Abstract
IFN regulatory factor 8 (IRF8) is a transcription factor that affects the differentiation and function of myeloid, dendritic, and B cells. Herein we report that IRF8 regulates the expression of Mdm2, a suppressor of p53-dependent and -independent apoptosis pathways, in germinal center (GC) B cells. In GC B cells of IRF8-deficient mice, Mdm2 transcripts were greatly down-regulated, and MDM2 protein was poorly expressed in GC of Irf8−/− mice. Small interfering RNA-induced repression of IRF8 in a GC-derived B cell line resulted in decreased expression of MDM2 at the protein level but increased expression of p53 and p21. We found that IRF8 binds to the Mdm2 P2 promoter, and that cotransfection of an IRF8 expression vector with an Mdm2 reporter construct stimulated significant increases in reporter activity. Additionally, transcripts of the p53 target Pmaip1 (Noxa) were significantly increased in IRF8-deficient GC B cells as well as in the IRF8 knockdown B cell line. Finally, cells deficient in IRF8 exhibited growth suppression and increased sensitivity to apoptosis induced by etoposide or IL-21. These results suggest that by regulating MDM2, IRF8 might allow GC B cells to tolerate physiological DNA breaks that otherwise would trigger apoptosis.
As a critical step in optimizing responses to specific pathogens, Ig genes undergo genomic sequence rearrangements in germinal center (GC)4 B cells, a process called Ig gene diversification, which includes somatic hypermutation (SHM) and class switch recombination (CSR). A hallmark for both SHM and CSR is the generation of DNA double-strand breaks (DSB) induced by activation-induced cytidine deaminase (1). In most somatic cells, the response to DSB is marked by the activation of p53, which in turn induces cell cycle arrest or apoptosis. By activating p21 to induce cell cycle arrest, p53 provides cells with the opportunity to repair DNA DSB before completing the cell cycle (2– 4). The activation of p53 can also result in cell death, caused by induction of proapoptotic genes (5), indicating that pathways controlling p53 activity must be exquisitely monitored. GC B cells, however, must be able to tolerate these physiologic DSB without experiencing apoptosis or cell cycle arrest.
Several studies have indicated that BCL6, a gene critical to the GC reaction, may function in various ways to protect GC B cells from growth arrest and apoptosis. These include direct transcriptional repression of both p53 (6) and the programmed cell death gene PDCD2 (7), as well as suppression of p21 transcription by interacting with MIZ1 bound to the p21 promoter (8). Recently, we reported that expression of BCL6 is regulated in part by the transcription factor IFN regulatory factor 8 (IRF8), also known as IFN consensus sequence binding protein (ICSBP) (9), suggesting the involvement of IRF8 in GC B cell physiology.
Tyrosine phosphorylation and dephosphorylation have been shown to control the activity of IRF8 (10, 11), and recent studies have identified contributions of ubiquitylation by TRIM21 and CBL to the function and degradation of the protein (12, 13). Although IRF8 has been shown to have major effects on the development and function of myeloid and dendritic cells (14, 15), it is also expressed in B lineage cells. Expression can be detected from the earliest stages of commitment to the lineage (16), is at highest levels in GC B cells, and is virtually extinguished in plasma cells (9, 17, 18).
To further understand the role of IRF8 in GC B cell physiology, we performed an exploratory experiment comparing the gene expression profiles of a GC lymphoma-derived B cell line with small interfering RNA (siRNA)-induced suppression of IRF8 vs unsuppressed cells. Among genes regulated by IRF8, Mdm2 was of particular interest. Mdm2 encodes a regulator of p53, which inhibits p53 function in two distinct ways. It blocks p53 transcriptional activity by binding directly to the transcriptional activation domain of p53 (19, 20). Additionally, as an E3 ligase, MDM2 ubiquitinates p53, inducing its nuclear export and proteosomal degradation (21, 22). MDM2 also directly inhibits p21 function by promoting p21 degradation via a ubiquitin-independent pathway (23). MDM2 is overexpressed in a significant portion of different types of cancers (24 –27), in which it acts as an oncogene by suppressing p53 activity, thereby permitting the uncontrolled growth of tumor cells.
In this study, we present results that show Mdm2 is a direct transcriptional target of IRF8. We also show that B cells deficient in IRF8 exhibit increased sensitivity to cell death induced by DNA DSB or by IL-21, a cytokine involved in several aspects of B cell survival and differentiation (28, 29). IRF8 may thus facilitate the expansion of Ag-specific B cells during the course of a GC response. These results also suggest that IRF8 may contribute to lymphomagenesis through its regulation of MDM2 and by collaborating with BCL6 to disable the p53 tumor suppressor pathway.
Materials and Methods
Mice, splenic GC B cells, and B cell lines
Normal C57BL/6J (The Jackson Laboratory) and C57BL/6-Irf8−/− mice described previously (30) were studied at 6–8 wk of age under a protocol (LIP-4) approved by the National Institute of Allergy and Infectious Diseases Animal Care and Use Committee. IRF8 conditional knockout mice (IRF8-CKO) were generated by CD19-driven Cre-mediated excision of Exon 2 of Irf8, which was flanked by LoxP sites (methods will be described elsewhere). Portions of spleens taken at necropsy were frozen in OCT and processed for immunohistochemical analysis. To isolate GC B cells, spleen cells from wild-type (WT) and Irf8−/− mice immunized i.p. 12 days previously with 100 μg of keyhole limpet hemocyanin in alum were stained with anti-GL7-FITC and peanut lectin (agglutinin) (PNA)-biotin followed by streptavidin-allophycocyanin GC cells (PNA+, GL7+) were sorted using a FACSAria (BD Biosciences).
The mouse NFS-202 B cell line that expresses IRF8 at high levels and NFS-202 clones expressing either of two Irf8-suppressive siRNAs or an inactive siRNA were described previously (9). The mouse NFS-203 cell line was derived from a late-stage B cell lymphoma of an NFS.V+ mouse and was found to express little or no IRF8 protein. MPC-11, a mouse plasmacytoma cell line that does not express IRF8, was obtained from Dr. M. Potter (National Cancer Institute, National Institutes of Health). Cells were cultured in DMEM containing 10% FBS, 2 mM glutamine, 2-mecaptoethanol, nonessential amino acids, and insulin/transferrin in a 5% CO2 atmosphere.
