Abstract
Background and Aims
Although the clinical phenotype of Lynch syndrome (also known as Hereditary Nonpolyposis Colorectal Cancer) has been well described, little is known about disease in PMS2 mutation carriers. Now that mutation detection methods can discern mutations in PMS2 from mutations in its pseudogenes, more mutation carriers have been identified. Information about the clinical significance of PMS2 mutations is crucial for appropriate counseling. Here, we report the clinical characteristics of a large series of PMS2 mutation carriers.
Methods
We performed PMS2 mutation analysis using long range PCR and MLPA for 99 probands diagnosed with Lynch syndrome-associated tumors showing isolated loss of PMS2 by immunohistochemistry. Penetrance was calculated using a modified segregation analysis adjusting for ascertainment.
Results
Germline PMS2 mutations were detected in 62% of probands (n = 55 monoallelic; 6 biallelic). Among families with monoallelic PMS2 mutations, 65.5% met revised Bethesda guidelines. Compared with the general population, in mutation carriers, the incidence of colorectal cancer was 5.2 fold higher and the incidence of endometrial cancer was 7.5 fold higher. In North America, this translates to a cumulative cancer risk to age 70 of 15–20% for colorectal cancer, 15% for endometrial cancer, and 25–32% for any Lynch syndrome-associated cancer. No elevated risk for non-Lynch syndrome-associated cancers was observed.
Conclusions
PMS2 mutations contribute significantly to Lynch syndrome but the penetrance for monoallelic mutation carriers appears to be lower than that for the other mismatch repair genes. Modified counseling and cancer surveillance guidelines for PMS2 mutation carriers are proposed.
Keywords: PMS2, Lynch syndrome, HNPCC, penetrance
Background
Lynch Syndrome (LS; also known as Hereditary Nonpolyposis Colorectal Cancer) is the most common form of hereditary colorectal cancer and accounts for 2.2% of newly diagnosed colorectal cancers and 2.3% of newly diagnosed endometrial cancers.1–3 Mutations in one of four mismatch repair genes: MLH1, MSH2, MSH6, and PMS2 cause LS and studies suggest that there may be differences in cancer risks for mutation carriers depending on the gene that is mutated.4, 5 Most recent valid estimates of cancer risks to age 70 suggest that individuals with LS have an increased risk of colorectal cancer (22–74%), endometrial cancer (32–42%), and a number of other cancers (such as those of the stomach, ovary, small intestine, biliary tract, and urinary tract).6–9 However, previous studies did not often include PMS2 analysis and therefore little is known about the cancer risks for PMS2 monoallelic mutation carriers. This is primarily due to the limited number of carriers described to date, which can be attributed to the historic difficulty in the identification of PMS2 mutations given the presence of a large family of pseudogenes located on the same chromosome.10–13 Most of the complications have recently been overcome by using long range PCR to preferentially amplify the PMS2 gene rather than its pseudogenes.14 With these methods, mutation testing by sequencing is greatly simplified and accurate except for exons 13–15 in which pseudogene-related conversion may confound the analysis.15–17
Traditionally, families most likely to have LS have been identified by using previously published classification systems known as the Amsterdam Criteria and the less stringent Bethesda Guidelines.18–21 However, many medical centers have adopted the practice of screening colorectal and endometrial tumors for LS at the time of surgical diagnosis, often using immunohistochemistry (IHC) to assess the presence or absence of the mismatch repair proteins. This approach allows for the identification of individuals at an increased risk of having LS regardless of their age or family history and facilitates the search for mutations by pinpointing the likely mutated gene based on the absence of its protein in the tumor.22 IHC showing isolated loss of PMS2 protein is suggestive of a germline PMS2 mutation. Two large studies have shown that loss of PMS2 protein and retention of MLH1, MSH2, and MSH6 mismatch repair proteins occurs in 4.3% of colorectal tumors exhibiting a high degree of microsatellite instability (95% CI 2.7% – 6.4%),23 and 11.5% of colorectal tumors with abnormal mismatch repair IHC (95% CI 6.7% – 18.0%).24 Baudhuin et al. found loss of PMS2 protein in <1% of colorectal tumors with abnormal IHC from cases unselected for family history of cancer and 4% of colorectal tumors with abnormal IHC that were classified as “moderate to high risk patients” based on “being referred for routine clinical molecular testing for Lynch syndrome at the Mayo Clinic.”25 Our unpublished data from a population-bases series of 483 colorectal cancer cases in central Ohio identified two probands with absence of PMS2 on IHC (0.4%).
There have been multiple reports of individuals with biallelic germline PMS2 mutations.13, 26–35 These individuals typically present with malignancy at a strikingly young age and have a distinct phenotype that is quite different from classically described LS, often consisting of gastrointestinal and hematologic malignancies, brain tumors, and Neurofibromatosis Type I features including café au lait spots.
Few reports of individuals heterozygous for PMS2 gene mutations currently exist and these have been limited due to small numbers of probands with deleterious mutations.36, 37 Here, we report clinical data for a large series of heterozygous PMS2 mutation carriers.
Patients and Methods
We tested 99 probands who had a LS-associated tumor (91 colorectal, 5 endometrial, 1 transitional cell of the renal pelvis, 1 small intestinal, and 1 gastric) that demonstrated isolated absence of PMS2 protein on IHC. Of these, 55 were enrolled in research studies approved by the institutional review board at The Ohio State University that allowed for PMS2 gene testing in our research laboratory. The remaining 44 samples were received anonymously through research collaborations with investigators in Newfoundland (n=3) and Sweden (n=6) and a formal collaboration with the NCI-funded Colon Cancer Family Registry, which provided 35 anonymous samples from four sites: Australasia, Seattle, Mayo Clinic, and Ontario. Personal and family medical history information was collected from all probands and cancer diagnoses among relatives were confirmed with pathology reports whenever possible.
Testing for PMS2 mutations
Sequence Analysis
Mutations specific to the PMS2 locus were characterized following a previously described method based on long range PCR,14 with the following modifications. Exons 6, 7, 8, and 10 were individually amplified directly from genomic DNA, and PCR2 was reduced to a smaller product (1618bp) using the following primers: 5′-ttgcttgtaatctgccagatgtggt-3′ and 5′-atctactttctcccttggttgacat-3′. When possible, relatives were tested for the mutation identified in the proband.
Multiplex Ligation-Dependent Probe Amplification (MLPA)
MLPA was carried out according to the manufacturer’s instructions (MRC Holland, Amsterdam, Netherlands). Briefly, 125ng of genomic DNA was denatured and hybridized for 18 hours at 60°C with the probe mix. Samples were then treated with ligase-65 at 54°C for 15 minutes. Subsequent PCR reactions were performed using a FAM-labeled primer. PCR products were resolved using an ABI 3700 sequencer and the results analyzed with Genotyper® software (Applied Biosystems). Deletions were suspected when the peak height was 60% or less than that of the controls. Due to the high degree of homology of the PMS2 pseudogene sequences, the MLPA kit is only reliable for exons 1, 2, 5, 6, 7, 8, 9, 10, 11 and 12.
