·12 | 0·311 Vc* | 30·81 | FSC: 0·252 |
| Total | 174 | 139·05 | 1·009 |
* P < 0·01
Table 4.
Pairwise comparisons of FST between populations of Hymenaea stigonocarpa based on the psbC/trnS3 region of plastid DNA
| Western group |
Central group |
Eastern group |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SMC | ITC | PIC | FAC | CVC | JTC | TOC | TUC | FUC | ADC | DIC | FBC | PEC | CHC | MCC | RPC | MUC | |
| ITC | 0·848 | 0·000 | |||||||||||||||
| PIC | 0·022 | 0·735 | 0·000 | ||||||||||||||
| FAC | 0·182 | 0·674 | 0·204 | 0·000 | |||||||||||||
| CVC | − 0·077 | 0·843 | − 0·269 | 0·256 | 0·000 | ||||||||||||
| JTC | 0·200 | 0·654 | 0·000 | 0·227 | 0·269 | 0·000 | |||||||||||
| TOC | 0·472 | 0·685 | 0·426 | 0·411 | 0·534 | 0·388 | 0·000 | ||||||||||
| TUC | − 0·059 | 0·751 | 0·062 | 0·200 | 0·007 | 0·198 | 0·458 | 0·000 | |||||||||
| FUC | 0·361 | 0·605 | 0·398 | 0·399 | 0·449 | 0·371 | 0·462 | 0·397 | 0·000 | ||||||||
| ADC | 1·000 | 0·940 | 0·848 | 0·812 | 0·922 | 0·841 | 0·841 | 0·904 | 0·227 | 0·000 | |||||||
| DIC | 1·000 | 0·925 | 0·831 | 0·790 | 0·914 | 0·816 | 0·818 | 0·892 | 0·197 | 0·000 | 0·000 | ||||||
| FBC | 0·778 | 0·806 | 0·734 | 0·704 | 0·803 | 0·698 | 0·727 | 0·762 | 0·255 | 0·205 | 0·167 | 0·000 | |||||
| PEC | 0·840 | 0·867 | 0·779 | 0·756 | 0·838 | 0·763 | 0·784 | 0·809 | 0·254 | 0·033 | 0·006 | 0·189 | 0·000 | ||||
| CHC | 0·793 | 0·819 | 0·738 | 0·708 | 0·809 | 0·700 | 0·724 | 0·769 | 0·209 | − 0·112 | − 0·040 | 0·134 | 0·039 | 0·000 | |||
| MCC | 0·692 | 0·713 | 0·715 | 0·.695 | 0·757 | 0·673 | 0·696 | 0·718 | 0·450 | 0·602 | 0·575 | 0·580 | 0·619 | 0·574 | 0·000 | ||
| RPC | 0·747 | 0·731 | 0·763 | 0·733 | 0·817 | 0·691 | 0·727 | 0·772 | 0·483 | 0·689 | 0·654 | 0·634 | 0·691 | 0·617 | 0·324 | 0·000 | |
| MUC | 0·649 | 0·694 | 0·674 | 0·658 | 0·712 | 0·636 | 0·655 | 0·675 | 0·361 | 0·522 | 0·496 | 0·512 | 0·543 | 0·494 | 0·218 | 0·263 | 0·000 |
Values given in bold are not significant at P > 0·05.
Haplotypes HA and HR (found only in the outgroups) were closest to H2, the most frequent haplotype in the western group (Fig. 2). Eastern populations were more diverse, as indicated by haplotype and nucleotide diversity indices (Table 1), followed by populations from the western and central groups. The central group includes two monomorphic populations with only the H1 haplotype, whereas all populations except SMC in the western group exhibited two haplotypes each (SMC was monomorphic). Most of the individuals sampled from the central and western groups had haplotypes H1 (71·4 %) and H2 (76·1 %), respectively, indicating a low degree of variation in these populations. In the eastern group, all of the haplotypes were directly linked to the H8 haplotype, whereas all of the central and western haplotypes were linked to the H1 and H2 haplotypes, respectively (Fig. 2).