Overexpression of IRF8 in NFS-203 cells
To overexpress IRF8 in NFS-203 cells, an expression vector containing a full-length Irf8 cDNA clone, pCDNA3.1-IRF8 (a gift from Dr. K. Ozato, National Institute of Child Health and Human Development, National Institutes of Health), was transfected into NFS-203 cells. Cells transfected with an empty plasmid served as controls.
Oligonucleotide microarray and quantitative real-time RT-PCR (qPCR)
Oligonucleotide microarrays were used to profile the transcriptome of B cells from Irf8−/− mice and Irf8 knockdown B cells as described previously (31). Briefly, mRNA was isolated from cells using TRIzol, and cDNA was prepared using a SuperScript III reverse transcriptase kit (Invitrogen). cDNA from samples and from a reference pool derived from mouse hematopoietic cell lines were labeled with Cy5 and Cy3, respectively, and hybridized to a 70-mer oligonucleotide microarray chip manufactured by the Microarray Research Facility at National Institute of Allergy and Infectious Diseases, National Institutes of Health. Microarray data are available under accession no. GSE1908 (www.ncbi.nlm.nih.gov/projects/geo/query/). For qPCR, total RNA was isolated and cDNA was prepared as above. The mixture of cDNA, primers, and Power SYBR Green PRC Master mix (Applied Biosystems) was run on an ABI 7900 real-time PCR system (Applied Biosystems). The primer sequences for Irf8, Mdm2, p21, p53, Pmaip1, pre-miR34a, and Gapdh are listed in supplemental Table I.5 The ΔΔCt value (cycle of threshold) was calculated using Gapdh as the reference gene.
Immunohistochemical analyses
Immunohistochemical staining of mouse spleen tissue was performed as described previously (9, 32). Briefly, serial frozen sections were incubated with Abs specific to MDM2 (R&D Systems) or BCL6 (Santa Cruz Biotechnology), or with PNA followed by anti-PNA Ab (Vector Laboratories). Subsequently, the sections were incubated with appropriate secondary Abs. The signals were developed using diaminobenzidine tetrahydrochloride (Sigma-Aldrich) as chromogen.
Western blot analysis
Proteins were extracted from mouse spleen or cultured cells using a buffer containing 1% Triton X-100, 400 mM NaCl, 10% glycerol, and 1× protease inhibitor (Roche). Equal amounts of protein samples were loaded onto a 4–12% gradient SDS-PAGE and run at a constant current. Subsequently, proteins were transferred to a nitrocellulose membrane. The membrane was blocked with 4% fat-free milk powder in PBS, followed by incubation with appropriate primary and secondary Abs. The primary Abs used were: anti-IRF8 (a gift from Dr. K. Ozato), anti-MDM2 (AF1244; R&D Systems), anti-p53 (clone 1C12; Cell Signaling Technology), and anti-p21 (F-5; Santa Cruz Biotechnology). Protein signals were detected using ECL reagents (Pierce).
Oligonucleotide pull-down assay
To test the in vitro binding of IRF8 to the Mdm2 promoter, we used an oligonucleotide pull-down assay as described by Slack et al. (33). Briefly, biotinylated double-stranded 34-mer oligonucleotides were synthesized (IDT DNA Technologies) that represent the Mdm2 P2 promoter sequence at +185 to +218 that contains an IRF/Ets composite site (IECS)-like element (see Fig. 2A) (34). The sequences for WT, mutant, and scrambled oligonucleotides are listed in supplemental Table II. The oligonucleotides were bound to streptavidin-conjugated magnetic beads (Dynal M280; Invitrogen) and added to a reaction mixture containing protein extracts, poly(dI-dC), and salmon sperm DNA. The mixture was incubated for 2 h at 4°C with constant rotation, followed by washing with ice-cold binding buffer. Proteins bound to the oligonucleotides were eluted with SDS loading buffer for 20 min at 85°C, and were loaded on a 4–12% gradient SDS-PAGE for Western blot analysis.
FIGURE 2.
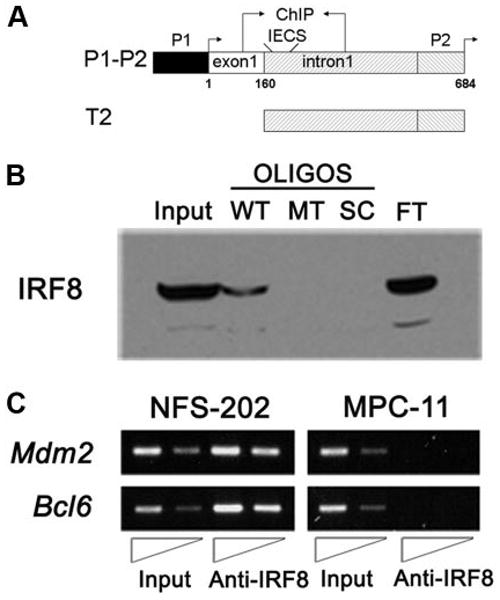
IRF8 binds sequences from the Mdm2 5′ regulatory region in vitro and in vivo. A, Schematic drawing of the Mdm2 promoter and reporter constructs P1–P2 and T2. Shown are the locations of the basal (P1) and p53-responsive promoters (P2) with their associated transcription start sites (arrows) and exon 1. The location of an IECS-like element and the region examined in ChIP analyses are also shown. The start site of exon 1 is indicated as +1. B, Western blot analyses of oligonucleotide pull-downs using the wild-type IECS-like (WT; core -GAAAAGAGGGAA-, italic denotes the two core elements), mutant (MT; core -GCAGAGAGTGTG-, underscore denotes the mutation), and scrambled (SC; core -GGAGAG GAGCGG-) oligonucleotides (OLIGOS). Also shown are control lanes with 10% of input nuclear protein (Input) and flow-through unbound protein (FT). C, ChIP analysis for in vivo binding of IRF8 to mouse Mdm2 P2 promoter. Anti-IRF8 Ab was used to precipitate fragments of genomic DNA from NFS-202 cells. The binding of IRF8 to Mdm2 P2 promoter is shown, together with its binding to Bcl6 promoter as a positive control. The same experiments were performed using DNA from MPC-11 cells that express little if any IRF8.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed as described previously (35). Cells (1 × 108) were cross-linked with 1% formaldehyde for 20 min at room temperature and then lysed. The lysates were sonicated for 30 s for 12 times to generate chromatin fragments with an average length of ∼200–600 bp. The chromatin was immunoprecipitated with goat anti-IRF8 Ab (C19; Santa Cruz Biotechnology). A 5-μl or 1-μl aliquot out of the 75 μl solution of DNA recovered from the immunoprecipitate or input DNA (1/60) was used for PCR, and the products were analyzed on a 2% agarose gel after 32 cycles of amplification. The following primers were used for PCR: mouse Mdm2 promoter (sense, 5′-CCGTGAAGGGTCGGAAGATG-3′, antisense, 5′-CACCCCCC–ACTCCCACCCAC-3′) and mouse Bcl6 promoter (sense, 5′-GTGCCTAATACTCT–AGCTGGAAGGAG-3′, antisense, 5′-GCTCGGCCTCTGGAATTCT-3′) as a positive control. The expected amplicon for the Mdm2 gene is a 220-bp fragment and for the Bcl6 gene is a 250-bp fragment (see Fig. 2A).