Testing for BRAF mutations
A standard PCR and sequencing approach was used to screen for activating mutations within exon 15 of BRAF. Briefly, 25–50ng of tumor DNA was amplified in a 15ul PCR reaction using Promega’s GoTaq® master mix and the following primer pair: Forward; 5′-cttcataatgcttgctctgatagga-3′ and Reverse; 5′-tttctagtaactcagcagcatctca-3′. Following successful amplification, PCR products were treated with ExoSAP-IT (USB Corporation) and sequenced with the forward primer. Products were analyzed using an ABI3700 machine.
Statistical Analysis
In order to estimate the age-specific cumulative cancer risk (penetrance) and relative incidence of the variant in monoallelic carriers compared to that for the population (hazard ratio, HR), the log likelihood was calculated for each pedigree under a segregation analysis model modified to take into account measured genotypes. To account for ascertainment, the likelihood for each population-based pedigree (case unselected for family history) was then conditioned on the PMS2 mutation status and cancer status of the proband and age at diagnosis. The likelihood for each clinic-based pedigree (ascertained via a family cancer clinic presumably because of a family history of cancer) was conditioned on the PMS2 mutation status of the proband and the cancer status and age of diagnosis of all relatives. Estimates were obtained by maximizing the sum of the logs of the conditioned likelihoods. Data were analyzed with a model in which the mean HR was estimated using region- and age-specific population incidence data as described by Antoniou et al.38 Separate hazard ratios were simultaneously estimated for the following cancer groups: colorectal cancer; endometrial cancer; less frequently reported LS-associated cancers (small bowel, stomach, kidney, ureter, ovary, and brain); and other (all other cancer sites) as in Quehenberger et al.7 Age was recorded at diagnosis of first cancer, death, or last known age (or polypectomy or hysterectomy when estimating colorectal or endometrial cancer risk respectively), whichever occurred first. Age-specific population incidences were obtained for 2002 from Globocan for North America, Australia and Sweden, being the regions from which the families in the penetrance analysis were recruited.39 For population-based families all relatives in generations with complete ascertainment were included irrespective of cancer status. For clinic-based families all available relatives were included in the penetrance analysis. Relatives with missing ages were given age 20 years if they had offspring, or one year more than their age of cancer diagnosis, or age 0 years otherwise.
The log HR, and hence the age-specific cumulative risk, of each cancer group was estimated under a dominant model by maximizing the total sample log-likelihood, equal to the sum of the family-specific ascertainment-conditioned log-likelihoods (families were assumed to be independent). The log HR were first assumed independent of age and sex, but were subsequently allowed to vary with age and sex. These analyses were performed with the pedigree analysis program MENDEL.40 Estimates were based on the most parsimonious model, as chosen by forward model selection, and the asymptotic likelihood ratio test was used to compare the goodness-of-fit of nested models. The population allele frequency was assumed to be 0.0001. Due to the rarity of mutations, the cancer incidences in non-carriers were assumed to equal those of the general population. To restrict risk estimation to monoallelic carriers, potential and known biallelic siblings of biallelic probands were excluded from the analysis.
Separately from the segregation analyses we also calculated carrier probabilities (Table 4) with a modified version of MENDEL 3.2 based on known genotypes and family relationships only; in particular they were not based on disease status. This calculation assumed Mendelian laws of inheritance, Hardy-Weinberg equilibrium and the allele frequency above.
Table 4.
Effective family size for the 39 families with a PMS2 mutation used in the penetrance analysis by recruitment region
| Region | Population-based | Clinic-based | ||||||
|---|---|---|---|---|---|---|---|---|
| families | Expected number of carriers. (sum of carrier probabilities)* | Individuals with probability of being a carrier >= 0.25 * | Number of relatives tested for family PMS2 mutation^ | families | Expected number of carriers. (sum of carrier probabilities)* | Individuals with probability of being a carrier>= 0.25 * | Number of relatives tested for family PMS2 mutation^ | |
| N. America | 17 | 106 | 255 | 13 | 12 | 117 | 221 | 22 |
| Australia | 7 | 52 | 147 | 1 | 1 | 6 | 10 | 0 |
| Sweden | 0 | 0 | 0 | 0 | 2 | 6 | 10 | 2 |
| Total | 24 | 158 | 402 | 14 | 15 | 129 | 241 | 24 |
Based on genetic relatedness to known or inferred carriers. Count does not include probands.
within two degrees of relatedness to known carriers.
Results
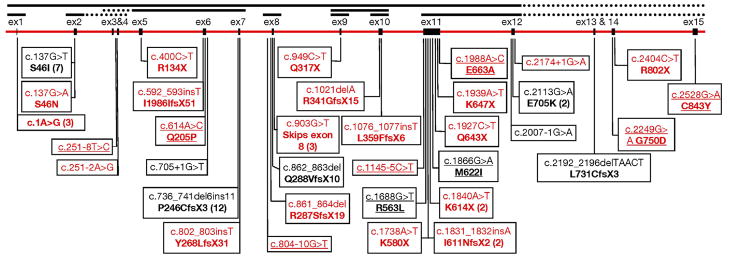
Of the 99 probands tested for PMS2 mutations using sequencing and MLPA, 61 (62%) were found to have deleterious mutations in the PMS2 gene. Another 10 probands had missense variants for which the significance has yet to be determined (Figure 1 shows the distribution of identified mutations). In all, 34 different deleterious PMS2 mutations were identified, 11 of which were seen in more than one proband. Five mutations occurred more than twice. The most notable of the five exceptions was an insertion/deletion frameshift mutation in exon 7, which was seen in 12 ostensibly unrelated probands. In an attempt to determine if this was either a founder mutation or one that arises frequently de novo, we conducted genotyping and haplotype analyses and found evidence of a shared haplotype suggesting that this mutation arose in the first millennium, possibly in Sweden or England, and is presently widespread in the world.41 The other four mutations, c.137G>T, an exon 10 deletion, c.903G>T, and c.1A>G, which occurred in 7, 3, 3, and 3 probands, respectively, are also prominent candidates for additional founder mutations within the PMS2 locus. However, unlike for the mutation within exon 7, we are presently unable to make significant conclusions regarding the status of these four additional mutations due to the relatively low number of affected probands and family members currently identified. With continued screening for PMS2 mutations we are hopeful that sufficient samples will eventually be identified so as to make conclusions regarding the origin of these four mutations and consequently their prevalence within the population. Finally, we note that as many as 6 other different mutations occurred in two unrelated probands each suggesting that further founder mutations may be present in the population.