DISCUSSION
The psbC-trnS region (Demesure et al., 1995) has been used in PCR-RFLP studies (Caron et al., 2000; Dutech et al., 2000; Heuertz et al., 2004); in the present study, using sequences of this region, H. stigonocarpa populations exhibited similar levels of genetic divergence (FST = 0·692) when compared with the values observed for other species of angiosperms with plastid DNA (median value of GST = 0·646; Petit et al., 2005). The plastid DNA sequences showed a subdivision of the geographic distribution of the H. stigonocarpa populations into three genetically differentiated groups (eastern, central and western), which exhibited high frequencies of haplotypes H8, H1 and H2, respectively. The high genetic differentiation between groups (FCT = 0·588) was concordant with the analysis with the Barrier software, which suggests the existence of barriers to gene flow. According to coalescence theory, H1 might be a more ancestral haplotype, since it is found in a more central position in the network (Posada and Crandall, 2001). Furthermore, H1 gave rise to H2 and H8 haplotypes that were found in populations that experienced demographic expansions in the eastern and western groups, as suggested by the star-shaped network around them. However, the H2 haplotype could also be considered an old haplotype since it is found in high frequency in western populations.
In the last taxonomic review of Hymenaea, Lee and Langenheim (1975) described three varieties of H. stigonocarpa: var. stigonocarpa, var. pubescens and var. brevipetiolata. According to these authors, var. stigonocarpa shares its wide distribution range with var. pubescens, and var. brevipetiolata, although collected only in west of Minas Gerais and Mato Grosso, could have a wider distribution area in cerrado. Due to the similarity in the geographic distribution of these varieties, the three genetic geographical groups found in the present study could not be explained by the occurrence of different sub-specific taxa.
The subdivision of the range of H. stigonocarpa populations into three genetically differentiated areas can be associated with climatic and vegetation changes within the region. After reviewing palynological records of tropical South America in the Late Quaternary, Behling and Hooghiemstra (2001) suggested temporal and spatial changes in the distribution of savanna vegetation. During the last glacial period, savannas, both north and south of the equator, expanded, reflecting markedly drier conditions (Behling, 2002). Other records indicated that the southern portion of the present-day cerrado region might have been reduced in area due to strong cold fronts, which moved across the Brazilian highlands far to the north during glacial times (Behling and Hooghiemstra, 2001). In the period approx. >48000 to approx. 18000 radiocarbon years before present (YBP), the landscape of Catas Altas (20°05′S, 43°22′W) was characterized by subtropical grasslands with small areas of subtropical gallery forests containing Araucaria (Behling and Lichte, 1997). Increase in rainfall and greatly reduced annual average temperatures in this region favoured the expansion of Araucaria forests, vegetation typical of southern Brazil today, in areas presently covered by cerrado vegetation. Subtropical grassland vegetation expanded, replacing the cerrado regions far to the north in the highlands of south-eastern Brazil, from present-day latitudes of about 28–27°S to about 20°S (Catas Altas). This also suggested that the temperature in the last glacial maximum was 5–7 °C lower than observed today (Behling, 1998). The expansion of the subtropical grassland into the cerrado region may have reduced typical cerrado vegetation, thus isolating populations and decreasing the gene flow. This would explain lower haplotype and nucleotide diversity values observed in the populations from the western and central groups.
During glacial times, maritime influences could have determined different climates between central and eastern Brazil. Several lines of evidence show that the arid climate was more extreme in the central Brazil region. The milder climate towards the Atlantic Ocean and the lower latitude allowed cerrado vegetation to spread eastwards to the coast (Behling and Hooghiemstra, 2001). In addition, the pollen records from the period 10990–10540 YBP of sand dunes in the middle São Francisco River region (10°24′S, 43°13′W, in north-eastern Brazil) show the presence of taxa that are today found in the Amazon and Atlantic rain forests, including species found in mountain regions, thus suggesting humid climatic conditions (De Oliveira et al., 1999). These facts could be a possible cause for the greater diversity found in the eastern populations (RPC, MCC and MUC). In these areas, the relatively higher temperatures and humidity (compared with the central and south areas of cerrado) could have resulted in the maintenance of larger populations, retaining genetic diversity. However, in the eastern group most of the haplotypes are private to one population, suggesting low gene flow. In this region only three populations were analysed, and these are geographically distant. Thus, for a more conclusive interpretation of the evolutionary history of this species in the eastern region, it would be necessary to analyse more populations.