Luciferase reporter assay
A murine IRF8 expression vector (pCDNA3.1-IRF8) was provided by Dr. K. Ozato. The reporter plasmid (800 ng) was used for transfection of HeLa cells grown in 12-well plates together with the pRL-SV40 Renilla luciferase vector (50 ng) (Promega) as an internal control. To test dosage effects, 100, 200, and 300 ng of the IRF8 expression vector were adjusted to a total 300 ng of plasmid with pCDNA3.1 empty vector. The Mdm2 luciferase reporter gene plasmids P1–P2 and T2 were provided by Dr. H. Wu (University of California at Los Angeles) (36). Luciferase activities were measured 22 h after transfection using the dual-luciferase reporter assay kit (Promega) according to the manufacturer's protocol. All samples were tested in triplicate.
Cell proliferation assay
NFS-202 B cell clones expressing IRF8-specific siRNAs no. 2 or no. 5, or an inactive siRNA control were cultured at 1000 cells/well for 24, 48, or 72 h. The number of viable cells in each well was measured using a Cell-Titer-Glo luminescent cell viability assay kit (Promega) based on the manufacturer's instructions.
Apoptosis rate measured by caspase-3 activity
NFS-202 or NFS-203 cells, cultured in 6-well plates at 1 × 106/well, were treated with 5 μM etoposide (Sigma-Aldrich) for 6 h or with 200 ng/ml IL-21 (R&D Systems) for 20 h. Cells were harvested and washed twice with cold PBS. Cell lysates were prepared and the activity of caspase-3 was measured based on hydrolysis of the peptide substrate acetyl-Asp-Glu-Val-Asp p-nitroanilide, which was determined colorimetrically (CASP-3-C; Sigma-Aldrich).
BrdU in vivo labeling assay
IRF8-CKO and control IRF8Flox/+CD19Cre/+ mice were immunized with sheep RBC i.p. for 12 days. Mice were injected i.p. with 1.5 mg of BrdU in PBS for 40 min. Spleens were taken and the splenocytes were stained with GL-7-FITC and anti-FAS-PE, followed by anti-BrdU-allophycocyanin using an allophycocyanin BrdU flow kit (BD Biosciences). Cells were analyzed by FACSCalibur.
Statistical analysis
Experimental data were analyzed using the general linear model procedure of SAS software (SAS Institute).
Results
Suppression of IRF8 expression in vitro and in vivo is associated with reduced levels of MDM2 transcripts and protein
We used oligonucleotide microarrays to compare transcriptional profiles of GC-origin NFS-202 mouse B lymphoma cells stably transfected with control or IRF8-specific siRNAs. We found that the levels of Mdm2 transcripts in cells deficient in IRF8 were decreased by 80% compared with levels in the control cells (data not shown). This effect was confirmed by qPCR. In NFS-202 cells with siRNA-induced knockdown of IRF8, the levels of Irf8 transcripts were reduced by 77% while the levels of Mdm2 transcripts were reduced by 72% (both p < 0.01) (Fig. 1A). Western blot analyses of protein extracts from control and Irf8 knockdown cells showed that the levels of IRF8 and MDM2 proteins were similarly reduced (Fig. 1B). To determine whether IRF8 is also involved in regulating MDM2 expression in vivo, we isolated GC B cells from WT and Irf8 knockout mice and found that the level of Mdm2 transcripts significantly decreased in Irf8−/− B cells compared to the WT B cells (p < 0.05) (Fig. 1A). We also performed immunohistochemical studies of frozen sections from spleens of WT and Irf8 knockout mice. In WT mice, GC B cells defined by binding of PNA expressed higher levels of MDM2 than did surrounding follicular B cells or other splenic elements. In contrast, in Irf8 knockout mice little if any MDM2 protein was expressed by the GC cells (Fig. 1C). These data strongly suggest that IRF8 is involved in regulating the expression of MDM2 in GC B cells.
FIGURE 1.