Figure 1. Schematic of the PMS2 genomic region along with the mutations identified in this study.
Mutations in black were described previously (Clendenning et al. 2006; Nakagawa et al 2004). Underlined mutations currently have an unconfirmed disease causing status. Complete exon deletions identified by MLPA are represented by black lines above the exons, with dashed regions representing possible extensions to the deleted regions that cannot be confirmed by the MLPA kit. Numbers in parentheses indicate the number of times each mutation was observed if observed more than once.
Six probands with a clinically distinct phenotype were found to have biallelic mutations in the PMS2 gene. Because these probands have a drastically different clinical presentation, we have chosen to describe the results of the monoallelic and biallelic mutation carriers separately.
The mutations were classified as being deleterious because a premature stop codon was introduced, either directly by a nonsense mutation or as a result of a frameshift mutation. Others were deleterious splice site mutations. The three mutations which did not meet these criteria (c.137G>T, c.137G>A and c.2113G>A) were classified as being deleterious based on previous studies. 12, 42
BRAF mutations
We performed BRAF analysis on tumor DNA from 47 probands. Tumor DNA was unavailable for the other 52 probands. The p.V600E mutation, which has been associated with sporadic colorectal cancers showing microsatellite instability in the context of somatic MLH1 promoter hypermethylation,43 was identified in one proband found to have an exon 10 deletion in PMS2, 2 probands who have the c.736_741del6ins11 mutation, and in 2 other probands that did not have detectable PMS2 mutations.
Monoallelic PMS2 mutation carriers
Table 1 shows that of the 55 probands found to have monoallelic germline PMS2 gene mutations, 47 had colorectal cancer, 5 had endometrial cancer, 1 had transitional cell carcinoma of the renal pelvis, 1 had cancer of the small intestine, and 1 had gastric cancer as their first cancer diagnosis. The mean age of cancer diagnosis was 50 years with a range of 23–77 years, which is similar to the mean age of diagnosis for the 28 probands who had neither a deleterious mutation nor an unclassified missense variant (mean age was 51 years; range, 31–84). Of the mutation carrying probands, 1 had metachronous colorectal cancer, 2 had synchronous colorectal cancers, and 4 had more than one type of LS-associated tumor. Five of the monoallelic mutation carrying probands (9.1%) had a family history of cancer that met the modified Amsterdam criteria;19 36 (65.5%) met the revised Bethesda guidelines;21 and 14 (25.5%) met none of the published clinical criteria for LS. Excluding the biallelic mutation carrying probands, of the probands who were diagnosed with their first LS-associated tumor before age 50, 57.4% (27/47) were found to have deleterious PMS2 gene mutations. Among probands diagnosed over 50 years of age, 61% (28/46) were positive for deleterious PMS2 gene mutations (Table 2). In this series, there were 19 monoallellic mutation carrying probands (34.5%) who had a parent with at least one LS-associated tumor. Among these 19 affected parents, there were 11 colorectal cancers, 5 gastric cancers, 3 endometrial cancers, and 2 pancreatic cancers. Parental mutation status confirmation was only feasible in 3 of the 19 affected parents. Samples from at least one additional family member were available for 13 of 55 probands with monoallelic PMS2 mutations and one proband was found to have a de novo PMS2 mutation as both of his parents were negative for his mutation and non-paternity was excluded.
Table 1.
Monoallelic PMS2 mutation carriers
| Patient | Mutation | Tumor Type | Age of Dx | 2nd Primary (Age) | Affected parent (Y/N)* | # Affected sibs (total sibs) | # Affected 2DRs (total 2DRs) |
|---|---|---|---|---|---|---|---|
| 1 | c.736_741del6ins11 (P246CfsX3) | TCC | 45 | Rectum (50) | N | 0(1) | 0(6) |
| 2 | c.736_741del6ins11 (P246CfsX3) | Ascending | 54 | Y^ | 0(1) | U | |
| 3 | c.736_741del6ins11 (P246CfsX3) | Descending | 51 | N^ | 0(2) | 0(3) | |
| 4 | c.736_741del6ins11 (P246CfsX3) | Ascending | 74 | Y^ | 1(2) | U | |
| 5 | c.736_741del6ins11 (P246CfsX3) | Splenic flexure | 47 | Y^ | 0(9) | 0(11) | |
| 6 | c.736_741del6ins11 (P246CfsX3) | Ascending | 44 | N | 1(12) | 6(23) | |
| 7 | c.736_741del6ins11 (P246CfsX3) | Rectal | 29 | N | 0(1) | 3(8) | |
| 8 | c.736_741del6ins11 (P246CfsX3) | Ascending | 58 | N | 0(2) | 2(12) | |
| 9 | c.736_741del6ins11 (P246CfsX3) | Cecum | 57 | N | 0(3) | 1(7) | |
| 10 | c.736_741del6ins11 (P246CfsX3) | Cecum | 51 | N | 0(5) | 0(7) | |
| 11 | c.736_741del6ins11 (P246CfsX3) | Stomach | 67 | Y^ | 2(8) | 1(11) | |
| 12 | c.736_741del6ins11 (P246CfsX3) | Cecum | 48 | N | 0(2) | 2(10) | |
| 13 | exon 8 deletion | Transverse | 28 | N^ | 0(1) | 1(5) | |
| 14 | c.1840A>T(K614X) | Sigmoid | 74 | N^ | 2(5) | 0(11) | |
| 15 | c.1840A>T(K614X) | Endometrial | 56 | Y^ | 1(3) | 0(4) | |
| 16 | c.862_863del (Q288VfsX10) | Ascending | 54 | N | 0(3) | 3(15) | |
| 17 | c.592_593insT (I198IfsX51) | Colon | 42 | N^ | 0(4) | 2(8) | |
| 18 | c.861_864del (R287SfsX19) | Rectal | 39 | N | 0(3) | 0(10) | |
| 19 | c.1076_1077insT (L359FfsX6) | Endometrial | 49 | Cecum (57) | Y | 0(1) | 2(8) |
| 20 | c.903G>T (Skips exon 8) | Endometrial | 61 | Y | 0(1) | 0(15) | |
| 21 | c.903G>T (Skips exon 8) | Ascending | 54 | N | 0(3) | 0(16) | |
| 22 | c.903G>T (Skips exon 8) | Colon | 54 | Y | 0(2) | 2(3) | |
| 23 | Exon 1 deletion | Ascending | 56 | N | 0(3) | U | |
| 24 | c.1927C>T (Q643X) | Descending | 54 | N | 0(3) | 1(11) | |
| 25 | c.400C>T (R134X) | Ascending | 66 | N | 0(1) | 1(5) | |