The present-day distribution of H. stigonocarpa reaches São Paulo State at its southern limit. Considering that the temperature in the last glacial maximum in south-eastern and central Brazil was 5–7 °C lower that it is today (Behling, 1998), the occurrence of this species may have been restricted to regions with a mild climate, closer to the coast or at lower latitudes. In the glacial period the temperature in mid-western and south-eastern Brazil was similar to present-day temperatures in southern Brazil, where H. stigonocarpa does not occur. Silberbauer-Gottsberger et al. (1977) showed a clear relationship between the degree of frost damage of species from cerrado and their geographical distribution. These authors concluded that frost seems to be one of the selective factors influencing the floristic composition of the cerrado at its southern limit. Due to the colder climatic conditions during the glacial time, the frost-sensitive cerrado vegetation must have also remained in the northern part of south-eastern Brazil, where frosts were rare or absent (Behling, 1998). After savanna vegetation and climatic conditions were re-established (5000–4000 YBP; Behling and Hooghiemstra, 2001), the species returned to the southern part of the present-day cerrado distribution. The southward re-colonization could explain the presence of the haplotypes H1, H2 and H8 in the FUC population, suggesting that this population may have originated from different lineages from eastern, central and western groups. Similar data have been observed in a Brazilian cerrado tree species, Caryocar brasiliense (Collevatti et al., 2003). The phylogeographic study of that species suggested that the population from western São Paulo State, the south-western limit of the cerrado geographical distribution, originated from multiple lineages of populations from Goiás (GO) and Mato Grosso (MT).
We suggest that a large polymorphic population of H. stigonocarpa covered most of the studied region, and that during the glacial periods it was reduced to small isolated populations, mainly in the central and western sites. The reduction of population size (bottleneck) would cause a depletion of genetic diversity due to genetic drift, which is more pronounced with plastid DNA markers, since its effective size is equal to one-half that of nuclear markers (Birky et al., 1983). The restricted distribution of haplotypes was maintained through limited seed dispersal during the expansion of the species. According to Avise (2000), a starburst phylogeographic pattern, as observed in this study, particularly considering the separate geographic groups (Fig. 2), is an expected signature for a species that has expanded its population and geographic range from a small number of founders. The parallel radiation from north to south with the maintenance of three distinct longitudinal haplotype groups could also have a relationship with geographic barriers that run in a north–south direction similarly to the observed haplotype groups. The Espinhaço mountain range may have contributed to the isolation of the easternmost of the central groups, and the Espigão Mestre the central ones of the western groups. Another important event, the extinction of megafauna in the Quaternary (approx. 10000 years ago), could have influenced the genetic structure found in H. stigonocarpa, as is also suggested for Caryocar brasiliense populations (Collevatti et al., 2003). Current dispersion of H. courbaril seeds is mainly by agoutis (Asquith et al., 1999), but Hallwachs (1986) suggested that this role was ‘inherited’ from large Pleistocene mammals. It is known that seed dispersal by large mammals was more effective, since they probably had greater dispersion capacities. It is possible that, due to the great similarity between the fruits and seeds of H. courbaril and H. stigonocarpa, seed dispersal in these two species may have been made by the same agents. With the megafauna extinction, dispersal of seeds and gene flow could have been reduced, thus increasing isolation and differentiation between populations. However, it must also be considered that human migration could have led to seed dispersal of H. stigonocarpa. This could explain some of our results, e.g. the CHC population has a haplotype typical of the central group but is geographically nearer to the eastern group.
This study provides information about the natural history of H. stigonocarpa and infers that climatic changes during the Quaternary helped shape the distribution and genetic structure of the species. Accompanied by palynological records, the phylogeographic data suggest that during glacial times the low temperatures resulted in extinction of H. stigonocarpa populations in most parts of the southern present-day cerrado area. Milder climatic conditions in the north and eastern portions of the cerrado resulted in the maintenance of populations. Following the postglacial climate amelioration, most parts of the present-day southern cerrado were re-colonized from three different lineages from the northern parts of this biome. Phylogeographic studies using plastid DNA data of species occurring in the Brazilian cerrado are still scarce. It is apparent that more phylogeographic studies with other species from the cerrado are needed to obtain a better understanding of the influence of Quaternary climatic changes on the evolutionary history of the flora of this biome.
ACKNOWLEDGEMENTS
This study was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq/Brazil). We also thank the Instituto Brasileiro de Meio Ambiente for providing facilities, Reinaldo M. Silva, Renan Milagres, Luciana C. Resende and Juliano Leal for technical assistance in this study, Rodrigo Redondo and Leandro M. Freitas for assistance with computing the analyses, and Rosangela L. Brandão, Alba L. Fonseca and Elder A. Paiva for their help in sample collection. A. C. S. Ramos received a PhD fellowship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES/Brazil). J. P. Lemos Filho and F. R. Santos received research fellowships from CNPq/Brazil.