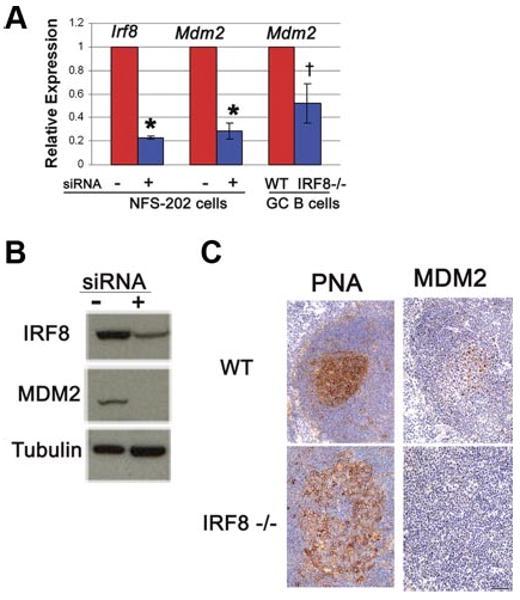
IRF8 regulates expression of MDM2 in vitro and in vivo. A, qPCR analyses of Irf8 and Mdm2 transcript levels in NFS-202 cells bearing an inactive (−) or an IRF8-suppressive siRNA (+, clone no. 5). Also shown are the Mdm2 transcript levels in GC B cells isolated from WT and Irf8−/− mice. The values were normalized to that in the control cells (†, p < 0.05; *, p < 0.01). B, Western blot analyses of IRF8 and MDM2 protein levels in NFS-202 cells expressing an inactive (−) or an IRF8-suppressive siRNA (+, clone no. 5). Tubulin expression was used as a loading control. C, Immunohistochemical analyses of serial frozen sections from WT and Irf8 knockout (IRF8−/−) mouse spleens for PNA binding and MDM2 protein expression.
IRF8 binds an IRF8 target sequence in the Mdm2 promoter in vitro and in vivo
To determine whether IRF8 might be directly involved in the transcriptional regulation of MDM2, we first examined DNA sequences 5′ of the Mdm2 start site for potential IRF8 binding sites. Regulation of Mdm2 expression is complex, involving two promoters, P1 and P2, that govern transcripts with different translational potentials (Fig. 2A) (36). Analysis of this region revealed a sequence on the minus strand in the P2 promoter (-GAAAAGAGGGAA-) similar to a recently identified IECS (15). An in vitro oligonucleotide pull-down assay using a magnetic bead-conjugated 34-bp oligonucleotide containing this motif was used to “fish” proteins from cell lysates of NFS-202 cells. Proteins eluted from the beads were examined by Western blotting using an IRF8-specific Ab to probe the bound proteins. The results showed that IRF8 bound to the WT motif from the Mdm2 promoter, but not to a probe with mutations in the IECS-like sequence or a scrambled form of the oligonucleotide (supplemental Table II and Fig. 2B).
To investigate the physiological relevance of this binding, we used ChIP assays to see if IRF8 was bound to 5′ regulatory sequences of Mdm2 in vivo. Previous studies showed that IRF8 is expressed at high levels in GC B cells but little, if any, in terminally differentiated plasma cells (9). To take advantage of this difference, we examined the genomic sequence containing the IECS-like motif from the GC-origin NFS-202 cells and, as a negative control, from the mouse plasmacytoma cell line MPC-11 (37). Study of cross-linked DNA precipitated with IRF8-specific Abs demonstrated that IRF8 occupied the promoter region in the GC cell line but not in the plasmacytoma cell line (Fig. 2C). Parallel analyses of the Bcl6 promoter region served as a positive control for recognition of target sequences by IRF8 (Fig. 2C).
IRF8 increases the transcriptional activity of the Mdm2 promoter
To further examine the cis-acting elements responsible for IRF8-mediated activation of Mdm2 transcription, we performed promoter reporter assays in HeLa cells (Fig. 3) using a luciferase gene linked to genomic DNA fragments of the Mdm2 promoter region: P1–P2, which contains both the P1 and P2 promoters; and T2, which contains the P2 promoter (Fig. 2A). HeLa cells were cotransfected with the reporter constructs and either an IRF8 expression vector or an empty vector. The results showed that the luciferase activities of P1–P2 and T2 were increased 4.6- and 3.8-fold, respectively, in cells overexpressing IRF8 (Fig. 3). These results indicated that sequences in the Mdm2 P2 promoter region were responsive to transcriptional activation by IRF8.
FIGURE 3.
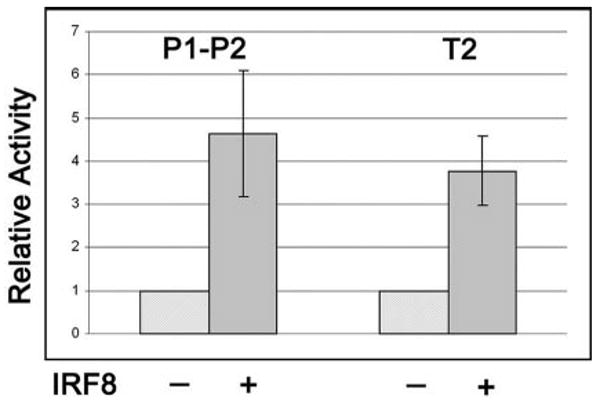
Promoter-reporter analyses of IRF8 regulation of Mdm2 transcription. HeLa cells were cotransfected with Mdm2 luciferase reporter constructs (see Fig. 2A) and either empty vector (−) or an Irf8 expression vector (+). Data from each reporter construct are normalized to the activity of the empty vector. Data shown are from three independent experiments (mean ± SD).
Suppression of IRF8 expression increases p53 and p21 protein levels
It is well known that MDM2 is a negative regulator of the functions of both p53 and p21 as a result of its activity as an E3 ligase as well as by E3 ligase-independent activities (20, 38). To determine the relationships among IRF8, MDM2, p53, and p21 levels in GC B cells, we used Western blot analyses to examine protein levels in extracts prepared from NFS-202 cells expressing inactive or IRF8-specific siRNAs. We also examined extracts prepared from NFS-203 cells that express negligible amounts of IRF8 transcript or protein, and NFS-203 cells bearing an IRF8 expression vector. The results showed that the levels of p53 and p21 proteins were significantly increased in cells with reduced expression of IRF8 (Fig. 4A). In contrast, levels of both p53 and p21 were substantially reduced in NFS-203 cells carrying an IRF8 expression vector (Fig. 4A). To determine whether the effects of siRNA-induced suppression of IRF8 on p53 and p21 expression were mediated at the transcriptional or posttranscriptional levels, we quantitated the levels of p53 and p21 transcripts in control and Irf8 knockdown cells by qPCR (Fig. 4B). The results showed that the transcript levels for both genes were similar in the control and Irf8 knockdown cells. Parallel studies showed that the levels of p53 and p21 transcripts in purified GC B cells from IRF8 knockout and WT mice were equivalent (data not shown). These data indicated that the observed regulation of p53 and p21 protein expression was mediated posttranscriptionally.