| 26 | c.1831_1832insA (I611NfsX2) | Cecum | 60 | N | 0(3) | 1(13) | |
| 27 | c.1831_1832insA (I611NfsX2) | Colon | 39 | Y | 0(2) | 1(7) | |
| 28 | Complete gene deletion | Colon | 54 | Y | 2(3) | 0(4) | |
| 29 | Complete gene deletion | Sigmoid | 67 | N | 3(4) | 1(12) | |
| 30 | Exon 9 deletion | Transverse | 49 | N | 0(3) | 1(13) | |
| 31 | c.137G>T (S46I) | Cecum | 32 | N^ | 0(1) | 0(5) | |
| 32 | c.137G>T (S46I) | Cecum | 47 | N | 0(4) | U | |
| 33 | c.137G>T (S46I) | Sigmoid | 44 | N^ | 0(4) | 2(15) | |
| 34 | c.137G>T (S46I) | Small intestine | 46 | N | 0(1) | 0(2) | |
| 35 | c.137G>T (S46I) | Transverse | 43 | N | 0(0) | 1(13) | |
| 36 | c.137G>T (S46I) | Sigmoid | 62 | Ascending (62) | N | 0(3) | 0(15) |
| 37 | c.2113G>A (E705K) | Colon | 61 | Y^ | 0(0) | U | |
| 38 | c.2113G>A (E705K) | Hepatic flexure | 50 | Y | 0(1) | 3(13) | |
| 39 | c.705+1G>T (Loss of splice donor site) | Endometrial | 34 | Colon (58) | N | 0(1) | 2(14) |
| 40 | c.2007−1G>A (Loss of splice acceptor site) | Rectosigmoid | 77 | N | 0(3) | U | |
| 41 | c.2174+1G>A (Loss of splice donor site) | Colon | 71 | Colon (71) | Y | 0(10) | U |
| 42 | c.1738A>T (K580X) | Transverse | 49 | N | 0(9) | U | |
| 43 | c.1939A>T (K647X) | Sigmoid | 42 | N | 0(1) | 3(11) | |
| 44 | deletion of exons 5, 6, 7 | Cecum | 50 | N | 0(1) | 0(7) | |
| 45 | deletion of exons 5, 6, 7 | Ascending | 37 | N | 0(5) | 1(10) | |
| 46 | deletion exon 2 | Cecum | 46 | N | 0(5) | 0(10) | |
| 47 | c.1021delA (R341GfsX15) | Transverse | 49 | Y | 0(2) | 2(11) | |
| 48 | c.2404C>T (R802X) | Colon | 36 | Rectum (37) | Y | 0(2) | U |
| 49 | c.949C>T (Q317X) | Ascending | 39 | N | 0(2) | 0(6) | |
| 50 | c.802_803insT (Y268LfsX31) | Sigmoid | 59 | Y | 0(1) | 2(15) | |
| 51 | Exon 10 deletion | Endometrial | 30 | Cecum (44) | N | 0(2) | 1(15) |
| 52 | Exon 10 deletion | Ascending | 60 | N | 0(2) | 0(5) | |
| 53 | Exon 10 deletion | Rectal | 45 | Y | 0(2) | 0(11) | |
| 54 | Exon 11-12 deletion | Ascending | 51 | N | 0(2) | U | |
| 55 | c.2192_2196delTAACT (L731CfsX3) | Transverse | 23 | N | New mutation |
Affected with any of the following cancers: colorectal, endometrial, stomach, ovarian, pancreatic, ureter and renal pelvis, biliary tract, brain tumors, and small bowel. Dx = diagnosis; TCC = transitional cell carcinoma; U = unknown;
= de novo mutation excluded
Table 2.
Number of PMS2 mutation carriers by age at diagnosis and family history in probands with isolated loss of PMS2 in their tumor by IHC.
| Total | PMS2 mutation carrier (%) | |
|---|---|---|
| Age <50 | 47 | 27 (57.4) |
| and affected FDR and/or SDR | 27 | 17 (63) |
|
| ||
| Age >50 | 46 | 28 (60.9) |
| and affected FDR and/or SDR | 26 | 15 (57.7) |
FDR = first degree relative, SDR = second degree relative. Probands were considered mutation positive only when a deleterious mutation was identified. Patients with uncertain missense variants were considered negative. Biallelic mutation carriers (n=6) were excluded from this table.
In total, we identified 55 monoallelic mutation carriers among the relatives of probands. We obtained colorectal cancer screening information at the time of ascertainment for 29 (53%) of these relatives. Of these, 10 had not undergone colonoscopy; 8 had polyps removed previously (histology unknown); and 11 had normal colonoscopies. Colon cancer screening status was not reported for the remaining 26 (47%) identified monoallelic mutation carriers however 8 of these mutation carriers reported a history of colorectal cancer.
Biallelic PMS2 mutation carriers
Six probands were found to have biallelic PMS2 gene mutations. Five of these presented with colorectal cancer as their first tumor and one presented with medulloblastoma at age 7. Four of six biallelic mutation carriers had multiple colonic polyps at the time of ascertainment. The mean age of gastrointestinal tumor diagnosis was 21 years (range, 14–28 years). One of the five biallelic mutation carrying probands was homozygous for a PMS2 mutation and her parents were consanguineous. Four of the 5 probands had at least one sibling with a malignancy before age 40 (range of sibling malignancy, 6–38 years). IHC results from these probands’ tumor analyses demonstrated loss of PMS2 expression in their tumors as well as adjacent unaffected tissue. A summary of these clinical findings can be found in Table 3.
Table 3.
Biallelic PMS2 gene mutation carriers
| Pt | PMS2 mutation (a) | PMS2 mutation (b) | Personal History (age of dx) | Family History (age of dx) |
|---|---|---|---|---|
| 56 | c.2249G>A (G750D) | Complete gene deletion | Rectal cancer (22) Brain tumor (23) |
Brother: CRC (21) |
| 57 | c.1A>G (5′ truncation) | Deletion exons 9 and 10 | Synchronous descending colon cancer (28) | 2 great, great uncles with CRC (48, 56) |
| 58 | c.1A>G (5′ truncation) | c.614A>C (Q205P) | Colon cancer (20) Duodenal cancer (41) Lymphoma |
Brother: brain tumor (38) Sister: brain tumor (31) |
| 59* | c.137G>T (S46I) | c.137G>A (S46N) | Sigmoid cancer (14) | Noncontributory |
| 60 | c.1A>G (5′ truncation) | c.251-2A>G (Loss of splice acceptor site) | Rectal cancer (24) Endometrial cancer (35) Glioma (35) |
Brother: CRC (26), glioblastoma (34) Brother: glioma (24) |
| 61*^ | c.949C>T (Q317X) | c.949C>T (Q317X) | Meduloblastoma (7) Ampullary adenocarcinoma (16) |
Brother: T-cell lymphoma (6) |
Documented features of neurofibromatosis;51 CRC = colorectal cancer; Dx = diagnosis
Parents of this proband are first cousins
Penetrance
Table 4 shows the effective size and ascertainment of the 39 PMS2 mutation-carrying families used in the penetrance analysis. On average (not including the proband) each of the 24 population-based families consisted of approximately 6.6 expected carriers – compared to 9.0 for the 15 clinic-based families.