LITERATURE CITED
- Asquith NM, Terborgh J, Arnold AE, Riveros CM. The fruits the agouti ate: Hymenaea courbaril seed fate when its disperser is absent. Journal of Tropical Ecology. 1999;15:9–235. [Google Scholar]
- Avise JC. Molecular markers: natural history and evolution. New York, NY: Chapman and Hall; 1994. [Google Scholar]
- Avise JC. Phylogeography: the history and formation of species. Harvard, MA: Harvard University Press; 2000. [Google Scholar]
- Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, et al. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annual Review of Ecology and Systematics. 1987;18:489–522. [Google Scholar]
- Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Behling H. Late quaternary vegetational and climatic changes in Brazil. Review of Paleobotany and Palynology. 1998;99:143–156. [Google Scholar]
- Behling H. South and southeast Brazilian grasslands during Late Quaternary times: a synthesis. Palaeogeography, Palaeoclimatology, Palaeoecology. 2002;177:19–27. [Google Scholar]
- Behling H, Hooghiemstra H. Neotropical savanna environments in space and time: late Quaternary interhemispheric comparison. In: Markraf V, editor. Interhemispheric climate linkages. New York, NY: Academic Press; 2001. pp. 307–323. [Google Scholar]
- Behling H, Lichte M. Evidence of dry and cold climatic conditions at glacial times in tropical southeastern Brazil. Quaternary Research. 1997;48:348–358. [Google Scholar]
- Belahbib N, Pemonge MH, Ouassou A, Sbay H, Kremer A, Petit RJ. Frequent cytoplasmic exchanges between oak species that are not closely related: Quercus suber and Q. ilex in Morocco. Molecular Ecology. 2001;10:2003–2012. doi: 10.1046/j.0962-1083.2001.01330.x. [DOI] [PubMed] [Google Scholar]
- Birky CW, Maruyama T, Fuerst P. An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics. 1983;103:513–527. doi: 10.1093/genetics/103.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci SJ, Goldstein G, Meinzer FC, Franco AC, Campanello P, Scholz FG. Mechanisms contributing to seasonal homeostasis of minimum leaf water potential and predawn disequilibrium between soil and plant water potential in Neotropical savanna trees. Trees. 2005;19:296–304. [Google Scholar]
- Caron H, Dumas S, Marque G, Messier C, Beou E, Petit RJ, Kremer A. Spatial and temporal distribution of chloroplast DNA polymorphism in a tropical tree species. Molecular Ecology. 2000;9:1089–1098. doi: 10.1046/j.1365-294x.2000.00970.x. [DOI] [PubMed] [Google Scholar]
- Cavers S, Navarro C, Lowe AJ. Chloroplast DNA phylogeography reveals colonization history of neotropical tree, Cedrela odorata L., in Mesoamerica. Molecular Ecology. 2003;12:1451–1460. doi: 10.1046/j.1365-294x.2003.01810.x. [DOI] [PubMed] [Google Scholar]
- Clark CM, Wentworth TR, O'Malley DM. Genetic discontinuity revealed by chloroplast microsatellites in eastern North American Abies (Pinaceae) American Journal of Botany. 2000;87:774–782. [PubMed] [Google Scholar]
- Collevatti RG, Grattapaglia D, Hay JD. Evidences for multiple maternal lineages of Caryocar brasiliense populations in the Brazilian cerrado based on the analysis of chloroplast DNA sequences and microsatellite haplotype variation. Molecular Ecology. 2003;12:105–115. doi: 10.1046/j.1365-294x.2003.01701.x. [DOI] [PubMed] [Google Scholar]
- Crestana CSM, Dias IS, Mariano G. Ecologia de polinização de Hymenaea stilbocarpa Hayne, o Jatobá. Silvicultura em São Paulo. 1985;17/19:31–37. [Google Scholar]
- Demesure B, Sodzi N, Petit RJ. A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Molecular Ecology. 1995;4:129–131. doi: 10.1111/j.1365-294x.1995.tb00201.x. [DOI] [PubMed] [Google Scholar]
- De Oliveira PE, Barreto AMF, Suguio K. Late Pleistocene/Holocene climatic and vegetational history of the Brazilian caatinga: the fossil dunes of the middle São Francisco River. Palaeogeography, Palaeoclimatology, Palaeoecology. 1999;152:319–337. [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1987;12:13–15. [Google Scholar]