FIGURE 4.
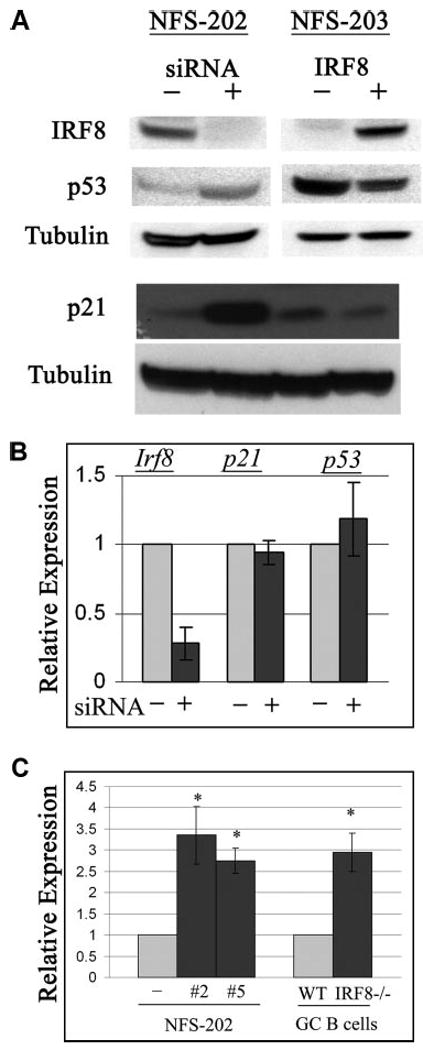
Regulation of p21 and p53 in mouse B cells by IRF8. A, Protein extracts from cell lines including NFS-202 cells bearing an inactive (−) or an IRF8-suppressive siRNA (+, clone no. 5) and NFS-203 cells bearing empty vector (−) or an IRF8 expression vector (+) were tested by Western blotting with Abs specific for IRF8, p53, p21, and tubulin. B, qPCR analyses of Irf8, p21, and p53 transcripts in NFS-202 cells bearing an inactive (−) or an IRF8-suppressive siRNA (+, clone no. 5). C, qPCR analysis of Pmaip1 transcript in NFS-202 cells bearing an inactive (−) or two IRF8-suppressive siRNAs (clone nos. 2 and 5) as well as in splenic B cells from WT or Irf8 knockout mice (IRF8−/−). Data shown are from three independent experiments (mean ± SD) (*, p < 0.01).
It would be expected that increased levels of p53 protein in IRF8-deficient B cells would be associated with increased expression of p53 target genes besides p21. To examine this issue, we measured the transcript levels of Pmaip1, a direct target of p53 that encodes the proapoptotic protein PMAIP1/NOXA, in NFS-202 cells with IRF8 knockdown as well as in GC B cells isolated from Irf8 knockout mice. The results showed that the levels of Pmaip1 transcripts were increased significantly in both cell populations (p < 0.01) (Fig. 4C).
Suppression of IRF8 slows proliferation and enhances apoptosis of B lymphocytes
To examine the functional importance of IRF8-dependent regulation of MDM2, p53, p21, and PMAIP1/NOXA we first compared the proliferation of NFS-202 cells bearing either of two IRF8-specific siRNAs (clone nos. 2 and 5) or an inactive siRNA (control) during a 72-h period. The results showed that the cells deficient in IRF8 grew much more slowly than the control cells (Fig. 5A). As noted previously, MDM2 functions to regulate the proapoptotic activity of p53 in a variety of cell types. To determine whether this is also the case for mature B cells, we first treated NFS-202 cells bearing inactive or IRF8-specific siRNAs with etoposide to induce DNA DSB and measured the extent of caspase-3 activation. Caspase-3 activity was significantly increased in NFS-202 cells treated with etoposide, while the levels in treated IRF8-deficient cells were >2-fold higher than the levels in treated control cells (p < 0.001) (Fig. 5B). Treatment with etoposide also stimulated substantial caspase-3 activity in NFS-203 cells that express little IRF8, but this increase was completely abrogated in cells bearing the IRF8 expression vector (Fig. 5C). These results indicated that the sensitivity of B cells to apoptosis caused by etoposide-induced DNA DSB was inversely correlated with levels of IRF8 expression.
FIGURE 5.
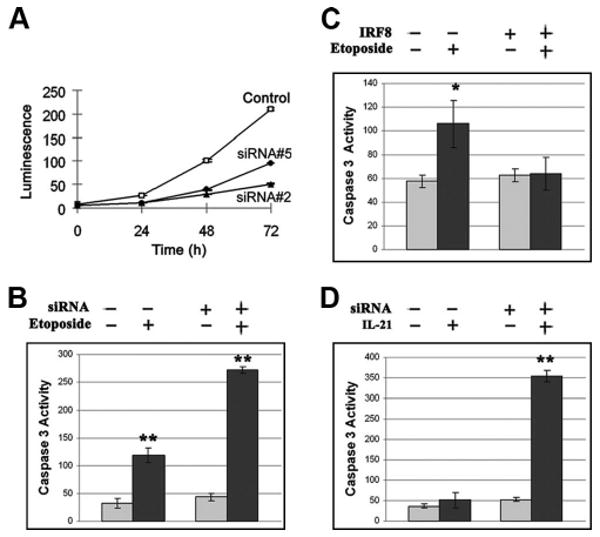
Regulation of proliferation and apoptosis in mouse B cells by IRF8. A, Proliferation of NFS-202 cells expressing an inactive (control) or either of two IRF8-suppressive siRNAs (clone nos. 2 and 5). Proliferation of cells with IRF8-suppressive siRNAs was significantly reduced (p < 0.001). Data shown are from three independent experiments (mean ± SD). B, Caspase-3 activity in NFS-202 cells expressing an inactive (−) or an IRF8-suppressive siRNA (+, clone no. 5) cultured in the presence or absence of etoposide (5 mM) for 6 h. C, Caspase-3 activity in NFS-203 cells bearing an empty vector (−) or an IRF8 expression vector (+) cultured in the presence or absence of etoposide for 6 h. D, Caspase-3 activity in NFS-202 cells bearing an inactive (−) or an IRF8-suppressive siRNA (+, clone no. 5) cultured in the presence or absence of IL-21 (200 ng/ml) for 20 h. For panels B–D, data shown are from three independent experiments (mean ± SD). *, p < 0.05; **, p < 0.001.