Table 5 shows that in the 39 families with PMS2 mutations used for penetrance analysis, there were 34 cases of colorectal cancer with a mean age of 59 years in men and women who were within 2 degrees of relationship of a known mutation carrier (not including the probands). There were 9 relatives diagnosed with endometrial cancer with an average age of diagnoses of 50 years, and 22 cases of cancer classified as less frequent LS cancers with an average age of diagnosis of 56 years.
Table 5.
Number and mean (standard error) age of diagnosis of first cancers in individuals with a carrier probability of >=0.25 (excluding probands) reported in the 39 PMS2 mutation-carrying families used in penetrance analysis by ascertainment.
| Population-based (24 families) | Clinic-based (15 families) | ||||
|---|---|---|---|---|---|
| Cancer site | Sex | Number of cases | Mean age at diagnosis (SE) | Number of cases | Mean age at diagnosis (SE) |
| Colorectal | Males | 7 | 61.0 (7.0) | 9 | 47.3 (7.1) |
| Females | 10 | 65.7 (5.5) | 8 | 62.7 (3.4) | |
| Endometrial | Females | 6 | 49.2 (5.9) | 2 | 50.0 (26.0) |
| Less frequent Lynch cancers | Males | 5 | 57.8 (9.1) | 7 | 61.0 (5.4) |
| Females | 4 | 59.7 (13.8) | 5 | 54.8 (7.4) | |
| Ovary | Females | 2 | 42 (20.0) | 0 | |
| Stomach | Males | 2 | 70.0 (17.0) | 3 | 72.7 (1.3) |
| Females | 2 | 77.5 (10.5) | 4 | 60.7 (5.8) | |
| Small intestine | Males | 1 | 59 | 0 | |
| Females | 0 | 0 | |||
| Kidney (renal pelvis) | Males | 0 | 3 | 57 (6.2) | |
| Females | 0 | 0 | |||
| Ureter | Males | 0 | 0 | ||
| Females | 0 | 0 | |||
| Brain | Males | 2 | 45 (15.0) | 1 | 38 |
| Females | 0 | 1 | 31 | ||
| Other Cancers* | Males | 21 | 66.8 (3.8) | 11 | 52.6 (5.9) |
| Females | 26 | 57.0 (2.9) | 10 | 62.7 (4.6) | |
| Totals | 79 | 52 | |||
Breast (n=19), lung (n=13), prostate (n=8), skin (n=7), non-Hodgkin’s lymphoma (n=7), leukemia (n=3), pharynx (n=2), liver (n=2), pituitary gland (n=2), pancreas (n=1), testis (n=1), thyroid (n=1), unspecified (n=2).
The increased risk of colorectal cancer for male carriers, did not differ from that for female carriers (p = 0.3) and when combined, the overall increased risk was 5.2 fold (95% CI 2.8 – 9.7). Based on the incidence rates in the North American population, this increased risk would result in a cumulative risk to age 70 of approximately 20% (11%–34%) for male carriers and 15% (8%–26%) for female carriers. Female carriers were at approximately 8 times the general population risk of endometrial cancer resulting in a 15% (6%–35%) risk to age 70 in North America. There was some evidence for increased risk of the less common LS-associated cancers but the increase was not statistically significant for males or females (both p = 0.3). There was no evidence of increased risk for cancers not previously associated with LS (p >0.2). Overall, based on the hazard ratio estimates above and the region specific cancer incidence rates in North America, 25% (16% – 48%) of male PMS2 mutation carriers and 32% (21% – 53%) of female PMS2 mutation carriers will be diagnosed with any LS-associated cancer by age 70. This compares with about 6.5% of males, and 7.2% of females, in the general North American population. There was no evidence that the hazard ratio for any cancer group varied with age (p ≥ 0.1) or by country of recruitment (p ≥ 0.4), or by whether the family was ascertained on a population-basis or from a family cancer clinic (p ≥ 0.5) (Table 6).
Table 6.
Penetrance as hazard ratio and risk to age 70 for cancer based on sex for North America, Australia, and Sweden.
| Cancer | Sex | Hazard ratio (95% CI) | Cumulative risk (95% CI) | ||
|---|---|---|---|---|---|
| to age 50 | to age 60 | to age 70 | |||
| Colorectal | Male | 5.2 (2.8–9.7) | 2% (1%–4%) | 8% (4%–14%) | 20% (11% – 34%) |
| Female | 5.2 (2.8–9.7) | 2% (1%–4%) | 6% (3%–10%) | 15% (8% – 26%) | |
| Endometrial | Female | 7.5 (2.8–20.0) | 3% (1%–8%) | 9% (3%–21%) | 15% (6% – 35%) |
| Less frequent Lynch cancers* | Male | 2.5 (0.4–16.2) | 1% (0%–8%) | 3% (0%–17%) | 6% (1% – 33%) |
| Female | 2.5 (0.5–12.6) | 1% (0%–7%) | 3% (1%–13%) | 6% (1% – 25%) | |
| Other cancers | Male | 0.9 (0.3–2.3) | 4% (1%–9%) | 10% (4%–24%) | 24% (10% – 51%) |
| Female | 1.5 (0.8–3.1) | 8% (4%–15%) | 16% (8%–29%) | 27% (15% – 48%) | |
| Any Lynch Syndrome-associated cancer | Male | 4% (2%–10%) | 10% (6%–24%) | 25% (16%–48%) | |
| Female | 6% (4%–14%) | 16% (10%–31%) | 32% (21%–53%) | ||
Kidney (renal pelvis), stomach, ovary, small bowel, ureter, brain
Discussion
This is the largest series of PMS2 mutation carriers reported to date and probands were identified both by population-based screening of colorectal and/or endometrial tumors and through ascertainment in high risk specialty clinics. This study illustrates that PMS2 gene mutations account for many cases of LS and perhaps have historically been overlooked and underestimated given the technical difficulty of identifying them. In a previous population-based study of colorectal and endometrial cancers, we found that 6 of 44 (13.7%) probands with LS had PMS2 mutations (these 6 mutation carriers are included in this analysis).1, 2
In the current study, we found deleterious PMS2 gene mutations in 62% of probands selected on the criterion that their tumors showed isolated loss of PMS2 protein by IHC. In addition, we found 10 probands with variants of uncertain significance in PMS2 that could be deleterious. In contrast, we previously screened 43 probands whose tumors showed loss of both MLH1 and PMS2 proteins on IHC (MLH1 and PMS2 form a heterodimer) and we detected no deleterious PMS2 mutations in these probands. It should be noted that the possibility of gene conversion events between the 3′ end of PMS2 and the PMS2-CL pseudogene 16 means that in a proportion of probands we may have screened exons 13, 14, and 15 from the pseudogene rather than the true PMS2 locus. Our analysis identified 2 deleterious mutations and 2 variants of uncertain significance in these exons (figure 1). In addition, the inability of the MLPA kit to detect deletions of exons 3, 4, 13, 14, and 15 means that it is possible that some of the 38 individuals who tested negative (10 of whom had missense variants of unknown significance) have PMS2 gene mutations that we could not identify with the current technique. Overall, PMS2 mutations contribute significantly to LS, yet clinical testing for PMS2 gene mutations is not currently available in the United States.