- Dumolin-Lapègue S, Demesure B, Fineschi S, Le Corre V, Petit RJ. Phylogeographic structure of white oaks throughout the European continent. Genetics. 1997;146:1475–1487. doi: 10.1093/genetics/146.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupanloup I, Schneider S, Excoffier L. A simulated annealing approach to define the genetic structure of populations. Molecular Ecology. 2002;11:2571–2581. doi: 10.1046/j.1365-294x.2002.01650.x. [DOI] [PubMed] [Google Scholar]
- Dutech C, Maggia L, Joly HI. Chloroplast diversity in Vouacapoua americana (Caesalpiniaceae), a neotropical forest tree. Molecular Ecology. 2000;9:1427–1432. doi: 10.1046/j.1365-294x.2000.01027.x. [DOI] [PubMed] [Google Scholar]
- Eiten G. The cerrado vegetation of central Brazil. Botanical Review. 1972;38:201–341. [Google Scholar]
- El Mousadik A, Petit RJ. Chloroplast DNA phylogeography of the argan tree of Morocco. Molecular Ecology. 1996;5:547–555. doi: 10.1111/j.1365-294x.1996.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Ennos RA. Estimating the relative rates of pollen and seed migration among plant populations. Heredity. 1994;72:250–259. [Google Scholar]
- Ewing B, Green P. Basecalling of automated sequencer traces using Phred II: error probabilities. Genome Research. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendi M, Green P. Basecalling of automated sequencer traces using Phred I: accuracy assessment. Genome Research. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3·0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Excoffier LP, Smouse E, Quattro JM. Analysis of molecular variance inferred from metric distances among haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira ME, Grattapaglia D. Introdução ao uso de marcadores moleculares em análise genética. Brasília: EMBRAPA/CENARGEN; 1995. [Google Scholar]
- Ferris C, Oliver RP, Davy AJ, Hewitt GM. Native oak chloroplasts reveal an ancient divide across Europe. Molecular Ecology. 1993;2:337–344. doi: 10.1111/j.1365-294x.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- Forster P, Bandelt HJ, Rohl A, et al. Cambridge: Fluxus Technology Ltd; 2000. NETWORK 3·1·1·0. Software free available at: www.fluxus-engineering.com . [Google Scholar]
- Gibbs PE, Oliveira PE, Bianchi MB. Postzygotic control of selfing in Hymenaea stigonocarpa (Leguminosae-Caesalpinioideae), a bat-pollinated tree of the Brazilian cerrados. International Journal of Plant Sciences. 1999;160:1–7. [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Research. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Gugerli F, Senn J, Anzidei M, Madaghiele A, Büchler U, Sperisen C, et al. Chloroplast microsatellites and mitochondrial nad1 intron 2 sequences indicate congruent phylogenetic relationships among Swiss stone pine (Pinus cembra), Siberian stone pine (Pinus sibirica), and Siberian dwarf pine (Pinus pumila) Molecular Ecology. 2001;10:1489–1497. doi: 10.1046/j.1365-294x.2001.01285.x. [DOI] [PubMed] [Google Scholar]
- Hallwachs W. Agouti (Dasyprocta punctata): the inheritors of guapinol (Hymenaea coubaril: Leguminosae) In: Estrada A, Fleming TH, editors. Frugivores and seed dispersal. Dordrecht: W. Junk Publishers; 1986. pp. 285–304. [Google Scholar]
- Hamilton MB. Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Molecular Ecology. 1999;8:521–523. [PubMed] [Google Scholar]
- Heuertz M, Fineschi S, Anzidei M, Pastorelli R, Salvini D, Paule L, et al. Chloroplast DNA variation and postglacial recolonization of common ash (Fraxinus excelsior L.) in Europe. Molecular Ecology. 2004;13:3437–3452. doi: 10.1111/j.1365-294X.2004.02333.x. [DOI] [PubMed] [Google Scholar]
- Hopper SD, Gioia P. The Southwest Australian floristic region: evolution and conservation of a global hot spot of biodiversity. Annual Review of Ecology, Evolution and Systematics. 2004;35:623–650. [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lee Y-T, Langenheim JH. Systematics of the genus Hymenaea L. (Leguminosae, Caesalpinioidae, Detarieae) University of California Publications in Botany. 1975;69:1–109. [Google Scholar]