Follicular Th cells are critically involved in determining the fate of B cells involved in GC responses to T-dependent Ags due, in part, to their expression of the type I cytokine IL-21 (39, 40). IL-21 induces apoptosis of resting and anti-IgM-stimulated B cells, but promotes the maturation of CD40-stimulated B cells into Ig class-switched memory cells and plasma cells (28, 29). To determine whether IRF8 might be involved in the cell fate decisions made by GC B cells exposed to IL-21, we examined NFS-202 cells bearing inactive or IRF8-specific siRNAs for their responses to stimulation with IL-21. Although treatment with IL-21 had no effect on the levels of caspase-3 activity in control cells, the levels were significantly increased in cells deficient in IRF8 (p < 0.001) (Fig. 5D). These results strongly suggest that IRF8 enables GC B cells to tolerate apoptotic signals induced by exposure to IL-21.
IRF8 deficiency reduces proliferation of GC B cells in vivo
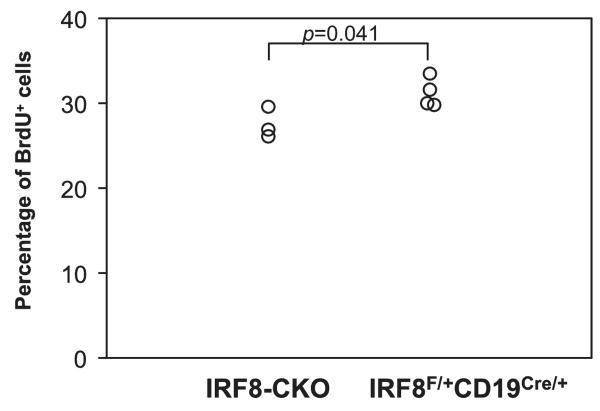
Although GC centroblasts are among the most rapidly dividing cells in the body, the mechanisms governing their high proliferative rate are poorly understood. This is perhaps best exemplified by the fact that GC B cells do not express MYC at either the protein or the transcriptional level (41), whereas both transcripts and protein are present at elevated levels in the highly proliferative transient-amplifying compartment of intestinal crypts (42). Proliferative cells of the GC are readily detected at the peak of a primary response by evaluating mice treated with BrdU from the time of immunization for the presence of BrdU-containing GC cells. To determine whether IRF8 might influence GC B cell proliferation in vivo, we examined mice immunized with SRBC 12 days previously for the frequencies of GL-7+FAS+ cells that stained for BrdU. For this study, we compared GC B cells from mice homozygous (F/F) and heterozygous (F/+) for a conditional allele of IRF8 that had been crossed with mice expressing CD19-Cre to selectively deplete B cells of IRF8. These studies showed that the frequency of BrdU+ GC B cells from F/F CD19-Cre/+ (CKO) mice was 12% lower than that of B cells from F/+ CD19-Cre controls (p < 0.05) (Fig. 6). These data indicate that IRF8 has a relatively minor role in governing GC B cell proliferation in response to a strong antigenic stimulus.
FIGURE 6.
Regulation of proliferation in GC B cells by IRF8. IRF8-CKO and IRF8Flox/+CD19Cre/+ control mice were immunized with SBBC for 12 days. Mice were injected i.p. with BrdU for 40 min. Splenocytes were stained with GL-7-FITC and anti-FAS-PE, followed by fixation and permeabilization and stained with an anti-BrdU-allophycocyanin Ab. BrdU+ GC B cells (GL-7+FAS+) were analyzed by FACS. Each circle represents one mouse. Data represent two independent experiments with similar results.
Discussion
B lymphocytes engaged in the GC response uniquely experience physiologic DNA DSB resulting from activation-induced cytidine deaminase-induced SHM and CSR. In most cells, DNA DSB induces a stress response featured by p53 activation leading to growth arrest or apoptosis. This stands in striking contrast to the relative immunity of GC B cells to these downstream consequences of DNA breakage. The results of this and other recent studies indicate that the protein products of two genes, IRF8 and BCL6, expressed together at high levels in mature GC B cells synergize to suppress p53-mediated responses. IRF8 mediates the direct transcriptional activation of the gene encoding MDM2, which promotes p53 degradation, while BCL6 represses p53 transcription by binding directly to sequences within the p53 promoter region (6). IRF8 also directly regulates the transcriptional activity of Bcl6 (9). These activities provide a basis for understanding the near complete absence of p53 transcripts or protein in GC B cells (6). Additionally, BCL6 was also reported to repress p21 expression through its interaction with another transcription factor, MIZ1 (8). p21 plays an essential role in cell-cycle arrest after DNA damage (2– 4). Because of the physiological occurrence of DNA DSB in GC B cells, inhibition of p21 activity is probably critical to the highly proliferative state of GC centroblasts. Taken together, the results of this and previous studies indicate that BCL6 and MDM2 play complementary roles in suppressing the expression of p53 and p21 in GC B cells: BCL6 by acting through transcriptional repression, and MDM2 by promoting protein degradation. IRF8 contributes to these regulatory circuits by activating both BCL6 and MDM2 at the transcriptional level (Fig. 7).
FIGURE 7.
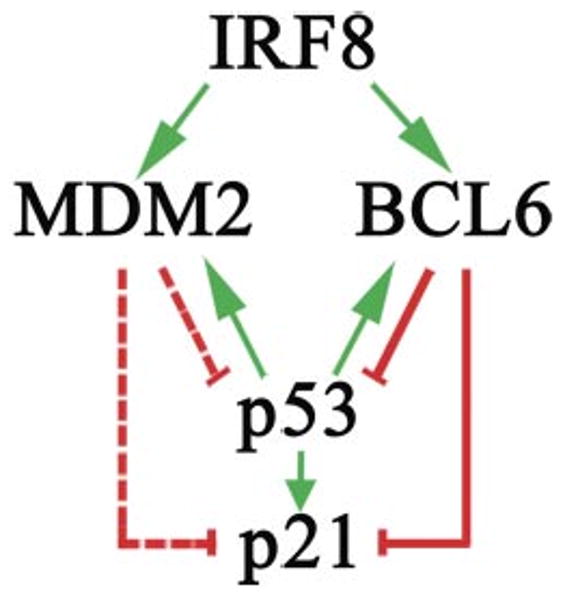
IRF8 contributes to coordinate regulation of p53 and p21 by BCL6 and MDM2 in GC B cells. Green arrows and red solid lines indicate transcriptional activation and repression, respectively. Red dashed lines indicate inhibition by induction of protein degradation. Regulatory loops between p53 and BCL6 or MDM2 are shown.