Our data predict that approximately one in four male carriers of PMS2 mutations and one in three female carriers of PMS2 mutations will be diagnosed with a LS-associated cancer by age 70. These risks appear to be lower than the respective cancer risks for MLH1 and MSH2 mutation carriers. For example, in a Dutch series of clinic-based families with MLH1 and MSH2 mutations the risks to age 70 for LS-associated cancers were estimated to be 38% for males and 54% for females.7 In Australian families ascertained from a population-based series of CRC diagnosed before age 45, the corresponding risks were 75% and 86%.9 In a series of Scottish families ascertained from a population-based series of CRC diagnosed before age 35 the risks for CRC and endometrial cancers alone were 70% for males and 72% for females.6 However, due to the substantial size of the 95% confidence intervals of our estimates, we cannot definitively conclude that the penetrance for PMS2 mutations is lower than for mutations in other mismatch repair genes. There have been numerous other studies estimating penetrance for LS-associated cancers for MLH1 and MSH2 mutation carriers to be even higher, however the majority were based on families that were ascertained because they had striking clinical histories and as penetrance estimates were not conditioned on this ascertainment, the previous estimates were upwardly biased.9, 44, 45
If the risks of cancer for PMS2 mutation carriers are lower than that for carriers of mutations in other mismatch repair genes, which is consistent with the observed data, the molecular explanation for this is unclear. It has been hypothesized that MLH1 can form a heterodimer with MLH3 or PMS1 in the absence of functional PMS2, which may compensate for the MutLα heterodimer in the mismatch repair process.46 This may also explain why the cancer risks for MSH6 mutation carriers are also lower than the risks for MLH1 and MSH2 mutations carriers (although still higher than for the PMS2 mutation carriers) since MSH2 can also form a heterodimer with MSH3 in the absence of functional MSH6.
Given the relatively low estimates of penetrance, published guidelines for ascertaining patients with LS including the Amsterdam criteria and the revised Bethesda guidelines may not be adequate for identifying patients with PMS2 mutations. If clinicians relied solely on the Amsterdam criteria to identify the mutation carriers in this series, 50/55 (90.9%) mutation carriers would not have been identified. If clinicians relied on revised Bethesda guidelines alone, 14/55 (25%) mutation carriers would have been missed. The above limitations have been described for ascertaining patients with mutations in the other mismatch repair genes as well. In a population based study, 7 of 10 (70%) of endometrial cancer patients and 5 of 23 (22%) of colorectal cancer patients who had LS did not meet either set of published criteria.1, 2 In a population-based series of colorectal cancer patients diagnosed under age 45, 50% of LS cases would have been missed using Amsterdam criteria alone.22
The argument could be made that given the low penetrance in monoallelic PMS2 mutation carriers, identification of probands without a family history of LS-associated tumors is not of great importance. However, we have shown that PMS2-associated LS also demonstrates highly variable clinical characteristics. The age of first LS-associated tumor varied widely among our molonallelic mutation-carrying probands with a range of 23–77 years (mean, 50 years). Roughly half (49.1%) of all of the monoallelic mutation carriers were diagnosed with cancer before age 50 (Table 7). Thus, general population screening recommendations for colon cancer surveillance beginning at age 50 would not be sufficient for families with a PMS2 gene mutation. However, the standard LS cancer screening guidelines (colonoscopy every 1–2 years beginning at age 20–25)47 for carriers of MLH1 or MSH2 mutations may be extreme given the lower penetrance. Our data suggest that PMS2 mutation carriers should probably follow an intermediate screening regimen such as beginning colonoscopy every 1–2 years at 30, as was recommended by Lindor et al. for individuals with MSH6 mutations.48 It should be noted, though, that three monoallelic mutation carriers (5% of all mutations identified) were diagnosed with their first cancer before age 30.
Table 7.
Age distribution of monoallelic PMS2 gene mutation carrying probands at diagnosis
| Age range (years) | # of mutation carrying probands (%) |
|---|---|
| 20–29 | 3 (5) |
| 30–39 | 8 (15) |
| 40–49 | 16 (29) |
| 50–59 | 16 (29) |
| 60–69 | 8 (15) |
| 70–79 | 4 (7) |
It is worth noting that in this series the typical V600E mutation in BRAF was present in the tumors of 3 probands out of 28 with monoallelic PMS2 mutations tested for BRAF. Therefore, the proposed use of BRAF analysis to exclude LS, which is based on results from MLH1 and MSH2 mutation carriers, 49, 50 may not be applicable in the case of PMS2 mutation carriers.
Our results show that PMS2 mutations are common (>60%) in patients whose tumors do not stain for PMS2 by IHC. However, when the staining for MLH1 is also defective, PMS2 mutations are not found (n=43). This suggests that in practice, searching for PMS2 mutations by molecular methods can be limited to cases where PMS2 alone is abnormal by IHC.
This study shows that identifying PMS2 gene mutations is just as important as identifying mutations in other LS-associated mismatch repair genes so that targeted screening recommendations can be made and predictive testing offered to at-risk family members. As we learn more about the cancer risks and spectrums of cancers through future studies, counseling and surveillance recommendations for those with PMS2 gene mutations may be further modified.
Supplementary Material
Supplemental Figure 1 PMS2 immunohistochemistry in colonic tissue from a patient with a monoallelic PMS2 mutation.
There is absence of PMS2 staining in the tumor tissue only (lower left).
Acknowledgments
Grant Support: This work was supported by the National Cancer Institute, National Institutes of Health grants CA67941, CA16058, and RFA # CA-95-011 and through cooperative agreements: Australasian Colorectal Cancer Family Registry (U01 CA097735); Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01 CA074800); Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783); Seattle Colorectal Cancer Family Registry (U01 CA074794); grant 04-0570 from the Swedish Cancer Society; grant 06-1252 from The Stockholm Cancer Center Foundation; and The National Health and Medical Research Council, Australia.