- Lorenz-Lemke AP, Muschner VC, Bonatto SL, Cervi AC, Salzano FM, Freitas LB. Phylogeographic inferences concerning evolution of Brazilian Passiflora actinia and P. elegans (Passifloraceae) based on ITS (nrDNA) variation. Annals of Botany. 2005;95:799–806. doi: 10.1093/aob/mci079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni CR, Ducousso A, Caron H, Petit RJ, Kremer A. Chloroplast DNA variation of Quercus rubra L. in North America and comparison with other Fagaceae. Molecular Ecology. 2005;14:513–524. doi: 10.1111/j.1365-294X.2005.02400.x. [DOI] [PubMed] [Google Scholar]
- Manni F, Guerard E, Heyer E. Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier's algorithm. Human Biology. 2004;76:173–190. doi: 10.1353/hub.2004.0034. [DOI] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorites. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Latouche-Halle C, Wagner DB. Geographic structure of chloroplast DNA polymorphisms in European oaks. Theoretical and Applied Genetics. 1993;87:122–128. doi: 10.1007/BF00223755. [DOI] [PubMed] [Google Scholar]
- Petit RJ, El Mousadik A, Pons O. Identifying populations for conservation on the basis of genetic markers. Conservation Biology. 1998;12:844–855. [Google Scholar]
- Petit RJ, Aguinagalde I, de Beaulieu J-L, Bittkau C, Brewer S, Cheddadi R, et al. Glacial refugia: hotspots but not melting pots of genetic diversity. Science. 2003;300:1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Molecular Ecology. 2005;14:689–701. doi: 10.1111/j.1365-294X.2004.02410.x. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Intraspecific gene genealogies: trees grafting into networks. Trends in Ecology and Evolution. 2001;16:37–45. doi: 10.1016/s0169-5347(00)02026-7. [DOI] [PubMed] [Google Scholar]
- Ratter JA, Bridgewater S, Ribeiro JF. Biodiversity patterns of the woody vegetation of the Brazilian cerrado. In: Pennington RT, Lewis GP, Ratter JA, editors. Neotropical savannas and seasonally dry forests: plant diversity, biogeography and conservation. Boca Raton, FL: CRC Press; 2006. pp. 31–66. [Google Scholar]
- Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM. Rapid diversification of a species-rich genus of neotropical rain forest trees. Science. 2001;293:2242–2245. doi: 10.1126/science.1061421. [DOI] [PubMed] [Google Scholar]
- Rizzini CT. Árvores e madeiras úteis do Brasil; manual de dendrologia brasileira. São Paulo: Edgard Blucher; 1971. [Google Scholar]
- Rozas J, Sánches-Delbarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analysis by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Salgueiro F, Felix D, Caldas JF, Margis-Pinheiro M, Margis R. Even population differentiation for maternal and biparental gene markers in Eugenia uniflora, a widely distributed species from the Brazilian coastal Atlantic rain forest. Diversity Distribution. 2004;10:201–210. [Google Scholar]
- Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA. Phylogeographic studies in plants: problems and prospects. Molecular Ecology. 1998;7:465–474. [Google Scholar]
- Schierenbeck KA, Symonds VV, Gallagher KG, Bell J. Genetic variation and phylogeographic analyses of two species of Carpobrotus and their hybrids in California. Molecular Ecology. 2005;14:539–547. doi: 10.1111/j.1365-294X.2005.02417.x. [DOI] [PubMed] [Google Scholar]
- Silberbauer-Gottsberger I, Morawetz W, Gottsberger G. Frost damage of cerrado plants in Botucatu. Biotropica. 1977;9:253–261. [Google Scholar]
- Silva MR, Silva MS, Martins KA, Borges S. Studies on the use of jatoba flour in biscuits as a source of dietary fibre containing no added simple sugars. Ciência e Tecnologia de Alimentos. 2001;21:176–82. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;24:4876–4887. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weising K, Gardner RC. A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome. 1999;42:9–19. [PubMed] [Google Scholar]
- Zhang Q, Chiang TY, George M, Liu JQ, Abbott RJ. Phylogeography of the Qinghai-Tibetan Plateau endemic Juniperus przewalskii (Cupressaceae) inferred from chloroplast DNA sequence variation. Molecular Ecology. 2005;14:3513–3524. doi: 10.1111/j.1365-294X.2005.02677.x. [DOI] [PubMed] [Google Scholar]