The finding that IRF8 regulates the expression of MDM2 as well as BCL6 suggests a wider role for this transcription factor in GC development than was appreciated previously. GC centroblasts express IRF8 and BCL6 at the highest levels among any B cell subset and have the shortest doubling times of almost any eukaryotic cell type. A mechanistic or functional explanation for this has not been fully developed, but the fact that IRF8 and BCL6 together suppress p53-dependent as well as p53-independent growth arrest and apoptosis pathways suggests the importance of this phenotype. The contributions of IRF8 to this biology would appear to extend beyond regulation of BCL6 and MDM2, as microarray and qPCR analyses of the NFS-202 cells suggest that IRF8 may also suppress transcription of Cdkn1b (p27) and Cdkn2c (p18) (data not shown). The high proliferative rate of GC B cells may be required to generate the number of cells needed to survive negative selection as centrocytes for generation of effective T-dependent responses.
The regulation of MDM2 by IRF8 also has implications for lymphomagenesis. Aberrant constitutive expression of BCL6 due to chromosomal translocations and/or activation-induced cytidine deaminase-dependent mutations in 5′ noncoding regulatory sequences is a feature of the great majority of human diffuse large B cell lymphomas (43, 44). Coexpression of IRF8 with BCL6, even at physiologic levels, could markedly impair the ability of cells to repair the types of DNA damage that could contribute to disease progression. Thus, by synergizing with BCL6 to regulate p53 and p21, IRF8 may promote the proliferative phase of the normal GC reaction but also the proliferative expansion of B cell lymphomas through its regulation of MDM2. It will be important to determine whether IRF8 and BCL6 function similarly in other cells types, such as macrophages and dendritic cells, that express both genes at high levels (45, 46).
Acknowledgments
We thank Jonathan Kuo for technical help. We are indebted to Dr. Keiko Ozato (National Institutes of Health) and Dr. Hong Wu (University of California at Los Angeles) for providing research reagents.
Footnotes
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Abbreviations used in this paper: GC, germinal center; SHM, somatic hypermutation; CSR, class switch recombination; DSB, double-strand break; IRF, IFN regulatory factor; siRNA, small interfering RNA; WT, wild type; PNA, peanut lectin (agglutinin); IECS, IRF/Ets composite site; qPCR, quantitative real-time RT-PCR; ChIP, chromatin immunoprecipitation.
The online version of this article contains supplemental material.
Disclosures: The authors have no financial conflicts of interest.
References
- 1.Maizels N. Immunoglobulin gene diversification. Annu Rev Genet. 2005;39:23–46. doi: 10.1146/annurev.genet.39.073003.110544. [DOI] [PubMed] [Google Scholar]
- 2.Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 3.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 4.Dulic V, Kaufmann WK, Wilson SJ, Tlsty TD, Lees E, Harper JW, Elledge SJ, Reed SI. p53-Dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 5.Shen Y, White E. p53-Dependent apoptosis pathways. Adv Cancer Res. 2001;82:55–84. doi: 10.1016/s0065-230x(01)82002-9. [DOI] [PubMed] [Google Scholar]
- 6.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 7.Baron BW, Anastasi J, Thirman MJ, Furukawa Y, Fears S, Kim DC, Simone F, Birkenbach M, Montag A, Sadhu A, et al. The human programmed cell death-2 (PDCD2) gene is a target of BCL6 repression: implications for a role of BCL6 in the down-regulation of apoptosis. Proc Natl Acad Sci USA. 2002;99:2860–2865. doi: 10.1073/pnas.042702599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 9.Lee CH, Melchers M, Wang H, Torrey TA, Slota R, Qi CF, Kim JY, Lugar P, Kong HJ, Farrington L, et al. Regulation of the germinal center gene program by IFN regulatory factor 8/IFN consensus sequence binding protein. J Exp Med. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang W, Saberwal G, Horvath E, Zhu C, Lindsey S, Eklund EA. Leukemia-associated, constitutively active mutants of SHP2 protein tyrosine phosphatase inhibit NF1 transcriptional activation by the interferon consensus sequence binding protein. Mol Cell Biol. 2006;26:6311–6332. doi: 10.1128/MCB.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unlu S, Kumar A, Waterman WR, Tsukada J, Wang KZ, Galson DL, Auron PE. Phosphorylation of IRF8 in a pre-associated complex with Spi-1/PU.1 and non-phosphorylated Stat1 is critical for LPS induction of the IL1B gene. Mol Immunol. 2007;44:3364–3379. doi: 10.1016/j.molimm.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong HJ, Anderson DE, Lee CH, Jang MK, Tamura T, Tailor P, Cho HK, Cheong J, Xiong H, Morse HC, III, Ozato K. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol. 2007;179:26–30. doi: 10.4049/jimmunol.179.1.26. [DOI] [PubMed] [Google Scholar]
- 13.Xiong H, Li H, Kong HJ, Chen Y, Zhao J, Xiong S, Huang B, Gu H, Mayer L, Ozato K, Unkeless JC. Ubiquitin-dependent degradation of interferon regulatory factor-8 mediated by Cbl down-regulates interleukin-12 expression. J Biol Chem. 2005;280:23531–23539. doi: 10.1074/jbc.M414296200. [DOI] [PubMed] [Google Scholar]