The work on samples from the Colon Cancer Family Registries was completed through cooperative agreements with the Ontario Familial Colorectal Cancer Registry, Australasian Colorectal Cancer Family Registry, Mayo Colorectal Cancer Registry, and the Seattle Familial Colorectal Cancer Family Registry. The authors would like to thank Soledad Fernandez, PhD, Maureen Mork, Jennifer Panescu, Shuying Sun, and Michael Walsh for their contributions to this work and our colleagues throughout the world who contributed samples from their patients to this research.
Abbreviations used in this manuscript
- CRC
colorectal cancer
- HR
hazard ratio
- IHC
immunohistochemistry
- LS
Lynch syndrome
- MSI
microsatellite instability
Footnotes
Conflicts of Interest: No conflicts of interest exist
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–60. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 2.Hampel H, Frankel W, Panescu J, et al. Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res. 2006;66:7810–7. doi: 10.1158/0008-5472.CAN-06-1114. [DOI] [PubMed] [Google Scholar]
- 3.Hampel H, Panescu J, Lockman J, et al. Comment on: Screening for Lynch Syndrome (Hereditary Nonpolyposis Colorectal Cancer) among Endometrial Cancer Patients. Cancer Res. 2007;67:9603. doi: 10.1158/0008-5472.CAN-07-2308. [DOI] [PubMed] [Google Scholar]
- 4.Goecke T, Schulmann K, Engel C, et al. Genotype-phenotype comparison of German MLH1 and MSH2 mutation carriers clinically affected with Lynch syndrome: a report by the German HNPCC Consortium. J Clin Oncol. 2006;24:4285–92. doi: 10.1200/JCO.2005.03.7333. [DOI] [PubMed] [Google Scholar]
- 5.Hendriks YM, Wagner A, Morreau H, et al. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: impact on counseling and surveillance. Gastroenterology. 2004;127:17–25. doi: 10.1053/j.gastro.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 6.Dunlop MG, Farrington SM, Carothers AD, et al. Cancer risk associated with germline DNA mismatch repair gene mutations. Hum Mol Genet. 1997;6:105–10. doi: 10.1093/hmg/6.1.105. [DOI] [PubMed] [Google Scholar]
- 7.Quehenberger F, Vasen HF, van Houwelingen HC. Risk of colorectal and endometrial cancer for carriers of mutations of the hMLH1 and hMSH2 gene: correction for ascertainment. J Med Genet. 2005;42:491–6. doi: 10.1136/jmg.2004.024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hampel H, Stephens JA, Pukkala E, et al. Cancer risk in hereditary nonpolyposis colorectal cancer syndrome: later age of onset. Gastroenterology. 2005;129:415–21. doi: 10.1016/j.gastro.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Jenkins MA, Baglietto L, Dowty JG, et al. Cancer risks for mismatch repair gene mutation carriers: a population-based early onset case-family study. Clin Gastroenterol Hepatol. 2006;4:489–98. doi: 10.1016/j.cgh.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Nicolaides NC, Carter KC, Shell BK, et al. Genomic organization of the human PMS2 gene family. Genomics. 1995;30:195–206. doi: 10.1006/geno.1995.9885. [DOI] [PubMed] [Google Scholar]
- 11.Nicolaides NC, Kinzler KW, Vogelstein B. Analysis of the 5′ region of PMS2 reveals heterogeneous transcripts and a novel overlapping gene. Genomics. 1995;29:329–34. doi: 10.1006/geno.1995.9997. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa H, Lockman JC, Frankel WL, et al. Mismatch repair gene PMS2: disease-causing germline mutations are frequent in patients whose tumors stain negative for PMS2 protein, but paralogous genes obscure mutation detection and interpretation. Cancer Res. 2004;64:4721–7. doi: 10.1158/0008-5472.CAN-03-2879. [DOI] [PubMed] [Google Scholar]
- 13.De Vos M, Hayward BE, Picton S, et al. Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am J Hum Genet. 2004;74:954–64. doi: 10.1086/420796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clendenning M, Hampel H, LaJeunesse J, et al. Long-range PCR facilitates the identification of PMS2-specific mutations. Hum Mutat. 2006;27:490–5. doi: 10.1002/humu.20318. [DOI] [PubMed] [Google Scholar]
- 15.Niessen RC, Kleibeuker JH, Jager PO, et al. Getting rid of the PMS2 pseudogenes: mission impossible? Hum Mutat. 2007;28:414. doi: 10.1002/humu.20447. [DOI] [PubMed] [Google Scholar]
- 16.Hayward BE, De Vos M, Valleley EM, et al. Extensive gene conversion at the PMS2 DNA mismatch repair locus. Hum Mutat. 2007;28:424–30. doi: 10.1002/humu.20457. [DOI] [PubMed] [Google Scholar]
- 17.Clendenning M, de la Chapelle A. Response to: Getting Rid of the PMS2 Pseudogenes: Mission Impossible? Hum Mutat. 2007;28:415. doi: 10.1002/humu.20447. [DOI] [PubMed] [Google Scholar]
- 18.Vasen HF, Mecklin JP, Khan PM, et al. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34:424–5. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 19.Vasen HF, Watson P, Mecklin JP, et al. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–6. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez-Bigas MA, Boland CR, Hamilton SR, et al. A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst. 1997;89:1758–62. doi: 10.1093/jnci/89.23.1758. [DOI] [PubMed] [Google Scholar]
- 21.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–8. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Southey MC, Jenkins MA, Mead L, et al. Use of molecular tumor characteristics to prioritize mismatch repair gene testing in early-onset colorectal cancer. J Clin Oncol. 2005;23:6524–32. doi: 10.1200/JCO.2005.04.671. [DOI] [PubMed] [Google Scholar]
- 23.Gill S, Lindor NM, Burgart LJ, et al. Isolated loss of PMS2 expression in colorectal cancers: frequency, patient age, and familial aggregation. Clin Cancer Res. 2005;11:6466–71. doi: 10.1158/1078-0432.CCR-05-0661. [DOI] [PubMed] [Google Scholar]
- 24.Truninger K, Menigatti M, Luz J, et al. Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer. Gastroenterology. 2005;128:1160–71. doi: 10.1053/j.gastro.2005.01.056. [DOI] [PubMed] [Google Scholar]
- 25.Baudhuin LM, Burgart LJ, Leontovich O, et al. Use of microsatellite instability and immunohistochemistry testing for the identification of individuals at risk for Lynch syndrome. Fam Cancer. 2005;4:255–65. doi: 10.1007/s10689-004-1447-6. [DOI] [PubMed] [Google Scholar]
- 26.Nicolaides NC, Papadopoulos N, Liu B, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 27.De Rosa M, Fasano C, Panariello L, et al. Evidence for a recessive inheritance of Turcot’s syndrome caused by compound heterozygous mutations within the PMS2 gene. Oncogene. 2000;19:1719–23. doi: 10.1038/sj.onc.1203447. [DOI] [PubMed] [Google Scholar]
- 28.Trimbath JD, Petersen GM, Erdman SH, et al. Cafe-au-lait spots and early onset colorectal neoplasia: a variant of HNPCC? Fam Cancer. 2001;1:101–5. doi: 10.1023/a:1013881832014. [DOI] [PubMed] [Google Scholar]
- 29.Agostini M, Tibiletti MG, Lucci-Cordisco E, et al. Two PMS2 mutations in a Turcot syndrome family with small bowel cancers. Am J Gastroenterol. 2005;100:1886–91. doi: 10.1111/j.1572-0241.2005.50441.x. [DOI] [PubMed] [Google Scholar]
- 30.De Vos M, Hayward BE, Charlton R, et al. PMS2 mutations in childhood cancer. J Natl Cancer Inst. 2006;98:358–61. doi: 10.1093/jnci/djj073. [DOI] [PubMed] [Google Scholar]
- 31.Felton KE, Gilchrist DM, Andrew SE. Constitutive deficiency in DNA mismatch repair. Clin Genet. 2007;71:483–98. doi: 10.1111/j.1399-0004.2007.00803.x. [DOI] [PubMed] [Google Scholar]
- 32.Felton KE, Gilchrist DM, Andrew SE. Constitutive deficiency in DNA mismatch repair: is it time for Lynch III? Clin Genet. 2007;71:499–500. doi: 10.1111/j.1399-0004.2007.00801.x. [DOI] [PubMed] [Google Scholar]
- 33.Kruger S, Kinzel M, Walldorf C, et al. Homozygous PMS2 germline mutations in two families with early-onset haematological malignancy, brain tumours, HNPCC-associated tumours, and signs of neurofibromatosis type 1. Eur J Hum Genet. 2008;16:62–72. doi: 10.1038/sj.ejhg.5201923. [DOI] [PubMed] [Google Scholar]
- 34.Poley JW, Wagner A, Hoogmans MM, et al. Biallelic germline mutations of mismatch-repair genes: a possible cause for multiple pediatric malignancies. Cancer. 2007;109:2349–56. doi: 10.1002/cncr.22697. [DOI] [PubMed] [Google Scholar]
- 35.Will O, Carvajal-Carmona LG, Gorman P, et al. Homozygous PMS2 deletion causes a severe colorectal cancer and multiple adenoma phenotype without extraintestinal cancer. Gastroenterology. 2007;132:527–30. doi: 10.1053/j.gastro.2006.11.043. [DOI] [PubMed] [Google Scholar]
- 36.Worthley DL, Walsh MD, Barker M, et al. Familial mutations in PMS2 can cause autosomal dominant hereditary nonpolyposis colorectal cancer. Gastroenterology. 2005;128:1431–6. doi: 10.1053/j.gastro.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Hendriks YM, Jagmohan-Changur S, van der Klift HM, et al. Heterozygous mutations in PMS2 cause hereditary nonpolyposis colorectal carcinoma (Lynch syndrome) Gastroenterology. 2006;130:312–22. doi: 10.1053/j.gastro.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 38.Antoniou AC, Pharoah PD, McMullan G, et al. Evidence for further breast cancer susceptibility genes in addition to BRCA1 and BRCA2 in a population-based study. Genet Epidemiol. 2001;21:1–18. doi: 10.1002/gepi.1014. [DOI] [PubMed] [Google Scholar]
- 39.Ferlay JBF, Pisani P, Parkin DM. Globocan 2002: Cancer Incidence, Mortality and Prevalence Worldwide: IARC CancerBase No. 5. IARCPress; Lyon: 2004. version 2.0. [Google Scholar]
- 40.Lange K, Cantor R, Horvath S, et al. Mendel version 4.0: A complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet. 2001;69:A1886. [Google Scholar]
- 41.Clendenning M, Senter L, Hampel H, et al. A frame-shift mutation of PMS2 is a widespread cause of Lynch syndrome. J Med Genet. 2008 doi: 10.1136/jmg.2007.056150. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kadyrov FA, Dzantiev L, Constantin N, et al. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 43.Wang L, Cunningham JM, Winters JL, et al. BRAF mutations in colon cancer are not likely attributable to defective DNA mismatch repair. Cancer Res. 2003;63:5209–12. [PubMed] [Google Scholar]
- 44.Lin KM, Shashidharan M, Thorson AG, et al. Cumulative incidence of colorectal and extracolonic cancers in MLH1 and MSH2 mutation carriers of hereditary nonpolyposis colorectal cancer. J Gastrointest Surg. 1998;2:67–71. doi: 10.1016/s1091-255x(98)80105-4. [DOI] [PubMed] [Google Scholar]
- 45.Carayol J, Khlat M, Maccario J, et al. Hereditary non-polyposis colorectal cancer: current risks of colorectal cancer largely overestimated. J Med Genet. 2002;39:335–9. doi: 10.1136/jmg.39.5.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boland CR, Koi M, Chang DK, et al. The biochemical basis of microsatellite instability and abnormal immunohistochemistry and clinical behavior in Lynch Syndrome: from bench to bedside. Fam Cancer. 2007 doi: 10.1007/s10689-007-9145-9. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Network NCC. NCCN Clinical Practice Guidelines in Oncology. 2007;2007 In: http://www.nccn.org/professionals/physician_gls/PDF/colorectal_screening.pdf.
- 48.Lindor NM, Petersen GM, Hadley DW, et al. Recommendations for the care of individuals with an inherited predisposition to Lynch syndrome: a systematic review. JAMA. 2006;296:1507–17. doi: 10.1001/jama.296.12.1507. [DOI] [PubMed] [Google Scholar]
- 49.Loughrey MB, Waring PM, Tan A, et al. Incorporation of somatic BRAF mutation testing into an algorithm for the investigation of hereditary non-polyposis colorectal cancer. Fam Cancer. 2007;6:301–10. doi: 10.1007/s10689-007-9124-1. [DOI] [PubMed] [Google Scholar]
- 50.Bessa X, Balleste B, Andreu M, et al. A prospective, multicenter, population-based study of BRAF mutational analysis for Lynch syndrome screening. Clin Gastroenterol Hepatol. 2008;6:206–14. doi: 10.1016/j.cgh.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 51.Jackson C, Holter S, Pollett A, et al. Cafe’-au-lait macules and pediatric malignancy caused by biallelic mutations in the DNA mismatch repair (MMR) gene PMS2: Case report and review of the literature. Pediatric Blood and Cancer. 2008 doi: 10.1002/pbc.21514. (in press) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 PMS2 immunohistochemistry in colonic tissue from a patient with a monoallelic PMS2 mutation.
There is absence of PMS2 staining in the tumor tissue only (lower left).