- 14.Tamura T, Ozato K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J Interferon Cytokine Res. 2002;22:145–152. doi: 10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- 15.Tamura T, Thotakura P, Tanaka TS, Ko MS, Ozato K. Identification of target genes and a unique cis element regulated by IRF-8 in developing macrophages. Blood. 2005;106:1938–1947. doi: 10.1182/blood-2005-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Lee CH, Qi C, Tailor P, Feng J, Abbasi S, Atsumi T, Morse HC., III IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 2008;112:4028–4038. doi: 10.1182/blood-2008-01-129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cattoretti G, Shaknovich R, Smith PM, Jack HM, Murty VV, Alobeid B. Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. J Immunol. 2006;177:6930–6939. doi: 10.4049/jimmunol.177.10.6930. [DOI] [PubMed] [Google Scholar]
- 18.Martinez A, Pittaluga S, Rudelius M, Davies-Hill T, Sebasigari D, Fountaine TJ, Hewitt S, Jaffe ES, Raffeld M. Expression of the interferon regulatory factor 8/ICSBP-1 in human reactive lymphoid tissues and B-cell lymphomas: a novel germinal center marker. Am J Surg Pathol. 2008;32:1190–1200. doi: 10.1097/PAS.0b013e318166f46a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 20.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 21.Brooks CL, Gu W. Dynamics in the p53-Mdm2 ubiquitination pathway. Cell Cycle. 2004;3:895–899. [PubMed] [Google Scholar]
- 22.Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin Y, Lee H, Zeng SX, Dai MS, Lu H. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 2003;22:6365–6377. doi: 10.1093/emboj/cdg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bueso-Ramos CE, Yang Y, deLeon E, McCown P, Stass SA, Albitar M. The human MDM-2 oncogene is overexpressed in leukemias. Blood. 1993;82:2617–2623. [PubMed] [Google Scholar]
- 25.Cordon-Cardo C, Latres E, Drobnjak M, Oliva MR, Pollack D, Woodruff JM, Marechal V, Chen J, Brennan MF, Levine AJ. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 1994;54:794–799. [PubMed] [Google Scholar]
- 26.Ladanyi M, Cha C, Lewis R, Jhanwar SC, Huvos AG, Healey JH. MDM2 gene amplification in metastatic osteosarcoma. Cancer Res. 1993;53:16–18. [PubMed] [Google Scholar]
- 27.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 28.Mehta DS, Wurster AL, Whitters MJ, Young DA, Collins M, Grusby MJ. IL-21 induces the apoptosis of resting and activated primary B cells. J Immunol. 2003;170:4111–4118. doi: 10.4049/jimmunol.170.8.4111. [DOI] [PubMed] [Google Scholar]
- 29.Ozaki K, Spolski R, Feng CG, Qi CF, Cheng J, Sher A, Morse HC, III, Liu C, Schwartzberg PL, Leonard WJ. A critical role for IL-21 in regulating immunoglobulin production. Science. 2002;298:1630–1634. doi: 10.1126/science.1077002. [DOI] [PubMed] [Google Scholar]
- 30.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, Lou J, Knobeloch KP, Gabriele L, Waring JF, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 31.Zhang JQ, Okumura C, McCarty T, Shin MS, Mukhopadhyay P, Hori M, Torrey TA, Naghashfar Z, Zhou JX, Lee CH, et al. Evidence for selective transformation of autoreactive immature plasma cells in mice deficient in Fasl. J Exp Med. 2004;200:1467–1478. doi: 10.1084/jem.20041575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi CF, Xiang S, Shin MS, Hao X, Lee CH, Zhou JX, Torrey TA, Hartley JW, Fredrickson TN, Morse HC., III Expression of the cyclin-dependent kinase inhibitor p27 and its deregulation in mouse B cell lymphomas. Leuk Res. 2006;30:153–163. doi: 10.1016/j.leukres.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 33.Slack A, Chen Z, Tonelli R, Pule M, Hunt L, Pession A, Shohet JM. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci USA. 2005;102:731–736. doi: 10.1073/pnas.0405495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanno Y, Levi BZ, Tamura T, Ozato K. Immune cell-specific amplification of interferon signaling by the IRF-4/8-PU.1 complex. J Interferon Cytokine Res. 2005;25:770–779. doi: 10.1089/jir.2005.25.770. [DOI] [PubMed] [Google Scholar]
- 35.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang CJ, Freeman DJ, Wu H. PTEN regulates Mdm2 expression through the P1 promoter. J Biol Chem. 2004;279:29841–29848. doi: 10.1074/jbc.M401488200. [DOI] [PubMed] [Google Scholar]
- 37.Smith GP. Sequence of the full-length immunoglobulin κ-chain of mouse myeloma MPC11. Biochem J. 1978;171:337–347. doi: 10.1042/bj1710337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Wang H, Li M, Agrawal S, Chen X, Zhang R. MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J Biol Chem. 2004;279:16000–16006. doi: 10.1074/jbc.M312264200. [DOI] [PubMed] [Google Scholar]
- 39.Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 40.Vinuesa CG, Tangye SG, Moser B, Mackay CR. Follicular B helper T cells in antibody responses and autoimmunity. Nat Rev Immunol. 2005;5:853–865. doi: 10.1038/nri1714. [DOI] [PubMed] [Google Scholar]
- 41.Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci USA. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bettess MD, Dubois N, Murphy MJ, Dubey C, Roger C, Robine S, Trumpp A. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol Cell Biol. 2005;25:7868–7878. doi: 10.1128/MCB.25.17.7868-7878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasqualucci L, Migliazza A, Basso K, Houldsworth J, Chaganti RS, Dalla-Favera R. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101:2914–2923. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- 44.Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 45.Toney LM, Cattoretti G, Graf JA, Merghoub T, Pandolfi PP, Dalla-Favera R, Ye BH, Dent AL. BCL-6 regulates chemokine gene transcription in macrophages. Nat Immunol. 2000;1:214–220. doi: 10.1038/79749. [DOI] [PubMed] [Google Scholar]
- 46.Pantano S, Jarrossay D, Saccani S, Bosisio D, Natoli G. Plastic downregulation of the transcriptional repressor BCL6 during maturation of human dendritic cells. Exp Cell Res. 2006;312:1312–1322. doi: 10.1016/j.yexcr.2005.12.020. [DOI] [PubMed] [Google Scholar]